Abstract
Priming effects (PEs) are defined as short-term changes in the turnover of soil organic matter (SOM) caused by the addition of easily degradable organic compounds to the soil. PEs are ubiquitous but the direction (acceleration or retardation of SOM decomposition) and magnitude are not easy to predict. It has been suggested that the ratio between the amount of added PE-triggering substrate to the size of initial soil microbial biomass is an important factor influencing PEs. However, this is mainly based on comparison of different studies and not on direct experimentation. The aim of the current study is to examine the impact of glucose-to-microbial biomass ratios on PEs for three different ecosystems. We did this by adding three different amounts of 13C-glucose with or without addition of mineral N (NH4NO3) to soils collected from arable lands, grasslands and forests. The addition of 13C-glucose was equivalent to 15%, 50% and 200% of microbial biomass C. After one month of incubation, glucose had induced positive PEs for almost all the treatments, with differences in magnitude related to the soil origin and the amount of glucose added. For arable and forest soils, the primed C increased with increasing amount of glucose added, whereas for grassland soils this relationship was negative. We found positive correlations between glucose-derived C and primed C and the strength of these correlations was different among the three ecosystems considered. Generally, additions of mineral N next to glucose (C:N = 15:1) had little effect on the flux of substrate-derived C and primed C. Overall, our study does not support the hypothesis that the trigger-substrate to microbial biomass ratio can be an important predictor of PEs. Rather our results indicate that the amount of energy obtained from decomposing trigger substrates is an important factor for the magnitude of PEs.
Introduction
Priming effects (PEs) are defined as short-term changes in the turnover of soil organic matter (SOM) caused by the input of easily degradable organic compounds (e.g. plant residues, root exudates, excretes of soil animals) to the soil [1]. So far, a reliable prediction of the direction (acceleration or retardation of SOM decomposition) and magnitude of PEs in response to organic carbon additions cannot be given. Several environmental factors influence PEs, such as the amount and chemical structure of added substrates [2–4], the inorganic nutrient availability [1,5] and the microbial biomass and community structure [6–8]. A meta-analysis [6] by Blagodatskaya and Kuzyakov (2008) indicated that the magnitude and direction of PEs are dependent on the ratio of the amount of added substrate to the size of the microbial biomass. Their analysis revealed a linear increase in PEs with increasing amount of trigger compounds as long as the added C substrate is less than 15% of the size of the microbial biomass. In contrast, an exponential decrease in PEs was found when the amount of trigger compounds was more than 50% of the size of the soil microbial biomass. This decrease in PEs with high amount of triggering compounds is suggested to be due to the so called preferential microbial substrate utilization [9,10] where soil microbes switch to utilize added easily degradable C sources instead of native soil C.
The meta-analysis of Blagodatskaya and Kuzyakov (2008) considered all publications on PEs with information on microbial biomass C. However, it is known that the structure and functioning of microbial communities and quality of soil organic matter greatly differ among ecosystems [11,12] and that this can have a strong impact on PEs [13]. Moreover, the studies taken into consideration in the meta-analysis included both single and multiple applications of the triggering substrates. Single and multiple applications will differently affect soil microbial biomass as well as community structure. These differences can have an impact on PEs [14–23].
The effects of concentrations of triggering compounds on PEs was recently tested [24]. In this study different amounts of the same trigger compound (glucose) were added to different soil ecosystems collected along an elevation gradient. The doses of added glucose were based on the initial size of the soil microbial biomass and they used multiple additions rather than a single one to resemble the temporal dynamics of labile C input in the field. The main finding was that PEs are increasing with higher amounts of trigger substrates and, therefore, not strongly related to the initial size of the soil microbial biomass. Yet, the magnitude of increase of PEs with increasing amount of glucose varied among the ecosystems included in the study.
The current study was already started when the paper of Liu et al. (2017) [24] appeared and had basically the same purpose: to investigate if the effects of trigger substrate to microbial biomass ratios on PEs are in agreement with the outcome of the meta-analysis of Blagodatskaya and Kuzyakov (2008). In particular, we were interested in this since most studies on PEs use an amount of trigger substrate that is at least 45% of soil microbial biomass [2,4,25,26], which is far higher than recommended by Blagodatskaya and Kuzyakov (2008). Based on the meta-analysis of Blagodatskaya and Kuzyakov (2008), we hypothesized that the trigger-substrate to microbial biomass ratio is an important predictor of PEs. To test the general validity of this hypothesis, we included three different ecosystems in our study, namely arable fields, grasslands and forests. Like most other studies, we have used single trigger substrate additions, whereas the study of Liu et al, (2017) used multiple applications.
Concurrently with addition of different amounts of a PE-triggering substrate (13C-glucose) we added ammonium nitrate (NH4NO3) to study the effect of nitrogen on PEs, as the availability of N can also influence the magnitude of PEs [5]. According to the “microbial nitrogen mining” hypothesis microbes use labile C as an energy source to decompose recalcitrant organic matter in order to obtain mineral N [27]. Hence, N addition may reduce mining for N and consequently SOM decomposition [28]. However, the N mining theory has been challenged as simultaneous addition of C and N was shown to stimulate rather than decrease priming [2,29,30]. Stimulation of decomposition can be driven by the stoichiometry of substrates, with the highest decomposition rates observed when the ratios of supplies of C and N to microbes match their demands [31,32]. Hence, simultaneous addition of C and N compounds can alleviate the stoichiometric constraint [28,33], causing a better match with N demands necessary for microbial growth and enzyme production and, consequently, resulting in higher PEs [2]. Based on these considerations and our previous results [2] we hypothesized that addition of N will not have a negative effect on PEs.
Materials and methods
Soil sampling and processing
In August 2015, soil (0–10 cm) was collected from three different ecosystems in the central part of the Netherlands [34], i.e. arable fields, beech forests and natural grasslands developed on abandoned arable fields. The arable fields were planted with maize (Zea mays L.), the dominant plant species in the forests was beech (Fagus syslvatica L.) and the natural grasslands were dominated by grasses such as common bent (Agrostis capillaris L.), tufted grass (Holcus lanatus L.) and forbs such as narrow-leave plantain (Plantago lanceolata L.) [34]. For each ecosystem type, we collected soils from four separate sites that were about 1 km apart, representing four ecosystem replicates [34]. In the laboratory, fresh soil from each plot was sieved (4 mm) and homogenized, removing fine roots and other plant debris. Field-moist soil was then stored at 4 °C until further use.
Measurements of biotic and abiotic soil properties
Before the start of the experiments from each of our soil samples we collected a random subsample to determine biotic and abiotic soil conditions [34].
Soil chemical properties
Descriptions of the methods used for the evaluation of soil chemical properties are reported in [35]. Soil pH (soil: H2O,1:2 w:v), gravimetric moisture content (dried at 105°C, to constant mass) and maximum water holding capacity (WHC) was measured in fresh soil samples. Total C and N in all soil samples were measured by a combustion method using an elemental analyser (Thermo flash EA 1112, Thermo Fisher Scientific Inc.). Mineral N was extracted by shaking 10 g dry weight soil with 50 ml 1 M KCl for two hours. Concentration of N-NH4+ and N-NO3- in the KCl extract were determined using an AutoAnalyzer (SEAL QuAAtro Segmented Flow Analysis system). The orthophosphate fraction from the soils was extracted in a 1:20 (w/v) ratio with a 0.5 molar solution of NaHCO3 at pH 8.5. Concentration of P-(PO4) in the extracts was determined by an AutoAnalyzer (SEAL QuAAtro Segmented Flow Analysis system). Chemical properties of the soils are listed in Table 1.
Table 1. Mean values (±SE) of chemical soil properties for each of the three soil types (arable, grassland, forest).
| Soil | pH | C:N ratio | N-NO3 [mg kg-1 dw soil] |
N-(NH4) [mg kg-1 dw soil] |
P-(PO4) [mg kg-1 dw soil] |
|---|---|---|---|---|---|
| Arable | 5.7 ± 0.3 | 17.4 ± 0.4 | 31.5 ± 5.4 | 0.2 ± 0.1 | 231.5 ± 15.9 |
| Grassland | 5.5 ± 0.1 | 17.7 ± 0.9 | 5.1 ± 0.5 | 0.3 ± 0.3 | 99.7 ± 6.8 |
| Forest | 3.9 ± 0.3 | 24.1 ± 0.4 | 2.3 ± 0.4 | 1.6 ± 0.1 | 18.6 ± 1.8 |
DNA extractions and quantitative PCRs
To determine bacterial biomass we extracted DNA from soils using the PowerSoil DNA Isolation Kit (MOBIO Laboratories, Carlsbad, California, USA) according to the manufacturer’s instruction with some modifications: after adding solution C1 (causing cell lysis), samples were incubated at 60 °C for 30 min; after adding solution C6 (releasing DNA from spin filter), samples were incubated at 30 °C for 10 min. Total DNA was quantified using a NanoDrop ND-1000 Spectrophotometer (Bio-Rad Laboratories Inc.).
Briefly, each qPCR reaction for bacterial quantification (total volume 15 μl) consisted of 7.5 μl of Sybergreen (iTaq Universal SYBR Green Supermix), 0.6 μl of forward primer (Eub 338, 10 pmol μl-1) [36], 0.6 μl of reverse primer (Eub 518, 10 pmol μl-1) [36], 0.6 μl of reverse primer (Eub 518, 10 pmol μl-1)[37], 3.3 μl Nucleic acid free water (Sigma) and 3 μl of DNA. Plasmid Ter331 (Collimonas 16S) was used as a standard for the quantification. The PCR program consisted of an initial denaturation step at 95 °C for 2 min, followed by 40 cycles of 95 °C for 10 sec, 53 °C for 10 sec and 72 °C for 25 sec. The qPCRs were performed with a Rotor-Gene RG-3000 (Corbett research). For each template DNA we analysed four biological replicates in duplicate. The qPCR results, expressed as 16S rRNA gene copy numbers g-1 of dry weight soil, were used to calculate the bacterial cell numbers using a conversion factor of 4.1 copies per cell [38] and bacterial biomass C as described by [39].
Fungal biomass
Ergosterol, a sterol found in fungal membranes, was used as a biomarker for fungal biomass. We used the protocol described by [40]. Briefly, 4 g of moist soil was shaken with 6 ml of methanol in the presence of glass beads, to disrupt the fungal mycelium and to release the ergosterol into the extractant. After centrifugation and filtration, ergosterol was measured on a 1260 Bio-inert LC coupled with a 6460 QQQ (Agilent, Santa Clara, USA). The results obtained, expressed as mg kg-1, were used to calculate the fungal biomass using a conversion factor of 5.4 Conversion factors of 5.4 mg ergosterol g-1 biomass C [41].
Mesocosm set-up
Glass bottles (500 ml) were filled with moist soil equivalent to 80 g of dry weight and the soil was pre-incubated in a climate chamber for an acclimatization period of 15 days at 20°C. The acclimatization period was based on the results of a previous pilot experiment (data not shown), where soil CO2 efflux rates were regularly checked to confirm stabilization of the soil microbial activity.
Soil samples were mixed with single additions of three different aqueous solutions of uniformly labelled 99 atom% 13C-glucose (Campro Scientific GmbH). The amount of glucose-C added was equivalent to 15%, 50%, and 200%, of the microbial biomass carbon (Table 2) [6]. Besides the addition of the three different glucose solutions, half of the mesocosms also received an aqueous solution of NH4NO3 to establish a final C-glucose to N ratio of 15:1 [8]. Controls consisted of soils without addition. The solutions were stirred into the soils to ensure a homogeneous mixture. Each treatment included four soil replicates per ecosystem type. Soils were incubated at 20 °C in the dark for 30 days. The soil moisture was maintained at 60% of the water holding capacity throughout the incubation period by weighing the microcosms once a week and watering with deionized water when needed. After 4 days (time period based on the development of total CO2 respiration) and 30 days (end of the experiment) of incubation, soil was sampled from each treatment using an ethanol- cleaned tweezer to minimize contamination. These soil samples were used for microbial biomass measurements. The samples were frozen (-20 °C) prior to DNA and ergosterol extractions.
Table 2. Mean values (±SE) of initial fungal and bacterial biomass (mg C g-1 dw soil) of the sampled soils and amount of 13C-glucose added (mg C g-1 dw soil) to the soils in a quantity of C equal to 15%, 50%, and 200% of the microbial biomass carbon.
| Fungal biomass | Bacterial biomass | Tot Microbial biomass | 13C-glucose (mg C g-1 dw soil) | |||
|---|---|---|---|---|---|---|
| Soil type | (mg C g-1 dw soil) | 15% | 50% | 200% | ||
| Arable | 0.19 ± 0.01 a | 0.21 ± 0.04 a | 0.40 ± 0.05 a | 0.06 ± 0.01 | 0.20 ± 0.03 | 0.81 ± 0.10 |
| Forest | 0.52 ± 0.04 b | 0.17 ± 0.04 a | 0.69 ± 0.05 c | 0.10 ± 0.10 | 0.34 ± 0.03 | 1.38 ± 0.11 |
| Grassland | 0.28 ± 0.03 a | 0.30 ± 0.02 a | 0.59 ± 0.05 ab | 0.09 ± 0.09 | 0.29 ± 0.03 | 1.17 ± 0.10 |
Statistically significant differences (P < 0.05) are marked with different letters. Fungal and bacterial biomass are estimated on basis of ergosterol (fungal) and qPCR-16SrDNA (bacteria).
CO2 measurements
For CO2 efflux measurements, the bottles containing the soils were tightly closed 24 hours before sampling. Headspace CO2 was sampled through the lid septa and directly injected into 5.9 mL evacuated Exetainer vials (Labco Ltd., Buckinghamshire, UK). We sampled at 0.2 (5 hours), 1, 2, 3, 4, 5, 7, 9, 11, 15, 21, 30 days of incubation (12 sampling times in total). An aliquot of the gas samples (250 μl for each vial) was injected using an auto-sampler in the Ultra GC gas chromatograph (Interscience, Breda, The Netherlands) equipped with a flame ionization detector (FID) and a Rt-QBOND (30 m, 0.32 mm, ID) capillary column. Helium was used as carrier gas and the oven temperature was kept at 50 °C with a flow of 5 ml. For the analysis of the 13C-CO2 we used a Thermo Scientific gaschromatograph with combustion interface (Conflo III) system connected to a Thermo Scientific Delta V Plus isotope ratio mass spectrometer (Thermo Scientific, Bremen, Germany). A second aliquot of the gas samples (250 μl) was injected into the split injector (split ratio 1:10) and eluted with helium (5 ml/min) on Rt-QBOND (30 m, 0.32 mm, ID) capillary column at 31°C. The reference gas was calibrated with Methane δC (VPDB) -38.25 (Arndt Schimmelmann, Indiana University, Bloomington, USA). CO2 concentrations of 12 air samples per bottle were used to calculate the cumulative respiration for the whole incubation period. Cumulative fluxes were calculated by linear interpolation between measuring times.
Priming effect calculations
The percentage of respired CO2 derived from 13C substrate was calculated for all treatments and sampling times according to the formula:
| (1) |
Where δC is the δ13C value of the respired CO2 from control soils, δT is the δ13C value in respired CO2 from treated soils and δL is the δ13C value of the labelled substrate [42]. Data were expressed in μg C g−1 dry weight soil. PE was then calculated as the total respired CO2 in treated soils minus the substrate-derived CO2 and minus the respired CO2 in the control and expressed in μg C g−1 dry weight soil [13]:
| (2) |
Statistical analyses
To test how the amount of added glucose, nitrogen addition and their interactions affected glucose-derived CO2, PEs and microbial biomass (measured at fourth and thirtieth day of incubation) per soil type, we used two-way ANOVA. Data grouped per soil type with or without N addition were analysed with a one-way ANOVA, followed by post-hoc Tukey’s test, to determine differences between glucose additions. In case of unequal variances among treatments, statistical comparisons were performed by Tamhane’s test. We used one-sample T-tests to test whether PEs were different from zero. We used a regression analysis to test the relationship between the amount of added 13C-glucose (15%, 50%, and 200% of the microbial biomass C) and PEs. We used Pearson’s correlation coefficient to test for the relationships between substrate derived C and total respired C, and between substrate derived C and primed C. Statistical analyses were performed using IBM SPSS Statistics 22.
Results
CO2 effluxes and priming effect
The highest CO2 effluxes were observed in the arable and forest soils that received the highest amount of glucose (C-glucose equal to 200% of the microbial biomass carbon) (P < 0.05). In contrast, the lowest levels of CO2 evolution were seen for grassland soils amended with the highest amount of glucose (S1 Fig). Almost none of the additions of C-glucose equal to 15% and 50% of the microbial biomass carbon resulted in significant extra total CO2 evolution as compared to the control (P > 0.05). For grassland soils there were no significant differences in CO2 evolution for any of the carbon and nitrogen additions (S1 Fig) (P > 0.05).
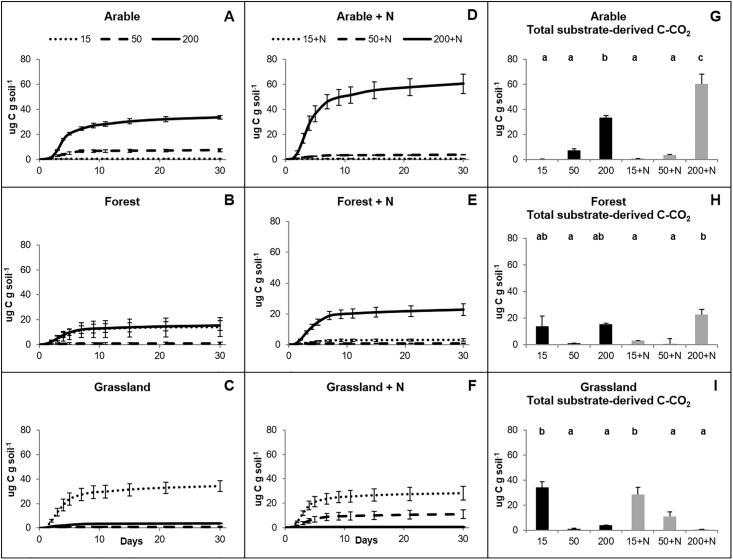
The CO2 evolution pattern was generally the same as compared to the release of substrate-derived C (13CO2, released from labeled glucose) (Fig 1). The positive relationship between substrate-derived CO2 and total soil respiration is confirmed by correlation analysis (R2 = 0.4577; P < 0.001; S2A Fig). Yet, the strength of the relationship between substrate derived CO2 and total respiration decreased for the different soils following this order: arable soils (R2 = 0.6977; P < 0.0001) > grassland soils (R2 = 0.3576; P = 0.002) > forest soils (R2 = 0.2938; P = 0.006). The highest amounts of substrate-derived C were observed for arable and forest soils that received C-glucose equal to 200% (P < 0.05; Fig 1G and 1H). On the contrary, for the grasslands soils the amount of substrate-derived C were highest in the treatments with the lowest amount of added glucose (P < 0.05; Fig 1I).
Fig 1. Substrate-derived CO2 (μg C-CO2 g dw soil-1) in soils from three ecosystems as induced by three different amounts of 13C-glucose (15%, 50%, and 200% of the microbial biomass carbon).
A-F: Cumulative accumulation of substrate-derived CO2 over 30 days of incubation. G-H: Total substrate-derived CO2 after 30 days of incubation. N: NH4NO3. Statistically significant differences (P < 0.05) are marked with different letters. NS: no significant differences. Error bars represent standard errors (n = 4). Two-way ANOVA results are reported in S1 Table.
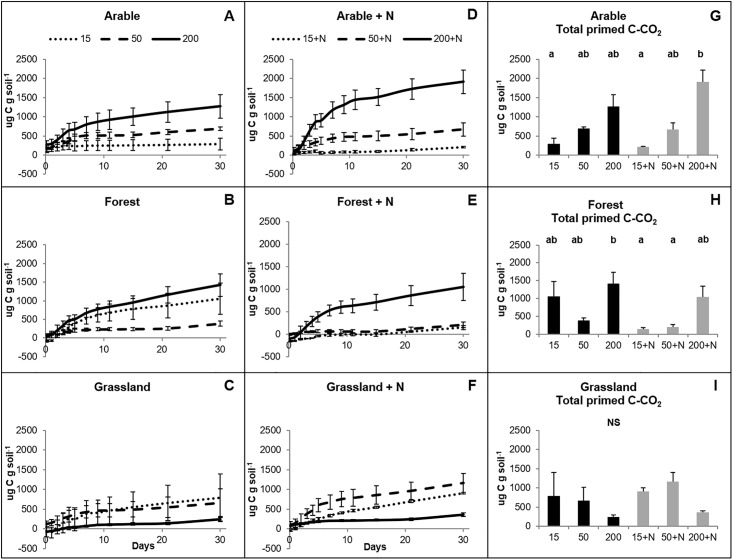
After 30 days of incubation, glucose had induced a positive PE (increase of respiration of unlabeled C) for almost all the soils (Fig 2; S2 Table). Primed C in arable soils increased with increasing amount of glucose added (Fig 2G) (P < 0.05). In forest soils this pattern was less clear although the highest amounts of primed C were also seen for the highest glucose additions (Fig 2H). The responses of the grassland soils to the different amount of glucose added were not significantly different in magnitude from each other in all treatments (P > 0.05). However, PE becomes significantly different from zero when glucose additions were combined with mineral N additions (S1 Table). For several additions in the forest and grassland soils we observed an initial (< 10 days) negative PE (Fig 2).
Fig 2. Primed CO2 (μg C-CO2 g dw soil-1) in soils from three ecosystems as induced by three different amounts of 13C-glucose (15%, 50%, and 200% of the microbial biomass carbon) after 30 days of incubation.
A-F: Cumulative accumulation of primed CO2 over 30 days of incubation. G-H: Total primed CO2 after 30 days of incubation. N: NH4NO3. Statistically significant differences (P < 0.05) are marked with different letters. NS: no significant differences. Error bars represent standard errors (n = 4). Two-way ANOVA results are reported in S1 Table.
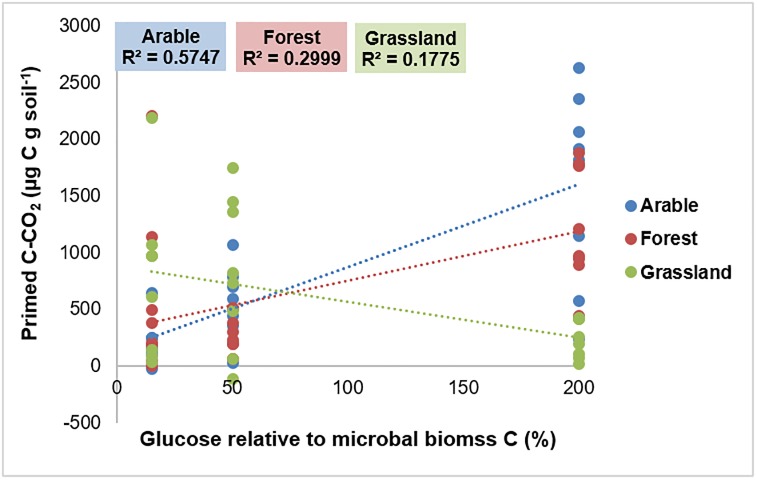
The regression analysis showed a significant positive linear relationship between the amount of the added C expressed as % of the microbial biomass C and primed C for arable soils (R2 = 0.5747; P < 0.0001) and forest soils (R2 = 0.2999; P = 0.006), respectively (Fig 3). For the grassland soils this was a negative relationship (R2 = 0.1775; P = 0.04).
Fig 3. Regression analysis for the amount of added C-glucose expressed as % of microbial biomass (15%, 50% and 200%) versus primed C-CO2.
Treatments are grouped together according to the ecosystems from which the soils were obtained.
We found a positive correlation between substrate-derived CO2 and PEs for the different ecosystems (S2B Fig). In this case the strength of the positive relationship between substrate derived CO2 and PEs decreased from the arable soils (R2 = 0.72; P < 0.0001) to forest soils (R2 = 0.2702; P = 0.009) and grassland soils (R2 = 0.1856; P = 0.03).
The ratio between primed C to substrate-derived C was highest for the arable soils amended with the lowest amount of C-glucose combined with N (P < 0.05; S3 Fig). This relative strong impact of low doses of glucose on PEs was not seen for the soils originating from natural ecosystems (S3B and S3C Fig).
In general, addition of N had no significant effect on glucose-derived C and PEs (P = 0.347 and 0.581, respectively; S1 Table). It stimulated higher substrate derived respiration only in arable soils amended with C-glucose equal to 200% of the microbial biomass carbon (P < 0.01) and had the same tendency for primed C (P = 0.007) (Figs 1G and 2G).
Microbial biomass
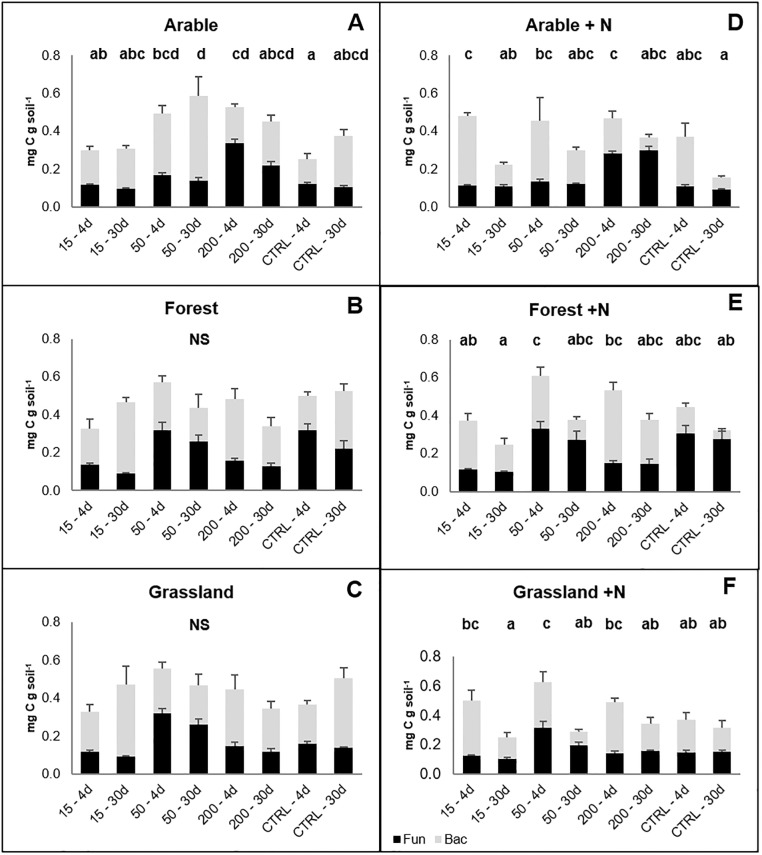
Initial microbial biomass was highest for forest soils, followed by grassland and arable soils (P < 0.05, Table 2). In addition, we found the highest initial fungal biomass in forest soils (P < 0.05), whereas there were no differences between arable and grassland soils. The three ecosystems did not differ in terms of soil bacterial biomass (P > 0.05, Table 2). After 4 days and 30 days of incubation, soils were sampled from each treatment to estimate the microbial biomass carbon. Generally, the microbial biomass remained constant throughout the incubation period, showing no particular differences (P < 0.05) among arable soils and no significant differences among forest and grassland treatments (Fig 4, S3 Table). The proportion of fungal and bacterial biomass fluctuated between treatments but without a consistent pattern. Yet, we observed a trend for all the treatments amended with nitrogen, namely a decrease in total microbial biomass over time (Fig 4D, 4E and 4F). This appeared to be mainly caused by a decrease in bacterial biomass (S4 Fig).
Fig 4. Microbial biomass (fungi + bacteria, mg C g dw soil-1) measured at 4 and 30 days of incubation of glucose-amended soils obtained from three ecosystems.
Fungal and bacterial biomass are estimated on basis of ergosterol (fungal) and qPCR-16SrDNA (bacteria). 15, 50 and 200 indicate the quantity of glucose-C added representing 15%, 50%, and 200% of the initial microbial biomass carbon. 4d: fourth day of incubation. 30d: thirtieth day of incubation. CTRL: control treatment. N: NH4NO3. Statistically significant differences (P < 0.05) are marked with different letters. NS: no significant differences. Error bars represent standard errors (n = 4). Black columns: fungal biomass. Grey columns: bacterial biomass. Two-way ANOVA results are reported in S3 Table.
Discussion
Amount of trigger compound and priming effects
Linear regression between the amount of added C (expressed % of the initial microbial biomass) and primed C revealed contrasting results for the different ecosystems. The meta-analysis by Blagodatskaya and Kuzyakov (2008) has indicated that additions of trigger compounds up to 15% of microbial biomass C induce a linear increase in PEs. In contrast, when the added amount of trigger compounds is higher than 50% of the microbial biomass C, an exponential decrease in PEs was seen. Our study could not confirm their findings and therefore the ratio between the amount of added C to the size of the soil microbial biomass does not appear to be a universal predictor for soil organic matter PEs. A similar conclusion has recently been drawn [24].
In contrast to our observations, Liu and colleagues (2017) found a positive linear relationship between increasing C additions and PEs in the natural ecosystems they studied, including grassland and forest ecosystems. These contrasting results might be related to different composition and structure of the soil microbial communities investigated in the two studies [43]. In our case, the strongest positive relationship of PEs with the amount of added glucose was seen for arable soils. A common agricultural practice is to amend soils with different organic matter residues such as manure and compost. Part of these materials are easily degradable as indicated by initial high respiration rates after addition to arable soils. Therefore, microbes in agricultural soils may be better adapted to receive high inputs of easily degradable organic matter and this could be the reason why they can cope better with temporary high organic carbon additions than soil microbes in natural ecosystems [44–46].
We found a positive correlation between substrate-derived C (13C) and primed C (12C) albeit that the strength of this correlation was different for the three ecosystems included. Arable soils showed the strongest correlation, followed by forest and grassland soils. These results indicate that the amount of energy obtained from decomposing trigger substrates is an important factor that defines the magnitude of PEs [47]. A strong positive correlation between substrate derived CO2 and primed CO2 was recently also observed [25]. Yet, the accumulation curves between substrate-derived C and primed C differed (Figs 1 and 2). Substrate-derived C accumulation was completed during the first week of incubation whereas primed C increased until the end of the experiment (30 days). This has been reported before and indicates that the initial microbial activation by the added glucose continued for a longer period [6,48].
Arable soils showed a different pattern of PE stimulation than natural soils as the lowest amount of glucose resulted in the highest ratio between primed C to substrate-derived C, in particular when glucose was combined with N (S3 Fig). This indicates a relatively stronger impact of low trigger substrate concentrations on PEs, which may be ascribed to the higher temporal heterogeneity of availability of energy sources in arable soils due to agricultural management practices such as fallow followed by addition of organic fertilizers. In such managed arable soils, microbes may have a strategy to become highly activated when easily degradable substrates become available [49,50].
It is largely assumed that lack of N in soils will induce high PEs since soil microbes are triggered to mine SOM to acquire this nutrient [5,24]. Following this so-called microbial N mining theory, a less strong PE is expected when C substrate is added together with N. In line with our hypothesis, we found that N addition had no or little effect on PEs. Arable and grassland soils did not differ in C:N ratio, while forest soils had the highest one. Hence, according to the N mining theory, the strongest negative effect of N addition on PEs could have been expected for the forest soils. This was not the case, indicating that other SOM properties are probably more important for describing the effect of N on PEs [51]. Several other studies did also show that simultaneous addition of C and N can increase or have no effect on PEs [2,29,30,34]. In line with these studies N mining theory was recently challenged [25] since in its current form it does not contribute to an explanation for PEs.
In grassland and forest soils, amended with the lowest glucose input, we observed a temporary decrease in the decomposition of SOM after the first few days of incubation. Negative PE was previously shown in the early phases after C addition to soils [2,24,52]. The shift of microbes from SOM decomposition to uptake of added C and N substrates is indicated to be the underlying mechanism of negative PEs [9,48]. Liu et al. (2017) found an overall negative PE for the whole incubation period with multiple low C additions and attributed this to a minimum amount of energy (threshold) needed to overcome N limitation. Qiao et al. (2016) indicated that different mechanisms can be responsible for negative PEs depending on the intrinsic C:N ratios of soil organic matter and C:N ratios of the trigger substrates. In our case, negative PE is followed by positive PE and is therefore more likely to be the result of initial activation of microbes using internal reserve material (negative apparent PE) [6].
The lack of respiration response (S1 Fig) and the very low PEs that we have seen in the grassland soils that received high C substrate additions might be due to a glucose oversaturation of microorganisms. Microbes present in the grassland samples receiving the highest amount of 13C-glucose started using this substrate but due to possible osmotic stress the total activity subsequently declined [53].
The discrepancy in the results between our work and other studies investigating PEs might be due to the differences we had in the experimental approaches, such as the ecosystems under investigation, the length of the experiments, the simultaneous amendments of C and N versus only C additions and the frequency of C input (single versus repeated C additions). With respect to the latter, single versus multiple additions might influence PE differently [13] as they have different impacts on the ability of microbes to invest energy in the synthesis of SOM-degrading enzymes [7,24,25]. Single substrate applications represent short-term pulses of easily accessible and degradable substrates that produce hotspots of microbial activity that induce accelerated decomposition process rates [54].
Microbial biomass response
In our study the microbial biomass remained overall constant during the whole incubation period of the experiment although we found differences in PEs among soils. Generally, glucose did not stimulate the growth of fungi and bacteria differently, and both microbial groups appeared to play a role in PEs. The addition of the trigger compound could have served as energy source for the soil microbial community, stimulating the production of extracellular enzymes with subsequent increase in the decomposition of SOM [55].
We observed a decrease in microbial biomass in all treatments amended with N. Reduction of bacterial and fungal biomass in response to N fertilization is consistent with previous studies [56–58]. The addition of glucose and N to the soil mesocosms might have altered the demands necessary for microbial growth [59]. Furthermore, the effects of N fertilization can depend on the soil conditions prior to fertilization [56].
Conclusions and perspectives
In this study we investigated the effect of different ratios of the amount of organic trigger compounds to initial soil microbial biomass on PEs. We did not find the contrasting effects of low and high ratios as predicted by a previous meta-analysis [6]. Our results support those of Liu et al. (2017) with respect to the lack of predictability of trigger substrate to microbial biomass ratios but only partly with respect to a consistent increase of positive PEs with increasing concentrations of trigger substrates. In arable and forest soils the primed C increased with an increasing amount of added glucose, but this was not the case for grassland soils. Hence, the proposed predictive value of trigger-substrate concentrations for explaining PEs [24] requires more investigations. Yet, we observed a significant relationship between the mineralization of the added trigger substrate and PEs for all soil types, indicating the importance of energy obtained from trigger-substrates for PEs. N additions together with glucose had little or no effect on PEs, rejecting the N mining theory. To be able to explain mechanisms of PEs and integrate PEs in global soil carbon models, more studies of soils with different properties are needed. Furthermore, it is important to understand how the quality and quantity of added compounds affect PEs in arable soils in order to improve the management of soil organic carbon dynamics.
Supporting information
A-F: Cumulative accumulation of total CO2 over 30 days of incubation. G-H: total CO2 after 30 days of incubation. CTRL: control treatment. Statistically significant differences (P < 0.05) are marked with different letters. NS: no significant differences. N: NH4NO3. Error bars represent standard errors (n = 4).
(TIF)
Treatments are grouped together according to the soil type.
(TIF)
N: NH4NO3. Statistically significant differences (P < 0.05) are marked with different letters. NS: no significant differences. Error bars represent standard errors (n = 4).
(TIF)
15, 50 and 200 represent the quantity of C added as equal to 15%, 50%, and 200% of the initial microbial biomass carbon. 4d: fourth day of incubation. 30d: thirtieth day of incubation. CTRL: control treatment. N: NH4NO3.
(TIF)
(DOCX)
15, 50 and 200 represent the quantity of C added as equal to 15%, 50%, and 200% of the microbial biomass carbon. N: NH4NO3. *: Significant values (P < 0.05).
(DOCX)
(DOCX)
Acknowledgments
We thank Machiel Bosch of Natuurmonumenten for permission to sample the grassland soils and Derks family for sampling the arable soils. We are thankful to Stijn van Gils, Jan Geert Bruggink and Iris Chardon for their practical help in the field and lab. This is publication number 6729 of the NIOO-KNAW Netherlands Institute of Ecology. Raw data are available on: https://dataverse.nl/dvn/dv/nioo-me.
Data Availability
Raw data are available on: https://dataverse.nl/dvn/dv/nioo-me.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Kuzyakov Y, Friedel JK, Stahr K. Review of mechanisms and quantification of priming effects. Soil Biol Biochem. 2000;32: 1485–1498. 10.1016/S0038-0717(00)00084-5 [DOI] [Google Scholar]
- 2.Di Lonardo DP, De Boer W, Klein Gunnewiek PJA, Hannula SE, Van der Wal A. Priming of soil organic matter: Chemical structure of added compounds is more important than the energy content. Soil Biol Biochem. 2017;108: 41–54. 10.1016/j.soilbio.2017.01.017 [DOI] [Google Scholar]
- 3.Guenet B, Neill C, Bardoux G, Abbadie L. Is there a linear relationship between priming effect intensity and the amount of organic matter input? Appl Soil Ecol. 2010;46: 436–442. 10.1016/j.apsoil.2010.09.006 [DOI] [Google Scholar]
- 4.Whitaker J, Ostle N, McNamara NP, Nottingham AT, Stott AW, Bardgett RD, et al. Microbial carbon mineralization in tropical lowland and montane forest soils of Peru. Front Microbiol. 2014;5: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dijkstra F a, Carrillo Y, Pendall E, Morgan J a. Rhizosphere priming: a nutrient perspective. Front Microbiol. 2013;4: 216 10.3389/fmicb.2013.00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blagodatskaya E, Kuzyakov Y. Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: critical review. Biol Fertil Soils. 2008;45: 115–131. 10.1007/s00374-008-0334-y [DOI] [Google Scholar]
- 7.Fontaine S, Mariotti A, Abbadie L. The priming effect of organic matter: a question of microbial competition? Soil Biol Biochem. 2003;35: 837–843. 10.1016/S0038-0717(03)00123-8 [DOI] [Google Scholar]
- 8.Fontaine S, Henault C, Aamor A, Bdioui N, Bloor JMG, Maire V, et al. Fungi mediate long term sequestration of carbon and nitrogen in soil through their priming effect. Soil Biol Biochem. 2011;43: 86–96. 10.1016/j.soilbio.2010.09.017 [DOI] [Google Scholar]
- 9.Kuzyakov Y. Review: Factors affecting rhizosphere priming effects. J Plant Nutr Soil Sci. 2002;165: 382. [Google Scholar]
- 10.Cheng W, Kuzyakov Y. Root Effects on Soil Organic Matter Decomposition. 2005; 119–144. [Google Scholar]
- 11.Blagodatskaya E, Khomyakov N, Myachina O, Bogomolova I, Blagodatsky S, Kuzyakov Y. Microbial interactions affect sources of priming induced by cellulose. Soil Biol Biochem. 2014;74: 39–49. 10.1016/j.soilbio.2014.02.017 [DOI] [Google Scholar]
- 12.Schimel JP, Schaeffer SM. Microbial control over carbon cycling in soil. Front Microbiol. 2012;3: 348 10.3389/fmicb.2012.00348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiao N, Xu X, Hu Y, Blagodatskaya E, Liu Y, Schaefer D, et al. Carbon and nitrogen additions induce distinct priming effects along an organic-matter decay continuum. 2016;6: 19865 10.1038/srep19865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Nobili M, Contin M, Mondini C, Brookes P. Soil microbial biomass is triggered into activity by trace amounts of substrate. Soil Biol Biochem. 2001;33: 1163–1170. 10.1016/S0038-0717(01)00020-7 [DOI] [Google Scholar]
- 15.Blagodatskaya EV, Blagodatsky SA, Anderson T-H, Kuzyakov Y. Priming effects in Chernozem induced by glucose and N in relation to microbial growth strategies. Appl Soil Ecol. 2007;37: 95–105. 10.1016/j.apsoil.2007.05.002 [DOI] [Google Scholar]
- 16.Mondini C, Cayuela ML, Sanchez-Monedero M a., Roig a., Brookes PC. Soil microbial biomass activation by trace amounts of readily available substrate. Biol Fertil Soils. 2006;42: 542–549. 10.1007/s00374-005-0049-2 [DOI] [Google Scholar]
- 17.Luna-Guido M., Beltrán-Hernández R., Dendooven L. Dynamics of 14 C-labelled glucose in alkaline saline soil. Soil Biol Biochem. 2001;33: 707–719. [Google Scholar]
- 18.Hamer U, Marschner B. Priming effects in different soil types induced by fructose, alanine, oxalic acid and catechol additions. Soil Biol Biochem. 2005;37: 445–454. 10.1016/j.soilbio.2004.07.037 [DOI] [Google Scholar]
- 19.Perelo LW, Munch JC. Microbial immobilisation and turnover of 13 C labelled substrates in two arable soils under eld and laboratory conditions. Soil Biol Biochem. 2005;37: 2263–2272. 10.1016/j.soilbio.2005.02.039 [DOI] [Google Scholar]
- 20.Santruckova H, Picek T, Tykva R, Šimek M, Bohumil P. Short-term partitioning of 14C-[U]-glucose in the soil microbial pool under varied aeration status. Biol Fertil Soils. 2004;40: 386–392. 10.1007/s00374-004-0790-y [DOI] [Google Scholar]
- 21.Conde E, Cardenas M, Ponce-Mendoza a., Luna-Guido ML, Cruz-Mondragón C, Dendooven L. The impacts of inorganic nitrogen application on mineralization of 14C-labelled maize and glucose, and on priming effect in saline alkaline soil. Soil Biol Biochem. 2005;37: 681–691. 10.1016/j.soilbio.2004.08.026 [DOI] [Google Scholar]
- 22.Falchini L, Naumova N, Kuikman PJ, Bloem J, Nannipieri P. CO2 evolution and denaturing gradient gel electrophoresis profiles of bacterial communities in soil following addition of low molecular weight substrates to simulate root exudation. Soil Biol Biochem. 2003;35: 775–782. 10.1016/S0038-0717(03)00105-6 [DOI] [Google Scholar]
- 23.Shen J, Bartha R. Priming effect of glucose polymers in soil-based biodegradation tests. Soil Biol Biochem. 1997;29: 1195–1198. 10.1016/S0038-0717(97)00031-X [DOI] [Google Scholar]
- 24.Liu X-JA, Sun J, Mau RL, Finley BK, Compson ZG, van Gestel N, et al. Labile carbon input determines the direction and magnitude of the priming effect. Appl Soil Ecol. 2017;109: 7–13. 10.1016/j.apsoil.2016.10.002 [DOI] [Google Scholar]
- 25.Mason-Jones K, Schmücker N, Kuzyakov Y. Contrasting effects of organic and mineral nitrogen challenge the N-Mining Hypothesis for soil organic matter priming. Soil Biol Biochem. 2018;124: 38–46. 10.1016/j.soilbio.2018.05.024 [DOI] [Google Scholar]
- 26.Sullivan BW, Hart SC. Evaluation of mechanisms controlling the priming of soil carbon along a substrate age gradient. Soil Biol Biochem. 2013;58: 293–301. 10.1016/j.soilbio.2012.12.007 [DOI] [Google Scholar]
- 27.Moorhead DL, Sinsabaugh RL. A theoretical model of litter decay and microbial interaction. Ecol Monogr. 2006;76: 151–174. 10.1890/0012-9615(2006)076[0151:ATMOLD]2.0.CO;2 [DOI] [Google Scholar]
- 28.Chen R, Senbayram M, Blagodatsky S, Myachina O, Dittert K, Lin X, et al. Soil C and N availability determine the priming effect: microbial N mining and stoichiometric decomposition theories. Glob Chang Biol. 2014;20: 2356–2367. 10.1111/gcb.12475 [DOI] [PubMed] [Google Scholar]
- 29.Tian Q, Yang X, Wang X, Liao C, Li Q, Wang M, et al. Microbial community mediated response of organic carbon mineralization to labile carbon and nitrogen addition in topsoil and subsoil. Biogeochemistry. 2016;128: 125–139. 10.1007/s10533-016-0198-4 [DOI] [Google Scholar]
- 30.Chowdhury S, Farrell M, Bolan N. Priming of soil organic carbon by malic acid addition is differentially affected by nutrient availability. Soil Biol Biochem. 2014;77: 158–169. 10.1016/j.soilbio.2014.06.027 [DOI] [Google Scholar]
- 31.Hessen D, Ågren G, Anderson T. Carbon sequestration in ecosystems: the role of stoichiometry. Ecology. 2004;85: 1179–1192. Available: http://www.esajournals.org/doi/abs/10.1890/02-0251 [Google Scholar]
- 32.Sterner RW, Elser JJ. Ecological stoichiometry: the biology of elements from molecules to the biosphere. Princeton University Press; 2002. [Google Scholar]
- 33.Drake JE, Darby BA, Giasson MA, Kramer MA, Phillips RP, Finzi AC. Stoichiometry constrains microbial response to root exudation- insights from a model and a field experiment in a temperate forest. Biogeosciences. 2013;10: 821–838. 10.5194/bg-10-821-2013 [DOI] [Google Scholar]
- 34.Di Lonardo DP, Manrubia M, De Boer W, Zweers H, Veen GF, Van der Wal A. Relationship between home-field advantage of litter decomposition and priming of soil organic matter. Soil Biol Biochem. 2018;126: 49–56. 10.1016/j.soilbio.2018.07.025 [DOI] [Google Scholar]
- 35.van der Wal A, van Veen J a., Smant W, Boschker HTS, Bloem J, Kardol P, et al. Fungal biomass development in a chronosequence of land abandonment. Soil Biol Biochem. 2006;38: 51–60. 10.1016/j.soilbio.2005.04.017 [DOI] [Google Scholar]
- 36.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56: 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muyzer G, De Waal E. C, Uitterlinden A. G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Env Microbiol. 1993;59: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santelli CM, Orcutt BN, Banning E, Bach W, Moyer CL, Sogin ML, et al. Abundance and diversity of microbial life in ocean crust. Nature. 2008;453: 653–6. 10.1038/nature06899 [DOI] [PubMed] [Google Scholar]
- 39.Bloem J, Veninga M, Shepherd J. Fully automatic determination of soil bacterium numbers, cell volumes, and frequencies of dividing cells by confocal laser scanning microscopy and image analysis. Appl Env Microbiol. 1995;61: 926–936. Available: http://aem.asm.org/content/61/3/926.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Ridder-Duine AS, Smant W, van der Wal A, van Veen J a., de Boer W. Evaluation of a simple, non-alkaline extraction protocol to quantify soil ergosterol. Pedobiologia (Jena). 2006;50: 293–300. 10.1016/j.pedobi.2006.03.004 [DOI] [Google Scholar]
- 41.Klamer M, Baath E. Estimation of conversion factors for fungal biomass determination in compost using ergosterol and PLFA 18:2ω6,9. Soil Biol Biochem. 2004;36: 57–65. 10.1016/j.soilbio.2003.08.019 [DOI] [Google Scholar]
- 42.Nottingham AT, Turner BL, Chamberlain PM, Stott AW, Tanner EVJ. Priming and microbial nutrient limitation in lowland tropical forest soils of contrasting fertility. Biogeochemistry. 2012;111: 219–237. 10.1007/s10533-011-9637-4 [DOI] [Google Scholar]
- 43.Ushio M, Wagai R, Balser TC, Kitayama K. Variations in the soil microbial community composition of a tropical montane forest ecosystem: Does tree species matter? Soil Biol Biochem. 2008;40: 2699–2702. 10.1016/j.soilbio.2008.06.023 [DOI] [Google Scholar]
- 44.Lupwayi NZ, Larney FJ, Blackshaw RE, Kanashiro DA, Pearson DC. Phospholipid fatty acid biomarkers show positive soil microbial community responses to conservation soil management of irrigated crop rotations. Soil Tillage Res. 2017;168: 1–10. 10.1016/j.still.2016.12.003 [DOI] [Google Scholar]
- 45.Bowles TM, Acosta-Martínez V, Calderón F, Jackson LE. Soil enzyme activities, microbial communities, and carbon and nitrogen availability in organic agroecosystems across an intensively-managed agricultural landscape. Soil Biol Biochem. 2014;68: 252–262. 10.1016/j.soilbio.2013.10.004 [DOI] [Google Scholar]
- 46.Arcand MM, Levy-Booth DJ, Helgason BL. Resource legacies of organic and conventional management differentiate soil microbial carbon use. Front Microbiol. 2017;8: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wild B, Schnecker J, Alves RJE, Barsukov P, Bárta J, Čapek P, et al. Input of easily available organic C and N stimulates microbial decomposition of soil organic matter in arctic permafrost soil. Soil Biol Biochem. 2014;75: 143–151. 10.1016/j.soilbio.2014.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuzyakov Y, Bol R. Sources and mechanisms of priming effect induced in two grassland soils amended with slurry and sugar. Soil Biol Biochem. 2006;38: 747–758. 10.1016/j.soilbio.2005.06.025 [DOI] [Google Scholar]
- 49.Butler JL, Williams M a, Bottomley PJ, Myrold DD. Microbial community dynamics associated with rhizosphere carbon flow microbial community dynamics associated with rhizosphere carbon flow. Appl Environ Microbiol. 2003;69: 6793–6800. 10.1128/AEM.69.11.6793-6800.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu Y, Murase J, Watanabe A, Sugimoto A, Kimura M. Linking microbial community dynamics to rhizosphere carbon flow in a wetland rice soil. FEMS Microbiol Ecol. 2004;48: 179–186. 10.1016/j.femsec.2004.01.004 [DOI] [PubMed] [Google Scholar]
- 51.Schmidt MWI, Torn MS, Abiven S, Dittmar T, Guggenberger G, Janssens I a., et al. Persistence of soil organic matter as an ecosystem property. Nature. 2011;478: 49–56. 10.1038/nature10386 [DOI] [PubMed] [Google Scholar]
- 52.Wang H, Boutton TW, Xu W, Hu G, Jiang P, Bai E. Quality of fresh organic matter affects priming of soil organic matter and substrate utilization patterns of microbes. Sci Rep. 2015;5: 10102 10.1038/srep10102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989;53: 121–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuzyakov Y, Blagodatskaya E. Microbial hotspots and hot moments in soil: Concept & review. Soil Biol Biochem. 2015;83: S29–S36. 10.1016/j.soilbio.2015.01.025 [DOI] [Google Scholar]
- 55.Schimel J, Weintraub MN. The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: a theoretical model. Soil Biol Biochem. 2003;35: 549–563. 10.1016/S0038-0717(03)00015-4 [DOI] [Google Scholar]
- 56.Demoling F, Ola Nilsson L, Bååth E. Bacterial and fungal response to nitrogen fertilization in three coniferous forest soils. Soil Biol Biochem. 2008;40: 370–379. 10.1016/j.soilbio.2007.08.019 [DOI] [Google Scholar]
- 57.Knorr M, Frey SD, Curtis PS. Nitrogen additions and litter decomposition: A meta analysys. Ecology. 2005;86: 3252–3257. 10.1890/05-0150 [DOI] [Google Scholar]
- 58.Ramirez KS, Craine JM, Fierer N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob Chang Biol. 2012;18: 1918–1927. 10.1111/j.1365-2486.2012.02639.x [DOI] [Google Scholar]
- 59.Mooshammer M, Wanek W, Zechmeister-Boltenstern S, Richter A. Stoichiometric imbalances between terrestrial decomposer communities and their resources: Mechanisms and implications of microbial adaptations to their resources. Front Microbiol. 2014;5: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A-F: Cumulative accumulation of total CO2 over 30 days of incubation. G-H: total CO2 after 30 days of incubation. CTRL: control treatment. Statistically significant differences (P < 0.05) are marked with different letters. NS: no significant differences. N: NH4NO3. Error bars represent standard errors (n = 4).
(TIF)
Treatments are grouped together according to the soil type.
(TIF)
N: NH4NO3. Statistically significant differences (P < 0.05) are marked with different letters. NS: no significant differences. Error bars represent standard errors (n = 4).
(TIF)
15, 50 and 200 represent the quantity of C added as equal to 15%, 50%, and 200% of the initial microbial biomass carbon. 4d: fourth day of incubation. 30d: thirtieth day of incubation. CTRL: control treatment. N: NH4NO3.
(TIF)
(DOCX)
15, 50 and 200 represent the quantity of C added as equal to 15%, 50%, and 200% of the microbial biomass carbon. N: NH4NO3. *: Significant values (P < 0.05).
(DOCX)
(DOCX)
Data Availability Statement
Raw data are available on: https://dataverse.nl/dvn/dv/nioo-me.