Abstract
Mass spectrometry (MS) is one of the most effective techniques for high-throughput, high-resolution characterization of glycan structures. Although many software applications have been developed over the last decades for the interpretation of MS data of glycan structures, only a few are capable of dealing with the large data sets produced by glycomics analysis. Furthermore, these applications utilize databases that can lead to redundant glycan annotations and do not support post-processing of the data within the software or by third party applications. To address the needs, we present GRITS Toolbox, a freely-available, platform-independent software application capable of storing and processing glycomics MS data along with associated metadata. GRITS Toolbox automatically annotates MS data using an integrated glycan identification module that references manually curated databases of mammalian glycans (provided with the software) or any user-defined databases. Extensive display routines are provided to post-process the data and refine the automated annotation using expert knowledge of the user. The software also allows side by side comparison of annotations from different MS runs or samples and exporting of annotations into Excel format.
Keywords: free software, glycomics, glycomics mass spectrometric data, mass spectrometry annotation, mass spectrometry data processing
Introduction
Glycans, nucleic acids (DNA/RNA), proteins and lipids constitute major classes of biomolecules required for the survival of all living organisms (Marth 2008). The systematic study of glycans (Glycomics), including their structures, functions and interactions with other molecules, has led to significant advances in our understanding of the biological mechanisms underlying adaptation, development and disease (Ohtsubo and Marth 2006; Cummings and Pierce 2014). Mass spectrometry (MS) is the most widely used technology for the identification and quantification of glycan structures. Continuing improvements in the accuracy and sensitivity of this technique have resulted in rapid growth in the amount and throughput of glycomics data that is being generated. This trend calls for new software tools capable of processing, interpreting and storing large volumes of glycomics data.
Many software tools have been developed to assist in the identification of glycan structures from MS data. Several web-based tools, such as GlycoFragment (Lohmann and von der Lieth 2004), GlycoPeakfinder (Maass et al. 2007) or GlycoMod (Cooper et al. 2001), suggest compositional or structural annotations of spectral data that is submitted via the internet. The main limitation of these tools is that they accept and process one MS or MSn spectrum at a time, which makes these web-based programs unsuited for the annotation of high-throughput data consisting of hundreds or even thousands of MS/MS spectra. Furthermore, most of these software applications cannot manipulate or recall annotations once the browser session has expired; this limitation requires the data to be resubmitted for regeneration of the annotations.
In addition to web-based tools, several standalone software applications have been developed for local installation and data analysis. Two of the most advanced systems for the interpretation of data generated by MS analysis of released glycans are GlycoWorkbench (Ceroni et al. 2008; Damerell et al. 2012, 2015) and SimGlycan® (Apte and Meitei 2010). GlycoWorkbench is a freely available multiplatform Java application with advanced routines that facilitate the creation and graphical display of glycan structures and the annotation of MS profiling and MS/MS data with these structures and their fragments. MS data can be annotated with structures from multiple databases that are integrated into GlycoWorkbench, including CFG glycan database (Raman et al. 2006), CarbBank (Doubet et al. 1989; Doubet and Albersheim 1992), GLYCOSCIENCES.de (Lutteke et al. 2006) and GlycomeDB (Ranzinger et al. 2008, 2011). The program displays the annotated data in several different formats and supports interactive post-processing of the annotations and their export in Excel format. Although GlycoWorkbench is capable of annotating several MS spectra at once, each spectrum must be loaded individually, making the handling of large datasets cumbersome. The second standalone application used for the interpretation of MS data from free glycans is the commercial program SimGlycan®, which runs on Microsoft Windows® systems. This program supports the loading of standard, open source mzXML files (Pedrioli et al. 2004) or proprietary data files containing complete MS/MS runs and annotates these data with structures from the KEGG glycan database (Hashimoto et al. 2006). However, neither local post-processing nor export of the data for post-processing by other software tools is supported by this software. Both software tools display annotations in the commonly used graphical representation described in “Essentials of Glycobiology” (Varki et al. 2015) and allow data-processing sessions to be saved and reopened for further data manipulation and display.
Here, we present GRITS Toolbox, a freely available software system that we have developed for archiving, processing and interpreting analytical data with a focus on glycomics data generated by MS of N-linked and O-linked glycans released from glycoproteins or cells. GRITS Toolbox implements an extensive set of graphical user interface functions to visualize, review, manually modify and export experimental data and annotated MS data. GRITS Toolbox has been developed for the interpretation of data generated by analysis of free, released or labeled glycans. The analysis of intact glycoconjugates, such as glycopeptides or glyco-lipids is currently out-of-scope of the software. The software is in continuous development and new features and performance improvements are being added with each new version. Here, we present version 1.2 of our software and discuss future developments in the “Future Work” section. We chose the name GRITS as a recursive acronym for “GRITS Really Is The Solution,” an abbreviation that also reflects an aspect of the regional flavor of the southeastern United States, where this work was performed.
Results
GRITS Toolbox is an integrated, modular system that implements separate user interfaces for the entry, processing, visualization and export of data and metadata, as described in the following sections.
Project information and sample description
For a typical user, the initial interaction with the software starts with the creation of a project. In this context, a project is a digital container that allows information and data to be grouped together. At this stage, optional metadata can be attached to the project, specifying global information such as a general description of the project, information about collaborating partners (names, addresses, contact information and funding), and a list of user-defined keywords or tags. After a project is created, a list of samples (each called an analyte) which are studied or analyzed as part of the project needs to be generated before any experimental data can be loaded and attached. The user can describe each sample at the desired level of detail in human language or in tabular form representing the information using dictionary identifiers or ontology URIs that are readily indexed and standardized, as required for submission to databases and repositories. Similarly, GRITS Toolbox provides interfaces to associate experimental data with supporting information about sample preparation sufficient for experts and non-experts in glycoscience to understand the experiment that generated the data and to reproduce the experimental results. Initiatives such as Minimum Information Required for A Glycomics Experiment (MIRAGE) (Kolarich et al. 2013; York et al. 2014) have recognized the need for such information in order to understand, evaluate and reproduce glycomics experiments. Clearly organizing, storing and archiving all this diverse information along with the raw and annotated data are critical requirements for effective data sharing and utilization.
MS data
After creating project and sample descriptions, MS data can be attached to the corresponding sample. Even if the application is solely used for MS interpretation it requires the creation of projects and samples to maintain a consistent data model. Two forms of data can be loaded: the raw data file as provided by the instrument and the corresponding files in an XML standard format (mzXML (Pedrioli et al. 2004) or mzML (Martens et al. 2011)). GRITS Toolbox only works with data from the XML file but the instrument file, if provided, is also stored for archival purposes. If the instrument vendor software does not support data export to one of the XML formats, free conversion tools, such as msConvert (Chambers et al. 2012), can be used to generate the file in the appropriate format, allowing data files to be loaded regardless of the instrument used to generate the data. It is also possible to invoke msConvert within GRITS and convert the instrument files to mzXML/mzML files without opening another software. GRITS Toolbox is flexible and supports many different MS experiments commonly used for glycan identification, including MS profiling, Tandem MS/MS, Total Ion Mapping (TIM) and LC-MS/MS (Aoki et al. 2007). Once the files are copied into the GRITS project, each spectral scan in the file and the peaks it comprises can be browsed either in tabular format or visually as a graphically annotated spectrum.
Annotated MS data
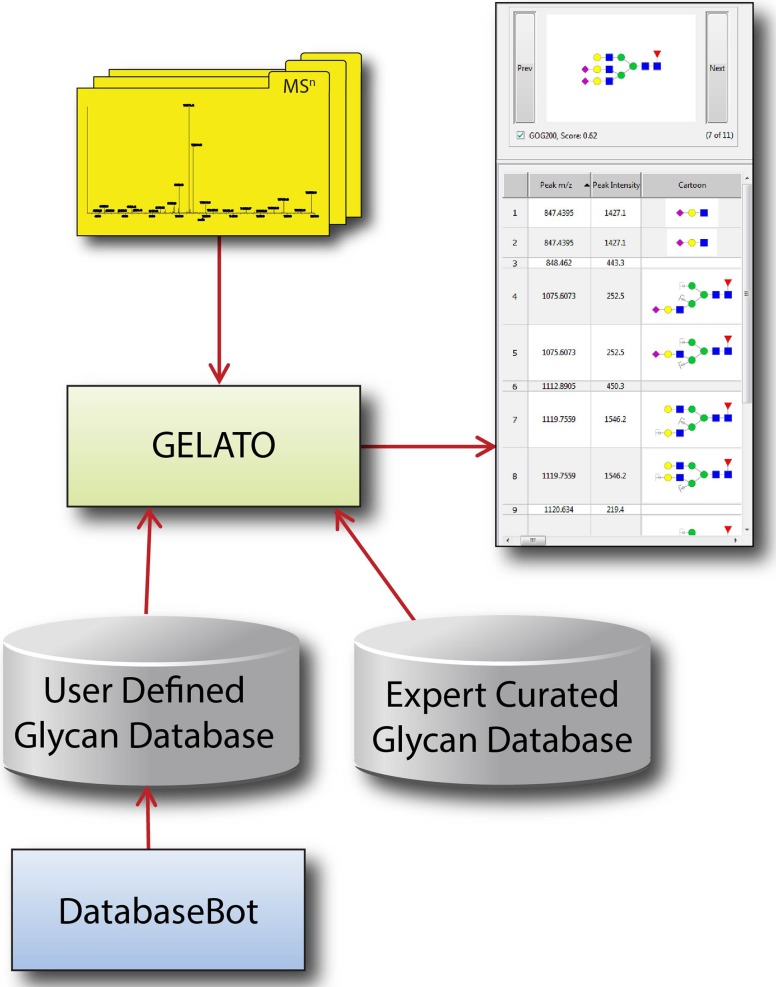
Uploaded MS data can be interpreted using the integrated annotation module, named Glycomics Elucidation and Annotation Tool (GELATO, described in (AlJadda et al. 2015)), which associates the spectral features of each mass spectrum with a specific glycan structure or set of structures (see Figure 1). The fragmentation and interpretation algorithm in GELATO is implemented using functions provided by the GlycoWorkbench Java library. These functions predict the fragmentation products of given glycan structures based on several user-specified settings: accuracy, possible adducts, possible cleavages, possible neutral exchanges, derivatization and reducing end modification. Table I shows an overview over the different options that can be used for the annotation. Although GELATO reuses many of the existing GlycoWorkbench functions, it also provides additional features that are not readily available in GlycoWorkbench. These include: the ability to specify different accuracy settings for MS1 and MSn spectra; application of different fragmentation settings for each MS level or ion-dissociation method; prediction of ions resulting from neutral loss; the ability to create new types of ion structures or adducts and ions formed by neutral exchange.
Fig. 1.
Annotation of experimental MSn data using GELATO module. Glycan structures that have been curated by experts using Qrator have been used to populate GRITS databases. Alternatively, the users can use DatabaseBot module of GRITS to create custom databases. Structures in these databases are used by GELATO to simulate quasi-molecular ions, which are compared to experimentally observed MS1 ions. Matching candidate structures are fragmented using the extended GlycoWorkbench fragmentation algorithm integrated in GELATO and the theoretical fragments are compared to fragment ions in the MS2 scan.
Table I.
Overview of the major settings in GELATO and a listing of possible assignments for these settings
| Annotation setting | Supported option |
|---|---|
| Accuracy | Any value in ppm or Dalton. Different accuracy settings for MS1 and MSn |
| Glycan derivatization | None, Per-methylation, Per-deuteromethylation, C13 Per-methylation, Per-Acetylation, Per-deuteroacetylation |
| Reducing end (modification) | Free reducing end, Reduced reducing end, Methylation, Deoxygenation, PA, 2AB, many other common labels, or user defined label |
| Cleavage types | A, B, C, X, Y, Z; number of cleavages can be chosen freely (different settings for different MS level or activation method possible) |
| Adducts | H+, Na+, H-, Cl-, Li+, K+, Ca++, or user defined adducts |
| Ion exchange | Any specified adduct except H |
| Neutral loss or gain | H2O, CH2, Sialic acid, or user defined neutral loss |
To provide candidate structures for use by the GELATO module, GRITS Toolbox is equipped with a set of integrated databases that have been curated by human experts using a web-based system called Qrator (Eavenson et al. 2015). For each type of glycan structure (N-glycan, O-glycan, glycosphingolipid glycans) in the Qrator system, a separate structure database has been created and integrated in GRITS Toolbox. Alternatively, the user can create a custom structure database using an integrated database builder, called DatabaseBot (described in Section Databases and Filtering). The structures in these databases are proposed as annotations for spectral features in experimental data sets if the experimental m/z is within the specified precursor tolerance of the theoretical m/z of the structure. If the experimental data comes from MS profiling experiments, the m/z values for all peaks in the MS spectra are compared to the theoretical quasi-molecular m/z values for the candidate structures. If the spectra were generated by an MS/MS, LC-MS/MS or TIM experiment, candidate structures are identified by comparing the precursor m/z values for each MS2 spectrum to the theoretical values for each structure. As with the precursor, if the m/z values of the ions produced by in silico fragmentation of a candidate structure are within the specified fragment tolerance of the observed m/z values of peaks in the MS2 spectrum, the peaks are annotated using the simulated fragments. The GELATO algorithm is recursive, repeating this procedure for MSn spectra with n > 2 and thereby facilitating the annotation of deep tandem MS data sets.
Table II shows the results of a benchmark analysis performed using four different data files (A through D) generated by tandem MS/MS experiments, each of which are annotated using two different glycan databases. The first database is the built-in N-Glycan database (1190 N-glycan structures), while the second one contains a subset (590 N-glycan structures) of the same glycans. For each dataset and each database, three different annotation runs were performed, varying the allowed cleavage types (run 1: up to two cleavages—only [B,Y]; run 2: up to two glycosidic cleavages [B,C,Y,Z]; run 3: up to two glycosidic cleavages [B,C,Y,Z] and up to one crossring cleavages [A,X]). All other annotation settings were kept the same (accuracy for MS1/MSn:600 ppm/300 ppm; up to four sodium adducts; no exchanges and no neutral losses). The analysis was performed on a 2015 iMac with an i7 processor and 16GB RAM. The number of MS2 scans annotated for data set A (209 MSn scans in total) is 64 using the full database (1,190 structures) and 54 using the smaller database (590 structures). The corresponding numbers are, respectively, 184 and 162 for data set B (409 MSn scans), 220 and 182 for data set C (2,000 MSn scans) and 251 and 200 for data set D (3,000 MSn scans). For each data set, the total time (hh:mm:ss, including that used to perform the annotation plus that used to organize and write the annotation results to the data files) is shown. A similar annotation run (N-glycans database and B-Y cleavages) was performed with a LC-MS/MS data set consisting of 37576 scans, with a total time of 2:45:47, resulting in 11,072 annotated MSn scans.
Table II.
Benchmark of annotation times relative to the number of scans
| Data set | N-Glycans Database (1190 Structures) | N-Glycans Database (590 Structures) | ||||
|---|---|---|---|---|---|---|
| Cleavage Types: B-Y | Cleavage Types: B-Y-C-Z | Cleavage Types: B-Y-C-Z-A-X | Cleavage Types: B-Y | Cleavage Types: B-Y-C-Z | Cleavage Types: B-Y-C-Z-A-X | |
| A— 209 scans | 00:03:25 | 00:03:31 | 00:06:54 | 00:01:42 | 00:01:48 | 00:03:23 |
| B—409 scans | 00:03:44 | 00:04:08 | 00:17:57 | 00:01:52 | 00:02:11 | 00:09:52 |
| C—2000 scans | 00:05:12 | 00:06:58 | 00:53:02 | 00:03:08 | 00:03:22 | 00:26:17 |
| D—3000 scans | 00:06:49 | 00:08:05 | 01:15:15 | 00:03:34 | 00:04:20 | 00:43:10 |
Annotation scoring
For each structure that is assigned to an experimental precursor ion in a Tandem MS/MS run, confidence scores are calculated. GELATO calculates two scores, a “counting score” and an “intensity score”. The counting score is the ratio of the number of peaks annotated by fragmentation of a candidate structure to the number of all peaks in the experimental MS/MS spectrum. Alternatively, the intensity score is the ratio of the total intensity of all annotated peaks to the total intensity of all peaks in the MS/MS spectra.
Annotation post-processing
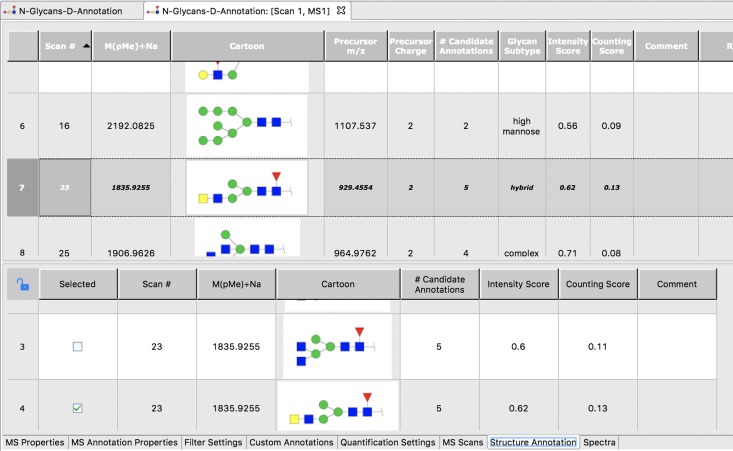
All matching candidate structures and their fragment ions are stored in a GRITS MS annotation file. These results are then presented to the user for post-processing (see Figure 2). Structural annotations of each peak in the MS profiling spectrum (which are used as MS2 precursor ions) are shown in the upper part of the editor. Alternative structures for each peak with the same m/z value are shown in the lower part of the editor. Each peak in the profiling spectrum can be individually selected and edited by the user, making it possible to refine annotation(s) by selecting or deselecting candidate structures based on score, expert knowledge and/or prior experience.
Fig. 2.
Screenshot of the results of an MS/MS annotation. The highest scoring candidate structure for each MS1 peak (i.e., MS2 precursor) identified by GELATO is shown as a row in the upper part of the screen. Clicking on one of these rows shows a list of alternative structures (lower part of the screen) for this precursor ion, allowing final annotation of each MS1 ion to be selected manually, based on the MS2 spectrum obtained by fragmenting the ion (Figures 3 and 4) or user expertise. Ions in the MS2 spectrum can be viewed and evaluated (Figure 3) by double-clicking on a row in the upper table. In this example, MS2 scan # 23 (precursor m/z 929.4554) has been selected (row with dark shading) and possible annotations for this precursor ion are shown in the bottom portion. Monosaccharide symbols follow the SNFG (Symbol Nomenclature for Glycans) system (Varki et al. 2015).
In addition to the information from the MS data file (e.g., MS2 scan number, m/z, intensity) and the annotations (e.g., representations of glycan sequence in different graphical formats including IUPAC notation (McNaught 1997) and the notation suggested by “Essentials of Glycobiology” (Varki et al. 2015)), the tables contain columns describing features of the glycan structure that can help the user sort and select structures with which to annotate the spectra. Data in these columns include the number of specific monosaccharides (e.g., sialic acids) in each structure, the presence of predefined motifs (e.g., Lewis type fucosylation patterns) and other specific structural features (e.g., the number of branches, core fucosylation, presence of LacNAc and LacDiNAc repeats, presence of bisecting residues, alternative sialic acids and core type for O-glycans). Scores for each annotation of the MS/MS data are calculated and included in the table as well (bottom portion of Figure 2).
Manual selection of structures for final annotation of each precursor ion will depend substantially on the annotation of MS2 with fragment ions, which can be viewed (see Figure 3) by double-clicking on a candidate structure (upper portion of Figure 2). All candidate structures are shown in the summary table (Figure 3) along with all of their fragments that can be assigned to peaks in the MS2 spectra. This information can be used to select/deselect candidate structures based on agreement of their predicted and observed fragments.
Fig. 3.
Fragment overview of a MS2 spectrum selected by double-clicking on a structure (dark shaded row) in the upper portion of Figure 2. The header of each column shows a candidate structure (corresponding to an item in the candidate list in the lower portion of Figure 2) for the precursor ion of the selected MS2 spectrum. Theoretical fragments of each of these structures that match m/z values observed in the MS2 spectrum are shown in rows below. Structures to be saved for the final annotation can be selected by checking the boxes above each structure in the header. MSn fragmentation (n > 2) can be displayed in similar fashion.
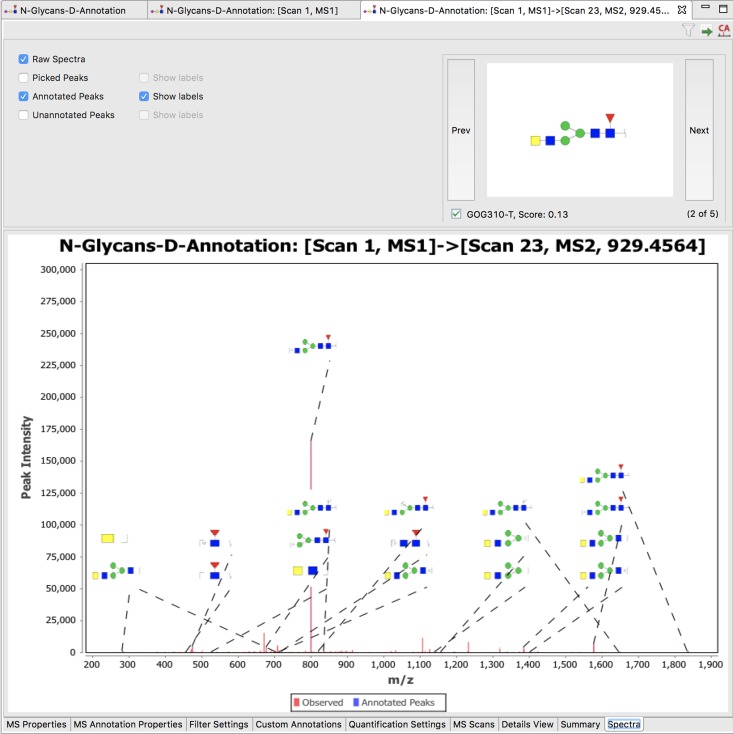
It is also possible to get a general overview of peak annotation by invoking a graphical representation of the spectrum with cartoon representations of the annotations rendered above each ion (see Figure 4).
Fig. 4.
Screenshot of an MS2 spectrum (bottom) (selected by clicking to the Spectra tab at the bottom of the page in Figure 3) annotated with the predicted fragments of a candidate structure (top right). The “Prev” and “Next” buttons allow the user to scroll through the candidate structures of the precursor ion and evaluate each based on the annotation of its fragment-ion spectrum. Structures that are deemed correct can be selected using the check box below the “Prev” button. Several options (top left) are available to control how various features of the spectrum are displayed.
All annotations selected by the user are stored in the GRITS MS annotation file, so this information can be accessed again when the MS data set is reopened. Both the overview table containing the detected m/z values, peak intensities and annotation information (Figure 2, upper part) and the summary page (Figure 3) can be exported to Excel for post-processing of the annotated data.
Databases and filtering
GRITS Toolbox comes with a selection of databases, as mentioned in the previous section, that are based on our manually curated Mammalia database. However, if the users are working on other types of samples (e.g., plant, worm, insect, bacteria) or on disease related samples, these databases provide little help since many sample-related structures are missing. Therefore, GRITS Toolbox provides a module (DatabaseBot) to create new custom databases and configure the GELATO module to annotate spectra using such custom databases rather than or in addition to the existing ones during annotation. A new custom database can be created in several ways: by supplementing one of the existing databases with new structures, by removing unwanted structures from an existing database, or by generating a new database altogether.
In order to help eliminate possible irrelevant or redundant annotations, GRITS Toolbox offers a filtering mechanism at several points in the process of MS data analysis. Composition and/or motif-based filtering can be applied while creating a custom database from an existing database, allowing the exclusion of irrelevant structures from the queried database. Filtering can also be applied during GELATO annotation. Based on user preferences, the GELATO module can ignore structures from a database, thereby reducing the search space to only those structures which pass the specified filter criteria. GRITS Toolbox also offers the ability to apply post-filtering after annotations are generated and presented to the user in the form of a table. Once the annotation results are shown in a table representation, the user can highlight certain structures based on filter criteria to facilitate the candidate selection process or automatically select final candidates when they match the given filter criteria. Score-based filtering is also available, which allows the user to automatically select top candidates based on their intensity or counting scores generated by GELATO annotation.
Comparison of results across samples or experimental conditions
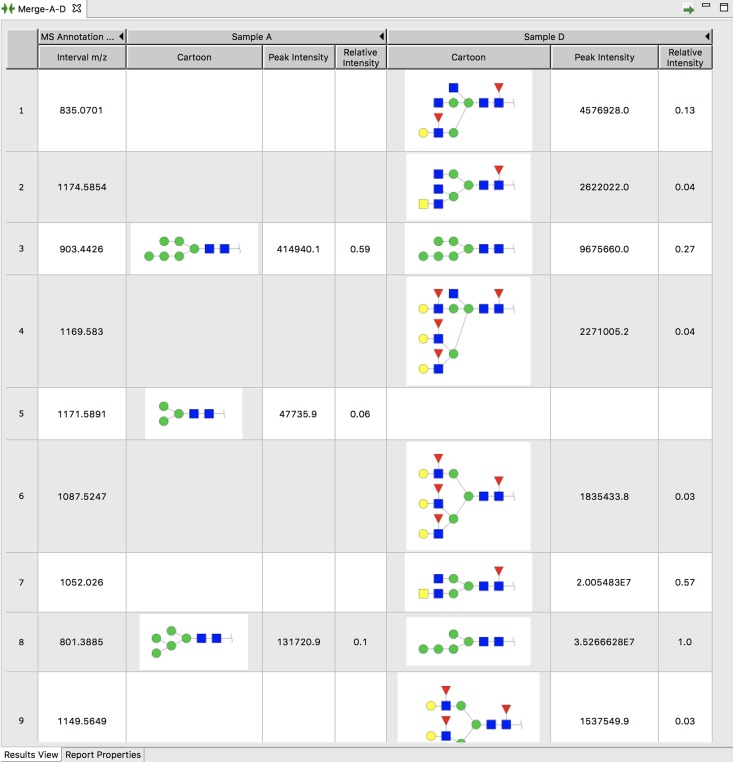
One of the most important aspects of glycomics research is the comparison of experiment results across samples or conditions. GRITS Toolbox provides a “merge” tool to compare annotation results from different samples side-by-side to more readily detect glycomic changes. The users can select two or more annotation results to create a merge report as shown in Figure 5. The merge report is interactive in that the user can double-click on any annotated structure to see or change the candidate annotations by going back to the original annotation page (Figure 2). The results can also be exported into Excel for further processing if necessary.
Fig. 5.
Screenshot of a merge report which is generated by selecting “Tools→MS Glycan Annotation Merge→New MS Glycan Annotation Report” from the menu and selecting two or more samples (Sample A and Sample D in this screenshot). Interval (first column) is obtained by looking at all samples and retrieving m/z values within the user provided tolerance interval (500 ppm is used for this example). For each selected annotation (Sample A and Sample D) the structure, intensity and relative intensity (ratio of peak intensity to most abundant peak) are shown if the peak was present in this sample.
Discussion
GRITS Toolbox is freely available platform independent software that was developed to allow processing and annotation of glycomics MS data, to capture and archive metadata associated with MS and non-MS data. The core functionality of the GRITS Toolbox resides in its ability to facilitate the elucidation of glycan structures based on MS data. This feature utilizes the GlycoWorkbench fragmentation algorithm, but has also been extended to provide more flexible and thorough annotation of high throughput MS data. GRITS Toolbox has also been designed to support new features that are not included in GlycoWorkbench or other currently available software tools. These novel features include, but are not limited to, the prediction of ions generated by neutral loss processes and the ability to specify custom ion structures (e.g., novel adducts). Furthermore, while most other annotation tools are only capable of handling limited amounts of MS data at a time, GRITS Toolbox is able to process and annotate thousands of MS spectra simultaneously. This makes the program well suited for the interpretation of large-scale data sets that will increasingly characterize the cutting edge of glycomics research. GRITS Toolbox offers extended display options that allow annotation of MSn data to be viewed and explored using different tabular or graphical representations assisting users in the manual post-processing of annotations.
A major advance toward robust, automated analysis of MS glycomics data, which is incorporated within the workflows supported by GRITS Toolbox, is the ease with which highly curated or otherwise customized databases can be invoked for MS data analysis. Other currently available software tools utilize various broadly available databases. Databases integrated into GlycoWorkbench include the CFG glycan database, CarbBank, GLYCOSCIENCES.de and GlycomeDB. However, reliance on these databases can result in degenerate annotation of an MS spectrum with different instances of the same structure. Such redundant annotation is usually due to the representation of the same structure in more than one database or the presence of incompletely specified structures in the same database. For example, a spectrum may be simultaneously annotated with several structures that differ only in the anomeric configuration of the reducing end or in the extent to which glycosidic linkage positions are specified. In many cases, these structures cannot be distinguished using MS alone. GlycoWorkbench partially addresses this problem by allowing users to create custom databases of limited scope and to use these databases for spectral annotation. Similar limitations, including the potential for redundant annotation, also apply to SimGlycan® software, although the “Enterprise Edition” allows users to edit existing glycan databases to enhance annotation. In contrast to the currently available annotation tools, the automatic annotation function in GRITS Toolbox utilizes manually curated mammalian databases developed through the Qrator project. These expert-curated databases are included within the GRITS Toolbox and provide a solid foundation for annotating MS glycan data. If these databases are insufficient or inappropriate (e.g., for work on non-mammalian samples), they can also be supplemented or replaced by creating or importing alternative glycan databases. Furthermore, if automatic annotation is insufficient, users can post-process the automatic annotations and refine them based on their specific expertise.
Future work
GRITS Toolbox (January 2019) was developed as a standalone annotation tool for free and released glycans in Tandem MS/MS experiments. However, the software also supports MS profiling, TIM and LC-MS/MS data. Besides the ongoing improvements and performance optimizations there are several major projects planned to increase the software’s function and usability. (1) Improved scoring—as described above GRITS Toolbox uses spectrum-based scores to help the user in the annotation. However, probabilistic scores and false discovery rates would be a much more useful tool especially for high throughput experiments. (2) Extension of the glycan databases—the databases provided with GRITS are manually curated databases of human and mammalian glycans. These databases are however not complete and an ongoing effort of our group is the extension of these databases with missing structures and topologies. (3) Improved data analysis for LC-MS/MS—many of the features implemented for Tandem MS/MS data processing are still not very well suited for LC-MS/MS datasets or high-throughput experiments. Additional display options are needed to allow user friendly post processing and manual verification of these annotations. (4) Database-less annotation—one of the limiting factors in GRITS Toolbox are the databases. The curated databases work well for human and mammalian samples but are not well suited for samples from other species. We are working on a module that annotates spectra with glycan compositions rather than structures from a database, which allows to easily use GRITS toolbox for any type of glycan samples without creating a database first.
In addition to the described efforts above, GRITS Toolbox is a platform that can be extended by third party plugins to add new functionality to the software. Notably there are two external ongoing efforts to extend GRITS functionality. A plugin for loading, processing and interpretation of glycan microarray data (manuscript in preparation) and a plugin for annotation of MS data from intact glycolipids (manuscript in preparation).
Acknowledgments
We would like to thank the developers of GlycoWorkbench for their collaborative spirit and for their efforts to develop an outstanding product that reshaped glycomic data analysis. The availability of this program library has been an invaluable resource that provided the authors with a solid foundation upon which we have developed the advanced functionality implemented within GRITS Toolbox.
Abbreviations
- DNA
deoxyribonucleic Acid
- MS
mass spectrometry
- RNA
ribonucleic Acid
- XML
extensible markup language
- URI
uniform resource identifier
Funding
This work was supported by the National Institute of General Medical Sciences [Grant No. 8P41GM103490].
Availability
The current version of the software system is freely available from our project website: http://www.grits-toolbox.org; last accessed April 2, 2019. The freely available Java JDK 1.8 (http://www.oracle.com/technetwork/java/javase/downloads/jdk8-downloads-2133151.html; last accessed April 2, 2019) framework is required for running the software. The software can be used on Windows, MacOS and Linux systems. A set of video tutorials has been created to help getting started (https://www.youtube.com/channel/UCH-K1KDIcru-GXFio0awO9Q; last accessed April 2, 2019). An example workspace with data to demonstrate the GRITS Toolbox features is available in the download section of the project website as well. As of January 2019, GRITS Toolbox has been downloaded by 195 individuals from 126 different institutions (27 companies, 15 research centers and 84 universities).
References
- AlJadda K, Ranzinger R, Porterfield MP, Weatherly B, Korayem M, Miller JA, Rasheed K, Kochut KJ, York WS. 2015. Gelato and sage: An integrated framework for ms annotation. CoRR. abs/1512.08451.
- Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M. 2007. Dynamic developmental elaboration of n-linked glycan complexity in the drosophila melanogaster embryo. J Biol Chem. 282(12):9127–9142. [DOI] [PubMed] [Google Scholar]
- Apte A, Meitei NS. 2010. Bioinformatics in glycomics: Glycan characterization with mass spectrometric data using simglycan. Methods Mol Biol. 600:269–281. [DOI] [PubMed] [Google Scholar]
- Ceroni A, Maass K, Geyer H, Geyer R, Dell A, Haslam SM. 2008. Glycoworkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J Proteome Res. 7(4):1650–1659. [DOI] [PubMed] [Google Scholar]
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J et al. 2012. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 30(10):918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CA, Gasteiger E, Packer NH. 2001. Glycomod—a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics. 1(2):340–349. [DOI] [PubMed] [Google Scholar]
- Cummings RD, Pierce JM. 2014. The challenge and promise of glycomics. Chem Biol. 21(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerell D, Ceroni A, Maass K, Ranzinger R, Dell A, Haslam SM. 2012. The glycanbuilder and glycoworkbench glycoinformatics tools: updates and new developments. Biol Chem. 393(11):1357–1362. [DOI] [PubMed] [Google Scholar]
- Damerell D, Ceroni A, Maass K, Ranzinger R, Dell A, Haslam SM. 2015. Annotation of glycomics ms and ms/ms spectra using the glycoworkbench software tool. Methods Mol Biol. 1273:3–15. [DOI] [PubMed] [Google Scholar]
- Doubet S, Albersheim P. 1992. Carbbank. Glycobiology. 2(6):505. [DOI] [PubMed] [Google Scholar]
- Doubet S, Bock K, Smith D, Darvill A, Albersheim P. 1989. The complex carbohydrate structure database. Trends Biochem Sci. 14(12):475–477. [DOI] [PubMed] [Google Scholar]
- Eavenson M, Kochut KJ, Miller JA, Ranzinger R, Tiemeyer M, Aoki K, York WS. 2015. Qrator: a web-based curation tool for glycan structures. Glycobiology. 25(1):66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Goto S, Kawano S, Aoki-Kinoshita KF, Ueda N, Hamajima M, Kawasaki T, Kanehisa M. 2006. Kegg as a glycome informatics resource. Glycobiology. 16(5):63R–70R. [DOI] [PubMed] [Google Scholar]
- Kolarich D, Rapp E, Struwe WB, Haslam SM, Zaia J, McBride R, Agravat S, Campbell MP, Kato M, Ranzinger R et al. 2013. The minimum information required for a glycomics experiment (mirage) project: Improving the standards for reporting mass-spectrometry-based glycoanalytic data. Mol Cell Proteomics. 12(4):991–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann KK, von der Lieth C-W. 2004. Glycofragment and glycosearchms: web tools to support the interpretation of mass spectra of complex carbohydrates. Nucleic Acids Res. 32(Web Server issue):W261–W266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutteke T, Bohne-Lang A, Loss A, Goetz T, Frank M, von der Lieth C-W. 2006. Glycosciences.De: an internet portal to support glycomics and glycobiology research. Glycobiology. 16(5):71R–81R. [DOI] [PubMed] [Google Scholar]
- Maass K, Ranzinger R, Geyer H, von der Lieth CW, Geyer R. 2007. “Glyco-peakfinder”--de novo composition analysis of glycoconjugates. Proteomics. 7(24):4435–4444. [DOI] [PubMed] [Google Scholar]
- Martens L, Chambers M, Sturm M, Kessner D, Levander F, Shofstahl J, Tang WH, Rompp A, Neumann S, Pizarro AD et al. 2011. Mzml--a community standard for mass spectrometry data. Mol Cell Proteomics. 10(1):R110 000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth JD. 2008. A unified vision of the building blocks of life. Nat Cell Biol. 10(9):1015–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaught AD. 1997. Nomenclature of carbohydrates (recommendations 1996). Adv Carbohydr Chem Biochem. 52:43–177. [PubMed] [Google Scholar]
- Ohtsubo K, Marth JD. 2006. Glycosylation in cellular mechanisms of health and disease. Cell. 126(5):855–867. [DOI] [PubMed] [Google Scholar]
- Pedrioli PG, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R et al. 2004. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 22(11):1459–1466. [DOI] [PubMed] [Google Scholar]
- Raman R, Venkataraman M, Ramakrishnan S, Lang W, Raguram S, Sasisekharan R. 2006. Advancing glycomics: Implementation strategies at the consortium for functional glycomics. Glycobiology. 16(5):82R–90R. [DOI] [PubMed] [Google Scholar]
- Ranzinger R, Herget S, von der Lieth CW, Frank M. 2011. Glycomedb--a unified database for carbohydrate structures. Nucleic Acids Res. 39(Database issue):D373–D376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranzinger R, Herget S, Wetter T, von der Lieth C-W. 2008. Glycomedb - integration of open-access carbohydrate structure databases. BMC Bioinformatics. 9:384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T et al. 2015. Symbol nomenclature for graphical representations of glycans. Glycobiology. 25(12):1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Agravat S, Aoki-Kinoshita KF, McBride R, Campbell MP, Costello CE, Dell A, Feizi T, Haslam SM, Karlsson N et al. 2014. Mirage: the minimum information required for a glycomics experiment. Glycobiology. 24(5):402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]