Abstract
During the height of the 2009 H1N1 swine-derived influenza pandemic, a clinical trial was conducted in which seven subjects were immunized using a monovalent, MF59®-adjuvanted vaccine, developed from an egg- passaged candidate vaccine virus (CVV), A/California/07/2009 X-181. Whole blood was collected prior to immunization and at 8, 22, and 202 days post-vaccination, and subjects’ serological responses were evaluated. Here, we reconstruct and examine the longitudinal, influenza-specific circulating B cell repertoire of one subject in that study. Genotypic analysis of 390 total subject-derived antibodies (Abs) revealed a total of 29 germline genes in use among immunoglobulin heavy chain variable regions (IgHV), with the majority of those sequences isolated representing memory recall responses and two major lineages dominating the early response. In vitro phenotyping showed a diverse set of binding epitopes on the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA), many of which are considered subdominant. Strong correlations were found between IgHV germline usage among non-related lineages and both binding epitope and neutralization breadth. Results here highlight the potential for Ab responses to be misdirected to egg-adaptive artifacts on CVVs while simultaneously stressing the ability to mount potent, broadly neutralizing responses to mostly novel antigens via recall of subdominant memory responses, as well as the need for evaluating alternative endpoint assays and anti-NA responses following clinical trials.
Keywords: Influenza, vaccination, antibody
Introduction
Vaccines protect from infection by establishing or modifying immune memory. In the case of influenza, these responses are directed primarily at the surface proteins hemagglutinin (HA)[1], which mediates entry into the host cell, and neuraminidase (NA), which releases newly formed virions from the host cell’s membrane. Eighteen genotypically distinct subtypes of HA have been described for influenza A, falling into two major subdivisions (Group 1 and Group 2), and two diverse lineages for influenza B have been described (Yamagata and Victoria). Within each subtype and lineage, influenza viruses are further subject to antigenic drift—constant, rapid viral evolution driven by mutations in the genes that encode the HA and NA. Because of this genetic and antigenic diversity, influenza-related deaths average nearly half a million people worldwide each year[2], with circulating influenza results in annual outbreaks; transfer from birds or swine, in new pandemics. In the case of the swine-derived H1N1 pandemic strain that emerged in 2009, estimates suggested that 20% of people worldwide were infected,[3] in large part because its antigenic properties were different from those of the concurrent, seasonal circulating H1N1.
In the immediate aftermath of the 2009 pandemic (pdm2009), several studies analyzed how immune repertoires had been affected by vaccination against that wildtype strain, A/California/07/2009. Most of those studies focused on B-cell repertoires and the breadth of reactivity of the antibodies produced. Certain subjects exhibited an unexpected antibody (Ab) binding phenotype specific for HA epitopes other than the immunodominant globular head, and this profile correlated with expanded heterotypic neutralization breadth.[4] It was further shown that these broadly-neutralizing, heterotypic, non-head Abs preferentially use the germline gene 1~69 to encode the variable region of their immunoglobulin heavy chains (lgHV)[5–9]. Most of these studies involved large patient cohorts and therefore could examine only snapshots of each subject’s repertoire. These snapshots were never intended to assess the composition of the flu-specific B cell repertoire throughout a single season.
In the current study, we took the opportunity to reconstruct and examine a longitudinal intra-seasonal antibody repertoire using samples, originally collected for another study, from a cohort of seven subjects who received an MF59-adjuvanted single-component pdm2009 subunit vaccine made from egg-passaged, A/California/07/2009-derived, reassortant CVV NYMC-X181 (hereafter, “X-181”).[10]. The first report focused on T-cell and polyclonal responses. However, the structure of that study also provided an opportunity to trace the subjects’ serological responses to pdm2009 in real time, to assess the presence of broadly protective heterotypic antibodies, and to determine which responses were converted into long-term memory. We provide here an analysis of the complete intra-seasonal antibody repertoire of a single individual in that study—the subject who exhibited the lowest pre-vaccination baseline titer (a 32-year-old male hereafter known as Subject 7). We examined this repertoire using paired heavy and light chain sequences from single cells and characterized each antibody’s epitope, neutralization breadth, clonal relationships, and IgHV germline usage. Thus, we were able to trace the rise and fall of expanded clonal lineages as well as orphan clones from non-expanded lineages at up to six months post-vaccination. We believe that these analyses are the most complete case study to date on the rapidly developing immune response following vaccination against influenza within a season.
Materials and Methods
Study design and sample preparation
The study design, a complete description of sample collection procedures, and statements of Ethics Committee approval and informed consent have all been previously described.[10]
Cell sorting
Cell sorting was completed at the Duke Human Vaccine Institute (DHVI) using methods previously published.[11–13] PBMCs from Day 8 were sorted for all plasmablasts in an unbiased fashion via FACS. Gating for plasmablasts was as described [11]. Antigen-specific sorting of memory B cells (MBCs) from Days 0, 22, and 202 was performed using a recombinant X-181 protein produced in insect cells via a baculovirus expression system by collaborators at Harvard Medical School [15]. The X-181 protein was labeled in two fluorochrome colors by N- hydroxysuccinimide ester chemistry. Antigen-specific MBCs were sorted as viable singlet events gated as CD3/14/16/235a negative, CD19 positive, surface IgD negative that were double-positive for the X-181 protein. Sample plots and gating for antigen-specific sorting are presented in Raymond et al., Figure S7 [16]. Single cell sorting, PCR isolation of IgHV, Vκ, and Vλ from sorted cells, and synthesis of linear expression cassettes were also completed at DHVI as previously published.[14] Cell culture supernatants from 293T cells transfected with paired heavy and light chain linear cassettes were harvested at DHVI and sent for characterization.
Clonal lineage clustering
Paired Ig heavy and light sequences from cell transfections were grouped into clonal lineages using Clonalyst software [17, 18] from single-cell, paired heavy- and light-chain sequences. Antibodies are designated by heavy- chain sequence number.
Antigen production
Monobulks:
Unblended, monovalent lots of subunit vaccine antigen were produced in embryonated chicken eggs by Novartis Vaccines (now Seqirus) in either pilot or engineering batches as previously described. [19]
Recombinant protein:
HA gene segments were cloned into pCMV-KM2l and featured a foldon trimerization domain and a 6x-His tag. NA gene segments were cloned into pRS5α featuring a tetramerization domain and a 6x-His tag. All vectors were sequence verified. Plasmid DNA was transfected into Expi293F cells (ThermoFisher, A14527) using the Expi293 Expression System (Life Technologies, A14524) according to the recommended protocol. Culture media were harvested three days post-transfection by centrifugation. Recombinant antigens (rAgs) were purified with HisTrap Excel columns (GE Healthcare, 17–3712-05) by FPLC (GE, AKTA Pure 25) according to the recommended protocol. Purified proteins were characterized by SDS-PAGE and SEC-HPLC.
Production of human monoclonal antibodies (mAbs)
Linear DNA cassettes for Ig VH, Vκ, and Vλ segments were synthesized as GBIocks at Integrated DNA Technologies and cloned into pRS5α expressing full-length IgG1 heavy, kappa light, or lambda light chains where appropriate, by Gibson assembly (New England Biolabs, E5510S) according to the recommended protocol. Variable regions of Ig chains were sequence verified. Plasmid DNA for paired Ig heavy and light chains were co-transfected into Expi293F cells using the Expi293 Expression System according to the recommended protocol. Culture media were harvested three days post-transfection by centrifugation. mAbs were purified with HiTrap MabSelect Columns (GE Healthcare, 28–4082-53) by FPLC according to the recommended protocol. Purified mAbs were characterized by SDS-PAGE and SEC-HPLC.
Binding Affinity by Simple ELISA
Monobulk antigens were diluted to 1.0 μg/ml in Dulbecco’s Phosphate Buffered Saline (DPBS; Lonza, 17–512F); rAgs to 1.5 μg/ml. Diluted antigen was coated onto 96-well (Nunc, 439454) or 384-well (Greiner Bio-One, 781097) plates and stored at 4 °C overnight. Plates were washed three times in DPBS with 0.05% Tween-20 (PBS-T; Omnipur, 9480) and blocked with DPBS+1% BSA (Rockland, BSA-30) for 1h at RT and washed three times. Patient-derived transfection supernatants (tfx sups) were used neat; mAbs were normalized to 15 μg/ml in DPBS. Serial dilutions were performed 1:2 in DPBS + 1% BSA + 0.1% Triton-X (ELISA Buffer; EMD Millipore, TX1568–1) in a separate plate (BD Falcon, 353077) before being added to the reaction plate. Plates were incubated 1h at RT washed three times. HRP-conjugated anti-human F(ab’)2 secondary Ab (Jackson ImmunoResearch, 109–035-097) was diluted 1:5000 in ELISA Buffer before being loaded onto plates. Plates were incubated 1h at RT washed three times. Colorimetric outcomes were developed with TMB Substrate (Rockland, TMBE-1000) for 5 min at RT and stopped with 2N sulfuric acid (BDH, BDH3500–1). Plates were incubated an additional 15 min at RT prior to being read for 450nm absorbance using a Tecan Infinite M200 NanoQuant. Statistical determination of EC50 concentrations was performed using Prism software (GraphPad Software, Inc.).
Rough Epitope Mapping by Competition ELISA
Control Titration.
Control mAbs of known binding epitope [5, 15, 16] were biotinylated using NHS-PEG4-Biotin, No-Weigh™ Format kit (Thermo, 21329) at 3 mg/ml in DPBS. Biotinylated mAbs (B-mAbs) were normalized to 15 μg/ml in DPBS, and 1:2 serial dilutions were performed in ELISA Buffer in a separate plate (BD Falcon, 353077) before being added to the reaction plate, prepared using monobulk antigen as described above. After blocking, one volume of ELISA Buffer was added to the reaction plate followed by one volume of B-mAb dilution. Plates were incubated lh with shaking at RT, then washed three times. HRP-conjugated Avidin D (Vector Labs, A-2004) was diluted 1:10,000 in ELISA Buffer and loaded onto plates. Plates were then processed as described above. Concentrations of respective B-mAbs at which elicited 450 nm absorbances were 2.0–3.0 (nominally, “EC75”) were selected for use in subsequent assays described below.
Experimental Competition.
Subject-derived mAbs were normalized to 100 μg/ml (>10x EC75of the B-mAb competitor) in DPBS. Serial dilutions were performed 1:2 in ELISA Buffer in a separate plate before being added to the reaction plate. Assays were carried out as described above with the substitution of one volume of mAb dilution for one volume of ELISA Buffer following the block step.
Microneutralization assay
Microneutralization assays were carried out in triplicate as previously described. [15]
Results
Analysis of individual clones
A total of 390 paired heavy and light chain sequences were isolated from memory B cells sorted for HA specificity (at Day 0, n = 27; Day 22, n = 47; and Day 202, n = 99) or from an unbiased harvest of all plasmablasts isolated at Day 8 (n = 217; see materials and methods for gating summary). Because the cell harvest at Day 8 was sorted for all plasmablasts regardless of antigen specificity, sampling of the repertoire at Day 8 is considerably more complete than at Days 0, 22, and 202. All 390 Abs isolated were assayed for binding by ELISA to the subunit vaccine antigen (monobulk) made from X-181, which predominantly contains HA but also contains NA. Monobulk-reactive clones were further assayed for binding to recombinant HA (rHA) and/or to recombinant NA (rNA), depending on harvest strategy. Non-reactive clones were not subjected to further analysis by ELISA. Among the 217 paired heavy and light chain isolated from the Day 8 repertoire, ELISA detected binding by 157 to the X-181 monobulk. Over 15% (24/157) of this reactive repertoire bound rNA and not rHA (Fig 1A). No Ab from the HA-specific MBC harvests at Days 0, 22, and 202 were texted for NA affinity. Using a rHA construct for the trimeric full-length soluble ectodomain (FLsE) and a rHA trimeric globular head-only construct (both sequence-verified to match the egg-adapted X-181 HA sequence) as ELISA antigens, we found that B cell receptors (BCRs) specific to the X-181 HA head dominated the immediate response (~75%, 97/133 at Day 8) before reaching a post-vaccination balance of approximately 60%/40%, head/non-head (Fig 1B). We did not detect any HA-reactive clones in the pre-vaccination repertoire. Nearly 97% of head-specific clones at Day 8 were representatives of expanded clonal lineages (Fig 1C). This percentage had declined by Day 22 (to 73%) before falling even more by Day 202 (to 21%), resulting in a circulating HA head-specific repertoire composed of 79% non-expanded orphan clones. By contrast, the non-head binding repertoire showed virtually no preference for expanded lineages vs. orphan clones at any time point assessed (Fig 1D). Although the Day 0, 22, and 202 repertoires had been sorted for antigen specificity, a substantial number of clones—27/27 at Day 0, 9/47 at Day 22, and 45/99 at Day 202-had no ELISA-detected reactivity. This phenomenon has been observed previously, and may be an artifact induced during sorting or potentially due to the different glycosylation patterns on HA based on the expression systems used to generate the antigens.[20]
Figure 1. Binding Profile of HA-Specific Patient Repertoire.

X-181 monobulk-reactive samples were assayed via ELISA using a panel of recombinant antigens. Antigens included a trimeric Full-Length soluble Ectodomain (FLsE) construct for the HA of X-181 (the vaccine strain), a truncated trimeric Head-only construct for the HA of X-181, and a tetrameric recombinant rNA FLsE from the A/Brevig Mission/1/1918 strain (commercially available). A) Specificity of surface proteins amongst X-181 monobulk-reactive plasmablast samples. Due to sorting constraints, repertoires at other time points could not be assessed for NA binding. B-D) HA-specific samples at each time point are shown. Samples which bound both the FLsE and the rHA Head-only construct are listed as being specific for the HA globular head. Clones which bound only to the FLsE but not to the Head-only construct are reported as binding elsewhere on the FLsE. B) Preference of HA binding epitope following vaccination. Percentages are based on the number of HA-reactive clones at each time point: Day 0, none detected; Day 8, n = 133; Day 22, n = 38; Day 202, n = 54. C) Percentage of HA Head-specific clones from expanded clonal lineages. Percentages are based on the number of HA-Head-specific clones at each time point: Day 8, n = 97; Day 22, n = 22; Day 202, n = 33. D) Percentage of HA Non-Head-specific clones from expanded clonal lineages. Percentages are based on the number of HA non-head-specific clones at each time point: Day 8, n = 36; Day 22, n = 16; Day 202, n = 21.
Analysis of clonal lineages
Computational analysis of the 390 paired heavy and light chain sequences uncovered several related clonal lineages (CLs), where a lineage is defined as a family of clones with identical HCDR3 regions, regardless of the collection date. Clones described as “orphans” are clones for which no other clone with an identical HCDR3 was detected at anytime point. Several CLs not detected pre-vaccination expanded quickly (Fig 2), with a total of 71 HA-specific CLs identified across the entire analysis: 45 CLs specific to the HA head and 26 CLs with binding to the stem (non-head) region of the HA ectodomain. Two lineages were present in circulation both pre- and post- vaccination. A total of 12 lineages were detected at multiple time points post-vaccination but not at Day 0. Of these, five (CLs 6515, 6649, 6708, 6741, and 6936) were detected in circulation at all three post-vaccination time points. Two of the five persistent lineages (CL6515 in blue and CL6649 in red) dominated (Fig 2). Although the prominence of CL6515 within the circulating repertoire began to wane by Day 22, CL6649 appeared to have continued to expand throughout the three-week period. An analysis of both the phenotype and crystal structure of CLs 6515 [16] and 6649 [15] have been published, and it was determined in those analyses that CL6515 was specific for an egg-adapted artifact within the receptor binding site (RBS) on the X-181 HA, while CL6649 bound a previously undescribed lateral patch at some distance from the RBS on the HA head.
Figure 2. Clonal Lineage Repertoires Show Robust Clonal Expansion.

Paired heavy and light chain sequences isolated from subject-drawn PBMCs were analyzed In slllco and grouped into related lineages—where a lineage is defined as a family of clones with identical HCDR3 regions. Lineages isolated in circulation at multiple time points are highlighted a specific color. Lineages present at only a single time point are shown in grayscale. Clonal lineages 6515[16] and 6649[15] are emphasized where present.
IgHV gene segment usage
The peripheral blood repertoire derived from a total of 29 different IgHV unmutated, common ancestor (UCA) germline genes, often represented by more than one allele. Of these, 10 were favored, with 1~69, 3~23, 4~30–4, 4~39, and 5~51 particularly prevalent (Fig 3A). We then expressed at least one clone from each lineage as a recombinant full-length IgG1, along with a representative sampling of orphan clones from each time point, for further phenotypic analysis by both microneutralization and competition ELISA using control mAbs with known epitopes. (Note: Sequence data for certain lineages contained irreconcilable ambiguities. These lineages were omitted from further analysis.) Epitope mapping by competition ELISA (Fig 3B) showed that lineages using VH1~69 accounted for nearly half of all NA-binding and HA non-head-binding lineages, while VH5~51 accounted for nearly half of all lineages specific for the sialic acid RBS on the X-181 HA globular head. No antibodies from lineages using VH1~69 bound HA at the RBS, and no antibody from a lineage using VH5~51 bound HA outside the RBS. Despite its abundance within the circulating repertoires (except at Day 8), VH4~30–4 yielded mostly lineages that recognized neither HA nor NA. Other, less prevalent, germline genes yielded lineages of more varied binding phenotypes.
Figure 3. IgHV Germline Usage.
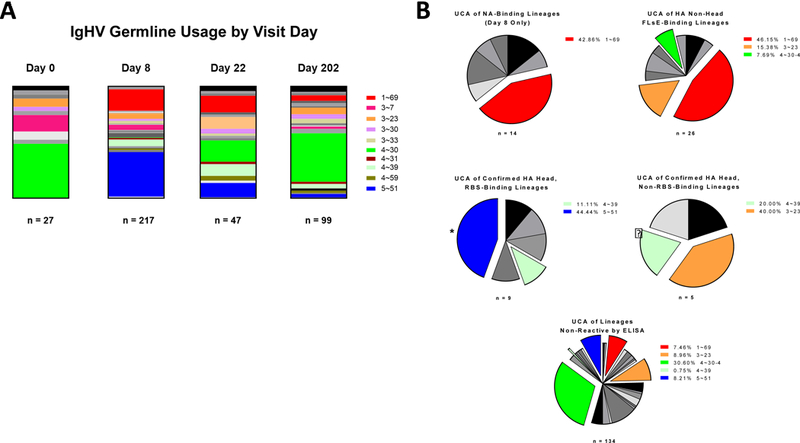
The NT sequence of the IgHV of each clone was assayed In silico for its unmutated common ancestor germline IgHV gene. Clones using the same IgHV germline gene are grouped together, regardless of lineage relationship. Select germline genes are highlighted specific colors. Remaining germline genes are presented in grayscale. A) IgHV germline usage by patient visit day. B) Clones were assessed for binding specificity by both simple and competition ELISA. Clones described as binding at the X-181 HA RBS competed directly with Ab6639, whose crystal structure is published.[16] Clones described as binding on the HA head but not at the RBS bound the trimeric head-only construct but did not compete directly with Ab6639. Clones described as binding the X-181 HA non-head FLsE did bind the Trimeric X-181 HA FLsE construct but not the X-181 HA head-only construct. Most competed directly with CR6261, whose HA-Stem-binding crystal structure is published.[29] Clones of a similar binding epitope were grouped based on their IgHV germline usage, regardless of lineage relationship. The most prominent germline genes are highlighted specific colors. Remaining germline genes are presented in grayscale
* Includes CL6515 [16]
? Includes CL6649 [15].
Neutralization phenotypes
Every Ab isolated was assayed against live A/California/07/2009 egg-passaged reassortant NYMC-X181 using tfx sups. Tfx sups with neutralizing activity against the vaccine strain were further assayed using the H1N1 components of the concurrent and previous TIV formulations from 2009: A/Brisbane/ 59/2007 and A/Solomon lslands/03/2006, respectively. Select Abs expressed as full-length IgG1 were further assayed against a panel of thirteen total viruses: seven H1N1 strains (including those mentioned above), one H1N2 strain, one H5N1 pseudovirus, two H3N2 strains, and one each of B-Yamagata and B-Victoria strains. Most of the Abs from expanded clonal lineages—especially those present in circulation at multiple time points—elicited a heterotypic target phenotype, neutralizing multiple H1N1 strains in that expanded virus survey panel (Fig 4). HA-specific lineages tested that used VH1~69 all had broad heterotypic responses across the group 1 viruses in the panel; lineages using VH5~51 had substantially less breadth, with only minimal cross-reactivity against heterologous strains; lineages using VH4~30–4 were non-reactive. Despite preferential expansion of heterotypic lineages, the homotypic phenotype of the predominant CL6515 lineage (using VH5~51)—due to its specificity for an egg- derived mutation in the RBS[16]—meant that homotypic plasmablasts dominated the overall early response.
Figure 4. Epitope Mapping and Phenotyping of Representative rlgG1 mAh Clones from each HA-reactive Lineage.

Members of each clonal lineage were expressed as full-length IgG1 and purified* and assessed by competition ELISA for binding epitope. Clones were then assessed for ability to neutralize a panel of influenza strains. Several lineages for which epitope binding data were inconclusive have been omitted from the table. Clones are grouped by IgHV germline usage, regardless of clonal relationship. Virus strains are H1N1 unless otherwise noted
*Whenever possible. Some sequences contained NT sequence ambiguities which would affect AA sequences. Calls were not made In these cases, and samples were omitted from further analysis.
Somatic hypermutation
Expanded clonal lineages (i.e. non-orphans) were further assessed for somatic hypermutation (SHM) and grouped by their neutralization phenotype (Fig 5A, 5B, and 5D) or binding preference in the case of NA-binders isolated from Day 8 plasmablasts (Fig 5C). Lineages with ≥97.5% identity to their IgHV UCA germlines were considered primary responses.[8, 21, 22] All HA-specific lineages with any detectable neutralizing activity were memory recall responses, as were 89% (8/9) of NA-specific lineages. 77% of expanded lineages with no detectable ELISA or microneutralization activity were primary responses, at or near 100% identity with their respective IgHV UCA germline gene segments. Of all lineages representing primary responses, 91% (10/11) were non-reactive, and none reacted to pandemic HA. Among orphan clones (data not shown), 54/125 nonneutralizing orphans, 6/27 orphans with limited breadth, 4/11 broadly-neutralizing orphans, and 0/5 NA-specific orphans had ≥97.5% identity with their respective IgHV UCA germline segments. In the absence of sister clones for clone phylogeny, it is difficult to determine whether such clones are indeed primary responses or are merely the product of sampling limitations.
Figure 5. SHM of Expanded Lineages.

Lineages for which ≥2 clones were detected were assessed for their homology to their respective germline IgHV segments using Clonalyst, with 100% being identical and unmutated. The dotted red line is drawn at 97.5% homology. Lineages at or above this threshold (≤2.5% SHM) are considered primary responses; lineages below this threshold (>2.5% SHM) are considered recall responses. Lineages are grouped by neutralization phenotype. Bars indicate mean +/− standard deviation
*The threshold for broadly neutralizing lineages was set at >1/3 of strains tested. Lineages which neutralized ≤1/3 of the strains are described as having “limited” breadth. This includes strain-specific clones.
Discussion
The analysis here is a longitudinal intra-seasonal case study of the post-vaccination B cell repertoire in a subject exposed to a novel, egg-adapted HA derived from pandemic H1N1 influenza. Our relatively extensive sampling has allowed us to follow the rise and fall of clonal lineages within the post-vaccination season and to assess which antigenic sites on the pandemic vaccine generated the most potent responses. Previous work has shown that both Ab and T cell responses arise rapidly in response to a single dose of adjuvanted vaccine and that recall of imprinted immune memory dominates.[10]
Specificity for the globular HA head dominates circulating plasmacyte responses to seasonal influenza vaccines administered before 2009.[23, 24] By contrast, post-pandemic studies have shown that vaccination against pdmHINI A/California/07/2009 induced above average HA-stem-binding responses.[25] Our data confirm those results. In total, we identified 71 HA-specific lineages across the analysis — 45 of which were specific for the HA head and 26 for the HA stem.
Among the 45 head-specific lineages, we isolated members of only 2 at all three post-vaccination time points and those of another 2 at two of the three time points. We found representatives of the remaining 41 at only a single time point: 6, 10, and 21 at Day 8, Day 22, and Day 202, respectively. Only two HA-head-specific lineages, both isolated only at Day 22, derived from VH1~69, and neither bound the HA RBS. Among the 26 stem-specific lineages, we isolated members of 3 at all three post vaccination time points and those of another 4 at two of the three time points. We found members of 8, 4, and 7 lineages at only Day 8, Day 22, and Day 202, respectively. VH1~69 accounted for 12 of the 26 stem-directed lineages and for nearly three-quarters of those isolated at multiple time points, consistent with published reports of the strong association of this heavy chain gene segment with the broadly neutralizing, stem-directed phenotype.
We have further shown that the VH1~69 germline gene segment also encodes nearly half of all anti-NA lineages isolated from Subject 7. This finding suggests that a more extensive analysis of immunodominant epitopes on NA is warranted, since VHl~69-encoded antibodies are often polyreactive. We suggest that a longitudinal analysis of IgHV preference in NA-binding lineages will be a worthwhile future study.
The germline gene segment VH5~51 does not appear in any of the previously published studies, despite accounting for nearly half of all anti-HA lineages specific for the RBS (including the predominant CL6515) from this donor. Whether VH5~51 has properties that favor selection for HA binding at the RBS, analogous to those that make VH1~69 a preferential stem-binder, remains to be determined.
The clonal lineage with the largest number of representatives, CL6515, was specific for an egg- adaptation in the RBS of X-181, at an epitope conserved at its center and partly conserved at its periphery across the antigenic shift from pre-pandemic, seasonal H1 viruses to the pdm2009 virus. We have previously shown that neutralization titers obtained using polyclonal sera collected from this patient are 4-fold higher against the egg-passaged X-181 strain used in vaccine manufacture when compared to the corresponding wildtype strain in circulation (see [16], Table S1). In conjunction with that previous analysis, the current study quantifies the extent of this egg-adapted misdirection in real-time by demonstrating that this family of high-affinity clones overwhelms the initial response, accounting for 33% of short-lived plasmablasts in this subject (likely accounting for the initial surge in head-directed specificity observed in Fig 1B) and maintains that prevalence with more than 10% of Ag-specific MBCs three weeks post-vaccination. It is likely that at least some of the lineages which competed with CL6515 for binding at the RBS are also specific for egg-adaptations, suggesting that this estimate is on the conservative side. Such findings present a strong case for the pursuit of methods of vaccine manufacture without the use of eggs or other avian tissue.
Clonal lineage CL6649 had the second largest number of representatives in the post-vaccination repertoire. Members of this lineage bound the globular head of HA but did not bind directly at the sialic acid receptor binding site. CL6649 nonetheless exhibited a broadly neutralizing phenotype with respect to H1N1. We have carried out a more complete study of this lineage and its epitope and shown that the epitope is a lateral patch on the HA head, conserved from 1977 through 2010.[15] Its conservation across the 2009 antigenic shift, like that of the RBS, may account for the robustness of the recall response to those two epitopes when boosted with the 2009 vaccine. The relevance of epitopes at some distance from the HA RBS illustrates how use of the standard hemagglutination inhibition (HI) assay—which detects only binding at the RBS—as the primary end point assay in clinical trials could potentially bias results by by neglecting regions on HA outside the RBS, like that of CL6649. More recently, altered properties of NA for some H3N2 viruses has resulted in NA-mediated hemagglutination, which can further complicate interpretations of HI data.[26] The contributions from the 6649 lineage epitope and from the HA stem in the B-cell repertoire analyzed here, in conjunction with the recent NA interference discovered in H3N2, underscore the importance of evaluating alternative end-point assays, such as microneutralization, that take the entire Ab response into account.
Conclusions
Findings reported here represent a longitudinal intra-seasonal B cell repertoire analysis of a subject immunized against pdm2009 using egg-passaged CVV X-181. Our data show that rapidly arising memory-recall responses dominate the immediate response to vaccination against pandemic HA and that substantial components of those responses may be directed at otherwise subdominant epitopes. While it is possible that the presence of MF59 in the vaccine formulation aided this process as has shown in cases of H5N1 vaccination,[27–29] the limitations of the study design prevent us from drawing that conclusion definitively. We have also shown that at least one in three plasmablasts in the early response to vaccination with an egg-derived influenza vaccine in this subject have been misdirected to egg-adaptations induced during egg-based vaccine manufacture.[30, 31] Egg-derived specificity can attenuate the protective capabilities of the response, even in a polyclonal setting.[16] However, we have shown that, despite this misdirection, potent and broadly neutralizing Abs arise, with epitopes on the HA head, the HA stem, and NA, with the VH1~69 gene segment encoding a disproportionately large subset of broadly-neutralizing stem-binding lineages and NA-binding lineages.
Highlights.
First longitudinal intra-seasonal antibody repertoire analysis traces rise and fall of related clonal lineages in response to monovalent MF59-adjuvanted vaccine against a mostly novel antigen
Memory recall dominates the overall response throughout the season
Homotypic memory recall Abs dominate immediate response but wane throughout the season
NA-specific antibodies are prominent
VH1~69 accounts for large proportion of NA-specific and heterotypic HA-specific antibodies
Acknowledgements:
This research was conducted using funding from the National Institutes of Health (P01 Grant #AI089618–02). The authors wish to acknowledge the intellectual and technical contributions of Aaron Schmidt, Shaun Stewart, Kendall Dionne, Xiuwen Ma, and Annette Ferrari.
Abbreviations:
- AA
amino acid
- Ab
antibody
- BCR
B cell receptor
- B-mAbs
biotinylated monoclonal antibodies
- BSA
bovine serum albumin
- CL
clonal lineage
- CVV
candidate vaccine virus
- DHVI
Duke Human Vaccine Institute
- DPBS
Dulbecco’s Phosphate-Buffered Saline
- ECX
effective concentration at which x% of the maximum is reached
- FACS
fluorescence-activated cell sorting
- FLsE
full-length soluble ectodomain
- HA
hemagglutinin
- HCDR3
third complimentarity-determining region on the immunoglobulin heavy chain
- HRP
horseradish peroxidase
- Ig
immunoglobulin
- IgHV
immunoglobulin heavy chain variable region
- mAb
monoclonal antibody
- MBC
memory B cell
- NA
neuraminidase
- NT
nucleotide
- PBMC
peripheral blood mononuclear cell
- PBS-T
DPBS + 0.05% Tween20
- pdm2009
H1N1 pandemic Influenza strain
- rAgs
recombinant antigens
- RBS
receptor-binding site
- rHA
recombinant hemagglutinin
- rNA
recombinant neuraminidase
- RT
room temperature
- SHM
somatic hypermutation
- tfx sups
transfection supernatants
- TMB
3,3’,5,5’-tetramethylbenzidine
- UCA
unmutated common ancestor
Footnotes
Conflict of Interest Statement
Authors JF, GP, GDG, PRD, ECS, and PS were employees of Seqirus (formerly Novartis Influenza Vaccines), which manufactures and sells influenza vaccines, at the time this work was performed. Authors GDG and PRD are currently employed by GlaxoSmithKline and Pfizer, respectively, which both manufacture and sell influenza vaccines. Remaining authors have no conflicts to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Skehel JJ, Wiley DC. Receptor Binding and Membrane Fusion in Virus Entry: The Influenza Hemagglutinin. Annual Review of Biochemistry. 2000;69:531–69. [DOI] [PubMed] [Google Scholar]
- [2].WorldHealthOrganization. Immunization, Vaccines and Biologicals: Influenza. 2008. [Google Scholar]
- [3].Van Kerkhove MD, Hirve S, Koukounari A, Mounts AW, group HNpsw. Estimating age-specific cumulative incidence for the 2009 influenza pandemic: a meta-analysis of A(H1N1)pdm09 serological studies from 19 countries. Influenza Other Respir Viruses. 2013;7:872–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A. 2012;109:9047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ekiert DC, Bhabba G, Elsliger M, Friesen RHE, Jongeneelen M, Throsby M, et al. Antibody Recognition of a Highly Conserved Influenza Virus Epitope. Science. 2009;324:246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, et al. A Neutralizing Antibody Selected from Plasma Cells That Binds to Group 1 and Group 2 Influenza A Hemagglutinins. Science. 2011;333:850–6. [DOI] [PubMed] [Google Scholar]
- [7].Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen L-m, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature structural & molecular biology. 2009;16:265–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med. 2015;7:316ra192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Faenzi E, Zedda L, Bardelli M, Spensieri F, Borgogni E, Volpini G, et al. One dose of an MF59 -adjuvanted pandemic A/H1N1 vaccine recruits pre-existing immune memory and induces the rapid rise of neutralizing antibodies. Vaccine. 2012;30:4086–94. [DOI] [PubMed] [Google Scholar]
- [11].Moody MA, Zhang R, Walter EB, Woods CW, Ginsburg GS, McClain MT, et al. H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One. 2011;6:e25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 2011;108:14216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A. 2013;110:264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liao H-X, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. Journal of Virological Methods. 2009;158:171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Raymond DD, Bajic G, Ferdman J, Suphaphiphat P, Settembre EC, Moody MA, et al. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc Natl Acad Sci U S A. 2018;115:168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Raymond DD, Stewart SM, Lee J, Ferdman J, Bajic G, Do KT, et al. Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain. Nat Med. 2016;22:1465–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kepler TB. Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Res. 2013;2:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kepler TB, Munshaw S, Wiehe K, Zhang R, Yu JS, Woods CW, et al. Reconstructing a B-Cell Clonal Lineage. II. Mutation, Selection, and Affinity Maturation. Front Immunol. 2014;5:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wen Y, Han L, Palladino G, Ferrari A, Xie Y, Carfi A, et al. Conformationally selective biophysical assay for influenza vaccine potency determination. Vaccine. 2015;33:5342–9. [DOI] [PubMed] [Google Scholar]
- [20].Kuraoka M, Schmidt AG, Nojima T, Feng F, Watanabe A, Kitamura D, et al. Complex Antigens Drive Permissive Clonal Selection in Germinal Centers. Immunity. 2016;44:542–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jackson KJ, Liu Y, Roskin KM, Glanville J, Hoh RA, Seo K, et al. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe. 2014;16:105–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tan Y-C, Blum LK, Kongpachith S, Ju C-H, Cai X, Lindstrom TM, et al. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clinical Immunology. 2014;151:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Knossow M, Gaudier M, Douglas A, Barrѐre B, Bizebard T, Barbey C, et al. Mechanism of Neutralization of Influenza Virus Infectivity by Antibodies. Virology. 2002;302:294–8. [DOI] [PubMed] [Google Scholar]
- [24].Wiley D, Wilson I, Skehel J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–8. [DOI] [PubMed] [Google Scholar]
- [25].Thomson CA, Wang Y, Jackson LM, Olson M, Wang W, Liavonchanka A, et al. Pandemic H1N1 Influenza Infection and Vaccination in Humans Induces Cross-Protective Antibodies that Target the Hemagglutinin Stem. Front Immunol. 2012;3:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lin YP, Gregory V, Collins P, Kloess J, Wharton S, Cattle N, et al. Neuraminidase receptor binding variants of human influenza A(H3N2) viruses resulting from substitution of aspartic acid 151 in the catalytic site: a role in virus attachment? J Virol. 2010;84:6769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Khurana S, Chearwae W, Castellino F, Manischewitz J, King L, Honorkiewicz A, et al. Vaccines with MF59 Adjuvant Expand the Antibody Repertoire to Target Protective Sites of Pandemic Avian H5N1 Influenza Virus. Science Translation Medicine. 2010;2:1–7. [DOI] [PubMed] [Google Scholar]
- [28].Khurana S, Suguitan AL, Jr., Rivera Y, Simmons CP, Lanzavecchia A, Sallusto F, et al. Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med. 2009;6:e1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ellebedy AH, Krammer F, Li GM, Miller MS, Chiu C, Wrammert J, et al. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci U S A. 2014;111:13133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Robertson JS, Bootman JS, Newman R, Oxford JS, Daniels RS, Webster RG, et al. Structural changes in the haemagglutinin which accompany egg adaptation of an influenza A(H1N1) virus. Virology. 1987;160:31–7. [DOI] [PubMed] [Google Scholar]
- [31].Yasuo Ito TS; Takada Ayato; Kawamoto Ayumi; Otsuki Koichi; Masuda Hiroyuki; Yamada Mika; Suzuki Takashi; Kida Hiroshi; Kawaoka Yoshihiro. Differences in Sialic Acid-Galactose Linkages in the Chicken Egg Amnion and Allantois Influence Human Influenza Virus Receptor Specificity and Variant Selection. Journal of Virology. 1997;71:3357–62. [DOI] [PMC free article] [PubMed] [Google Scholar]


