Abstract
A class of carbohydrate-modified proteins, heparan sulfate proteoglycans (HSPGs), play critical roles both in normal development and during disease. Genetic studies using a model organism, Drosophila, have been contributing to understanding the in vivo functions of HSPGs. Despite the many strengths of the Drosophila model for in vivo studies, biochemical analysis of Drosophila HS is somewhat limited, mainly due to the insufficient amount of the material obtained from the animal. To overcome this obstacle, we generated mutant cell lines for four HS modifying enzymes that are critical for the formation of ligand binding sites on HS, Hsepi, Hs2st, Hs6st and Sulf1, using a recently established method. Morphological and immunological analyses of the established lines suggest that they are spindle-shaped cells of mesodermal origin. The disaccharide profiles of HS from these cell lines showed characteristics of lack of each enzyme as well as compensatory modifications by other enzymes. Metabolic radiolabeling of HS allowed us to assess chain length and net charge of the total population of HS in wild-type and Hsepi mutant cell lines. We found that Drosophila HS chains are significantly shorter than those from mammalian cells. BMP signaling assay using Hs6st cells indicates that molecular phenotypes of these cell lines are consistent with previously known in vivo phenomena. The established cell lines will provide us with a direct link between detailed structural information of Drosophila HS and a wealth of knowledge on biological phenotypic data obtained over the last two decades using this animal model.
Keywords: cell culture, Drosophila, heparan sulfate, mutant
Introduction
A class of carbohydrate-modified proteins, heparan sulfate proteoglycans (HSPGs), regulate key biological processes, including growth factor signaling and cell adhesion. Importantly, HSPGs serve as co-receptors for a number of signaling ligands such as fibroblast growth factors (FGFs), bone morphogenetic proteins (BMPs), Wnt/Wingless (Wg) and Hedgehog family members, playing critical roles in development and tumorigenesis (Bishop et al. 2007; Kirkpatrick and Selleck 2007; Li and Kusche-Gullberg 2016).
HS biosynthesis is a complex, multi-step process (Lindahl and Li 2009; Kreuger and Kjellen 2012; Lindahl 2014; Li and Kusche-Gullberg 2016). After synthesis of a glucuronic acid–galactose–galactose–xylose tetrasaccharide linkage region and addition of an N-acetylglucosamine (GlcNAc) residue, the HS-copolymerase EXT1/EXT2 adds alternating units of glucuronic acid (GlcA) and GlcNAc to the growing HS chain. Concomitant with polymerization, the polysaccharide undergoes sequential modification events. The modification begins by N-deacetylation and N-sulfation of GlcNAc units catalyzed by HS N-deacetylase/N-sulfotransferases (NDSTs). This is followed by C5-epimerization of GlcA residues, converting them to iduronic acid (IdoA), catalyzed by HS C5-epimerase (Hsepi). O-sulfation then occurs at different positions of the hexuronic acid and GlcN units, catalyzed by O-sulfotransferases, including Hs2st, Hs6sts and Hs3sts. After these modifications in the Golgi compartment, HS can be further modified extracellularly by a family of extracellular HS 6-O endosulfatases, Sulfs, which remove a specific subset of 6-O-sulfate groups within HS chains (Dhoot et al. 2001; Ai et al. 2003, 2006). Heparanase, an endoglucuronidase that specifically cleaves HS chains, is expressed at low levels in most mammalian tissues (Zcharia et al. 2009; Li and Kusche-Gullberg 2016). Increased heparanase expression is observed in pathological conditions, such as in tumor progression and metastasis, inflammation and fibrosis. Since only a fraction of potential targets in the HS chain are modified at each modification step, these processes generate remarkable structural heterogeneity of the molecules. Thus, the Golgi and extracellular enzymes regulate the amount and pattern of sulfate groups, forming binding sites on HS for a variety of ligand proteins (Esko and Selleck 2002; Nakato and Kimata 2002). However, how HSPG biosynthesis is regulated in vivo and how specific modifications of HS selectively affect signaling events remain a major question.
Genetic studies using a model organism, Drosophila have helped define in vivo functions of HSPGs and HS modifying enzymes (Nakato and Li 2016). There are remarkable advantages in the Drosophila model to study the role of HSPGs in development. Drosophila has the complete set of HS biosynthetic and modifying enzymes found in mammalian species, with the exception of heparanase, and produces complex HS structures that are equivalent to mammalian HS (Nakato and Li 2016). Importantly, Drosophila has only one gene for each of the enzymes in HS biosynthesis, which overcomes the complexity of genetic redundancy. Furthermore, a number of genetic tools (mutations, RNAi transgenic animals and overexpression constructs) for a complete set of genes of the HS biosynthetic machinery have been generated. These tools in combination with sophisticated molecular genetic techniques in this model enable us to manipulate HSPGs in vivo in a temporally and spatially controlled manner (Kamimura et al. 2011; Takemura and Nakato 2015). Using these tools, essential roles of HSPGs in many developmental processes have been defined, including morphogen gradient formation (Cadigan 2002; Yan and Lin 2009; Nakato and Li 2016), stem cell control (Hayashi et al. 2009; Dejima et al. 2011; Levings et al. 2016), regeneration (Takemura and Nakato 2017) and tumor formation (Levings and Nakato 2017).
The Drosophila model is also used to study a feedback regulatory network controlling HS biosynthesis known as “HS sulfation compensation”. This phenomenon was first recognized in a Chinese hamster ovary cell mutant strain, which lacks Hs2st activity (Bai and Esko 1996). This cell line produced HS with significantly higher levels of N- and 6-O-sulfation. This phenomenon was also observed in the Hs2st mouse null mutant model (Merry et al. 2001). HS purified from Hs2st−/− mouse embryonic fibroblasts did not have 2-O sulfate groups (as expected), but this loss was compensated by increased N- and 6-O sulfation. In Drosophila, Hs2st and Hs6st mutations induce compensatory increases in sulfation at 6-O- and 2-O-positions, respectively, restoring a wild-type net charge on HS in both genotypes (Kamimura et al. 2006). Analysis of Hs6st null (both maternally and zygotically) mutants revealed that 40% of these mutant embryos die with defects in FGF-dependent tracheal formation. The remaining mutant animals survive to the adult stage. During Drosophila development, this compensation rescues the FGF, Wg and BMP signaling pathways in vivo, ensuring the robust developmental systems (Kamimura et al. 2006; Dejima et al. 2013). These observations suggest that mutant HS retains some activities to form a signaling complex by providing proper 3D distribution of negative charge, although clearly at a lower rate compared to wild-type HS. However, the mechanism by which cells sense the lack of a specific sulfation event and induce a compensatory reaction is unknown.
Despite the many strengths of the Drosophila model for in vivo studies, information on Drosophila HS structure is somewhat limited. Drosophila HS has been analyzed biochemically by only one method, HS RPIP-HPLC disaccharide analysis (Toyoda et al. 2000). This technique determines the disaccharide composition of the polysaccharide. However, it has been difficult to determine other features of HS structure, such as molecular size, net charge, domain organization, N-sulfation pattern and the amount/distribution of IdoA residues, all of which are required for understanding the molecular mechanisms of HS–protein interactions. This is mainly due to the difficulty of metabolic radio-labeling of HS in vivo using Drosophila animals.
To fill this gap, it is ideal to establish an in vitro system to study HS biosynthesis using Drosophila cell lines. Recently, an efficient genetic method for generating Drosophila continuous cell lines of a given genotype has been developed (Simcox, Mitra et al. 2008; Simcox, Austin et al. 2008; Simcox 2013). The method uses expression of RasV12, a constitutively activated form of the oncogene Ras, to induce embryonic primary culture cells to progress to permanent cell lines. In this study, using this technique, we established novel cell lines mutant for four HS modifying enzymes: Hsepi, Hs2st, Hs6st and Sulf1. All these enzymes are known to be critical to the formation of ligand binding sites on HS as well as to be involved in the HS sulfation compensation (Kamimura et al. 2001, 2006; Kleinschmit et al. 2010, 2013; Wojcinski et al. 2011; Dejima, Takemura et al. 2013; Dejima, Kleinschmit et al. 2013). Morphological and immunological analyses of the established lines suggest that they are derived from mesodermal origin. Disaccharide analyses of HS from each mutant cell line reveal, in addition to the lack of respective enzymatic activity, compensatory increases of sulfation at different positions. We also show that these lines are useful for metabolic radiolabeling of glycosaminoglycan (GAG) for structural analysis. These tools will offer an excellent system to study the mechanisms of HS biosynthesis and HS–protein interactions. Thus, Drosophila genetics can be combined with HS structural analysis, making the Drosophia model highly unique and powerful to understand the structure–function relationship of HS.
Results
Establishment of novel Drosophila cell lines mutant for HS modifying enzymes
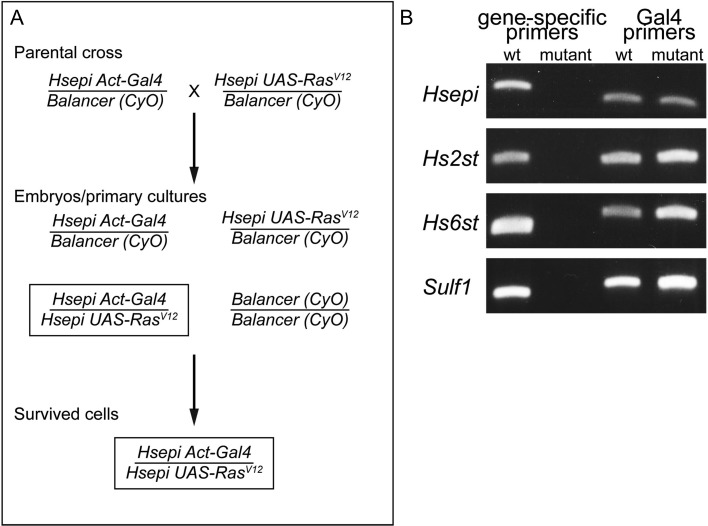
The Drosophila genome has single copies of homologs for Hsepi, Hs2st, Hs6st and Sulf1 genes. We have previously isolated Drosophila strains with null mutant alleles for each gene: Hsepid12 (Dejima et al. 2013), Hs2std267 (Kamimura et al. 2006), Hs6std770 (Kamimura et al. 2006) and Sulf1P1 (Kleinschmit et al. 2010). To establish mutant cell lines for these genes, we first generated recombinant chromosomes in which a null mutation for one of the four genes is combined with an Act5C-Gal4 transgene (a ubiquitous actin Gal4 driver) or a UAS-RasV12 (a constitutively active Ras) transgene (Figure 1A). Heterozygous flies carrying these chromosomes were crossed to obtain embryos with four different genotypes. These embryos with mixed genotypes were used to establish primary cultures according to the published protocol (Simcox, Mitra et al. 2008; Simcox, Austin et al. 2008; Simcox 2013). In case of Hsepi as an example shown in Figure 1A, homozygous mutant Hsepi embryos are the only cells expressing RasV12 in this culture (Act5C-Gal4/UAS-RasV12, boxed in Figure 1A). Since only RasV12-expressing cells can survive over passages, this cell line becomes homozygous null mutants for Hsepi. Thus, because of the proliferative advantage of the RasV12-expressing cells, this method does not require selection or cloning for a specific genotype. We used the same strategy for Hs2st, Hs6st and Sulf1 genes to establish mutant cell lines for respective genes.
Fig. 1.
Establishment of HS mutant cell lines. (A) Genetic crosses to obtain Hsepi mutant cell lines. A cross between Hsepi Act-Gal4 and Hsepi UAS-RasV12 will yield a mixture of embryos with four genotypes. Among these, only Hsepi homozygous mutant embryos bear both Act-Gal4 and UAS-RasV12 transgenes (boxed), thus expressing RasV12. After embryos with mixed genotypes are homogenized and primary cultures are plated, only Hsepi mutant cells survive during early passages. The same strategy was used for Hs2st, Hs6st and Sulf1 genes. (B) PCR analyses for genotyping. Genomic DNA samples were prepared from wild-type (wt) or indicated mutant cell lines (mutant), and used as templates. PCR primers were designed to specifically detect wild-type alleles for each of Hsepi, Hs2st, Hs6st and Sulf1 loci (left lanes). Control PCR was performed using primers that detect the Gal4 insertion in the genomic DNA (right lanes). In DNA samples isolated from mutant cell cultures, wild-type alleles were undetectable. This result shows that most cells in the established lines are homozygous for mutant alleles. This figure is available in black and white in print and in color at Glycobiology online.
To determine if we had obtained homozygous mutant cells, the genotype of each cell line was analyzed by PCR of genomic DNA using wild-type allele-specific primers (the sequences are described in “Materials and Methods”). The primer sequences are designed to specifically detect wild-type alleles for the genomic region of each gene, but not mutant alleles. We found that wild-type alleles became virtually undetectable by PCR after a few passages (4 weeks of culture), confirming the establishment of null mutant cell lines for these genes (Figure 1B, left lanes). All DNA samples show a band for Gal4 sequence in control PCR (Figure 1B, right lanes), consistent with the idea that all cell lines are heterozygous for Act-Gal4. Hereafter, the mutant cell lines are simply called Hsepi, Hs2st, Hs6st or Sulf1 cells, respectively. A previously established line, Ras-RMCE, which has Act5C-Gal4/UAS-RasV12 but no other mutation, was used as a control and referred to as “wild-type”.
Cell type of origins of the mutant cell lines
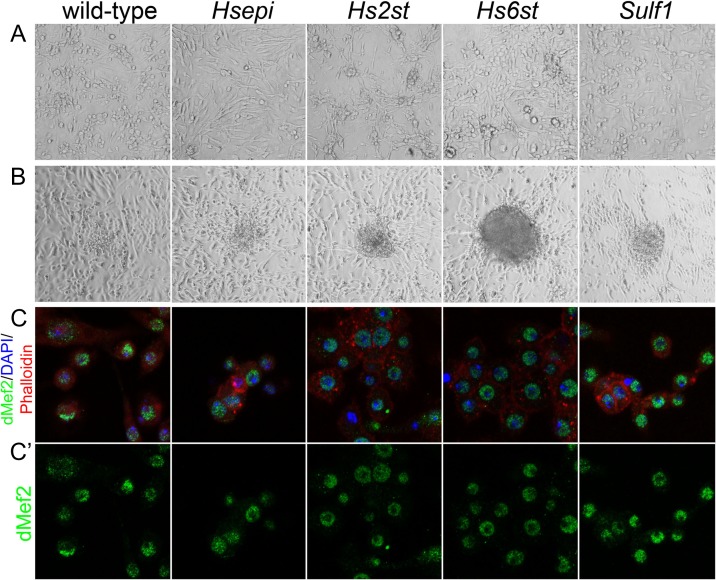
In this RasV12-dependent immortalization method, we do not select any specific cell types. For some reason, however, most cell lines established in previous studies were reported to be spindle-shaped cells of mesodermal origin (Simcox, Mitra et al. 2008; Simcox, Austin et al. 2008), suggesting the selective advantage of this cell type in the current protocol. This also appeared to be the case for our mutant cell lines. Cells looked morphologically heterogeneous for the first 30 days. However, after 3–4 rounds of passages, all lines showed spindle-shaped cells (Figure 2A), indistinguishable between different genotypes.
Fig. 2.
Morphological and immunological characterizations of the HS mutant cell lines. (A and B) Morphology of wild-type cells and established cell lines mutant for indicated genes. All cell lines show spindle-shaped morphology (A) and frequently form cell clumps characteristic of transformed cells (B). (C and C′) Confocal images showing the cell lines stained with anti-dMef2. High levels of nuclear dMef2 (green) were detected in all cell lines, suggesting they are of mesodermal origin. Nucleus and cell cortex are stained with DAPI (blue) and Phalloidin (red), respectively. Images in C′ show only green channel (dMef2) of the corresponding images in C. This figure is available in black and white in print and in color at Glycobiology online.
In addition to cell shape, growth behavior was also similar between the lines, which frequently form cell clumps on the top of adhered cells, piling up to form foci (Figure 2B). These cell clumps can be detached from the plate by pipetting without trypsin treatment. After re-seeding onto a new plate, the cells grow on the plate surface as spindle-shaped cells. These behaviors are characteristic of transformed cells, confirming the establishment of immortal cell lines.
To analyze the cellular origin of the established mutant cell lines, the cells were stained with a panel of antibodies. Staining with anti-dMef2 antibody, an established mesodermal marker, showed a high level of dMef2 expression in the nucleus of wild-type cells (Figure 2C and C′), confirming the previous observations (Simcox, Mitra et al. 2008; Simcox, Austin et al. 2008). We found that all established mutant cell lines also express nuclear dMef2 (Figure 2C and C′). No obvious signals were detected with anti-E-cadherin (an epithelial marker) and 22C10 (anti-Futsch, a neuronal marker) (data not shown). Together, these results indicated that the generated HS mutant cell lines are spindle-shaped cells of mesodermal origin, similar to most cell lines established by this protocol.
Disaccharide structures of HS from the mutant cell lines
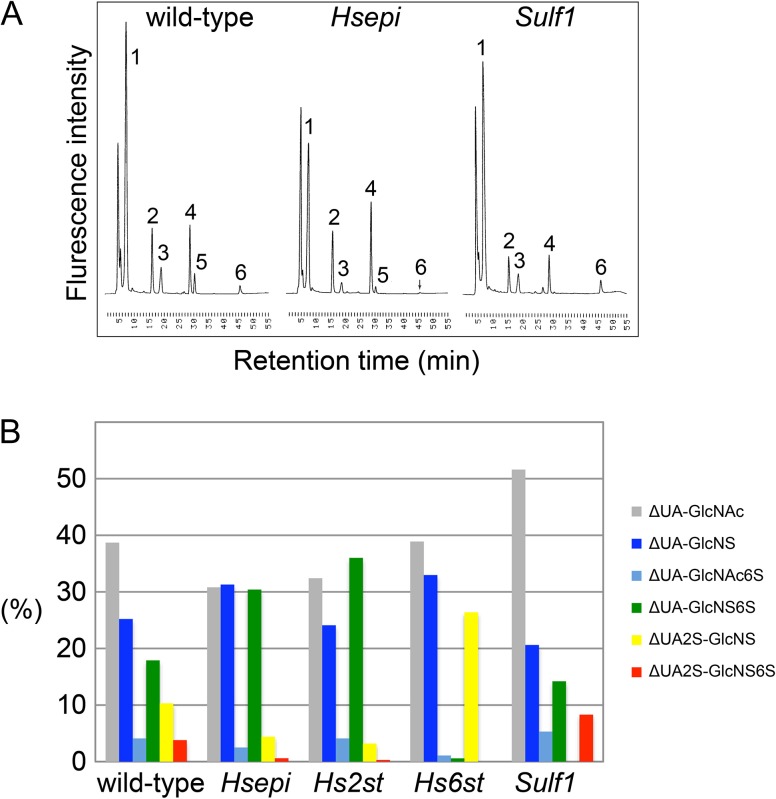
To determine the effects of the mutations on HS sulfation patterns, disaccharide analysis of HS isolated from each mutant cell line was performed. Briefly, HS samples isolated from wild-type, Hsepi, Hs2st, Hs6st and Sulf1 cells were completely digested into disaccharides by heparitinases. The resultant disaccharide species were separated and quantified by high performance liquid chromatography. HS isolated from the wild-type cell line (Ras-RMCE) showed a disaccharide composition closely related to that of Drosophila wild-type animals reported previously (Fig. S1; Toyoda et al. 2000).
Disaccharide composition of HS from Hsepi mutant cells showed a substantial reduction of 2-O-sulfate groups (Table I and Figure 3). This is caused by strong preference of Hs2st for IdoA as a substrate (Li et al. 2003; Dejima et al. 2013). The reduction of 2-O-sulfate-containing disaccharide units was accompanied by a compensatory increase in N- and 6-O-sulfate groups (ΔUA-GlcNS and ΔUA-GlcNS6S units). As a result, the proportion of non-sulfated disaccharide species (ΔUA-GlcNAc; 30.8%) decreased compared to wild-type (38.7%). Thus, this disaccharide pattern suggests that HS from Hsepi mutant cells has a higher charge density than that of wild-type, which is evidenced by analysis on ion-exchange chromatography (below). Total amount of HS and CS obtained from wild-type and Hsepi mutant cells is shown in Table II. We detected no major difference in the ratio of HS/CS and the disaccharide compositions of CS between these cell lines.
Table I.
Disaccharide analyses of HS from Hsepi, Hs2st, Hs6st and Sulf1 mutant cell lines
| HS (unsaturated disaccharide, %) | ||||||
|---|---|---|---|---|---|---|
| NAc | NS | NAc6S | NS6S | 2SNS | 2SNS6S | |
| Wild-type | 38.7 | 25.2 | 4.1 | 17.9 | 10.3 | 3.8 |
| Hsepi | 30.8 | 31.3 | 2.5 | 30.4 | 4.4 | 0.6 |
| Hs2st | 32.4 | 24.1 | 4.1 | 36.0 | 3.2 | 0.3 |
| Hs6st | 38.9 | 33.0 | 1.1 | 0.6 | 26.4 | N.D. |
| Sulf1 | 51.6 | 20.6 | 5.3 | 14.2 | N.D. | 8.3 |
Disaccharide composition of HS is shown for each respective genotype. The values are given as mol% of total disaccharides, and represent mean ± S.D. from three independent experiments. NAc, ΔUA-GlcNAc; NS, ΔUA-GlcNS; NAc6S, ΔUA-GlcNAc6S; NS6S, ΔUA-GlcNS6S; 2SNS, ΔUA2S-GlcNS; and 2SNS6S, ΔUA2S-GlcNS6S. N.D., not detectable. All results are shown as the average values from at least duplicate experiments. Graphical depiction of this result is shown in Figure 3B.
Fig. 3.
HS disaccharide profiling of wild-type and mutant cell lines. (A) Representative HPLC chromatograms of HS samples from wild-type (left), Hsepi (middle) and Sulf1 (right) cell lines. Each peak represents ΔUA-GlcNAc (1); ΔUA-GlcNS (2); ΔUA-GlcNAc6S (3); ΔUA-GlcNS6S (4); ΔUA2S-GlcNS (5); and ΔUA2S-GlcNS6S (6). (B) Graphical depiction of disaccharide composition of HS from each respective genotype. Bar graphs show percentages of the following disaccharides: ΔUA-GlcNAc (gray); ΔUA-GlcNS (blue); ΔUA-GlcNAc6S (light blue); ΔUA-GlcNS6S (green); ΔUA2S-GlcNS (yellow); and ΔUA2S-GlcNS6S (red). Results are shown as the average values from at least duplicate experiments (n = 2 or 3). This figure is available in black and white in print and in color at Glycobiology online.
Table II.
Total amount of HS and CS, and the disaccharide compositions of CS from wild-type and Hsepi mutant cells.
| Genotype | HS total amount (μg/mg dry cell) | CS total amount (μg/mg dry cell) | CS unsaturated disaccharide | ||
|---|---|---|---|---|---|
| 0S | 4S | 6S | |||
| Wild-type | 0.214 | 0.206 | 46.0 | 47.0 | 7.0 |
| Hsepi | 0.162 | 0.185 | 37.0 | 53.8 | 9.2 |
Similar to Hsepi mutant cells, HS from Hs2st mutant cells showed a significant loss of 2-O-sulfate-containing disaccharide units (Table I and Figure 3B). However, the compensation for reduced 2-O sulfation seems to be quite different in Hs2st mutant cells. Here, disulfated disaccharides containing N-sulfate and 6-O-sulfate groups (ΔUA-GlcNS6S) are substantially increased while the relative amount of disaccharides containing N-sulfate groups only (ΔUA-GlcNS) does not change. This is distinctly different from HS of Hsepi mutant cells, which bears significantly increased levels of both ΔUA-GlcNS and ΔUA-GlcNS6S units. This observation suggests an increased preference of Hs6st for N-sulfated disaccharides in the absence of Hs2st. Although we do not know the molecular basis for this difference, these observations are consistent with previous reports in Hsepi−/− mice (Li et al. 2003) and mutant flies (Kamimura et al. 2006; Dejima et al. 2013).
Disaccharide analysis of HS from Hs6st mutant cells revealed a significant reduction of 6-O-sulfate groups, as expected, leading to a loss of the tri-sulfated disaccharide unit (ΔUA2S-GlcNS6S) (Table I and Figure 3B). A massive elevation of the disaccharides, ΔUA-GlcNS and ΔUA2S-GlcNS, was observed, as reported in Hs6st mutant flies (Kamimura et al. 2006).
The disaccharide profile of Sulf1 mutant cells revealed abnormally high levels of the tri-sulfated disaccharide unit ΔUA2S-GlcNS6S (Table I and Figure 3). In addition, the ΔUA2S-GlcNS disaccharide was undetectable. On the other hand, there was no significant change in the amount of the 6-O-sulfated disaccharides, ΔUA-GlcNAc6S and ΔUA-GlcNS6S. These observations are consistent with the substrate specificity of Sulf1, which preferentially removes 6-O-sulfate groups from the tri-sulfated disaccharide units of HS (Ai et al. 2003, 2006; Kleinschmit et al. 2013).
Overall, the disaccharide profiles of HS from all cell lines closely reflect published in vivo data obtained from respective null mutant animals in all genotypes (Fig. S2; Kamimura et al. 2006; Kleinschmit et al. 2010; Dejima et al. 2013). We detected very small amounts of 2-O-sulfate-containing and 6-O-sulfate-containing units from Hs2st and Hs6st mutant cell lines, respectively. Their presence is probably due to residual enzyme activities derived from heterozygous cells remaining in the cultures. To test this idea, we compared disaccharide structures of HS samples from different passage points after the Hs2st mutant primary culture was seeded (Fig. S3). At an earlier passage (Passage #2), the mutant HS contained minor portions of 2SNS (4.2%) and 2SNS6S (0.7%) units. After cells had gone through additional passages (Passage #5), the 2SNS unit was reduced to 2.1% and the 2SNS6S unit became undetectable. This latter disaccharide composition is closely similar to that obtained from Hs2st null mutant animals. This observation supports the idea that these minor components were derived from heterozygous cells and that these residual activities will disappear after further passages. It also shows that the levels of these residual activities decrease during early passages (Passages #2–5), confirming that this methodology can establish homozygous mutant cell lines without selection or cloning.
Structural analysis of HS from wild-type Ras-RMCE cell line
One advantage of mutant cell lines over mutant animals is to obtain metabolically radiolabeled HS specimens which can be used for HS molecular structural analyses. One obstacle to obtain 35S-labeled GAGs from insect cells, however, is a high sulfate content in tissue culture media for insect cell lines. In general, these media, including Schneider’s insect medium and Shields and Sang M3 insect medium, contain extremely high levels of sulfate (e.g., 15.0 mM MgSO4 in M3 medium) (Schneider 1964), which makes it difficult to metabolically label the cells with 35S-sulfate. We therefore tested if we could radio-label Drosophila tissue culture cells using a mammalian cell culture medium (RPMI 1640 Medium, GlutaMAX™ Supplement, HEPES, ThermoFisher Scientific), which contains a lower level of sulfate (0.41 mM). We could successfully label GAG chains of S2 cells by incubating the cells with 35S-Na2SO4 for 16 h (data not shown). However, although the cells cultured in this medium looked healthy for a few days, they stopped dividing after a longer period of culture (>10 days), demonstrating that this medium was not optimal for the cells.
We therefore used a custom-ordered, M3 insect medium without MgSO4 (Life Technologies), to which we added a minimal amount of MgSO4 (final conc. 0.15 mM) (Kasevayuth and Yanagishita 2004). The growth and health of multiple Drosophila cell lines in this medium were indistinguishable from those using intact M3 medium (data not shown). Using these conditions, wild-type cell lines were incubated in the presence of 35S-sulfate for 6 h, and metabolically labeled macromolecules were isolated from conditioned media and cell lysates as described in “Materials and Methods” (Escobar Galvis et al. 2007; Jia et al. 2009; Dagalv et al. 2015).
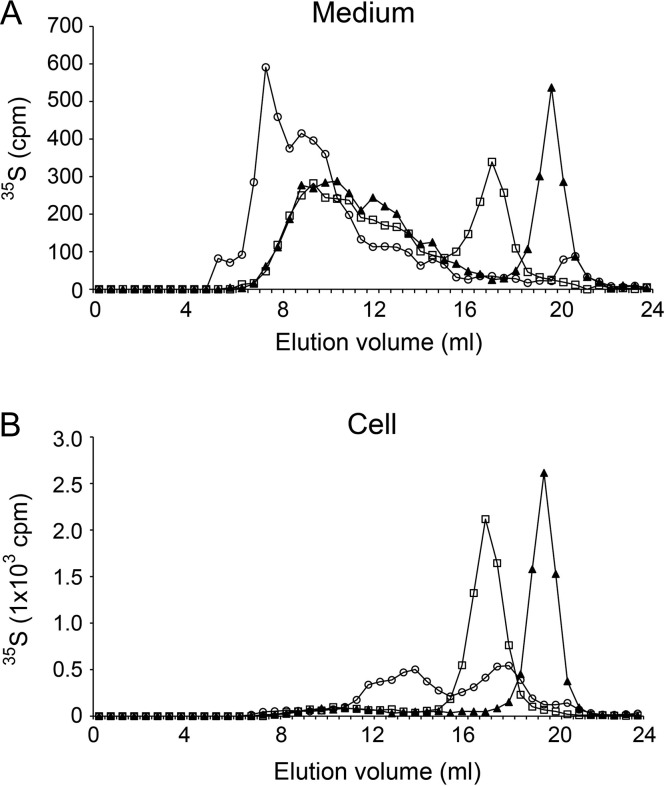
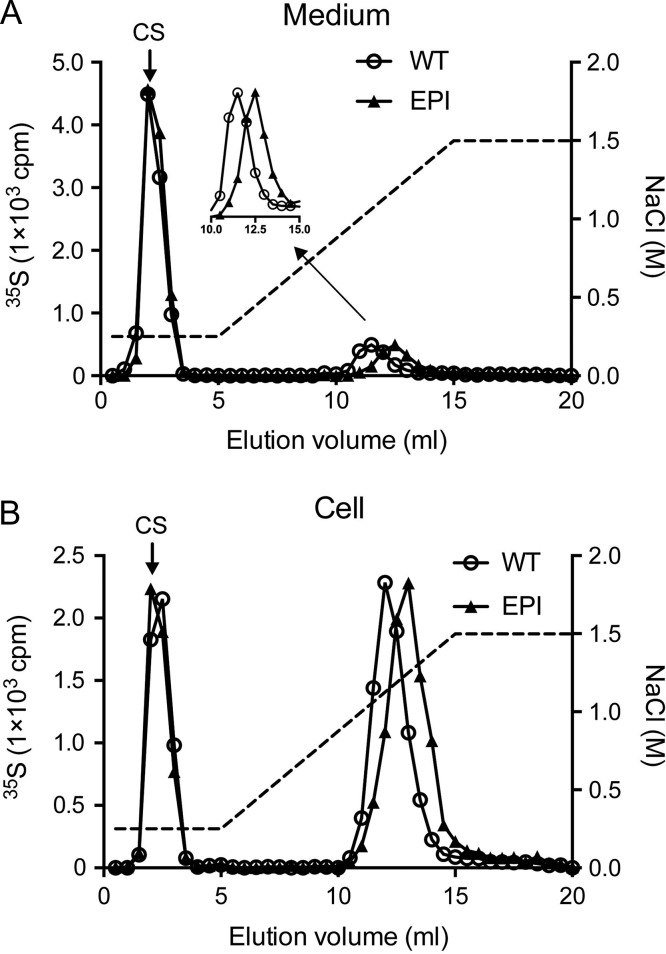
After purification by ion exchange chromatography, 35S-labeled macromolecules from cell lysate and medium of wild-type cells were analyzed by gel chromatography on a Superose 6 column (Figure 4). More than 90% of the 35S-labeled macromolecules isolated from cell fraction were susceptible to treatment with nitrous acid, demonstrating their HS nature. The untreated 35S-labeled cell lysate macromolecules were eluted in two peaks, where the first contained proteoglycans (susceptible to alkali treatment) while the second peak probably represented released HS-chains. The elution position of the single peak of 35S-labeled HS-chains obtained after alkali treatment corresponds to an apparent molecular weight of 10 kDa, calculated based on comparison with elution positions of standards of known molecular weight (Deligny et al. 2016). These HS-chains are considerably smaller than their mammalian counterparts. This is consistent with a previous observation that HS isolated from other Drosophila cell lines, Kc and S2 cells, was approximately 14 kDa (Kasevayuth and Yanagishita 2004). 35S-labeled HS-chains of similar size were also detected in the culture medium after alkali treatment (Figure 4A). However, in the medium only 35% of the 35S-macromolecules were HS (susceptible to treatment with nitrous acid). The remaining 65% were chondroitin sulfate (CS) proteoglycans and GAGs of higher apparent molecular weight.
Fig. 4.
Gel chromatography on Superose 6 of 35S-labeled proteoglycans/glycosaminoglycans from wild-type cells purified by DEAE ion exchange chromatography. Samples obtained from medium (A) or cell lysate (B) were analyzed directly (open circle), after alkali treatment (open square), or after treatment with both alkali and nitrous acid at pH 1.5 (filled triangle). Fractions of 0.5 mL were collected and analyzed by scintillation counting. This figure is available in black and white in print and in color at Glycobiology online.
Structural analysis of HS from Hsepi mutant cell line
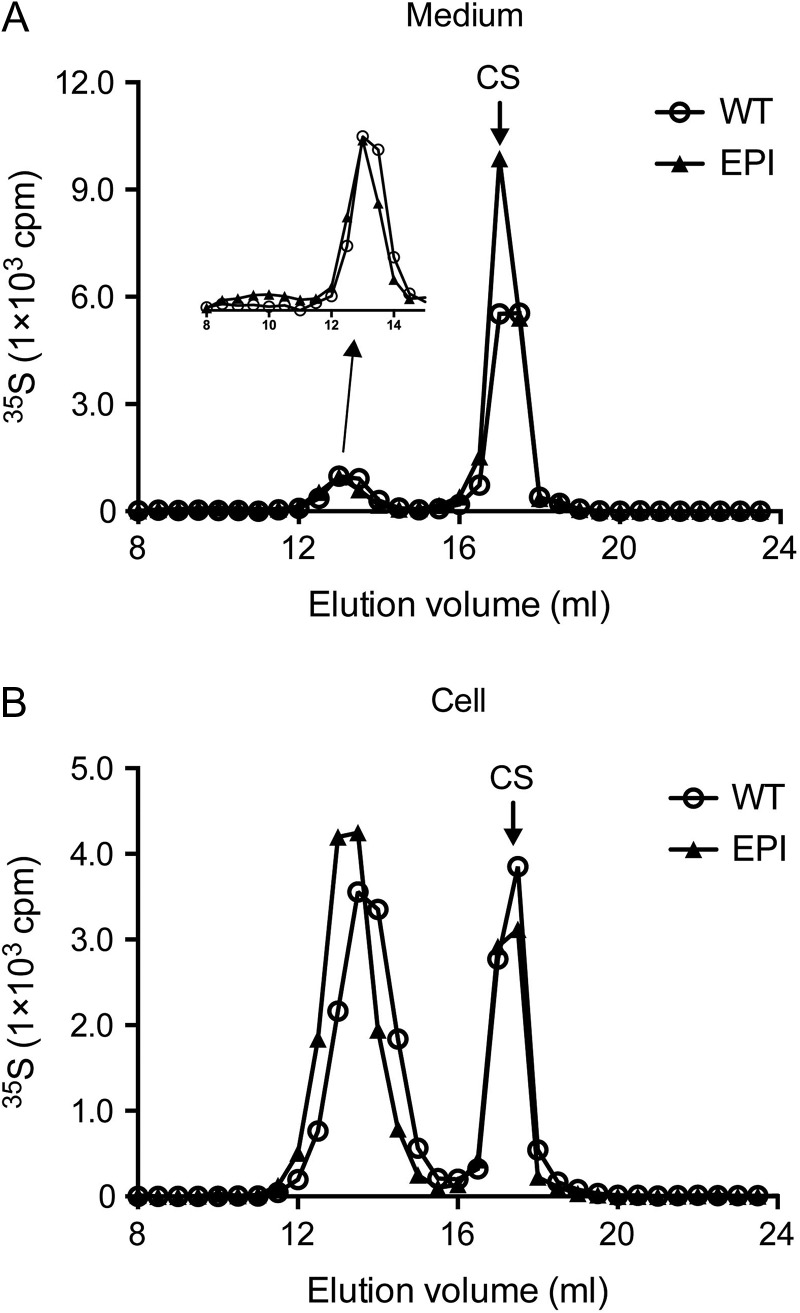
One of the cell lines, the Hsepi mutant cells, were further characterized to determine if the established mutant cell lines could be used as tools to study molecular phenotypes of biosynthesis enzyme deficiencies. In this experiment, we cultured wild-type and Hsepi mutant cells in the presence of 35S-sulfate for 24 h, and 35S-labeled GAGs were isolated from medium and cell fractions after alkali treatment (releasing the GAG chains) and chondroitinase treatment (degrading CS). Analysis by anion exchange chromatography revealed that HS isolated from both medium and cell fractions of Hsepi mutant cells was more retarded on a Mono Q column (Figure 5A and B), indicating a higher net negative charge density in Hsepi mutant HS compared to the wild-type polysaccharide. This resembles the phenotype of HS isolated from Hsepi mutant mouse tissues (Jia et al. 2009). As shown in Table I and Figure 3, HS from Hsepi mutant cells shows significantly elevated N- and 6-O-sulfation. Thus, the increased charge density predicted by disaccharide analysis was confirmed by the Mono Q analysis of the 35S-labeled HS.
Fig. 5.
Anion exchange chromatography of HS from wild-type and Hsepi cells. Metabolically 35S-labeled GAGs isolated from wild-type (WT, open circles) and Hsepi mutant (EPI, filled triangle) cells as described under “Materials and Methods” and were digested with Chondroitinase ABC prior to analysis. The samples (15,000 cpm each) from medium (A) and cell fractions (B) were applied to a Mono Q column eluted with a liner gradient of 0.25–1.5 M NaCl in NaAc buffer, pH 4.5. Eluted fractions (0.5 mL) were mixed with scintillation cocktail and assayed for radioactivity. “CS” indicates degraded chondroitin sulfate disaccharides. The dashed line indicates the salt gradient. This figure is available in black and white in print and in color at Glycobiology online.
It was recently shown that overexpression of mammalian Hsepi results in the production of longer HS chains, suggesting a possible role of this enzyme in chain length control (Fang et al. 2016). Therefore, we analyzed the HS samples from Hsepi cells on a Superose 12 column (Figure 6A and B). There was a small shift of the elution volume of the Hsepi mutant samples, which may indicate a slightly longer chain length of the mutant HS. We detected substantial shedding of CS in both cell lines, but it is more pronounced in the Hsepi cells.
Fig. 6.
Gel chromatography of HS chains from wild-type and Hsepi cells. GAG samples from metabolically 35S-labeled wild-type (WT, open circles) and Hsepi mutant (EPI, filled triangle) cells were prepared from medium (A) and cell (B) fractions. The samples (15,000 cpm each) were applied onto a Superose 12 column eluted in 50 mM HEPES buffer, pH 7.4, 1 M NaCl at a flow rate of 0.5 mL/min. Eluted fractions (0.5 mL) were mixed with scintillation cocktail and counted for radioactivity. “CS” indicates degraded chondroitin sulfate disaccharides. This figure is available in black and white in print and in color at Glycobiology online.
BMP signaling assay using Hs6st mutant cell line
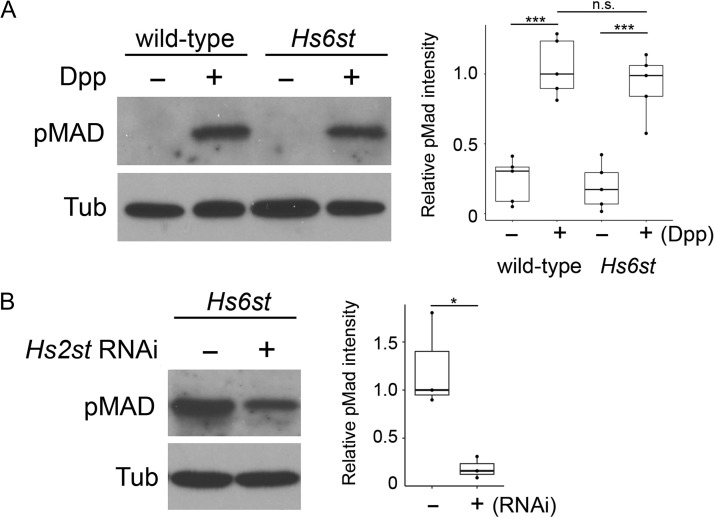
To examine if these cell lines are useful for biological assays, we performed a cell-based Decapentaplegic (Dpp) signaling assay (Akiyama et al. 2008; Dejima et al. 2011). Dpp is one Drosophila ortholog of BMPs and it is well established that Dpp signaling requires an HSPG co-receptor, Dally (Jackson et al. 1997; Fujise et al. 2003; Belenkaya et al. 2004; Akiyama et al. 2008; Hayashi et al. 2009; Dejima et al. 2011). Dpp signaling activity was monitored by phosphorylation of Mad protein, a direct readout of BMP signaling, using an antibody against phospho-Mad (pMad). Dpp-containing conditioned medium was prepared using Drosophila S2 cells as described in Materials and Methods.
We found that addition of relatively low levels of Dpp (equivalent to 2 × 10−9 M of a recombinant Dpp peptide, R&D Systems) can stimulate phosphorylation of Mad in wild-type cells (Figure 7A). In Hs6st mutant cells, this Dpp-mediated Mad phosphorylation was only partially impaired and the mutant cells retained significant activity to respond to Dpp (Figure 7A). This is consistent with a previous in vivo observation that the pMad levels were not significantly affected in the developing wing cells of Hs6st mutants (Dejima et al. 2013).
Fig. 7.
Dpp signaling assay using Hs6st mutant cell line. (A) Wild-type and Hs6st mutant cells were treated with Dpp-containing conditioned medium for 1 h at 25°C (+). Control cells (−) were treated with conditioned medium of S2 cells without a Dpp transgene. Dpp signaling was measured by immunoblot analysis of the cell lysate using anti-pMad antibody (pMad). α-Tubulin staining was used as an internal control (Tub). Quantification of the levels of pMad normalized with the α-Tubulin level is shown on the right. The intensity value in wild-type (+Dpp) sample was set to 1.0. The boxes represent the interquartile range and the line represents the median. The whiskers extend to the highest and lowest values. The results were obtained from five independent experiments. (B) The effect of Hs2st RNAi treatment on Dpp signaling in Hs6st cells. The pMad levels were measured two days after transfection of Hs2st dsRNA. Control cells (−) were treated in the same way without dsRNA. Quantification of Dpp signaling was obtained from three independent experiments. The intensity value in control sample (-dsRNA) was set to 1.0. P values are as follows: *P < 0.05; ***P < 0.001; N.S., not significant. This figure is available in black and white in print and in color at Glycobiology online.
This in vivo study also showed that the levels and patterns of pMad were severely disrupted in Hs2st; Hs6st double mutant cells, which suggested that HS sulfation compensation rescues Dpp signaling (Dejima et al. 2013). To analyze the contribution of the sulfation compensation to the moderate perturbation of Dpp signaling in Hs6st cells, we examined the effect of double-stranded (ds) RNAi knockdown of Hs2st in Hs6st mutant cells. We found that transfection of Hs2st dsRNA resulted in a substantial reduction in the level of pMad (Figure 7B). These results indicate that compensatory increase of 2-O sulfation rescues the ability of Hs6st mutant cells to respond to Dpp, consistent with the previous in vivo data (Dejima et al. 2013). Altogether, signaling profiles of these cell lines closely recapitulate previously known in vivo phenomena.
Discussion
The molecular mechanisms underlying regulation of HS biosynthesis and function remain largely unknown. Although all enzymes that polymerize and modify HS have been identified, it is still unknown how the size, charge and domain structures of HS are determined. Neither are the HS structures responsible for specific HS-dependent signaling and patterning events fully characterized. Addressing these questions requires better understanding of molecular basis for HS-ligand interactions (Xu and Esko 2014; Kjellen and Lindahl 2018). Over the last 2 decades, the Drosophila model has offered an excellent system to study how a specific change in a HSPG core-protein or HS modifying enzyme affects cellular and developmental phenomena (Nakato and Li 2016). On the other hand, structural information of Drosophila HS is still limited. Therefore, although we have accumulated a wealth of knowledge regarding morphological phenotypes of Drosophila mutants of HSPG/HS modifying enzyme genes, we have only been able to speculate about HS molecular phenotypes based on information from mammalian HS.
In this study, we established cell lines mutant for Hsepi, Hs2st, Hs6st and Sulf1, by immortalizing primary culture cells. In the EC rabbit endothelial cell line, a high level of overexpression of the EJ-ras gene resulted in reduced expression of a few HS modifying enzymes, leading to a significant decrease in O-sulfation (Lopes et al. 2006). This was not the case in the Drosophila mutant cell lines. HS disaccharide compositions of the mutant cells closely reflect published disaccharide patterns of HS from Drosophila animals with respective genotypes (Figures S1 and S2), showing no detectable effect of Ras-mediated transformation on the HS structures.
There are significant advantages to establish an in vitro system using Drosophila cells to analyze HS structures in detail. Most importantly, HS structural information obtained from these established Drosophila cell lines will be directly relevant to the in vivo analyses of Drosophila mutants. The established tools will allow us to link information regarding the roles of HS at molecular, cellular and animal levels. Indeed, a detailed picture of higher order structures of HS from these mutant cells would be extremely informative to define the range of HS structures that can (and cannot) mediate signaling in vivo.
Comparing structures of HS from mammals and Drosophila will provide clues to establish the fundamental properties of HS (both in structure and function). For example, the fact that HS chains isolated from Drosophila cells are significantly shorter than those of mammalian cells readily indicates that HS chains do not have to be as long as mammalian HS for its normal function in mediating cell signaling and regulating morphogenesis. A previous work conducted a structural analysis of HS prepared from wild-type Drosophila embryos (Kusche-Gullberg et al. 2012). In this study, HS was prepared from a large amount of specimen (15–30 g of wild-type embryos) and its domain organization was analyzed by selective deamination cleavage. The authors proposed that Drosophila HS from wild-type embryos shows unique structures with an extended NS-domain and the absence of internal N-acetylated sequences, similar to “low-sulfated heparin”. Future structural studies of HS from the established mutant cell lines will provide important information on the biological significance of the HS domain structures and fundamental rules of structure–function relationship of HS.
Mammalian genomes bear single genes for Hsepi and Hs2st but multiple genes for other classes of HS modifying enzymes. For example, three Hs6st genes (Hs6st-1, -2 and -3) show different expression patterns during development as well as different substrate preferences (Habuchi and Kimata 2010). Phenotypic comparison of Hs6st-1−/−, Hs6st-2−/− and double knockout mice indicated that the two genes have partially redundant functions in addition to their gene-specific roles. For Sulfs, two homologs (Sulf1 and Sulf2) are found in vertebrates. Although mouse Sulfs show differential expression patterns, they redundantly regulate neural innervation and enteric glial formation in the esophagus (Ai et al. 2007) and muscle regeneration (Langsdorf et al. 2007). Thus, these vertebrate paralogs of HS modifying enzymes have both paralog-specific functions and partially redundant roles. In contrast, there is no or minimal genetic redundancy in HS biosynthetic/modifying genes in Drosophila. Therefore, the biochemical phenotypes of these mutant cell lines represent the effects of a complete lack of each respective class of enzyme. This is reflected by sharp and clear HS sulfation compensation in mutants.
A previous in vivo study showed that FGF-dependent tracheogenesis occurs normally in a fraction of Hs2st or Hs6st null mutants (Kamimura et al. 2006). Disaccharide analysis of HS isolated from the mutant flies showed that the loss of 2-O-sulfate groups was compensated by an increase of 6-O-sulfation, and vise versa. These results suggest that mutant HS in both genotypes, which has a normal charge density but lacks the correct sulfation pattern, retains the ability to mediate FGF signaling, although the signaling efficiency is diminished. Given that HS chains show conformational flexibility by bending and twisting, Hs2st or Hs6st mutant HS chains may still position sulfate groups at appropriate three-dimensional locations required for protein binding at a certain efficiency (Kjellen and Lindahl 2018). The non-zero but reduced functionality of the mutant HS suggests that such HS has (1) fewer protein binding sites, (2) binding sites with lower affinities, or both. However, these issues still remain to be elucidated. Data set for detailed structural features, ligand binding, and in vivo activities of diverse HS from the mutant cells (with abnormal sulfation sequences but residual activities) will provide mechanistic insights into these questions.
Recently, a library of mouse lung endothelial cell lines has been generated by systematic deletion of HS biosynthetic and modifying genes (Qiu et al. 2018). As the authors stated, studies using such a library will lead to a better understanding of the structure–function relationships of HS. Our Drosophila mutant cell library will not only add more variations of HS structures, but also provide significant information on the “structure-in vivo function” relationship of HS, by directly linking structural features to abundant knowledge on morphological phenotypes obtained in this model. In addition, the comparative studies using the mouse and Drosophila libraries will highlight the commonalities and species-specific differences of HS structure and functions. Furthermore, in vitro and in vivo analyses in Drosophila can be performed in parallel, which provides a powerful platform to study the mechanisms of HS functions in specific biological phenomena.
Materials and Methods
Fly strains
The detailed information for fly strains used is described in Flybase. All flies were maintained at 25°C. The following strains were used as null mutants for each gene to establish cell lines: Hsepid12 (Dejima et al. 2013), Hs2std267 (Kamimura et al. 2006), Hs6std770 (Kamimura et al. 2006) and Sulf1P1 (Kleinschmit et al. 2010).
Primary cultures and cell lines
The following genetic crosses were made to generate the respective mutant cell lines: act5C-Gal4 Hsepid12/CyO X UAS-RasV12Hsepid12/CyO for Hsepi mutant cells; act5C-Gal4 Hs2std267/CyO X UAS-RasV12Hs2std267/CyO for Hs2st mutant cells; act5C-Gal4 Hs6std770/TM6B Tb X UAS-RasV12Hs6std770/TM6B Tb for Hs6st mutant cells; act5C-Gal4 Sulf1P1/TM6B Tb X UAS-RasV12Sulf1P1/TM6B Tb for Sulf1 mutant cells. Embryos were collected overnight from each cross and primary cultures were generated from the embryos by following an established protocol (Simcox, Mitra et al. 2008; Simcox, Austin et al. 2008; Simcox 2013) with some modifications. Briefly, eggs were dechorionated with 50% bleach and the surface of the dechorionated embryos was sterilized by rinsing with 70% ethanol. Approximately 100 μL of the packed volume of the embryos was used for a primary culture, and were homogenized with three gentle strokes using a 5 mL glass homogenizer. The homogenate was centrifuged at 1000 × g for 2 min at room temperature. The pellet was washed with fresh medium two more times. The washed embryos were suspended in 3 mL of M3 medium and cultured in a T-flask at 25°C. After 2 weeks, the first subculture was made. Confluent primary cell cultures were split in half and grown to confluent again. The doubling time was approximately 24 h both in wild-type and mutant cell lines. There was no difference in the growth rate between genotypes.
To determine the genotypes of primary culture cells, we detected wild-type chromosomes by PCR using primer sets that do not detect each of mutant alleles: Hsepid12, Hs2std267, Hs6std770, Sulf1P1. For example, PCR using Primers 1 and 2 produces a band of 574 bp from wild-type chromosomes, but not from Hsepid12. Similarly, Primer sets 3/4, 5/6 and 7/8 were used to detect wild-type chromosome bands of indicated lengths for Hsepi, Hs2st, Hs6st and Sulf1 loci, respectively.
Primer 1: (wild-type allele shows a 574 bp band with Primer 2 for Hsepi locus): 5′- CATTTGCTCATCTGTATCGCAGT-3′
Primer 2: 5′- CGCATATACACGCCCAGAGT-3′
Primer 3: (wild-type allele shows a 520 bp band with Primer 4 for Hs2st locus): 5′-GAATTTTGGCATTGCTTGGT-3′
Primer 4: 5′-TATCAGGATCAGCCAGTGGG-3′
Primer 5: (wild-type allele shows a 408 bp band with Primer 6 for Hs6st locus): 5′-CTGAAAGCCGAGAGACAAGG-3′
Primer 6: 5′-AGTATCCAAATCCGATGATGC-3′
Primer 7: (wild-type allele shows a 460 bp band with Primer 8 for Sulf1 locus): 5′-AGGATTCGGCTCCCCAGTAT-3′
Primer 8: 5′-AAGTGTTGTCCAGCTCTCCG-3′
The following primers were used for control PCR reaction to detect Gal4 sequence (a 500 bp band).
Primer 9: 5′-AAGAAAAACCGAAGTGCGCC-3′
Primer 10: 5′-CGTTTTCAGGAAGGGCAAGC-3′
Immunostaining of mutant cell lines
Immunostaining of the established cell lines was performed as previously described (Simcox 2013) with some modifications. Clean coverslips (12 mm Round, Warner Instruments) were prepared by washing in HCl overnight. Cells were seeded on the coverslips placed in a 35 mm culture dish and incubated at 25°C overnight. After the cells were washed once with PBS, the cells were fixed for 25 min in 4% paraformaldehyde. After washes with PBT (0.05% Triton X-100 in PBS), the fixed cells were blocked in 5% normal goat serum in PBT for 1 h at room temperature. Primary antibodies were applied in a 20 μL solution onto a coverslip and incubated for 2 h at 25°C in a moist chamber. The cells were washed with PBT, followed by incubation with secondary antibodies and Phalloidin-Alexa Fluor 633 (ThermoFisher Scientific). After extensive washes with PBT, the cells were mounted using VECTASHIELD (Vector Laboratory).
The following primary antibodies were used: rabbit anti-dMef2 (1:1000, a gift from B. Paterson), rat anti-E-cadherin (Developmental Studies Hybridoma Bank), mouse 22C10/Futsch (Developmental Studies Hybridoma Bank). Secondary antibodies were from the AlexaFluor series (1:500; ThermoFisher Scientific).
Disaccharide analysis
HS isolation and disaccharide composition analysis were carried out as previously described (Kamimura et al. 2006; Kleinschmit et al. 2010; Dejima et al. 2013). Approximately 5 mg of cells was used to isolate HS. The HS sample was digested with a heparitinase mixture (Seikagaku), and the resulting disaccharide species were separated using reversed-phase ion-pair chromatography. The effluent was monitored fluorometrically for post-column detection of HS disaccharides (Toyoda et al. 2000).
Metabolic 35S-labeling of HS
For metabolic labeling, cells were cultured in M3 medium without sulfate supplemented with 10% FBS and 0.15 mM MgSO4 to 95% confluency. After adding 50 μCi/ml Na235SO4 (specific activity, 1500 Ci/mmol; PerkinElmer Life Sciences), the cells were maintained in the same medium for the indicated periods of time.
Characterization of 35S-labeled HSPGs and HS from wild-type cells
After 6 h of incubation with 35S-sulfate, the medium was collected and the cells were washed with PBS followed by solubilization in 50 mM Tris–HCl, pH 7.4, 0.15 M NaCl, 1% Triton X-100 and Roche complete protease inhibitor cocktail at 4°C for 1 h under gentle shaking. 35S-Macromolecules in medium and cell lysate fractions were then purified by DEAE-ion exchange chromatography followed by gel chromatography on Superose 6 before or after treatment with alkali and HNO2 at pH 1.5 as previously described (Dagalv et al. 2015).
Comparison of 35S-labeled HS from Hsepi and wild-type cells
After 24 h of incubation with 35S-sulfate, the medium was collected and the cells were washed with PBS followed by lysis in extraction buffer (50 mM Tris–HCl, pH 7.4, 1% Triton X-100) at 4°C for 1 h under gentle shaking. The medium fractions were then mixed with equal volumes of 2 × extraction buffer (100 mM Tris–HCl, pH 7.4, 2% Triton X-100). Both cell lysate and medium fractions were then treated with NaOH at a final concentration of 0.5 M on ice overnight to release the GAG chains. After neutralization and centrifugation, the supernatants were applied to 2 ml DEAE-Sephacel columns (GE Healthcare), equilibrated with the extraction buffer. Following extensive washing with 50 mM NaAc, pH 4.5, 0.25 M NaCl, bound material was eluted with 50 mM NaAc, pH 4.5, 1.5 M NaCl. Eluates were desalted on a PD-10 column (GE Healthcare) and lyophilized. Products were dissolved in 0.05 M Tris–HCl, pH 8.0, 30 μM NaAc, 0.1 mg BSA/ml and treated with chondroitinase ABC (25 mU; Seikagaku) at 37°C overnight.
Dpp signaling assay
Cell-based Dpp signaling assay was performed as described previously (Akiyama et al. 2008; Dejima et al. 2011). Dpp-containing conditioned medium was prepared from Drosophila S2 cells expressing Dpp (transfected with pAW-HA-dpp). A recombinant Dpp peptide (R&D systems) was used to estimate Dpp amount in the conditioned media. Control conditioned medium was prepared from Drosophila S2 cells without pAW-HA-dpp transfection.
After wild-type and Hs6st cells were incubated with the Dpp-containing conditioned medium at 25°C for 1 h, cells were lysed in SDS sample buffer. Dpp signaling was assayed by immunoblot analysis using rabbit anti-pMad antibody (EP823Y, 1:2,000, Epitomics). α-Tubulin was stained with mouse anti-α-Tubulin antibody (DM1A, Sigma) and used as an internal control. The secondary antibodies used were anti-rabbit or mouse IgG conjugated with horseradish peroxidase (Sigma) and detected by ECL-Plus (GE Healthcare Life Sciences). Densitometric analysis for anti-pMad and anti-α-Tubulin staining was performed using ImageJ software.
For Hs2st RNAi treatment of Hs6st cells, the template for double-stranded RNA (dsRNA) synthesis was prepared by PCR with Hs2st-specific primers containing the T7 promoter sequence at the 5′ end (Primers 11 and 12 described below; underlined sequences are derived from Hs2st cDNA). The region corresponding to 261–560 bp of Hs2st cDNA was targeted. Double-stranded RNA was synthesized using a Megascript Kit (Ambion). A 500 ng of dsRNA was introduced into the cells using Effectene transfection reagents (Qiagen) in M3 medium. Control cells were treated in the same way without dsRNA. Dpp signaling was assayed 2 days after the dsRNA treatment.
Primer 11: 5′-TAATACGACTCACTATAGGGTTCAAGCCGCTTTCCAAG-3′
Primer 12: 5′-TAATACGACTCACTATAGGGCCATGTGGCCGTGATAAAG-3′
Supplementary Material
Acknowledgements
We are grateful to A. Simcox, B. Paterson, M. O’Connor, the Developmental Studies Hybridoma Bank, and the Bloomington Stock Center for fly stocks and reagents. We thank T. Su, S. Baker, M. Takemura and S. Takada for helpful discussions.
Funding
National Institute of General Medical Sciences [R01 GM115099 and R35 GM131688 to H.N.]; and VINNOVA (Sweden) [the Mobility Grant and to H.N and J.P.L.].
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Abbreviations
Dpp, decapentaplegic; GAG, glycosaminoglycan; GlcA, glucuronic acid; GlcNAc, N-acetylglucosamine; Hs2st, heparan sulfate 2-O sulfotransferase; Hs6st, heparan sulfate 6-O sulfotransferase; HS, heparan sulfate; Hsepi, glucuronyl C5 epimerase; HSPG, heparan sulfate proteoglycan; Sulf1, heparan sulfate endosulfatase 1.
References
- Ai X, Do AT, Kusche-Gullberg M, Lindahl U, Lu K, Emerson CP Jr. 2006. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J Biol Chem. 281:4969–4976. [DOI] [PubMed] [Google Scholar]
- Ai X, Do AT, Lozynska O, Kusche-Gullberg M, Lindahl U, Emerson CP Jr. 2003. QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J Cell Biol. 162:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai X, Kitazawa T, Do AT, Kusche-Gullberg M, Labosky PA, Emerson CP Jr. 2007. SULF1 and SULF2 regulate heparan sulfate-mediated GDNF signaling for esophageal innervation. Development. 134:3327–3338. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Kamimura K, Firkus C, Takeo S, Shimmi O, Nakato H. 2008. Dally regulates Dpp morphogen gradient formation by stabilizing Dpp on the cell surface. Dev Biol. 313:408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Esko JD. 1996. An animal cell mutant defective in heparan sulfate hexuronic acid 2-O-sulfation. J Biol Chem. 271:17711–17717. [DOI] [PubMed] [Google Scholar]
- Belenkaya TY, Han C, Yan D, Opoka RJ, Khodoun M, Liu H, Lin X. 2004. Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell. 119:231–244. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD. 2007. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 446:1030–1037. [DOI] [PubMed] [Google Scholar]
- Cadigan KM. 2002. Regulating morphogen gradients in the Drosophila wing. Semin Cell Dev Biol. 13:83–90. [DOI] [PubMed] [Google Scholar]
- Dagalv A, Lundequist A, Filipek-Gorniok B, Dierker T, Eriksson I, Kjellen L. 2015. Heparan sulfate structure: Methods to study N-sulfation and NDST action. Methods Mol Biol. 1229:189–200. [DOI] [PubMed] [Google Scholar]
- Dejima K, Kanai MI, Akiyama T, Levings DC, Nakato H. 2011. Novel contact-dependent bone morphogenetic protein (BMP) signaling mediated by heparan sulfate proteoglycans. J Biol Chem. 286:17103–17111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejima K, Kleinschmit A, Takemura M, Choi PY, Kinoshita-Toyoda A, Toyoda H, Nakato H. 2013. The role of Drosophila heparan sulfate 6-o-endosulfatase in sulfation compensation. J Biol Chem. 288:6574–6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejima K, Takemura M, Nakato E, Peterson J, Hayashi Y, Kinoshita-Toyoda A, Toyoda H, Nakato H. 2013. Analysis of Drosophila glucuronyl C-5 epimerase: Implications for developmental roles of heparan sulfate sulfation compensation and 2-O sulfated glucuronic acid. J Biol Chem. 288:34384–34393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deligny A, Dierker T, Dagalv A, Lundequist A, Eriksson I, Nairn AV, Moremen KW, Merry CLR, Kjellen L. 2016. NDST2 (N-Deacetylase/N-Sulfotransferase-2) enzyme regulates heparan sulfate chain length. J Biol Chem. 291:18600–18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhoot GK, Gustafsson MK, Ai X, Sun W, Standiford DM, Emerson CP Jr. 2001. Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 293:1663–1666. [DOI] [PubMed] [Google Scholar]
- Escobar Galvis ML, Jia J, Zhang X, Jastrebova N, Spillmann D, Gottfridsson E, van Kuppevelt TH, Zcharia E, Vlodavsky I, Lindahl U et al. 2007. Transgenic or tumor-induced expression of heparanase upregulates sulfation of heparan sulfate. Nat Chem Biol. 3:773–778. [DOI] [PubMed] [Google Scholar]
- Esko JD, Selleck SB. 2002. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 71:435–471. [DOI] [PubMed] [Google Scholar]
- Fang J, Song T, Lindahl U, Li JP. 2016. Enzyme overexpression—An exercise toward understanding regulation of heparan sulfate biosynthesis. Sci Rep. 6:31242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujise M, Takeo S, Kamimura K, Matsuo T, Aigaki T, Izumi S, Nakato H. 2003. Dally regulates Dpp morphogen gradient formation in the Drosophila wing. Development. 130:1515–1522. [DOI] [PubMed] [Google Scholar]
- Habuchi H, Kimata K. 2010. Mice deficient in heparan sulfate 6-O-sulfotransferase-1. Prog Mol Biol Transl Sci. 93:79–111. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Kobayashi S, Nakato H. 2009. Drosophila glypicans regulate the germline stem cell niche. J Cell Biol. 187:473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SM, Nakato H, Sugiura M, Jannuzi A, Oakes R, Kaluza V, Golden C, Selleck SB. 1997. dally, a Drosophila glypican, controls cellular responses to the TGF-beta-related morphogen, Dpp. Development. 124:4113–4120. [DOI] [PubMed] [Google Scholar]
- Jia J, Maccarana M, Zhang X, Bespalov M, Lindahl U, Li JP. 2009. Lack of L-iduronic acid in heparan sulfate affects interaction with growth factors and cell signaling. J Biol Chem. 284:15942–15950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura K, Fujise M, Villa F, Izumi S, Habuchi H, Kimata K, Nakato H. 2001. Drosophila heparan sulfate 6-O-sulfotransferase (dHS6ST) gene. Structure, expression, and function in the formation of the tracheal system. J Biol Chem. 276:17014–17021. [DOI] [PubMed] [Google Scholar]
- Kamimura K, Koyama T, Habuchi H, Ueda R, Masu M, Kimata K, Nakato H. 2006. Specific and flexible roles of heparan sulfate modifications in Drosophila FGF signaling. J Cell Biol. 174:773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura K, Maeda N, Nakato H. 2011. In vivo manipulation of heparan sulfate structure and its effect on Drosophila development. Glycobiology. 21:607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasevayuth K, Yanagishita M. 2004. Catabolism of heparan sulfate proteoglycans in Drosophila cell lines. Biochem Biophys Res Commun. 324:205–211. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick CA, Selleck SB. 2007. Heparan sulfate proteoglycans at a glance. J Cell Sci. 120:1829–1832. [DOI] [PubMed] [Google Scholar]
- Kjellen L, Lindahl U. 2018. Specificity of glycosaminoglycan–protein interactions. Curr Opin Struct Biol. 50:101–108. [DOI] [PubMed] [Google Scholar]
- Kleinschmit A, Koyama T, Dejima K, Hayashi Y, Kamimura K, Nakato H. 2010. Drosophila heparan sulfate 6-O endosulfatase regulates Wingless morphogen gradient formation. Dev Biol. 345:204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmit A, Takemura M, Dejima K, Choi PY, Nakato H. 2013. Drosophila heparan sulfate 6-O-endosulfatase Sulf1 facilitates wingless (Wg) protein degradation. J Biol Chem. 288:5081–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuger J, Kjellen L. 2012. Heparan sulfate biosynthesis: Regulation and variability. J Histochem Cytochem. 60:898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusche-Gullberg M, Nybakken K, Perrimon N, Lindahl U. 2012. Drosophila heparan sulfate, a novel design. J Biol Chem. 287:21950–21956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langsdorf A, Do AT, Kusche-Gullberg M, Emerson CP Jr., Ai X. 2007. Sulfs are regulators of growth factor signaling for satellite cell differentiation and muscle regeneration. Dev Biol. 311:464–477. [DOI] [PubMed] [Google Scholar]
- Levings DC, Arashiro T, Nakato H. 2016. Heparan sulfate regulates the number and centrosome positioning of Drosophila male germline stem cells. Mol Biol Cell. 27:888–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings DC, Nakato H. 2017. Loss of heparan sulfate in the niche leads to tumor-like germ cell growth in the Drosophila testis. Glycobiology. 28:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JP, Gong F, Hagner-McWhirter A, Forsberg E, Abrink M, Kisilevsky R, Zhang X, Lindahl U. 2003. Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking L-iduronic acid and in neonatal lethality. J Biol Chem. 278:28363–28366. [DOI] [PubMed] [Google Scholar]
- Li JP, Kusche-Gullberg M. 2016. Heparan sulfate: biosynthesis, structure, and function. Int Rev Cell Mol Biol. 325:215–273. [DOI] [PubMed] [Google Scholar]
- Lindahl U. 2014. A personal voyage through the proteoglycan field. Matrix Biol. 35:3–7. [DOI] [PubMed] [Google Scholar]
- Lindahl U, Li JP. 2009. Interactions between heparan sulfate and proteins—design and functional implications. Int Rev Cell Mol Biol. 276:105–159. [DOI] [PubMed] [Google Scholar]
- Lopes CC, Toma L, Pinhal MA, Porcionatto MA, Sogayar MC, Dietrich CP, Nader HB. 2006. EJ-ras oncogene transfection of endothelial cells upregulates the expression of syndecan-4 and downregulates heparan sulfate sulfotransferases and epimerase. Biochimie. 88:1493–1504. [DOI] [PubMed] [Google Scholar]
- Merry CL, Bullock SL, Swan DC, Backen AC, Lyon M, Beddington RS, Wilson VA, Gallagher JT. 2001. The molecular phenotype of heparan sulfate in the Hs2st-/- mutant mouse. J Biol Chem. 276:35429–35434. [DOI] [PubMed] [Google Scholar]
- Nakato H, Kimata K. 2002. Heparan sulfate fine structure and specificity of proteoglycan functions. Biochim Biophys Acta. 1573:312–318. [DOI] [PubMed] [Google Scholar]
- Nakato H, Li JP. 2016. Functions of heparan sulfate proteoglycans in development: Insights from drosophila models. Int Rev Cell Mol Biol. 325:275–293. [DOI] [PubMed] [Google Scholar]
- Qiu H, Shi S, Yue J, Xin M, Nairn AV, Lin L, Liu X, Li G, Archer-Hartmann SA, Dela Rosa M et al. 2018. A mutant-cell library for systematic analysis of heparan sulfate structure-function relationships. Nat Methods. 15:889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. 1964. Differentiation of larval Drosophila eye-antennal discs in vitro. J Exp Zool. 156:91–103. [DOI] [PubMed] [Google Scholar]
- Simcox A. 2013. Progress towards Drosophila epithelial cell culture. Methods Mol Biol. 945:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simcox AA, Austin CL, Jacobsen TL, Jafar-Nejad H. 2008. Drosophila embryonic ‘fibroblasts’: Extending mutant analysis in vitro. Fly (Austin Tex). 2:306–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simcox A, Mitra S, Truesdell S, Paul L, Chen T, Butchar JP, Justiniano S. 2008. Efficient genetic method for establishing Drosophila cell lines unlocks the potential to create lines of specific genotypes. PLoS Genet. 4:e1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura M, Nakato H. 2015. Genetic approaches in the study of heparan sulfate functions in Drosophila. Methods Mol Biol. 1229:497–505. [DOI] [PubMed] [Google Scholar]
- Takemura M, Nakato H. 2017. Drosophila Sulf1 is required for the termination of intestinal stem cell division during regeneration. J Cell Sci. 130:332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H, Kinoshita-Toyoda A, Selleck SB. 2000. Structural analysis of glycosaminoglycans in Drosophila and Caenorhabditis elegans and demonstration that tout-velu, a Drosophila gene related to EXT tumor suppressors, affects heparan sulfate in vivo. J Biol Chem. 275:2269–2275. [DOI] [PubMed] [Google Scholar]
- Wojcinski A, Nakato H, Soula C, Glise B. 2011. DSulfatase-1 fine-tunes Hedgehog patterning activity through a novel regulatory feedback loop. Dev Biol. 358:168–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Esko JD. 2014. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem. 83:129–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Lin X. 2009. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol. 1:a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zcharia E, Jia J, Zhang X, Baraz L, Lindahl U, Peretz T, Vlodavsky I, Li JP. 2009. Newly generated heparanase knock-out mice unravel co-regulation of heparanase and matrix metalloproteinases. PLoS One. 4:e5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.