Abstract
Birds, with their broad geographic ranges and close association with humans, have historically played an important role as carriers of human disease and as reservoirs for drug-resistant bacteria. Here, we examine scientific literature over a 15-year timespan to identify reported avian-bacterial associations and factors that may impact zoonotic disease emergence by classifying traits of bird species and their bacteria. We find that the majority of wild birds studied were migratory, in temperate habitats, and in the order Passeriformes. The highest diversity of bacteria was found on birds in natural habitats. The most frequently reported bacteria were Escherichia coli, Salmonella enterica, and Campylobacter jejuni. Of the bacteria species reported, 54% have shown pathogenicity toward humans. Percentage-wise, more pathogens were found in tropical (vs. temperate) habitats and natural (vs. suburban, urban, or agricultural) habitats. Yet, only 22% were tested for antibiotic resistance, and of those tested, 75% of bacteria species were resistant to at least one antibiotic. There were no significant patterns of antibiotic resistance in migratory versus non-migratory birds, temperate versus tropical areas, or different habitats. We discuss biases in detection and representation, and suggest a need for increased sampling in non-temperate zones and in a wider range of avian species.
Keywords: Microbial, Literature, Pathogen, Antibiotic resistance, Avian
Introduction
Zoonotic pathogens pose a significant public health threat, particularly with regard to bacterial diseases (Taylor et al. 2001; Jones et al. 2008). Birds have long played an important role in human disease, specifically in spreading microbial pathogens (Belshe 1998; Reed et al. 2003; Johnson et al. 2007; Moulin-Schouleur et al. 2007). This is likely due to several key avian traits. First, like humans, birds are found worldwide. Their ability to migrate long distances, colonize new areas, and withstand a range of environments allows for a global distribution (Fournier et al. 2000; Rappole et al. 2000; Humair 2002; Winker et al. 2007; Benskin et al. 2009; Altizer et al. 2011). Second, birds are prominent species in human-dominated habitat types. The close association of birds and humans in urban and agricultural settings facilitates zoonotic disease transfer (Waters et al. 1991; Marzluff 2001; Capua and Alexander 2002; McKinney 2002; Atterby et al. 2016). Third, birds and humans are host to some of the same bacteria species, many of which are pathogenic. While evidence for direct bacterial transmission from birds to humans is limited, several bird species have indirectly transmitted at least 12 genera of pathogenic bacteria through polluted water, ticks, and feces that lead to diarrhea, salmonellosis, Lyme disease, and other illnesses in humans (Tsiodras et al. 2008). Finally, both domestic agricultural and wild birds can contaminate shared spaces and cause human infections (Sacks et al. 1986; Graczyk et al. 2008; Bonnedahl et al. 2009; Ewers et al. 2009; Vincent et al. 2010; Bonnedahl and Järhult 2014). Bird-carried diseases are, therefore, of interest because of the threat not only toward birds, but also human health (Literák et al. 2010).
Changing environments, including those associated with urbanization, agriculture, and climate change, may affect the likelihood of birds acquiring pathogens. These novel environments can lead to new niches and evolutionary trajectories for birds (Darwin 1859; Lack 1940), which may affect the ecology of pathogens and their vectors (Dobson and Carper 1992; Holmes and Garnett 1994; Daszak et al. 2000; Harvell et al. 2002; Guernier et al. 2004; Harrus and Baneth 2005; Engering et al. 2013; Estrada-Peña et al. 2014; Rothernburger et al. 2017). As birds shift ranges to accommodate environmental changes, infected individuals could introduce novel pathogens into immunologically naïve populations (Hubálek 2004). With possible increased bacterial migration rates between individuals, antibiotic resistance is also forecasted to evolve and spread rapidly (Perron et al. 2007).
While zoonotic transmission of pathogens from birds to humans has been more difficult to quantify than conspecific transmission (Tsiodras et al. 2008), emerging infectious diseases are predicted to occur primarily through zoonotic transmission (Jones et al. 2008). This, coupled with the large percentage of bacterial pathogens (38%, Taylor et al. 2001), makes understanding associations of different bacteria and bird species valuable to public health efforts to combat infectious disease (Kruse et al. 2004; Vouga and Greub 2016). In a recent literature survey, 122 studies documented associations between wildlife and transmission of bacteria to our food chain (Greig et al. 2014). These data demonstrate the potential threat of wildlife-transmitted bacteria and influences on human health. The potentially urgent public health threat of bird-borne infectious diseases suggests that now is the right time to assess bird-bacteria associations.
Here we examine scientific literature over a 15-year timespan to quantify characteristics of the reported bacteria species involved in bird-bacteria relationships, and examine the potential zoonotic effects of these associations by classifying birds by specific habitats and bacteria in terms of pathogenicity and drug resistance. Specifically, we ask the following questions:
How are certain characteristics of birds such as taxonomic order, habitat, and migration status related to pathogenic bacteria species, specifically ones that are pathogenic toward humans?
What are the characteristics of bacteria that are found in birds with regard to taxonomy, pathogenicity, and antibiotic resistance?
Materials and Methods
Data Compiled from the Literature
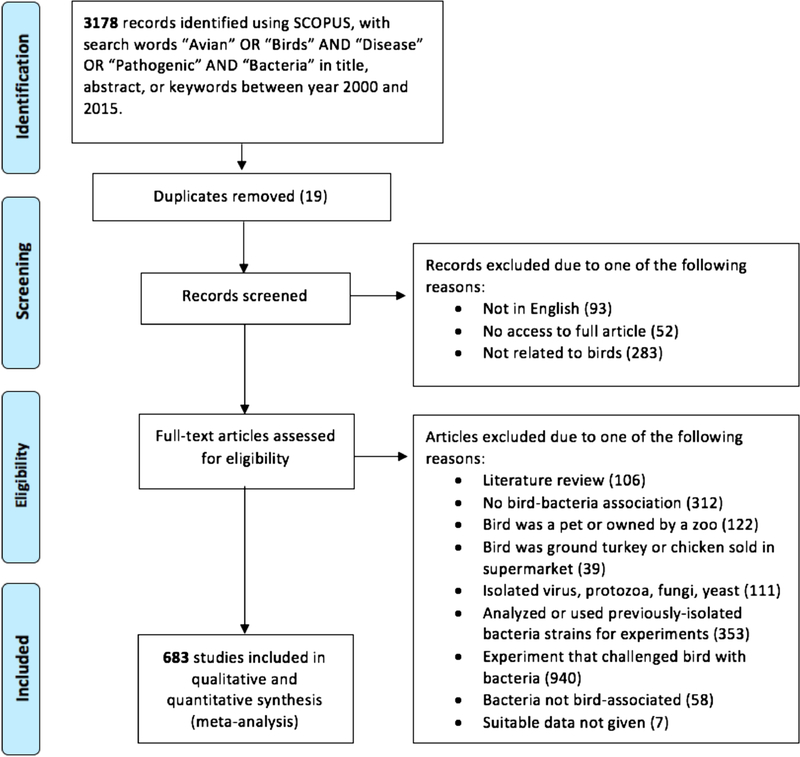
We conducted a search of papers published from 2000 to 2015 using SCOPUS, with the keywords “avian” or “birds,” and “disease” or “pathogenic,” and “bacteria” following PRISMA guidelines (Moher et al. 2009) (Fig. 1). Papers were limited to English primary sources that included birds with at least one bacterial association. Each bird-bacteria association represented one data point in our analysis. If a paper surveyed a single bird species with multiple bacteria species, we included each bird-bacteria association from that paper. For example, in one paper, researchers swabbed a herring gull and discovered 24 bacteria species (Bogomolni et al. 2008) for which we designated 24 separate data points.
Figure 1.
Screening process. PRISMA flowchart detailing the review and data selection process. Six hundred and eighty-three studies were ultimately used for analysis.
Type of Data Collected
For each bird-bacteria association, we made the following classifications: (1) geographic zone of study, (2) species, family, and order of bird, (3) whether the bird was wild or domestic, (4) migratory status of bird species, (5) habitat type, (6) species, family, and phylum of bacteria species associated with the bird, (7) whether the bacteria can cause human infection, (8) if bacteria were tested for antibiotic resistance, (9) if bacteria were resistant to any antibiotics, (10) antibiotics tested, and (11) antibiotics each bacteria species was resistant to. Further details of how we assessed these traits are provided below.
For geographic zone, we recorded whether a study took place in a tropical or temperate area. We noted the location where each species was found, and if a general area was given, such as “California,” we recorded that as the area. When coordinates were not given, we inputted the area into Google Earth Pro (Google Inc. 2015), scrolled to 100–200 m above ground, and used the coordinates at the automatically generated marker, usually in the centroid of the specified area. Tropical areas were bounded by the Tropic of Cancer (23.4371°N) and Tropic of Capricorn (23.4371°S).
If the bird species was not mentioned in an article, we recorded the family or order. Birds were considered “domestic” if they were raised solely for human consumption. Species were considered “migratory” if the “movement” section in the Handbook of the Birds of the World (HBW) (Hoyo et al. 1992–2013) indicated that the bird was a “migrant,” such as long-distance or altitudinal. If the section stated that a bird was “sedentary,” a “resident,” or “locally nomadic,” it was deemed “non-migratory.” We recorded “N/A” for any bird with “little or no information” about its movements, for any “domestic” bird, or if the bird was described in publications only as a family or order.
Birds found in “natural” habitats generally live in areas of non-agricultural flora and fauna (Blair 1996). Birds in “suburban” locations live in areas with both built cover and vegetation such as parks or gardens, and birds in “urban” habitats were located within a metropolis (Blair 1996). Additionally, habitats included “agricultural” for birds found in cultivated fields, “industrial livestock” for poultry houses or livestock farms, “multiple habitats” for studies taking place in multiple habitats, and “N/A” if the location was vague or unknown. We did not include pets. On a species level, if a bird was included in several studies with different habitats, it was placed into the “multiple habitats” category. Wild birds sampled around poultry houses were considered in “industrial livestock” habitats. “Domestic” birds lived in “industrial livestock” habitats, and were separated from wild bird analyses unless noted.
For bacteria species, we included species name, or if no species was listed, the genus. To determine pathogenicity toward humans, we used peer-reviewed journals to confirm if a species of bacteria has caused infections in humans. We recorded if bacteria species were tested for antibiotic resistance, the antibiotics used, and the results of those tests. Resistance was determined by the author of each study.
To detect bias in the characteristics of birds included in the bird-bacteria literature, we estimated the percentages of all birds in temperate versus tropical areas, migratory versus non-migratory, and natural versus agricultural versus suburban versus urban versus multiple habitat categories by randomly sampling 1000 birds from HBW (Hoyo et al. 1992–2013). We used chi square statistics to draw conclusions about patterns in the data. Data for wild birds living among domestic birds (in “industrial livestock” habitat) when shown were not analyzed statistically because of a low sample size for that group (n = 4).
Results
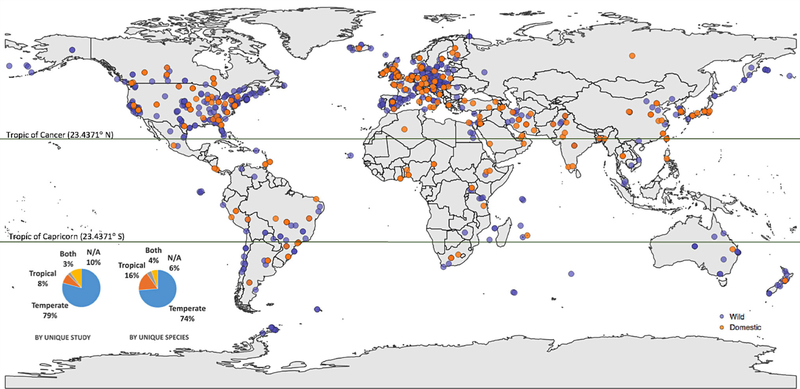
We analyzed 683 papers, which included 530 unique bird species, 11 phyla of bacteria representing at least 368 species, and 2289 unique bird-bacteria associations (Appendix Table 1, Appendix Table 2 in ESM). The samples in these studies were collected from all seven continents, with most samples from the USA and Europe (Fig. 2).
Figure 2.
Map of avian sampling locations. Each unique sampling location studied is represented by one plotted point on the map. Purple and orange dots are wild and domesticated species, respectively. Most of the sampling locations are in temperate locations (defined by areas that are above or below 23.4371 degrees latitude) compared with tropical locations. Inset pie charts show proportions of sampling locations by unique study (n = 683) and by unique species (n = 530).
Characteristics of Birds in this Study
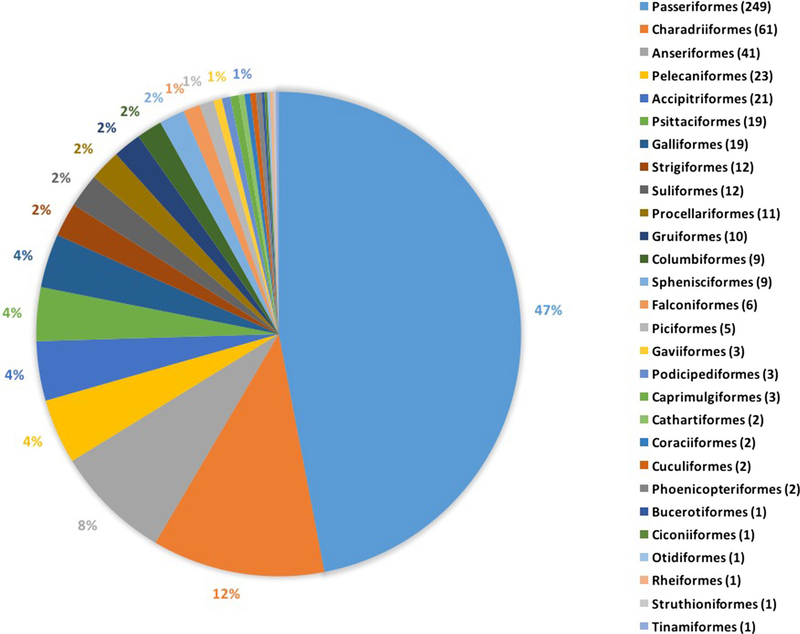
The 530 bird species included in the surveyed literature represent only 5% of 10,731 recognized bird species and an even smaller percent of the 18,000–20,000 estimated bird species (Barrowclough et al. 2016). Birds in this study spanned 29 of the 39 recognized bird orders (Gill and Donsker 2017) with a majority of species represented by three orders (Passeriformes 47% of 530 bird species; Charadriiformes 12%; Anseriformes, 8%) (Fig. 3). In some cases, this was an underrepresentation based on their abundance; for example, Passeriformes account for approximately 60% of all bird species. In other cases, there was overrepresentation: Charadriiformes account for 3% and Anseriformes less than 2% of all known bird species. Among the 683 studies, some species were overrepresented (Appendix Table 1, Appendix Figure 1 in ESM).
Figure 3.
Distribution of bird orders in the bird-bacteria literature over a 15-year timespan. We grouped the 530 unique bird species studied by their orders. In parentheses, the number of bird species in each order is shown. The most common orders were Passeriformes (47%), Charadriiformes (12%), and Anseriformes (8%). This chart includes 15 domestic species: 8 Galliformes, 5 Anseriformes, 1 Struthioniformes, 1 Rheiformes.
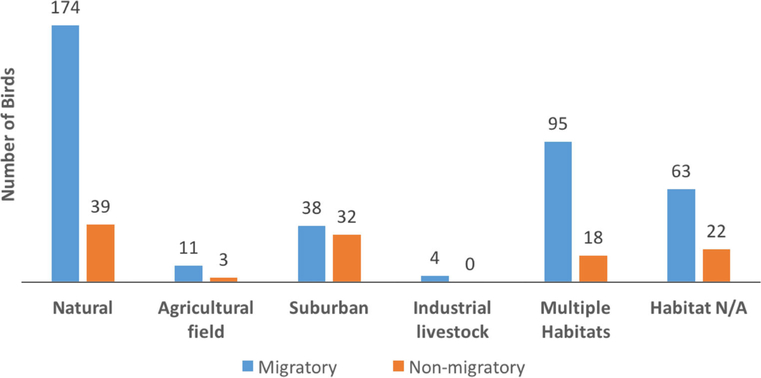
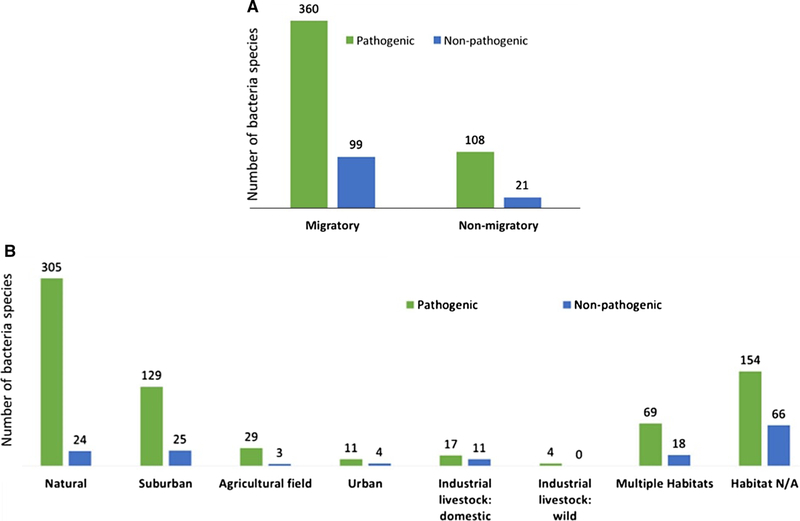
Most studies (75%) included birds that were migratory (Table 1). On a per-study level, 51% of studies focused on domestic birds in industrial livestock, 15% in natural habitats, 6% in suburban habitats, 3% in urban habitats, 2% in agricultural habitats, and 23% in multiple or unclearly defined habitats. On a per-species level, 4% of species studied were considered in domestic industrial livestock, 41% were only in natural habitats, 14% in suburban habitats, and 3% in agricultural habitats. No birds were found in only urban habitats (Table 1). Classification of birds in our analysis by migratory pattern and habitat showed that the migratory birds were significantly more common than non-migratory birds in all but suburban habitats (natural: = 85.56, P < 0.0001; suburban: = 0.51, P = 0.47; agricultural: = 4.57, P = 0.03; multiple habitats: = 52.47, P < 0.0001; Fig. 4).
Table 1.
Characteristics of Bird Species.
| Percent of species from literature-based analysis (of 515 species) (%) | Percent of species from HBW sample (of 1000 species) (%) | Comparison of literature survey to HBW sample | |
|---|---|---|---|
| Migration | |||
| Migratory | 75 | 32 | X2 = 223.59, df = 1, P < 0.0001 |
| Non-migratory | 22 | 58.6 | |
| Little or no information | 3 | 9.4 | |
| Location | |||
| Tropical | 16 | 72.8 | X2 = 594.0, df =2, P < 0.0001 |
| Temperate | 74 | 14.2 | |
| Tropical and temperate | 4 | 12.8 | |
| Little or no information | 6 | 0.2 | |
| Habitat | |||
| Natural | 42 | 72.9 | X2 = 338.92, df =5, P < 0.0001 |
| Agricultural | 3 | 3.5 | |
| Suburban | 14 | 17.4 | |
| Urban | 0 | 1.4 | |
| Industrial livestock (wild) | 0.8 | 0 | |
| Multiple habitats | 22.2 | 4.4 | |
| Little or no information | 18 | 0.4 | |
We characterized the migratory behavior, location, and habitat type of each of 515 unique wild species in the bird-bacteria literature from 2000 to 2015. To detect biases in the literature, we compared the characterizations of species in our literature-based analysis for the characterizations of 1000 species we randomly sampled from the Handbook of the Birds of the World (HBW).
Figure 4.
Bird migratory and habitat traits. Migration status of wild birds in different habitats, excluding 16 birds whose migration status was unknown (n = 499). A bird found only in one habitat in this literature-based analysis was listed under its respective habitat. Birds sampled in several different habitats were grouped into the “Multiple habitats” category.
We collected life history data from 1000 randomly selected species from HBW. Our sample reflected the makeup by order of the 10,731 known bird species (Gill and Donsker 2017). Starting with the most speciose orders, Passeriformes accounted for 65% of species (vs. 60% in Gill and Donsker 2017), Piciformes for 4.9% (vs. 4.1%), Caprimulgiformes for 4.9% (vs. 5.6%), Charadriiformes for 3.4% (vs. 3.6%), Columbiformes for 2.7% (vs. 3.2%), etc. Based on our estimated percentages using the HBW sample, the majority (73%) of the world’s bird species reside in tropical areas. The studies included in our analysis, however, were significantly biased toward temperate species (Fig. 2; Table 1). The bird-bacteria literature was also significantly biased toward migratory birds: 75% of the bird species studied migrate, while we estimate that 32% of the world’s species migrate (Table 1). The bias toward migratory birds could result from a bias toward temperate species; (temperate: 74% of species studied migrate vs. 32% of all birds: = 281.1, P < 0.0001; tropical: 36% of species studied migrate vs. 32% of all birds: = 0.21, P = 0.68). Finally, while most bird species are found in natural areas (about 73%), only 42% of species studied in the bird-bacteria literature lived there (Table 1). Overall, we found a significantly different distribution of species among habitats in the literature compared to distributions worldwide, driven by an underrepresentation of natural species and an over-abundance of species found in multiple or unclassified habitats (Table 1).
Characteristics of Bird-Associated Bacteria
The 368 bacteria species represented in our findings illustrate a small fraction of sequenced bacteria species (Schloss and Handelsman 2004), and even less of the estimated 109–1012 bacteria species in the world (Dykhuizen 1998; Locey and Lennon 2016; Larson et al. 2017). The most frequent bacteria species reported in the reviewed studies were Escherichia coli, Salmonella enterica, and Campylobacter jejuni (Appendix Figure 2 in ESM). Two or more of these bacteria species were often found in the same bird species (Appendix Table 1; Appendix Figure 2 in ESM). Per study, Pasteurella multocida and Borrelia burgdorferi were also commonly found in domestic and wild birds, respectively.
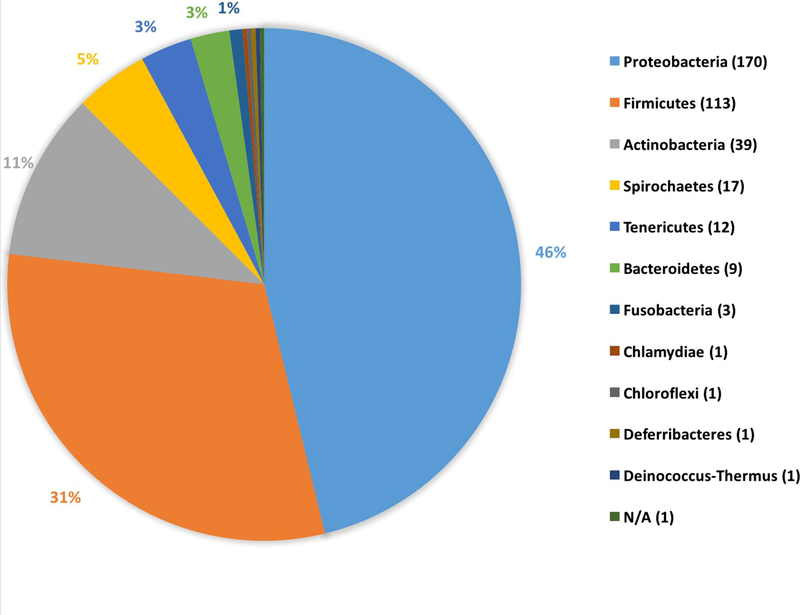
Out of 53 described bacteria phyla (Keller and Zengler 2004), 11 were reported in our analyses (21%). The majority of bacteria were from Proteobacteria (170 bacteria species out of 368; (46%), Firmicutes (31%), or Actinobacteria (11%)) (Fig. 5). One bacterium (Avispirillum sp.) has not been classified into a phylum yet (Waldenström 2006). Eleven phyla were found in migratory birds compared to seven phyla in non-migratory birds. Additionally, all 11 bacteria phyla were found in temperate habitats, but only eight phyla were represented in tropical habitats; temperate habitats had the additional phyla Chloroflexi, Deferribacteres, and Deinococcus-Thermus (Appendix Table 1, Appendix Table 2 in ESM). The greatest diversity of phyla was found in natural habitats (11 phyla), then industrial livestock (8), agricultural (6), suburban (6), and urban habitat (5).
Figure 5.
Distribution of bacteria phyla in the bird-bacteria literature over a 15-year timespan. We grouped 368 unique bacteria species studied by phyla. In parentheses, the number of bacteria species in each phylum is shown. The most common phyla were Proteobacteria (46%), Firmicutes (31%), and Actinobacteria (11%). There was one bacterium (Avispirillum sp.) that has not been classified yet (Waldenström 2006) and is in the phylum labeled, “N/A”.
Characteristics of Bird-Associated Bacteria that are Human Pathogens
Of the 538 identified bacteria species that are known human pathogens (Taylor et al. 2001), we found 199 in our study (37% of total bacterial pathogens). More pathogens were identified in temperate (193 species) compared to tropical habitats (48 species), but based on sampling effort as measured by the number of studies conducted, proportionately more pathogens were identified in tropical studies (48 species from 57 tropical studies, 193 species from 541 temperate studies ( = 15.97, P < 0.0001)). Thirty-five of 48 bacteria species found in tropical habitats areas were also found in temperate areas. Regarding habitat type, most pathogens (137 species) were found in 101 studies conducted in natural habitats, of which nearly half (49%) were not reported in other habitats. Suburban habitats had 38 species of pathogenic bacteria (from 39 studies), while agricultural and urban habitats had 18 and 13 species, from 12 and 20 studies, respectively. Over 350 studies in industrial livestock habitats identified 81 pathogens. Overall, the likelihood of detecting pathogenic bacteria relative to sampling effort differed significantly among habitats ( = 121.39, P < 0.0001), with the fewest pathogens, relative to the number of studies conducted, for domestic birds in industrial livestock habitats.
Relative to the total number of bacteria species found, migratory birds were not more likely to carry pathogenic bacteria than non-migratory birds ( = 1.42, P = 0.23; Fig. 6a). As a whole, however, pathogenic bacteria were significantly more frequent than non-pathogenic bacteria ( = 57.29, P < 0.0001), specifically in birds from natural, suburban, urban, and agricultural field habitats (natural: = 240.0, P < 0.0001; suburban: = 70.23, P < 0.0001; urban: = 21.12, P < 0.0001; agricultural: = 21.12, P < 0.0001; Fig. 6b). Domesticated industrial livestock carried nearly equal numbers of pathogenic and non-pathogenic bacteria ( = 1.29, P = 0.26; Fig. 6b). While the sample size for wild birds in industrial livestock habitats was too small to analyze, each of the four bacteria species detected in those birds was pathogenic (Fig. 6b), likely due to our own bias of placing “pathogen” within our search terms.
Figure 6.
Pathogenicity of bacteria in relation to bird characteristics. a Bacteria pathogenicity and migratory status of wild birds. Domestic birds were excluded from migratory counts. b Pathogenicity in each of the habitats. Bacteria on domestic species were included in the “Industrial livestock: domestic” category. Note: For both a and b, if multiple bacteria species were found on a bird, each bacteria species was counted.
Characteristics of Bacteria with Antibiotic Resistance
One hundred and nine bacteria species out of 368 (29.6%) were tested for antibiotic resistance, of which 75% (82 species) showed resistance to at least one antibiotic. Of the resistant bacteria in our analysis, 30 (37%) were found only in domesticated livestock. We documented 61 resistant bacteria species in migratory birds compared to 24 in nonmigratory birds, representing a roughly equal likelihood of identifying unique resistant bacteria based on sampling effort (61 from 48 studies testing resistance of bacteria from migratory birds; non-migratory: 24 from 23 studies; = 0.15, P = 0.70). Likewise, temperate and tropical birds harbored an equal diversity of resistant bacteria, with E. coli as the most resistant bacteria in both regions (Appendix Table 3 in ESM). Sixteen resistant bacteria species were found in 26 studies testing for resistance in bacteria from tropical birds, and 81 species were identified from 143 temperate studies ( = 0.004, P = 0.95). The diversity of resistant bacteria species detected did vary significantly among habitats based on relative sampling effort ( = 26.81, P < 0.0001). Forty resistant bacteria were detected from 19 studies in natural habitats (2.1 species/study), 2 species from 2 agricultural studies (1 species/study), 11 from 17 suburban studies (0.64 species/study), 1 species from 4 studies in urban habitats (0.25 species/study), and 44 from 105 industrial livestock studies (0.42 species/study). E. coli was resistant to the most antibiotics in every habitat except agricultural habitat, where E. coli antibiotic resistance was not assessed (Appendix Table 3 in ESM).
Patterns of Antibiotic Resistance
Of the 176 (26%) studies that tested for antibiotic resistance, the mean number of antibiotics tested was 11.5. In total, 125 antibiotics were tested among the studies, with the most common being tetracycline (71% of 176 studies), gentamicin (69%), and ampicillin (61%). The vast majority of the antibiotics tested, 111 of 125 (89%), were found to have at least one bacterium resistant to them (Table 2).
Table 2.
List of Antibiotics and Resistance.
| Mechanism/class | Antibiotic |
|---|---|
| Targets cell membrane | |
| Cationic peptides | Bacitracin* |
| Colistin* | |
| Lipopeptide | Daptomycin |
| Targets cell wall | |
| Carbapenems | Ertapenem |
| Imipenem* | |
| Meropenem* | |
| Cephalosporins (first generation) | Cefadroxil* |
| Cefazolin* | |
| Cefradine* | |
| Cephalothin* | |
| Cephalosporins (second generation) | Cefaclor* |
| Cefotetan* | |
| Cefotiam* | |
| Cefoxitin* | |
| Cefuroxime* | |
| Cephalexin* | |
| Cephalosporins (third generation) | Cefdinir |
| Cefixime* | |
| Cefoperazone* | |
| Cefotaxime* | |
| Cefpodoxime* | |
| Ceftazidime* | |
| Ceftibuten | |
| Ceftiofur* | |
| Ceftizoxime* | |
| Ceftriaxone* | |
| Cephalosporins (fourth generation) | Cefepime* |
| Cefpirome | |
| Cefquinome* | |
| Glycopeptides | Bleomycin |
| Teicoplanin* | |
| Vancomycin* | |
| Moenomycins | Flavomycin* |
| Monobactams | Aztreonam* |
| Penicillins | Amoxicillin* |
| Ampicillin* | |
| Carbenicillin* | |
| Cloxacillin* | |
| Mecillinam* | |
| Methicillin | |
| Mezlocillin* | |
| Oxacillin* | |
| Penicillin G* | |
| Piperacillin* | |
| Ticarcillin* | |
| Phosphonic acid derivatives | Fosfomycin* |
| Polypeptides | Enramycin* |
| Cephalosporin combination | Ceftazidime-clavulanic acid |
| Penicillin combination | Amoxicillin-clavulanic acid* |
| Ampicillin-sulbactam* | |
| Piperacillin-tazobactam* | |
| Ticarcillin-clavulanate* | |
| Targets DNA | |
| Aminocoumarin | Novobiocin* |
| Nitrofurans | Furazolidone* |
| Nitrofurantoin* | |
| Nitroimidazoles | Metronidazole* |
| Quinolones/Fluoro-quinolones | Ciprofloxacin* |
| Danofloxacin* | |
| Difloxacin* | |
| Enrofloxacin* | |
| Flumequine* | |
| Gatifloxacin* | |
| Levofloxacin* | |
| Marbofloxacin* | |
| Moxifloxacin* | |
| Nalidixic acid* | |
| Norfloxacin* | |
| Ofloxacin* | |
| Orbifloxacin* | |
| Oxolinic acid* | |
| Quinoxalin-di-N-oxides | Carbadox* |
| Targets folic acid synthesis | |
| Diaminopyrimidine | Trimethoprim* |
| Sulfonamides | Sulfachloropyridazine* |
| Sulfadiazine-trimethoprim* | |
| Sulfadimethoxine* | |
| Sulfamethazine* | |
| Sulfamethoxazole* | |
| Sulfamethoxazole-trimethoprim* | |
| Sulfathiazole* | |
| Sulfisoxazole* | |
| Sulphadimethoxine* | |
| Targets ribosomes | |
| Aminoglycosides | Amikacin* |
| Apramycin* | |
| Dihydrostreptomycin* | |
| Gentamicin* | |
| Kanamycin* | |
| Neomycin* | |
| Netilmicin* | |
| Paromomycin | |
| Spectinomycin* | |
| Streptomycin* | |
| Tobramycin* | |
| Chloramphenicol | Chloramphenicol* |
| Fusidane | Fusidic acid* |
| Glycylcyclines | Tigecycline* |
| Lincosamides | Clindamycin* |
| Lincomycin* | |
| Lincosamides, Aminoglycoside | Lincospectin* |
| Macrolides | Azithromycin*s |
| Clarithromycin* | |
| Erythromycin* | |
| Josamycin* | |
| Kitasamycin* | |
| Spiramycin | |
| Telithromycin* | |
| Tilmicosin* | |
| Tulathromycin | |
| Tylosin* | |
| Orthosomycins | Avilamycin* |
| Oxazolidinones | Linezolid* |
| Phenicols | Florfenicol* |
| Pleuromutilins | Tiamulin* |
| Streptogramin | Quinupristin-dalfopristin* |
| Streptogramin A* | |
| Streptogramin B* | |
| Virginiamycin* | |
| Tetracycline | Chlortetracycline* |
| Doxycycline* | |
| Minocycline* | |
| Oxytetracycline* | |
| Tetracycline* | |
| Targets RNA | |
| Mupirocin | Mupirocin |
| Rifamycin | Rifampicin* |
| Targets more than one area | |
| Combination drugs | Bacitracin-chloramphenicol |
| Penicillin-streptomycin | |
We classified antibiotics found in bird-bacteria literature from 2000 to 2015. Antibiotics are grouped by mechanism, on left. Combination antibiotics were considered as one unique antibiotic. An antibiotic with an asterisk (*) means that at least one bacterium has shown resistance to the respective antibiotic within our analysis.
The characteristics of the birds studied were not associated with significant differences in antibiotic resistance (migratory vs. non-migratory, temperate vs. tropic, and different habitats, X2, all P > 0.80). For example, bacteria from migratory birds were resistant to 79% (69/87) of the antibiotics tested compared to 85% (57/67) of antibiotics tested in non-migratory birds ( = 0.03, P = 0.87). The percent of antibiotics yielding resistance were 87% and 89% in temperate versus tropical birds, respectively, and among birds in different habitats, ranged from 71 to 95%, again with no significant patterns emerging.
In regard to bird taxonomy, chickens (Gallus gallus) carried bacteria found to be resistant to 96 antibiotics, the highest among domestic birds (Appendix Table 4 in ESM). Among migratory wild birds, the common buzzard (Buteo buteo) had the highest number of resistant antibiotics (20), while rock doves (Columba livia) and tawny owls (Strix aluco) had the highest number of antibiotic-resistant bacteria for non-migratory birds (24; Appendix Table 4 in ESM). Likewise, the bird orders Galliformes and Anseriformes had the most antibiotic-resistant bacteria for both domestic and wild birds (Appendix Table 5 in ESM).
Discussion
Zoonotic transmission of diseases is a clear public health issue (Jones et al. 2008; Cutler et al. 2010). Because diverse and abundant bird populations coexist with humans, and humans and birds are host to some of the same bacteria (da Costa et al. 2013), an investigation of bird-bacteria associations seems warranted. The goals of this literature-based analysis were to examine patterns of bird-bacteria associations and identify gaps in knowledge, specifically in determining understudied species, geographic locations, and habitat types. A 15-year timespan allowed us to obtain a snapshot of the current trends in the literature. A limitation could be that rare bacteria are overrepresented compared to common bacteria; however, the same common bacteria were sampled year after year (Appendix Table 1 in ESM).
We found a clear sampling bias toward domesticated birds, particularly chickens and turkeys, and other species closely associated with humans (e.g., rock doves). Further attention should be given to sampling bird species from orders underrepresented in bird-bacteria literature.
Additionally, temperate bird species are overrepresented compared to tropical bird species. Studies of tropical birds are particularly relevant in Asia and Africa, where numerous bacteria have evolved multi-drug resistance against antibiotics intended to treat tuberculosis, cholera, gonorrhea, salmonellosis, methicillin-resistant Staphylococcus aureus, and E. coli infections (Ndihokubwayo et al. 2013).
The overrepresentation of birds in multiple habitats possibly reflects an increase in urbanization (Luniak 2004). Additionally, the bias toward migratory birds can be partly explained by an oversampling of temperate species, which are largely migratory. It seems plausible that the literature should also skew toward subjects affecting humans: more pathogenic species, more domestic birds.
There is a lack of literature reporting bacterial diversity in temperate versus tropical birds. When only comparing bacterial diversity from soil and leaf litter samples in temperate and tropical regions, previous researchers found more bacterial diversity in temperate areas compared with the tropics (Kim et al. 2014; Tian et al. 2017), which reflects our findings as a whole. Previous studies have found that natural and agricultural habitats tend to have more bacteria phyla than urban habitats (Ibekwe et al. 2013; Jordaan 2015), and this finding is consistent with our results. The oversampling of migratory temperate birds may account for some of the apparent increase in bacterial diversity in migratory birds.
Had more bacteria species in birds been sampled, tropical zones would have proportionally more human pathogens than temperate zones. Likewise, natural habitats would have proportionally more pathogens than other habitats. Both of these findings are consistent with previous research (Guernier et al. 2004; Bradley and Altizer 2007). In wild birds, pathogenic bacteria were more common than non-pathogens. This could be because of selective isolation of known pathogens toward birds or a bias toward sampling dead birds (Benskin et al. 2009), which have a higher likelihood of bacterial infection. In our literature-based analysis, eight orders of birds had only human-pathogenic bacteria associated with them. It is likely that researchers have focused more on bacteria that are human pathogens in order to determine the threat from potential zoonotic reservoirs. One caveat is that we input “pathogen” or “disease” into our search terms, which inherently makes the results contain more pathogens. We statistically analyze human pathogens only, but our results also include bird pathogens.
In total, there was a high rate of resistance (75%) in tested bacteria. Worldwide, data regarding antimicrobial resistance are limited except for certain bacteria species of widespread concern, such as E. coli and S. aureus that have resistance rates of 50% or more in certain countries (WHO 2014). The high rate of resistance in our analysis can reflect the efforts of researchers focusing on certain pathogenic bacteria, or it can reflect the severity of resistance at hand. Interestingly, there were no significant differences in antibiotic resistance rates based on bird migratory status, geographic zone, or habitat type. Migratory birds are more likely to have antibiotic-resistant bacteria because they are exposed to antibiotics at higher rates due to traveling great distances and inhabiting a variety of environments, particularly in areas around humans (Allen et al. 2010). Additionally, industrial livestock and urban areas should have higher relative levels of resistant bacteria; because livestock are frequently treated with antibiotics, resistance develops more easily and can spread to the surrounding environment and to consumers in urban environments (Teuber 2001).
Domestic chickens were found to have the most antibiotic resistance, possibly as a reflection of the large number of studies that focused on chickens. It is also likely that chickens contain the most resistance because they are subject to numerous antibiotic therapies, and thus opportunities for resistance, in intensive farming situations (van den Bogaard et al. 2002; Muaz et al. 2018).
We suggest a better integration in the two areas of research of avian studies and microbiology. While it is unrealistic to expect those who conduct field studies in birds to add a microbial component, better communication and integration of field ornithology with microbial studies would allow for a less-biased understanding of bird-bacteria associations. More studies can utilize genomic methods and combine their findings with current bird microbiome studies to synthesize host-bacteria interactions. Some laboratories have already begun work in this direction (Taragel’ová et al. 2008; Literák et al. 2010; Oravcova et al. 2013). Finally, the significance of birds as vectors for pathogens, including viral pathogens that cause West Nile flu and H1N5 flu, makes understanding zoonotic transmission of diseases crucial to clinicians and to the field of public health, thus emphasizing the importance of multi-disciplinary studies in understanding bird-bacteria associations and their consequences.
Supplementary Material
Acknowledgements
We are grateful to Jaineet S. Chhabra and Marissa Schutte for help in data collection and Jeffrey G. Lee for technical assistance. We are also grateful for the data analyzation assistance provided by C. W. Chung. This research was supported by NIH/National Center for Advancing Translational Science (NCATS) UCLA CTSI Grant Number UL1TR001881 (PY). This work was also funded by a UCLA Faculty Career Development Award, a Hellman Fellowship (PY), and a Whitcome Undergraduate Research Scholarship (DC). Salary support for DS was provided by the National Institute of General Medical Science of the National Institutes of Health (NIH) under award number GM103427. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Compliance with Ethical Standards
Conflict of interest The authors declare that they have no conflict of interest.
Electronic supplementary material: The online version of this article (https://doi.org/10.1007/s10393-018-1342-5) contains supplementary material, which is available to authorized users.
References
- Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J (2010) Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology 8:251–259. 10.1038/nrmicro2312 [DOI] [PubMed] [Google Scholar]
- Altizer S, Bartel R, Han BA (2011) Animal migration and infectious disease risk. Science 331:296–302. 10.1126/science.1194694 [DOI] [PubMed] [Google Scholar]
- Atterby C, Ramey AM, Hall GG, Järhult J, Börjesson S, Bonnedahl J (2016) Increased prevalence of antibiotic-resistant E. coli in gulls sampled in Southcentral Alaska is associated with urban environments. Infection Ecology & Epidemiology 6 10.3402/iee.v6.32334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrowclough GF, Cracraft J, Klicka J, Zink RM (2016) How many kinds of birds are there and why does it matter? PLoS ONE 11(11):e0166307 10.1371/journal.pone.0166307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshe RB (1998) Influenza as a zoonosis: how likely is a pandemic? The Lancet 351:460–461. 10.1016/S0140-6736(98)22007-1 [DOI] [PubMed] [Google Scholar]
- Benskin CM, Wilson K, Jones K, Hartley IR (2009) Bacterial pathogens in wild birds: a review of the frequency and effects of infection. Biological Reviews 84:349–373. 10.1111/j.1469-185X.2008.00076.x [DOI] [PubMed] [Google Scholar]
- Blair RB (1996) Land use and avian species diversity along an urban gradient. Ecological Applications 6:506–519. 10.2307/2269387 [DOI] [Google Scholar]
- Bogomolni AL, Gast RJ, Ellis JC, Dennett MR, Pugliares KR, Lentell BJ, Moore MJ (2008) Victims or vectors: a survey of marine vertebrate zoonoses from coastal waters of the northwest atlantic. Diseases of Aquatic Organisms 81:13–38. 10.3354/dao01936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnedahl J, Drobni M, Gauthier-Clerc M, Hernandez J, Granholm S, Kayser Y, Melhus Å, Kahlmeter G, Waldenström J, Johansson A, Olsen B (2009) Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS one 4:e5958 10.1371/journal.pone.0005958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnedahl J, Järhult JD (2014) Antibiotic resistance in wild birds. Upsala Journal of Medical Sciences 119:113–116. 10.3109/03009734.2014.905663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley CA, Altizer S (2007) Urbanization and the ecology of wildlife diseases. Trends in Ecology & Evolution 22:95–102. 10.1016/j.tree.2006.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capua I, Alexander DJ (2002) Avian influenza and human health. Acta Tropica 83:1–6. 10.1016/S0001-706X(02)00050-5 [DOI] [PubMed] [Google Scholar]
- Cutler SJ, Fooks AR, van der Poel WHM (2010) Public health threat of new, reemerging, and neglected zoonoses in the industrialized world. Emerging Infectious Diseases 16:1–7. 10.3201/eid1601.081467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa PM, Loureiro L, Matos AJF (2013) Transfer of multidrug-resistant bacteria between intermingled ecological niches: the interface between humans, animals and the environment. International Journal of Environmental Research and Public Health 10:278–294. 10.3390/ijerph10010278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C (1859) On the origins of species by means of natural selection. London: Murray; 247. [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD (2000) Emerging infectious diseases of wildlife-threats to biodiversity and human health. Science 287:443–449. 10.1126/science.287.5452.443 [DOI] [PubMed] [Google Scholar]
- Dobson A, and Carper R (1992) Global warming and potential changes in host-parasite and disease-vector relationships. Global Warming and Biodiversity, Peters RL, Lovejoy RL (editors). New Haven, CT: Yale University Press, pp 201. [Google Scholar]
- Dykhuizen DE (1998) Santa Rosalia revisited: why are there so many species of bacteria? Antonie van Leeuwenhoek 73:25–33 [DOI] [PubMed] [Google Scholar]
- Engering A, Hogerwerf L, Slingenbergh J (2013) Pathogen-host-environment interplay and disease emergence. Emerging Microbes & Infections 2:e5 10.1038/emi.2013.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Peña A, Ostfeld RS, Peterson AT, Poulin R, de la Fuente J (2014) Effects of environmental change on zoonotic disease risk: an ecological primer. Trends in Parasitology 30:205–214. 10.1016/j.pt.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Ewers C, Antão E-M, Diehl I, Philipp H-C, Wieler LH (2009) Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Journal of Journal of Applied and Environmental Microbiology 75:184–192. 10.1128/AEM.01324-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier P-E, Tissot-Dupont H, Gallais H, Raoult D (2000) Rickettsia mongolotimonae: A rare pathogen in France. Emerging Infectious Diseases 6:290 10.3201/eid0603.000309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill F, Donsker D (2017) IOC world bird list (v 7.4). Accessed Oct 2017.
- Graczyk TK, Majewska AC, Schwab KJ (2008) The role of birds in dissemination of human waterborne enteropathogens. Trends in Parasitology 24:55–59. 10.1016/j.pt.2007.10.007 [DOI] [PubMed] [Google Scholar]
- Greig J, Rajić A, Young I, Mascarenhas M, Waddell L, LeJeune J (2014) A scoping review of the role of wildlife in the transmission of bacterial pathogens and antimicrobial resistance to the food chain. Zoonoses and Public Health 62:269–284. 10.1111/zph.12147 [DOI] [PubMed] [Google Scholar]
- Guernier V, Hochberg ME, Guégan J-F (2004) Ecology drives the worldwide distribution of human diseases. PLoS Biology 2:e141 10.1371/journal.pbio.0020141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrus S, Baneth G (2005) Drivers for the emergence and reemergence of vector-borne protozoal and bacterial diseases. International Journal for Parasitology 35:1309–1318. 10.1016/j.ijpara.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Harvell CD, Mitchell CE, Ward JR, Altizer S, Dobson AP, Ostfeld RS, Samuel MD (2002) Climate warming and disease risks for terrestrial and marine biota. Science 296:2158–2162. 10.1126/science.1063699 [DOI] [PubMed] [Google Scholar]
- Holmes EC, Garnett GP (1994) Genes, trees and infections: Molecular evidence in epidemiology. Trends in Ecology & Evolution 9:256–260. 10.1016/0169-5347(94)90291-7 [DOI] [PubMed] [Google Scholar]
- Hoyo JD, Elliott A, Sargatal J, and Christie DS (1992–2013) Handbook of the Birds of the World. 1–16. [Google Scholar]
- Hubálek Z (2004) An annotated checklist of pathogenic microorganisms associated with migratory birds. Journal of Wildlife Diseases 40:639–659. 10.7589/0090-3558-40.4.639 [DOI] [PubMed] [Google Scholar]
- Humair P-F (2002) Birds and borrelia. International Journal of Medical Microbiology 291:70–74. 10.1016/S1438-4221(02)80015-7 [DOI] [PubMed] [Google Scholar]
- Ibekwe AM, Leddy M, Murinda SE (2013) Potential human pathogenic bacteria in a mixed urban watershed as revealed by pyrosequencing. PloS one 8:e79490 10.1371/journal.pone.0079490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inc., Google (2015) Google Earth Pro Available: www.google.com/earth/.
- Johnson TJ, Kariyawasam S, Wannemuehler Y, Mangiamele P, Johnson SJ, Doetkott C, Skyberg JA, Lynne AM, Johnson JR, Nolan LK (2007) The genome sequence of avian pathogenic Escherichia coli strain o1: K1: H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. Journal of Bacteriology 189:3228–3236. 10.1128/JB.01726-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KE, Patel NG, Levy MA, Storeyard A, Balk D, Gittleman JL, Daszak P (2008) Global trends in emerging infectious diseases. Nature 7181:990–993. 10.1038/nature06536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordaan K (2015) Molecular profiling of microbial population dynamics in environmental water. Noordwes-Universiteit (doctoral dissertation). [Google Scholar]
- Keller M, Zengler K (2004) Tapping into microbial diversity. Nature Reviews Microbiology 2:141–150. 10.1038/nrmicro819 [DOI] [PubMed] [Google Scholar]
- Kim M, Kim W-S, Tripathi BM, Adams J (2014) Distinct bacterial communities dominate tropical and temperate zone leaf litter. Microbial Ecology 67:837–848. 10.1007/s00248-014-0380-y [DOI] [PubMed] [Google Scholar]
- Kruse H, Kirkemo A-M, Handeland K (2004) Wildlife as source of zoonotic infections. Emerging Infectious Diseases 12:2067–2072. 10.3201/eid1012.040707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack D (1940) Evolution of the Galapagos finches. Nature 146:324–327. 10.1038/146324a0 [DOI] [Google Scholar]
- Larson BB, Miller EC, Rhodes MK, Wiens JJ (2017) Inordinate fondness multiplied and redistributed: the number of species on earth and the new pie of life. The Quarterly Review of Biology 92:229–265. 10.1086/693564 [DOI] [Google Scholar]
- Literák I, Kulich P, Robesova B, Adamik P, Roubalova E (2010) Avipoxvirus in great tits (Parus major). European Journal of Wildlife Research 56:529–534. 10.1007/s10344-009-0345-5 [DOI] [Google Scholar]
- Locey KJ, Lennon JT (2016) Scaling laws predict global microbial diversity. Proceedings of the National Academy of Sciences of the United States of America 113:5970–5975. 10.1073/pnas.1521291113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luniak M (2004) Synurbization-adaptation of animal wildlife to urban development. Pages 50–55 in Proceedings of the 4th International Symposium of Urban Wildlife Conservation Tucson. [Google Scholar]
- Marzluff JM (2001) Worldwide urbanization and its effects on birds. Avian Ecology and Conservation in an Urbanizing World:19–47. [Google Scholar]
- McKinney ML (2002) Urbanization, biodiversity, and conservation the impacts of urbanization on native species are poorly studied, but educating a highly urbanized human population about these impacts can greatly improve species conservation in all ecosystems. BioScience 52:883–890. 10.1641/0006-3568(2002)052[0883:UBAC]2.0.CO;2 [DOI] [Google Scholar]
- Moher D, Liberati A, Tetzlaff J, Altman DG (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Medicine 6:e1000097 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin-Schouleur M, Répérant M, Laurent S, Brée A, Mignon-Grasteau S, Germon P, Rasschaert D, Schouler C (2007) Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. Journal of Clinical Microbiology 45:3366–3376. 10.1128/JCM.00037-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muaz K, Riaz M, Akhtar S, Park S, Ismail A (2018) Antbiotic residues in chicken meat: global prevalance, threats, and decontamination strategies: a review. Journal of Food Protection 81:619–627. 10.4315/0362-028X.JFP-17-086 [DOI] [PubMed] [Google Scholar]
- Ndihokubwayo JB, Yahaya AA, Dester A (2013) Antimicrobial resistance in the African region: issues, challenges and actions proposed. African Health Monitor 16:27–30 [Google Scholar]
- Oravcova V, Ghosh A, Zurek L, Bardon J, Guenther S, Cizek A, Literak I (2013) Vancomycin-resistant enterococci in rooks (corvus frugilegus) wintering throughout Europe. Environmental Microbiology 15:548–556. 10.1111/1462-2920.12002 [DOI] [PubMed] [Google Scholar]
- Perron GG, Gonzalez A, Buckling A (2007) Source-sink dynamics shape the evolution of antibiotic resistance and its pleiotropic fitness cost. Proceedings of the Royal Society B: Biological Sciences 274:2351–2356. 10.1098/rspb.2007.0640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappole JH, Derrickson SR, Hubálek Z (2000) Migratory birds and spread of West Nile virus in the western hemisphere. Emerging Infectious Diseases 6:319 10.3201/eid0604.000401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KD, Meece JK, Henkel JS, Shukla SK (2003) Birds, migration and emerging zoonoses: West Nile virus, lyme disease, influenza a and enteropathogens. Clinical Medicine & Research 1:5–12. 10.3121/cmr.1.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothernburger JL, Himsworth CH, Nemeth NM, Pearl DL, Jardine CM (2017) Environmental factors and zoonotic pathogen ecology in urban exploiter species. EcoHealth 14:630–641. 10.1007/s10393-017-1258-5 [DOI] [PubMed] [Google Scholar]
- Sacks JJ, Lieb S, Baldy LM, Berta S, Patton CM, White MC, Bigler WJ, Witte JJ (1986) Epidemic campylobacteriosis associated with a community water supply. American Journal of Public Health 76:424–428. 10.2105/AJPH.76.4.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J (2004) Status of the microbial census. Microbiology and Molecular Reviews 68:686–691. 10.1128/MMBR.68.4.686-691.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taragel’ová V, Koči J, Hanincová K, Kurtenbach K, Derdáková M, Ogden NH, Literák I, Kocianová E, Labuda M (2008) Blackbirds and song thrushes constitute a key reservoir of Borrelia garinii, the causative agent of borreliosis in central Europe. Journal of Journal of Applied and Environmental Microbiology 74:1289–1293. 10.1128/AEM.01060-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LH, Latham SM, Woolhouse MEJ (2001) Risk factors for human disease emergence. Philosophical Transactions of the Royal Society B: Biological Sciences 356:983–989. 10.1098/rstb.2001.0888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuber M (2001) Veterinary use and antibiotic resistance. Current Opinion in Microbiology 4:493–499. 10.1016/S1369-5274(00)00241-1 [DOI] [PubMed] [Google Scholar]
- Tian J, He N, Hale L, Niu S, Yu G, Liu Y, Blagodatskaya E, Kuzyakov Y, Gao Q, Zhou J (2017) Soil organic matter availability and climate drive latitudinal patterns in bacterial diversity from tropical to cold temperate forests. Functional Ecology 00:1–10. 10.1111/1365-2435.12952 [DOI] [Google Scholar]
- Tsiodras S, Kelesidis T, Kelesidis I, Bauchinger U, Falagas ME (2008) Human infections associated with wild birds. Journal of Infection 56:83–98. 10.1016/j.jinf.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bogaard AE, Willems R, London N, Top J, Stobberingh EE (2002) Antibiotic resistance of faecal enterococci in poultry, poultry farmers and poultry slaughterers. Journal of Antimicrobial Chemotherapy 49:497–505. 10.1093/jac/49.3.497 [DOI] [PubMed] [Google Scholar]
- Vincent C, Boerlin P, Daignault D, Dozois CM, Dutil L, Galanakis C, Reid-Smith RJ, Tellier PP, Tellis PA, Ziebell K (2010) Food reservoir for Escherichia coli causing urinary tract infections. Emerging Infectious Diseases 16:88–95. 10.3201/eid1601.091118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vouga M, Greub G (2016) Emerging bacterial pathogens: the past and beyond. Clinical Microbiology and Infection 22:12–21. 10.1016/j.cmi.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldenstrom J, Broman T, Carlsson I, Hasselquist D, Achterberd RP, Wagenaar JA, Olsen B (2002) Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Applied and Environmental Microbiology 68:5911–5917. 10.1128/AEM.68.12.5911-5917.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A, Higgins D, McCutchan T (1991) Plasmodium falciparum appears to have arisen as a result of lateral transfer between avian and human hosts. Proceedings of the National Academy of Sciences 88:3140–3144. 10.1073/pnas.88.8.3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winker K, McCracken KG, Gibson DD, Pruett CL, Meier R, Huettmann F, Wege M, Kulikova IV, Zhuravlev YN, Perdue ML (2007) Movements of birds and avian influenza from Asia into Alaska. Emerging Infectious Diseases 13:547 10.3201/eid1304.061072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (World Health Organization) (2014) Antimicrobial resistance: global report on surveillance, Geneva, Switzerland: World Health Organization [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.