Abstract
PCR-Restriction Fragment Length Polymorphism (RFLP) analyses targeting multiple nuclear genes were established for the simple and practical identification of Leishmania species without using expensive equipment. This method was applied to 92 clinical samples collected at 33 sites in 14 provinces of Ecuador, which have been identified at the species level by the kinetoplast cytochrome b (cyt b) gene sequence analysis, and the results obtained by the two analyses were compared. Although most results corresponded between the two analyses, PCR-RFLP analyses revealed distribution of hybrid strains between Leishmania (Viannia) guyanensis and L. (V.) braziliensis and between L. (V.) guyanensis and L. (V.) panamensis, of which the latter was firstly identified in Ecuador. Moreover, unexpected parasite strains having the kinetoplast cyt b gene of L. (V.) braziliensis and nuclear genes of L. (V.) guyanensis, L. (V.) panamensis, or a hybrid between L. (V.) guyanensis and L. (V.) panamensis were identified. This is the first report of the distribution of a protozoan parasite having mismatches between kinetoplast and nuclear genes, known as mito-nuclear discordance. The result demonstrated that genetically complex Leishmania strains are present in Ecuador. Since genetic exchanges such as hybrid formation were suggested to cause higher pathogenicity in Leishmania and may be transmitted by more species of sand flies, further country-wide epidemiological studies on clinical symptoms, as well as transmissible vectors, will be necessary.
Author summary
Leishmaniasis caused by intracellular protozoa of the genus Leishmania is a neglected tropical disease widely distributing worldwide, especially in tropical and subtropical areas. Approximately 20 species are known to be pathogenic to humans, of which eight species have been recorded as causative agents of cutaneous and mucocutaneous leishmaniases in Ecuador. Since infecting species are the major determinant of clinical outcomes, identification at the species level is important for the treatment and prognosis. The parasite species have been identified conventionally by multilocus enzyme electrophoresis (MLEE) and recently by genetic analysis such as sequencing and genotyping. In the present study, PCR-Restriction Fragment Length Polymorphism (RFLP) targeting multiple nuclear genes was employed, and the results were compared with those obtained by kinetoplast cytochrome b (cyt b) gene sequence analysis, which is widely applied to species identification. Although most results corresponded between the two analyses, PCR-RFLP revealed presence of unexpected genetically complex Leishmania strains having characteristics of hybrid and mito-nuclear discordance. Since hybrid strains of Leishmania were suggested to increase disease severity and may be transmitted by a wider range of sand fly species, careful epidemiological research, including clinical courses and vector research, will be needed.
Introduction
Leishmaniasis, caused by protozoan parasites of the genus Leishmania, is a neglected tropical disease widely distributed worldwide, especially in tropical and subtropical areas, affecting at least 12 million people in 96 countries [1]. Approximately 20 Leishmania species belonging to the subgenera Leishmania (Leishmania), Leishmania (Viannia) and Leishmania (Mundinia) are pathogenic to humans [1, 2]. Since infected parasite species is known to be the major determinant of clinical outcomes in leishmaniasis [1], identification of the causative parasite is important for appropriate treatment and prognosis.
Leishmania species have been classified conventionally by multilocus enzyme electrophoresis (MLEE) [3, 4]. Genetic analysis of kinetoplast and nuclear targets, such as cytochrome b (cyt b), cysteine protease (cpb), heat shock protein 70 (hsp70) genes and the internal transcribed spacer (ITS) regions of ribosomal RNA, has commonly been used for species identification due to its sensitivity, simplicity and reliability [5–13]. In addition, a simple PCR-Restriction Fragment Length Polymorphism (RFLP), which does not require costly equipment, was developed for species identification, and the ITS region and hsp70 gene are widely applied to epidemiological studies [11, 14–19].
In Ecuador, leishmaniasis is endemic in Pacific coast, Andean highland, and Amazonian areas, and eight species, Leishmania (Leishmania) mexicana, L. (L.) amazonensis, L. (L.) major-like, L. (Viannia) guyanensis, L. (V.) panamensis, L. (V.) braziliensis, L. (V.) naiffi, and L. (V.) lainsoni, have been recorded as causative agents of cutaneous leishmaniasis (CL) and mucocutaneous leishmaniasis (MCL) [8, 20, 21]. Of these, distribution of L. (L.) amazonensis and L. (L.) major-like have been reported to be localized, and infections by them have not been reported recently [8, 21]. Infection by L. (V.) guyanensis together with its closely-related species, L. (V.) panamensis, has been identified from CL patients in Pacific coast areas by MLEE [21–24]; however, our recent cyt b gene analysis revealed a wide range distribution of L. (V.) guyanensis, without detecting any L. (V.) panamensis in these areas [8]. These results suggest that endemic species may change, or the reported results may be caused by the discordance between the MLEE analysis and kinetoplast cyt b gene analysis employed for species identification. Recently, a countrywide epidemiological study was carried out based on the cyt b sequence analysis and it identified L. (V.) guyanensis and L. (V.) braziliensis widely in Pacific coast and Amazonian areas and L. (L.) mexicana in Andean high lands as current major causative species in Ecuador [8]. Additionally, L. (V.) naiffi and L. (V.) lainsoni were recently recorded in Amazonian areas [8, 20, 25].
In this study, a simple and practical method for the identification of Leishmania species in Ecuador was established on the basis of PCR-RFLP analyses targeting mannose phosphate isomerase (mpi) and 6-phosphogluconate dehydrogenase (6pgd) genes, and the result was compared with that obtained by the cyt b gene sequence analysis. This study demonstrated the presence of genetically complex Leishmania strains in Ecuador, and strongly suggested the importance of applying multiple target approaches to enhance the reliability of species identification and to characterize more detailed genetic properties of the parasite.
Methods
Parasite specimens and clinical samples
Frozen stocks of 24 parasite strains of five Leishmania species [L. (V.) guyanensis, L. (V.) panamensis, L. (V.) braziliensis, L. (L.) major-like, L. (L.) mexicana] that were isolated from CL patients in Ecuador and identified at the species level by MLEE [22–24] (Table 1) were spotted on an FTA Classic Card (Whatman, Newton Center, MA) and subjected to sequence analysis. Three strains of L. (V.) naiffi identified by cyt b gene analysis [25, 26] were also utilized (Table 1).
Table 1. Leishmania strains isolated in Ecuador.
| Species | Strains |
|---|---|
| L. (V.) guyanensis | MHOM/EC/05/EC4 |
| L. (V.) guyanensis | MHOM/EC/05/EC6 |
| L. (V.) guyanensis | MHOM/EC/05/EC7 |
| L. (V.) guyanensis | MHOM/EC/05/EC8 |
| L. (V.) guyanensis | MHOM/EC/05/EC9 |
| L. (V.) guyanensis | MHOM/EC/05/EC11 |
| L. (V.) guyanensis | MHOM/EC/05/EC12 |
| L. (V.) guyanensis | MHOM/EC/05/XPEA1 |
| L. (V.) guyanensis | MHOM/EC/05/LM3 |
| L. (V.) panamensis | MHOM/EC/87/G05 |
| L. (V.) panamensis | MHOM/EC/87/G06 |
| L. (V.) panamensis | MHOM/EC/87/G07 |
| L. (V.) panamensis | MHOM/EC/88/INH23 |
| L. (V.) braziliensis | MHOM/EC/00/Ppa20 |
| L. (V.) braziliensis | MHOM/EC/00/LASU22 |
| L. (V.) naiffi | 07tor |
| L. (V.) naiffi | 13tor1 |
| L. (V.) naiffi | 13tor2 |
| L. (L.) major-like | MHOM/EC/87/G09 |
| L. (L.) major-like | MHOM/EC/88/PT115 |
| L. (L.) mexicana | MHOM/EC/88/PT23 |
| L. (L.) mexicana | MHOM/EC/88/PT27 |
| L. (L.) mexicana | MHOM/EC/88/PT29 |
| L. (L.) mexicana | MHOM/EC/88/PT103 |
| L. (L.) mexicana | MHOM/EC/92/HU3 |
| L. (L.) mexicana | MHOM/EC/92/HU4 |
| L. (L.) mexicana | MHOM/EC/00/HU6 |
Most of the clinical samples employed in this study were collected from patients suspected of CL in the previous study [8, 20], and each 3 samples newly obtained from Provinces of Manabi and Santo Domingo de los Tsachilas, all of which were identified as L. (V.) guyanensis by the cyt b gene analysis, were included in this study. Leishmania parasites were identified on the basis of cyt b sequence analysis [8, 20]. The samples were collected at 33 sites in 14 provinces of Ecuador (S1 Fig). Residual tissue materials were spotted onto an FTA Classic Card, after taking scraped margin samples of active lesions for routine diagnosis. Two-mm-diameter disks of FTA card were punched out from each filter paper, washed three times with an FTA Purification Reagent (Whatman), and subjected to PCR amplification.
PCR and sequence analysis
PCR primers for amplification of cyt b, hsp70, mannose phosphate isomerase (mpi) and 6-phosphogluconate dehydrogenase (6pgd) gene fragments were designed based on the sequence regions conserved among species (Table 2). PCR amplification with a pair of outer primers was performed with 30 cycles of denaturation (95°C, 1 min), annealing (55°C, 1 min) and polymerization (72°C, 2 min) using Ampdirect Plus reagent (Shimadzu Biotech, Tsukuba, Japan). Each 0.5-μl portion of the PCR product was reamplified with inner primers under the same condition described above. The products were cloned into the pGEM-T Easy Vector System (Promega, Madison, WI) and sequences were determined on both strands by the dideoxy chain termination method using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Primers for amplification of a partial sequence of the kinetoplast cytochrome oxidase subunit II-NADH dehydrogenase subunit I region (COII-ND1) were also designed based on the sequences conserved among species (Table 2). The COII-ND1 sequences were determined on both strands by direct sequencing with inner primers, L.COII-2S and L.COII-2R. Restriction enzyme mapping was performed in silico by using BioEdit Sequence Alignment Editor to obtain species-specific RFLP patterns.
Table 2. Primer sequences used in this study.
| Target gene | Primer | Primer sequence (5’ to 3’) |
Expected amplicon size (bp) |
|
|---|---|---|---|---|
| cytochrome b | outer | L.cyt-AS | GCGGAGAGRARGAAAAGGC | 978 |
| (cyt b) | L.cyt-AR | CCACTCATAAATATACTATA | ||
| inner | L.cyt-S | GGTGTAGGTTTTAGTYTAGG | 866 | |
| L.cyt-R | CTACAATAAACAAATCATAATATRCAATT | |||
| cytochrome oxidase | outer | L.COII-1S | AACATAGTTCTCATTGCAGA | 954 |
| subunit II—NADH | L.COII-1R | ACAMCGRCCAGGTTCTCTAC | ||
| dehydrogenase subunit 1 | inner | L.COII-2S | AATGCAACATGCAGTTATWA | 736 |
| (COII-ND1) | L.COII-2R | AATGAATGTATAACATCAAC | ||
| heat shock protein 70 | outer | L.HSP-OS | GGGCACGACGTACTCGTGCG | 1,931 |
| (hsp70) | L.HSP-OR | AGTCGACCTCCTCGACCTTG | ||
| inner | L.HSP-IS2 | CCGTCGTACGTTGCGTTCAC | 1,735 | |
| L.HSP-IR2 | TGCTCTGGTACATCTTGGTC | |||
| outer* | L.HSP-Ty1S | GGCGAGCGCGCGATGACGAA | 847 | |
| L.HSP-OR | AGTCGACCTCCTCGACCTTG | |||
| inner* | L.HSP-Ty2S | CGTTCGACTTGTCCGGCATC | 468 | |
| L.HSP-IR2 | TGCTCTGGTACATCTTGGTC | |||
| mannose phosphate | outer | L.MPI-OS2 | GCCTGGGGCAAGRATGCCGC | 1,214 |
| isomerase | L.MPI-OR | CTCAAGTCGTTGGTCGACGC | ||
| (mpi) | inner | L.MPI-IS2 | CGTCCAGCTTCGTGGCRAAG | 1,130 |
| L.MPI-IR2 | GCCGTACGGYACCGCAAAGC | |||
| 6-phosphogluconate | outer | L.6PGD-OS | GAACGACCTCGGYATTATCG | 1,346 |
| dehydrogenase | L.6PGD-OR | GACACCAGCTGTCCGTACGG | ||
| (6pgd) | inner | L.6PGD-IS | GCCCTGAACATCGCCGAGAA | 1,272 |
| L.6PGD-IR | CGTGTACATGGCGTTGATGT | |||
*The primer sets were used for the PCR-RFLP analysis to differentiate L. (V.) guyanensis from L. (V.) panamensis.
PCR-Restriction Fragment Length Polymorphism (RFLP) analysis
Clinical samples spotted on FTA cards, in which parasites were identified by cyt b gene analysis in a previous study, were subjected to PCR-RFLP analysis. PCR amplifications targeting mpi and 6pgd were performed as described above using a high fidelity DNA polymerase, KOD plus (Toyobo, Osaka, Japan). The PCR products were digested by restriction enzymes HaeIII, HapI, and BstXI for the mpi gene and Bsp1286I and HinfI for the 6pgd gene, and resulting restriction fragment patterns were analyzed by 2% agarose gel electrophoresis. GeneRuler 100 bp Plus DNA Ladder (Thermo Fisher Scientific, Waltham, MA) was used as a DNA size marker. The gel was stained with GelRed Nucleic Acid Gel Stain (Biotium, Hayward, CA), and DNA fragments were visualized with UV transilluminator.
Differentiation between L. (V.) guyanensis and L. (V.) panamensis was performed by restriction enzyme-digestion of the hsp70 gene fragment [27]. Briefly, the hsp70 gene fragment was amplified by a nested PCR using sets of outer primers (L.HSP-Ty1S and L.HSP-OR) and inner primers (L.HSP-Ty2S and L.HSP-IR2) (Table 2). The amplicons were digested with a restriction enzyme, BccI, and resulting fragment patterns were analyzed by 3% agarose gel electrophoresis.
Ethics statement
Clinical samples were collected by local physicians and well-trained laboratory technicians of health centers of the Ministry of Health, Ecuador. For routine parasitological diagnosis, scratching smear samples of skin lesions were taken from suspected leishmaniasis patients at health centers. In this study, only residual tissue materials were collected after the routine procedure to minimize the burden on patients. Signed consent was obtained from the adult subjects and from the children’s parents or guardians, prior to the diagnostic procedures at each health center of the Ministry, providing information on the process of diagnosis and Leishmania species analysis, following the guidelines of the Ethics Committee of the Ministry. The subjects studied were volunteers in routine diagnosis/screening and treatment programs promoted by the Ministry. All routine laboratory examinations were carried out free of charge, and treatment with specific drug, meglumine antimoniate (Glucantime) was also offered free of charge at each health center. The study was approved by the ethics committee of the Graduate School of Veterinary Medicine, Hokkaido University (approval number: vet26-4) and Jichi Medical University (approval number: 17–080) [8].
Results
Sequence analysis of cyt b, hsp70, mpi and 6pgd genes from Leishmania strains
Leishmania cyt b, hsp70, mpi and 6pgd partial gene sequences were amplified from 27 strains of 6 species isolated in Ecuador. Sequences of these fragments showed high degrees of homology (88–100%, 82–100%, 83–100% and 94–100% in cyt b, mpi, 6pgd and hsp70 genes, respectively) with corresponding leishmanial genes registered in GenBank. The restriction enzyme mapping was performed in silico to see if species-specific enzyme sites could be found in cyt b, mpi, 6pgd and hsp70 gene fragments obtained in this study. Species-specific RFLP patterns could not be obtained for the cyt b gene because of intraspecies genetic variations through the sequences. On the hsp70 gene, restriction enzymes to differentiate Leishmania species were found; however, RFLP patterns including several smaller fragments (< 300 bp) were similar among species. Therefore, it seems difficult to identify the species based on RFLP patterns of hsp70 using agarose gel electrophoresis in some cases because of the resolution. On the other hand, restriction enzyme sites that can differentiate Leishmania species in Ecuador were identified in mpi and 6pgd genes, except for two very closely-related species, L. (V.) guyanensis and L. (V.) panamensis. Different RFLP patterns were obtained in L. (V.) guyanensis/L. (V.) panamensis, L. (V.) braziliensis/L. (V.) naiffi, L. (L.) major-like and L. (L.) mexicana for digested mpi gene fragments with a restriction enzyme HaeIII (Fig 1A). Although an RFLP polymorphism was observed in one (strain PT27) of seven L. (L.) mexicana strains, it did not affect species identification (Table 3). L. (V.) braziliensis and L. (V.) naiffi, showing the same RFLP patterns as HaeIII digestion, were differentiated by HpaI digestion (Table 3, Fig 1B). Although L. (V.) lainsoni, a recently reported species in the Ecuadorian Amazon [20], showed the same RFLP patterns as L. (V.) guyanensis/L. (V.) panamensis when digested with HaeIII and HpaI, BstXI-digestion successfully differentiated it from L. (V.) guyanensis/L. (V.) panamensis, as reported in Peruvian strains (S2 Fig) [28].
Fig 1. PCR-RFLP analyses of mpi gene fragments from 6 Leishmania species in Ecuador.
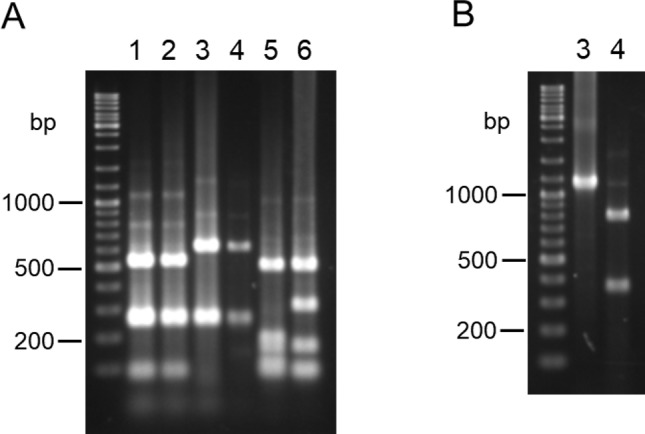
PCR amplification was performed with leishmanial mpi gene-specific primers, and PCR products were digested with (A) HaeIII and (B) HpaI, and resulting restriction fragment patterns were analyzed by agarose gel electrophoresis. 1. L. (V.) guyanensis, 2. L. (V.) panamensis, 3. L. (V.) braziliensis, 4. L. (V.) naiffi, 5. L. (L.) major-like, 6. L. (L.) mexicana.
Table 3. Fragment size of leishmanial mpi and 6pgd genes generated by digestion with selected restriction enzymes.
| Leishmania* | strain | mpi | 6pgd | ||
|---|---|---|---|---|---|
| HaeIII | HpaI | Bsp1286I | HinfI | ||
| L.guy | EC4 | 21, 87, 246, 259, 517 | 1130 | 90, 1182 | 107, 155, 200, 307, 503 |
| L.guy | EC6 | 21, 87, 246, 259, 517 | 1130 | 90, 1182 | 107, 155, 200, 307, 503 |
| L.guy | EC7 | 21, 87, 246, 259, 517 | 1130 | 90, 1182 | 107, 155, 200, 307, 503 |
| L.guy | EC8 | 21, 87, 246, 259, 517 | 1130 | 90, 1182 | 107, 155, 200, 307, 503 |
| L.guy | EC9 | 21, 87, 246, 259, 517 | 1130 | 90, 1182 | 107, 155, 200, 307, 503 |
| L.guy | EC11 | 21, 87, 246, 259, 517 | 1130 | 90, 1182 | 107, 155, 200, 307, 503 |
| L.guy | EC12 | 21, 87, 246, 259, 517 | 1130 | 90, 1182 | 107, 155, 200, 307, 503 |
| L.guy | XPEA1 | 21, 87, 246, 259, 517 | 1130 | 90, 1182 | 107, 155, 200, 307, 503 |
| L.guy | LM3 | 21, 87, 246, 259, 517 | 1130 | 90, 1182 | 107, 155, 200, 307, 503 |
| L.pan | G05 | 21, 87, 246, 259, 517 | 1130 | 90, 1182 | 107, 155, 200, 307, 503 |
| L.pan | G06 | 21, 87, 246, 259, 517 | 1130 | 90, 1182 | 107, 155, 200, 307, 503 |
| L.pan | G07 | 21, 87, 246, 259, 517 | 1130 | 90, 1182 | 107, 155, 200, 307, 503 |
| L.pan | INH23 | 21, 87, 246, 259, 517 | 1130 | 90, 1182 | 107, 155, 200, 307, 503 |
| L.bra | Ppa20 | 21, 246, 259, 604 | 1130 | 58, 90, 197, 927 | 107, 155, 200, 307, 503 |
| L.bra | LASU22 | 21, 246, 259, 604 | 1130 | 58, 90, 197, 927 | 6, 107, 149, 200, 307, 503 |
| L.nai | 07tor | 21, 246, 259, 604 | 352, 778 | 58, 90, 1124 | 155, 200, 414, 503 |
| L.nai | 13tor1 | 21, 246, 259, 604 | 352, 778 | 58, 90, 1124 | 155, 200, 414, 503 |
| L.nai | 13tor2 | 21, 246, 259, 604 | 352, 778 | 58, 90, 1124 | 155, 200, 414, 503 |
| L.maj | G09 | 81, 95, 116, 158, 187, 493 | 1130 | 529, 743 | 83, 169, 200, 406, 414 |
| L.maj | PT115 | 81, 95, 116, 158, 187, 493 | 1130 | 529, 743 | 83, 169, 200, 406, 414 |
| L.mex | PT23 | 81, 91, 162, 303, 493 | 1130 | 392, 880 | 32, 83, 200, 382, 575 |
| L.mex | PT27 | 81, 253, 303, 493 | 1130 | 392, 880 | 32, 83, 200, 382, 575 |
| L.mex | PT29 | 81, 91, 162, 303, 493 | 1130 | 392, 880 | 32, 83, 200, 382, 575 |
| L.mex | PT103 | 81, 91, 162, 303, 493 | 1130 | 392, 880 | 32, 83, 200, 382, 575 |
| L.mex | HU3 | 81, 91, 162, 303, 493 | 1130 | 392, 880 | 32, 83, 200, 382, 575 |
| L.mex | HU4 | 81, 91, 162, 303, 493 | 1130 | 392, 880 | 32, 83, 200, 382, 575 |
| L.mex | HU6 | 81, 91, 162, 303, 493 | 1130 | 392, 880 | 32, 83, 200, 382, 575 |
*L.guy: L. (V.) guyanensis, L.pan: L. (V.) panamensis, L.bra: L. (V.) braziliensis, L.nai: L. (V.) naiffi, L.maj: L. (L.) major-like, L.mex: L. (L.) mexicana
The nucleotide sequence data reported in this paper will appear in the DDBJ, EMBL and GenBank databases under the accession numbers LC468908-LC468956.
Digestion of the 6pgd gene with Bsp1286I resulted in distinct gene fragment patterns of L. (V.) guyanensis/L. (V.) panamensis, L. (V.) braziliensis, L. (V.) naiffi, L. (L.) major-like and L. (L.) mexicana; however, the patterns between L. (V.) guyanensis/L. (V.) panamensis and L. (V.) naiffi were similar and difficult to discriminate because of only about a 50 bp difference in a fragment of approximately 1 kbp (Fig 2A). The two species were successfully differentiated by digesting with HinfI (Fig 2B).
Fig 2. PCR-RFLP analyses of 6pgd gene fragments from 6 Leishmania species in Ecuador.
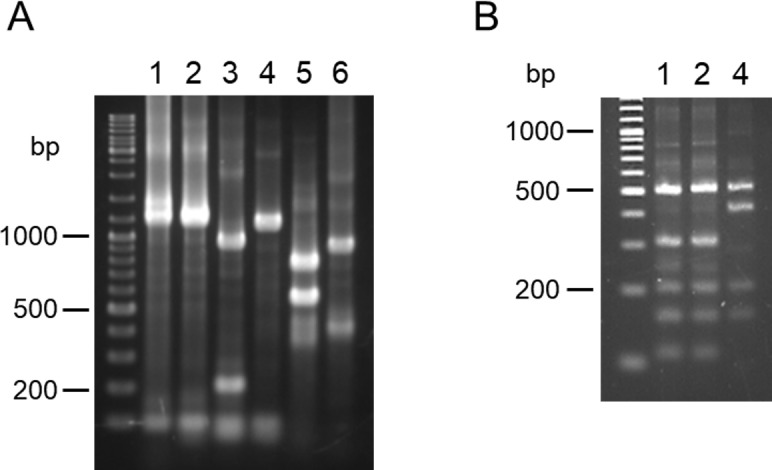
PCR amplification was performed with leishmanial 6pgd gene-specific primers, and PCR products were digested with (A) Bsp1286I and (B) HinfI, and resulting restriction fragment patterns were analyzed by agarose gel electrophoresis. 1. L. (V.) guyanensis, 2. L. (V.) panamensis, 3. L. (V.) braziliensis, 4. L. (V.) naiffi, 5. L. (L.) major-like, 6. L. (L.) mexicana.
Although L. (V.) guyanensis and L. (V.) panamensis were not discriminated by PCR-RFLP of mpi and 6pgd genes, PCR-RFLP of the hsp70 gene with a restriction enzyme, BccI, successfully differentiated the two species as reported previously (Fig 3) [27].
Fig 3. Differentiation between L. (V.) guyanensis and L. (V.) panamensis by PCR-RFLP of the hsp70 gene fragment.
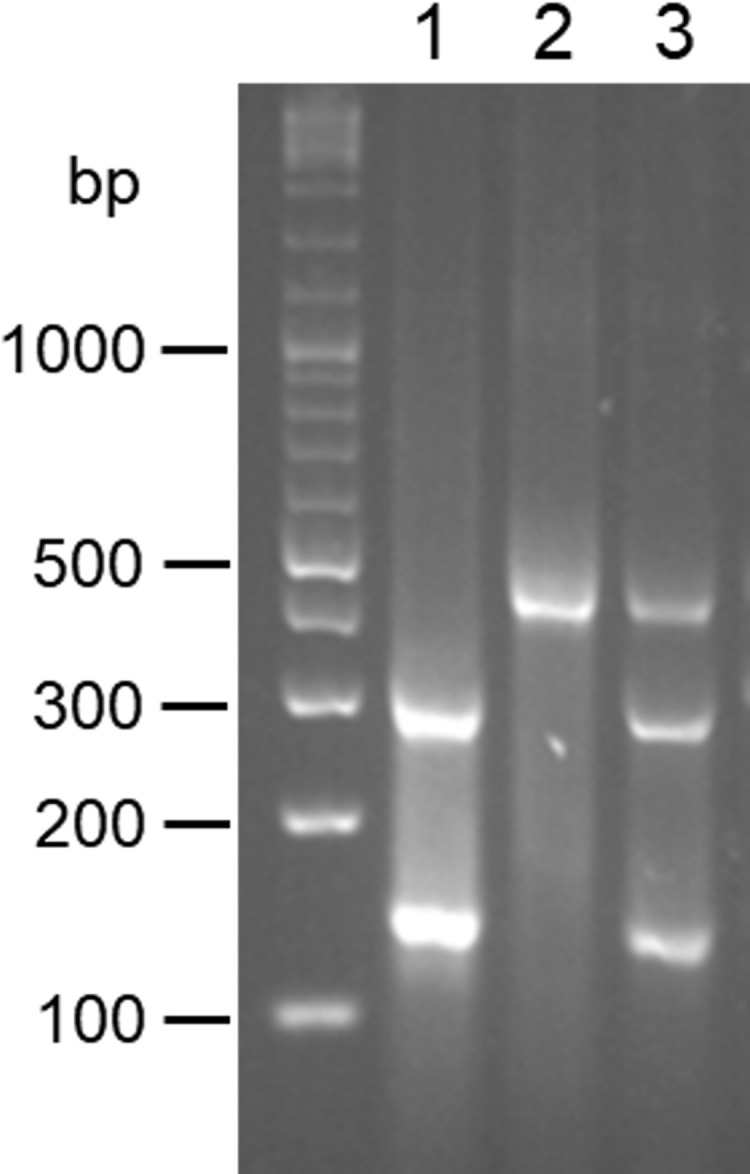
PCR amplification was performed with hsp70 gene-specific primers and the PCR products were digested with BccI. 1. L. (V.) guyanensis, 2. L. (V.) panamensis, 3. a hybrid of L. (V.) guyanensis and L. (V.) panamensis.
Identification of Leishmania species in clinical samples by PCR-RFLP
PCR-RFLP analyses of mpi gene with restriction enzymes, HaeIII and HpaI, and 6pgd gene with Bsp1286I and HinfI were applied to 92 clinical samples collected at 33 sites in 14 provinces of Ecuador. PCR-RFLP analysis of the hsp70 gene with a restriction enzyme, BccI, was used for differentiation between L. (V.) guyanensis and L. (V.) panamensis. The results obtained by PCR-RFLP analyses were compared with those obtained by the cyt b gene sequence analysis. The results of the species identification obtained by the two nuclear genes always agreed with each other. The identification by PCR-RFLP analyses completely matched with that obtained by the cyt b gene sequence analysis in all of L. (V.) naiffi (2 samples) and L. (L.) mexicana (3 samples) (Table 4). Of the 73 samples identified as L. (V.) guyanensis by cyt b gene analysis, 72 samples were identified as L. (V.) guyanensis by PCR-RFLP analyses, whereas one sample from a Pacific coast area showed a hybrid pattern of L. (V.) guyanensis and L. (V.) panamensis based on the PCR-RFLP of the hsp70 gene (Figs 3 and 4). The sequence of the hsp70 gene fragment was analyzed by direct sequencing, and a single nucleotide polymorphism was confirmed, showing “C” in L. (V.) guyanensis but “T” in L. (V.) panamensis, whereas a sample having a hybrid RFLP pattern had both “C” and “T” peaks at the corresponding position (S3 Fig), indicating the presence of a hybrid strain of L. (V.) guyanensis and L. (V.) panamensis in Ecuador. On the other hand, of the 14 samples identified as L. (V.) braziliensis by cyt b gene analysis, only 6 samples were identified as L. (V.) braziliensis by RFLP analyses (Table 4). In the other 8 samples identified as L. (V.) braziliensis by the cyt b gene analysis, three samples showed hybrid patterns in PCR-RFLP analyses of both the mpi and 6pgd genes (Fig 5A and 5B). The sequences of mpi and 6pgd gene fragments were analyzed by direct sequencing, and a single nucleotide polymorphism was confirmed, showing “C” in L. (V.) guyanensis but “T” in L. (V.) braziliensis of the mpi gene, and “T” in L. (V.) guyanensis but “C” in L. (V.) braziliensis of the 6pgd gene. On the other hand, the mpi and 6pgd genes from the three samples with hybrid RFLP patterns had both “C” and “T” peaks at the corresponding position (S4 Fig). From these results, the parasite species of these three samples were identified as a hybrid of L. (V.) braziliensis and L. (V.) guyanensis (Table 4, Fig 4). In the remaining 5 samples identified as L. (V.) braziliensis by sequence analysis of the cyt b gene, PCR-RFLP analyses showed that one sample from a Pacific coast area was L. (V.) guyanensis, three samples from the northern Pacific coast and Amazonian areas were L. (V.) panamensis, and one sample from a northern Pacific coast area had a hybrid pattern of L. (V.) guyanensis and L. (V.) panamensis (Table 4, Fig 4). The sequence analyses of mpi, 6pgd, and hsp70 gene fragments corresponded to PCR-RFLP analyses, indicating the presence of a mismatch between kinetoplast and nuclear genes, known as mito-nuclear discordance, in Leishmania distributing in Ecuador (Table 4, Fig 4). To further confirm the mito-nuclear discordance, partial sequences of the COII-ND1 region were analyzed as another target of kinetoplast genes in samples showing a mismatch between kinetoplast cyt b gene and nuclear mpi, 6pgd and hsp70 genes. The sequences were compared to each two corresponding sequences obtained from L. (V.) braziliensis and L. (V.) guyanensis in this study since this region has not been well-analyzed in subgenus Viannia species. The sequences from parasites with mito-nuclear discordance showed 98.9–99.1% and 98.5–98.9% identities with those of L. (V.) braziliensis and L. (V.) guyanensis, respectively (accession numbers: LC475135-LC475142). When partial COII gene sequences in the obtained COII-ND1 region sequences were analyzed on the GenBank database, the sequences from parasites with mito-nuclear discordance showed 99.5% and 98.9% identities with those of L. (V.) braziliensis and L. (V.) guyanensis, respectively. This result strongly suggested that the kinetoplast genes of these parasites originated from L. (V.) braziliensis, corresponding to the result of cyt b gene analysis.
Table 4. Comparison of Leishmania species identification in Ecuador between cyt b sequence analysis and PCR-RFLP analyses of nuclear DNAs.
|
Target gene (analysis) |
Identification* (numbers) | |||
|---|---|---|---|---|
|
cyt b (cloning and sequencing) |
L.g (73) | L.b (14) | L.n (2) | L.mex (3) |
|
mpi, 6pgd, and hsp70 (PCR-RFLP) |
L.g (72) L.g/L.p (1) |
L.b (6) L.g/L.b (3) L.g# (1) L.p# (3) L.g/L.p# (1) |
L.n (2) | L.mex (3) |
*L.g: L. (V.) guyanensis, L.p: L. (V.) panamensis, L.b: L. (V.) braziliensis, L.n: L. (V.) naiffi, L.mex: L. (L.) mexicana, L.g/L.p: a hybrid of L. (V.) guyanensis and L. (V.) panamensis, L.g/L.b: a hybrid of L. (V.) guyanensis and L. (V.) braziliensis
#mito-nuclear discordance
Fig 4. Geographic distribution of Leishmania species in Ecuador identified by PCR-RFLP analyses targeting multiple nuclear genes.
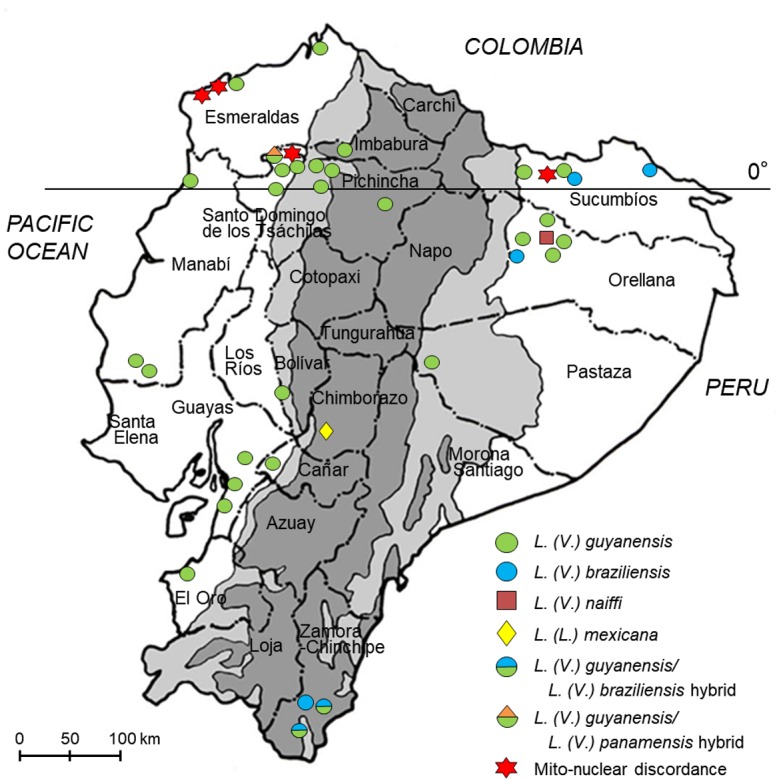
The dark gray areas show the Andean plateau (>1,000 m altitude), and the light gray areas show highland jungle or Andean slopes (400–1,000 m elevation). (Adapted from a map available at http://english.freemap.jp/).
Fig 5. Differentiation between L. (V.) guyanensis and L. (V.) braziliensis by PCR-RFLP of mpi and 6pgd gene fragments.
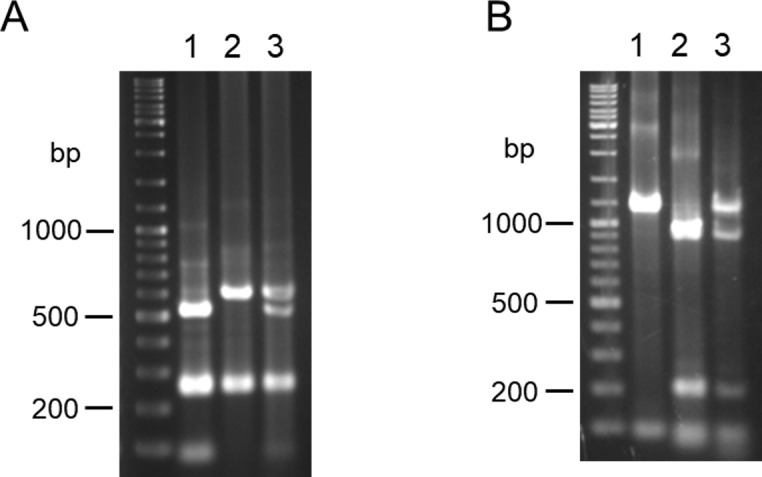
A, B. PCR amplification was performed with mpi gene- or 6pgd gene-specific primers and the PCR products were digested with HaeIII (A) or Bsp1286I (B), respectively. 1. L. (V.) guyanensis, 2. L. (V.) braziliensis, 3. a hybrid of L. (V.) guyanensis and L. (V.) braziliensis.
Discussion
In the present study, PCR-RFLP analyses were employed for the identification of Leishmania species distributing in Ecuador in order to develop a simple and practical way for species identification independent of expensive equipment such as a genetic analyzer. As a result, mpi and 6pgd genes, for which encoding enzymes have been widely used as the gold standard of species identification, were identified as suitable targets for this purpose in the tested samples. The results obtained by the PCR-RFLP analyses of multiple nuclear targets were compared to those of cyt b gene sequence analysis [7, 8, 29–36]. Although most results corresponded between the two analyses, PCR-RFLP revealed distribution of hybrid and mito-nuclear discordant Leishmania strains, which could not be identified only by cyt b gene sequence analysis. The results indicated that Leishmania strains distributing in Ecuador are genetically more complex than previously thought.
PCR-RFLP analysis has been employed for species identification of Leishmania species, and its utility is widely accepted [34]. The rRNA internal transcribed spacer 1 (ITS-1) region and hsp70 gene are mostly used as suitable target genes, of which the former is applied mainly in the Old World [6, 11, 12, 14, 17, 19, 27, 34, 37–41]. Although the hsp70 gene is one of the most valuable genetic markers for PCR-RFLP-based species identification, intraspecific polymorphism of RFLP patterns and very similar RFLP profiles among species, which affect species identification, have been reported in some Leishmania species [42]. In this study, other nuclear genes, mpi and 6pgd genes, for which encoding enzymes have been used for MLEE, were shown to be alternative useful targets for classification by PCR-RFLP analysis. Of these, the mpi gene was reported to be the only genetic marker that can distinguish two very closely-related species, L. (V.) braziliensis and L. (V.) peruviana [7, 43, 44]. In addition, a recent study demonstrated that PCR-RFLP of the shorter mpi gene fragment (approximately 500 bp) can differentiate 4 Leishmania species [L. (V.)braziliensis, L. (V.) peruviana, L. (V.) guyanensis, and L. (V.) lainsoni] and a hybrid of L. (V.) braziliensis and L. (V.) peruviana circulating in the Department of Huanuco, Peru [28]. In the present study, PCR-RFLP analyses of longer mpi and 6pgd gene fragments (>1000bp) were successfully established and applied to 92 clinical samples in Ecuador. Although a polymorphic RFLP pattern, which does not affect the identification, was detected in the mpi of one L. (L.) mexicana strain, the variant RFLP pattern was not detected in the present clinical samples identified as L. (L.) mexicana. Further sample analyses from different areas and different countries will be important to confirm the utility of this analysis, although polymorphic RFLP profiles may be detectable in these genes. Since polymorphism was also reported in the hsp70 gene of several Leishmania species [42], PCR-RFLP analyses of multiple target genes, rather than single nuclear or kinetoplast genes, will result in more accurate species identification and disclose more detailed genetic characteristics of the parasite.
Several samples showing hybrid RFLP patterns were identified as hybrid strains rather than mixed infection of different Leishmania species. It is due to the following reasons: 1) It is little or no chance to be infected by more than one parasite in a cutaneous lesion because the lesion is typically developed at the site bitten by a sand fly transmitting specific Leishmania species, 2) Even if mixed infection occurs, either parasite becomes dominant in the lesion, resulting in the presence of dominant allele by the genetic analysis. However, both alleles were comparably amplified as observed in the PCR-RFLP analysis, which is indicative of a putative hybrid strain. In addition, similar results were obtained on electrograms of the direct sequencing, showing comparable fluorescence intensities of polymorphic nucleotides derived from both species. 3) The presence of hybrid strain has been reported in the same area as described below [45]. Isolation of putative hybrid strains as a culture is necessary for further detailed characterization of these parasites.
Although multiple PCR-RFLP and cyt b sequence analyses showed corresponding results in most clinical samples, the present study revealed the distribution of several unexpected strains in Ecuador, including hybrid and mito-nuclear discordance strains. Since hybrid strains cannot be identified by the cyt b gene analysis after molecular cloning, this is another advantage of identifying parasite species by PCR-RFLP. Distribution of a hybrid strain of L. (V.) guyanensis/panamensis complex and L. (V.) braziliensis was reported in Zumba, a province of Zamora-Chinchipe in a southern part of Ecuador by using MLEE and random amplified polymorphic DNA (RAPD) [45]. The present study confirmed the presence of the hybrid strain in Zumba, and also in another area in the same province, Palanda. In addition, a hybrid of L. (V.) guyanensis and L. (V.) panamensis was detected in northern Pacific areas of Ecuador. This is the first report of the presence of a hybrid strain of L. (V.) guyanensis and L. (V.) panamensis in Ecuador. L. (V.) guyanensis and its closely related L. (V.) panamensis have been reported to be endemic in northern Pacific areas of Ecuador by MLEE; however, only L. (V.) guyanensis was identified in the same areas by cyt b gene analysis in recent studies [8, 21, 46]. The present study confirmed that L. (V.) guyanensis is dominantly present in these areas, suggesting that endemic species may change, or that there may be discordance between MLEE and genetic analysis. However, the identification of a hybrid of L. (V.) guyanensis and L. (V.) panamensis as a minor population suggests that parental L. (V.) panamensis may still be present in some of these areas. Another unexpected finding was identification of mito-nuclear discordant strains of Leishmania species in northern Pacific and Amazonian areas. Interestingly, mito-nuclear discordant strains were identified only in the species identified as L. (V.) braziliensis by cyt b gene analysis. This finding supports a recent study using cyt b gene analysis reporting increasing cases of L. (V.) braziliensis infection in Pacific coast areas when compared to previous studies using enzymatic MLEE analysis [8]. The hybrid strain of L. (V.) braziliensis and L. (V.) peruviana was suggested to increase disease severity when compared to parental species in an animal model [47]. Therefore, careful investigation is needed to clarify the presence of hybrid strains, including mito-nuclear discordance, and their effects on clinical courses. In addition, hybrid strains may increase the range of transmissible sand fly species if they have a potential to be transmitted by both vector species of parental parasites. Continuous vector research is important in these endemic areas, as well as parasitological and clinical studies. Further, basic parasitological research on how genetic exchange and mito-nuclear discordance occur among Leishmania species would be another interesting subject [48–51]. Mito-nuclear discordance is reported in various animals such as mammals, birds, reptiles, amphibians, fish and insects, and is inferred to result from various processes: 1) adaptive introgression of mitochondrial DNA, 2) demographic disparities, 3) sex-biased asymmetries, 4) hybrid zone movement, 5) an intracellular bacteria, Wolbachia infection in insects, and 6) human actions [52]. It provides deeper insights into the phylogenetic relationship, population structure, and evolutionary signature of these animals. Mito-nuclear discordance is also reported in helminth parasites: trematodes Schistosoma turkestanicum between populations [53], and cestodes Taenia solium between lineages [54], and between T. saginata and T. asiatica [55–57]. This is the first report of mito-nuclear discordance in protozoan parasites. Mito-nuclear discordance is speculated to be resulted from the similar process as hybridization of nuclear genes in protozoa. Further study is needed to disclose the mechanism of mito-nuclear discordance formation in protozoa. In addition, association of mito-nuclear discordance with the pathogenicity and vector competency of the parasites is important issues to be clarified. In this study, we established a novel PCR-RFLP-based genotyping approach to identify Leishmania species in Ecuador. Although the present PCR-RFLP analyses was shown to be practical for identification of Leishmania species in Ecuador, further study focusing on other Leishmania species and clinical samples from different countries will be needed to enhance the utility of this approach. PCR-RFLP analyses of clinical samples and subsequent comparison with kinetoplast cyt b sequence analysis revealed the distribution of genetically complex Leishmania strains having genetic characteristics of hybrid and mito-nuclear discordance. Although intraspecies genetic variation observed in the cyt b gene resulted in this gene as an unsuitable target for RFLP analysis, there is no doubt about the utility of cyt b gene sequence analysis for species identification and phylogenetic analysis since distinct interspecies genetic diversity of this gene overcomes the disadvantage of the intraspecies variation. However, the present study points to the importance of applying multiple target approaches as the combination of cyt b and the PCR-RFLP assays presented here, enhancing the reliability of species identification and characterization of genetic properties including hybrid and mito-nuclear discordance. Further studies are needed to reveal the parasitological characteristics of hybrid and mito-nuclear discordance, clinical outcomes caused by these parasites, and the range of vector species of these parasites. In addition, studies on mito-nuclear discordance in Leishmania and other protozoa may provide further insights into the mechanism of genetic exchanges of these parasites.
Supporting information
The dark gray areas show the Andean plateau (>1,000 m altitude), and the light gray areas show highland jungle or Andean slopes (400–1,000 m elevation). 1. San Lorenzo, 2. Esmeraldas, and 3. Atacames, Province of Esmeraldas; 4. Pedernales, 5. Montalvo, and 6. Pedro Pablo Gomez, Province of Manabi; 7. Cielo Verde, Province of Imbabura; 8. Puerto Quito, 9. Pedro Vicente Maldonado, 10. Los Bancos, 11. Nanegalito, 12. Pachijal, and 13. Quinche, Province of Pichincha; 14. Valle Hermoso, Province of Santo Domingo; 15. Balsapamba, Province of Bolivar; 16. Chanchan, Province of Chimborazo; 17. La Troncal, Province of Cañar; 18. El Triunfo, 19. Naranjal, and 20. Balao, Province of Guayas; 21. Santa Rosa, Province of El Oro; 22. Cascales, 23. Lago Agrio, and 24. Palma Roja, Province of Scumbios; 25. Coca, 26. Shangrila, 27. La Joya de los Sachas, 28. Pompeya, 29. Union Milagrena, and 30. Loreto, Province of Orellana; 31. Puyo, Province of Pastaza; 32. Palanda, and 33. Zumba, Province of Zamora-Chinchipe. (Adapted from a map available at http://english.freemap.jp/)
(TIF)
1. L. (V.) guyanensis, 2. L. (V.) panamensis, 3. L. (V.) lainsoni.
(TIF)
(TIF)
Direct sequence analysis showing a species-specific polymorphic site of Leishmania mpi gene (A) or 6pgd gene (B) fragments.
(TIF)
Acknowledgments
We would like to thank the following individuals for their invaluable support in the collection of FTA-cards from CL-suspected patients and for their technical assistance throughout the study: Enrique Bone (Coca, Orellana province), Edison Javier Torres Romero (Valle Hermoso, Santo Domingo de los Tsachilas province), Alexandra Narvaez (Palanda, Zamora-Chinchipe province), Gonzalo F. Shiguango (SNEM, Coca), Silvio V. Gonzales (SNEM, Coca), Kazue Hashiguchi (Universidad Catolica de Santiago de Guayaquil), and Flavio-Valeriano Zambrano (SNEM, Guayaquil).
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was financially supported by the Ministry of Education, Culture and Sports, Science and Technology (MEXT) of Japan (Grant Nos. 25257501 and 17H01685). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO/Department of Control of Neglected Tropical Diseases. Global leishmaniasis update, 2006–2015: A; turning point in leishmaniasis surveillance. WHO, Leishmaniasis. 2017; 92:557–572. [Google Scholar]
- 2.Paranaiba LF, Pinheiro LJ, Torrecilhas AC, Macedo DH, Menezes-Neto A, Tafuri WL, Soares RP. Leishmania enriettii (Muniz & Medina, 1948): A highly diverse parasite is here to stay. PLoS Pathog. 2017; 13:e1006303 10.1371/journal.ppat.1006303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rioux JA, Lanotte G, Serres E, Pratlong F, Bastien P, Perieres J. Taxonomy of Leishmania. Use of isoenzymes. Suggestions for a new classification. Ann Parasitol Hum Comp. 1990; 65:111–125. 10.1051/parasite/1990653111 [DOI] [PubMed] [Google Scholar]
- 4.Cupolillo E, Grimaldi G Jr, Momen H. A general classification of New World Leishmania using numerical zymotaxonomy. Am J Trop Med Hyg. 1994; 50:296–311. [DOI] [PubMed] [Google Scholar]
- 5.Laurent T, Van der Auwera G, Hide M, Mertens P, Quispe-Tintaya W, Deborggraeve S, De Doncker S, Leclipteux T, Bañuls AL, Büscher P, Dujardin JC. Identification of Old World Leishmania spp. by specific polymerase chain reaction amplification of cysteine proteinase B genes and rapid dipstick detection. Diagn Microbiol Infect Dis. 2009; 63:173–181. 10.1016/j.diagmicrobio.2008.10.015 [DOI] [PubMed] [Google Scholar]
- 6.da Silva LA, de Sousa Cdos S, da Graça GC, Porrozzi R, Cupolillo E. Sequence analysis and PCR-RFLP profiling of the hsp70 gene as a valuable tool for identifying Leishmania species associated with human leishmaniasis in Brazil. Infect Genet Evol. 2010; 10:77–83. 10.1016/j.meegid.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 7.Kato H, Cáceres AG, Mimori T, Ishimaru Y, Sayed AS, Fujita M, Iwata H, Uezato H, Velez LN, Gomez EA, Hashiguchi Y. Use of FTA cards for direct sampling of patients' lesions in the ecological study of cutaneous leishmaniasis. J Clin Microbiol. 2010; 48:3661–3665. 10.1128/JCM.00498-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato H, Gomez EA, Martini-Robles L, Muzzio J, Velez L, Calvopiña M, Romero-Alvarez D, Mimori T, Uezato H, Hashiguchi Y. Geographic distribution of Leishmania species in Ecuador based on the cytochrome b gene sequence analysis. PLoS Negl Trop Dis. 2016. a; 10:e0004844 10.1371/journal.pntd.0004844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talmi-Frank D, Nasereddin A, Schnur LF, Schönian G, Töz SO, Jaffe CL, Baneth G. Detection and identification of Old World Leishmania by high resolution melt analysis. PLoS Negl Trop Dis. 2010; 4:e581 10.1371/journal.pntd.0000581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Almeida ME, Steurer FJ, Koru O, Herwaldt BL, Pieniazek NJ, da Silva AJ. Identification of Leishmania spp. by molecular amplification and DNA sequencing analysis of a fragment of rRNA internal transcribed spacer 2. J Clin Microbiol. 2011; 49:3143–3149. 10.1128/JCM.01177-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraga J, Veland N, Montalvo AM, Praet N, Boggild AK, Valencia BM, Arévalo J, Llanos-Cuentas A, Dujardin JC, Van der Auwera G. Accurate and rapid species typing from cutaneous and mucocutaneous leishmaniasis lesions of the New World. Diagn Microbiol Infect Dis. 2012; 74:142–150. 10.1016/j.diagmicrobio.2012.06.010 [DOI] [PubMed] [Google Scholar]
- 12.Montalvo AM, Fraga J, Maes I, Dujardin JC, Van der Auwera G. Three new sensitive and specific heat-shock protein 70 PCRs for global Leishmania species identification. Eur J Clin Microbiol Infect Dis. 2012; 31:1453–1461. 10.1007/s10096-011-1463-z [DOI] [PubMed] [Google Scholar]
- 13.Chaouch M, Fathallah-Mili A, Driss M, Lahmadi R, Ayari C, Guizani I, Ben Said M, Benabderrazak S. Identification of Tunisian Leishmania spp. by PCR amplification of cysteine proteinase B (cpb) genes and phylogenetic analysis. Acta Trop. 2013; 125:357–365. 10.1016/j.actatropica.2012.11.012 [DOI] [PubMed] [Google Scholar]
- 14.Garcia L, Kindt A, Bermudez H, Llanos-Cuentas A, De Doncker S, Arevalo J, Wilber Quispe Tintaya K, Dujardin JC. Culture-independent species typing of neotropical Leishmania for clinical validation of a PCR-based assay targeting heat shock protein 70 genes. J Clin Microbiol. 2004; 42:2294–2297. 10.1128/JCM.42.5.2294-2297.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rotureau B, Ravel C, Couppié P, Pratlong F, Nacher M, Dedet JP, Carme B. Use of PCR-restriction fragment length polymorphism analysis to identify the main new world Leishmania species and analyze their taxonomic properties and polymorphism by application of the assay to clinical samples. J Clin Microbiol. 2006; 44:459–467. 10.1128/JCM.44.2.459-467.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spanakos G, Piperaki ET, Menounos PG, Tegos N, Flemetakis A, Vakalis NC. Detection and species identification of Old World Leishmania in clinical samples using a PCR-based method. Trans R Soc Trop Med Hyg. 2008; 102:46–53. 10.1016/j.trstmh.2007.05.019 [DOI] [PubMed] [Google Scholar]
- 17.Khanra S, Datta S, Mondal D, Saha P, Bandopadhyay SK, Roy S, Manna M. RFLPs of ITS, ITS1 and hsp70 amplicons and sequencing of ITS1 of recent clinical isolates of Kala-azar from India and Bangladesh confirms the association of L. tropica with the disease. Acta Trop. 2012; 124:229–234. 10.1016/j.actatropica.2012.08.017 [DOI] [PubMed] [Google Scholar]
- 18.Fraga J, Montalvo AM, Maes L, Dujardin JC, Van der Auwera G. HindII and SduI digests of heat-shock protein 70 PCR for Leishmania typing. Diagn Microbiol Infect Dis. 2013; 77:245–247. 10.1016/j.diagmicrobio.2013.07.023 [DOI] [PubMed] [Google Scholar]
- 19.Mouttaki T, Morales-Yuste M, Merino-Espinosa G, Chiheb S, Fellah H, Martin-Sanchez J, Riyad M. Molecular diagnosis of cutaneous leishmaniasis and identification of the causative Leishmania species in Morocco by using three PCR-based assays. Parasit Vectors. 2014; 7:420 10.1186/1756-3305-7-420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato H, Bone AE, Mimori T, Hashiguchi K, Shiguango GF, Gonzales SV, Velez LN, Guevara AG, Gomez EA, Hashiguchi Y. First human cases of Leishmania (Viannia) lainsoni infection and a search for the vector sand flies in Ecuador. PLoS Negl Trop Dis. 2016b; 10:e0004728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashiguchi Y, Velez LN, Villegas NV, Mimori T, Gomez EAL, Kato H. Leishmaniases in Ecuador: Comprehensive review and current status. Acta Trop. 2017; 166:299–315. 10.1016/j.actatropica.2016.11.039 [DOI] [PubMed] [Google Scholar]
- 22.Mimori T, Grimaldi G Jr, Kreutzer RD, Gomez EA, McMahon-Pratt D, Tesh RB, Hashiguchi Y. Identification, using isoenzyme electrophoresis and monoclonal antibodies, of Leishmania isolated from humans and wild animals of Ecuador. Am J Trop Med Hyg. 1989; 40:154–158. [DOI] [PubMed] [Google Scholar]
- 23.Calvopiña M, Armijos RX, Hashiguchi Y. Epidemiology of leishmaniasis in Ecuador: current status of knowledge—a review. Mem Inst Oswaldo Cruz. 2004; 99:663–672. [DOI] [PubMed] [Google Scholar]
- 24.Calvopina M, Armijos RX, Marco JD, Uezato H, Kato H, Gomez EA, Korenaga M, Barroso PA, Mimori T, Cooper PJ, Nonaka S, Hashiguchi Y. Leishmania isoenzyme polymorphisms in Ecuador: relationships with geographic distribution and clinical presentation. BMC Infect Dis. 2006; 6:139 10.1186/1471-2334-6-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato H, Calvopiña M, Criollo H, Hashiguchi Y. First human cases of Leishmania (Viannia) naiffi infection in Ecuador and identification of its suspected vector species. Acta Trop. 2013; 128:710–713. 10.1016/j.actatropica.2013.09.001 [DOI] [PubMed] [Google Scholar]
- 26.Kato H, Gomez EA, Yamamoto Y, Calvopiña M, Guevara AG, Marco JD, Barroso PA, Iwata H, Hashiguchi Y. Natural infection of Lutzomyia tortura with Leishmania (Viannia) naiffi in an Amazonian area of Ecuador. Am J Trop Med Hyg. 2008; 79:438–440. [PubMed] [Google Scholar]
- 27.Montalvo Alvarez AM, Nodarse JF, Goodridge IM, Fidalgo LM, Marin M, Van Der Auwera G, Dujardin JC, Bernal ID, Muskus C. Differentiation of Leishmania (Viannia) panamensis and Leishmania (V.) guyanensis using BccI for hsp70 PCR-RFLP. Trans R Soc Trop Med Hyg. 2010a; 104:364–367. [DOI] [PubMed] [Google Scholar]
- 28.Koarashi Y, Cáceres AG, Saca FMZ, Flores EEP, Trujillo AC, Alvares JLA, Yoshimatsu K, Arikawa J, Katakura K, Hashiguchi Y, Kato H. Identification of causative Leishmania species in Giemsa-stained smears prepared from patients with cutaneous leishmaniasis in Peru using PCR-RFLP. Acta Trop. 2016; 158:83–87. 10.1016/j.actatropica.2016.02.024 [DOI] [PubMed] [Google Scholar]
- 29.Luyo-Acero GE, Uezato H, Oshiro M, Takei K, Kariya K, Katakura K, Gomez-Landires E, Hashiguchi Y, Nonaka S. Sequence variation of the cytochrome b gene of various human infecting members of the genus Leishmania and their phylogeny. Parasitology. 2004; 128:483–491. [DOI] [PubMed] [Google Scholar]
- 30.Asato Y, Oshiro M, Myint CK, Yamamoto Y, Kato H, Marco JD, Mimori T, Gomez EA, Hashiguchi Y, Uezato H. Phylogenic analysis of the genus Leishmania by cytochrome b gene sequencing. Exp Parasitol. 2009; 121:352–361. 10.1016/j.exppara.2008.12.013 [DOI] [PubMed] [Google Scholar]
- 31.Kato H, Watanabe J, Mendoza Nieto I, Korenaga M, Hashiguchi Y. Leishmania species identification using FTA card sampling directly from patients' cutaneous lesions in the state of Lara, Venezuela. Trans R Soc Trop Med Hyg. 2011; 105:561–567. 10.1016/j.trstmh.2011.05.009 [DOI] [PubMed] [Google Scholar]
- 32.Leelayoova S, Siripattanapipong S, Hitakarun A, Kato H, Tan-ariya P, Siriyasatien P, Osatakul S, Mungthin M. Multilocus characterization and phylogenetic analysis of Leishmania siamensis isolated from autochthonous visceral leishmaniasis cases, southern Thailand. BMC Microbiol. 2013; 13:60 10.1186/1471-2180-13-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang BB, Chen DL, Chen JP, Liao L, Hu XS, Xu JN. Analysis of kinetoplast cytochrome b gene of 16 Leishmania isolates from different foci of China: different species of Leishmania in China and their phylogenetic inference. Parasit Vectors. 2013; 6:32 10.1186/1756-3305-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akhoundi M, Downing T, Votýpka J, Kuhls K, Lukeš J, Cannet A, Ravel C, Marty P, Delaunay P, Kasbari M, Granouillac B, Gradoni L, Sereno D. Leishmania infections: Molecular targets and diagnosis. Mol Aspects Med. 2017; 57:1–29. 10.1016/j.mam.2016.11.012 [DOI] [PubMed] [Google Scholar]
- 35.Bilbao-Ramos P, Dea-Ayuela MA, Cardenas-Alegría O, Salamanca E, Santalla-Vargas JA, Benito C, Flores N, Bolás-Fernández F. Leishmaniasis in the major endemic region of Plurinational State of Bolivia: Species identification, phylogeography and drug susceptibility implications. Acta Trop. 2017; 176:150–161. 10.1016/j.actatropica.2017.07.026 [DOI] [PubMed] [Google Scholar]
- 36.Al-Bajalan MMM, Al-Jaf SMA, Niranji SS, Abdulkareem DR, Al-Kayali KK, Kato H. An outbreak of Leishmania major from an endemic to a non-endemic region posed a public health threat in Iraq from 2014–2017: Epidemiological, molecular and phylogenetic studies. PLoS Negl Trop Dis. 2018; 12:e0006255 10.1371/journal.pntd.0006255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gadisa E, Genetu A, Kuru T, Jirata D, Dagne K, Aseffa A, Gedamu L. Leishmania (Kinetoplastida): species typing with isoenzyme and PCR-RFLP from cutaneous leishmaniasis patients in Ethiopia. Exp Parasitol. 2007; 115:339–343. 10.1016/j.exppara.2006.09.014 [DOI] [PubMed] [Google Scholar]
- 38.Montalvo AM, Fraga J, Monzote L, Montano I, De Doncker S, Dujardin JC, Van der Auwera G. Heat-shock protein 70 PCR-RFLP: a universal simple tool for Leishmania species discrimination in the New and Old World. Parasitology. 2010b; 137:1159–1168. [DOI] [PubMed] [Google Scholar]
- 39.Montalvo AM, Fraga J, El Safi S, Gramiccia M, Jaffe CL, Dujardin JC, Van der Auwera G. Direct Leishmania species typing in Old World clinical samples: evaluation of 3 sensitive methods based on the heat-shock protein 70 gene. Diagn Microbiol Infect Dis. 2014; 80:35–39. 10.1016/j.diagmicrobio.2014.05.012 [DOI] [PubMed] [Google Scholar]
- 40.Ben Abda I, de Monbrison F, Bousslimi N, Aoun K, Bouratbine A, Picot S. Advantages and limits of real-time PCR assay and PCR-restriction fragment length polymorphism for the identification of cutaneous Leishmania species in Tunisia. Trans R Soc Trop Med Hyg. 2011; 105:17–22. 10.1016/j.trstmh.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 41.Mosleh IM, Shönian G, Geith E, Al-Jawabreh A, Natsheh L. The Jordanian Mid Jordan Valley is a classic focus of Leishmania major as revealed by RFLP of 56 isolates and 173 ITS-1-PCR-positive clinical samples. Exp Parasitol. 2015; 148:81–85. 10.1016/j.exppara.2014.11.006 [DOI] [PubMed] [Google Scholar]
- 42.Espada CR, Ortiz PA, Shaw JJ, Barral AMP, Costa JML, Uliana SRB, Coelho AC. Identification of Leishmania (Viannia) species and clinical isolates of Leishmania (Leishmania) amazonensis from Brazil using PCR-RFLP of the heat-shock protein 70 gene reveals some unexpected observations. Diagn Microbiol Infect Dis. 2018; 91:312–318. 10.1016/j.diagmicrobio.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 43.Zhang WW, Miranda-Verastegui C, Arevalo J, Ndao M, Ward B, Llanos-Cuentas A, Matlashewski G. Development of a genetic assay to distinguish between Leishmania Viannia species on the basis of isoenzyme differences. Clin Infect Dis. 2006; 42:801–809. 10.1086/500326 [DOI] [PubMed] [Google Scholar]
- 44.Kato H, Cáceres AG, Hashiguchi Y. First evidence of a hybrid of Leishmania (Viannia) braziliensis/L. (V.) peruviana DNA detected from the phlebotomine sand fly Lutzomyia tejadai in Peru. PLoS Negl Trop Dis. 2016c; 10:e0004336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bañuls AL, Guerrini F, Le Pont F, Barrera C, Espinel I, Guderian R, Echeverria R, Tibayrenc M. Evidence for hybridization by multilocus enzyme electrophoresis and random amplified polymorphic DNA between Leishmania braziliensis and Leishmania panamensis/guyanensis in Ecuador. J Eukaryot Microbiol. 1997; 44:408–411. [DOI] [PubMed] [Google Scholar]
- 46.Gomez EA, Kato H, Torres-Romero EX, Velez LN, Villegas NV, Martillo VP, Zambrano FC, Kubo M, Hashiguchi K, Hashiguchi Y. Leishmaniasis caused by Leishmania (Viannia) guyanensis in north-central Pacific region of Ecuador: A clinico-epidemiological feature. Acta Trop. 2018; 185:204–211. 10.1016/j.actatropica.2018.05.016 [DOI] [PubMed] [Google Scholar]
- 47.Cortes S, Esteves C, Maurício I, Maia C, Cristovão JM, Miles M, Campino L. In vitro and in vivo behaviour of sympatric Leishmania (V.) braziliensis, L. (V.) peruviana and their hybrids. Parasitology. 2012; 139:191–199. 10.1017/S0031182011001909 [DOI] [PubMed] [Google Scholar]
- 48.Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, Lawyer P, Dobson DE, Beverley SM, Sacks DL. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science. 2009; 324:265–268. 10.1126/science.1169464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sadlova J, Yeo M, Seblova V, Lewis MD, Mauricio I, Volf P, Miles MA. Visualisation of Leishmania donovani fluorescent hybrids during early stage development in the sand fly vector. PLoS One. 2011; 6:e19851 10.1371/journal.pone.0019851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calvo-Álvarez E, Álvarez-Velilla R, Jiménez M, Molina R, Pérez-Pertejo Y, Balaña-Fouce R, Reguera RM. First evidence of intraclonal genetic exchange in trypanosomatids using two Leishmania infantum fluorescent transgenic clones. 2014; PLoS Negl Trop Dis 8:e3075 10.1371/journal.pntd.0003075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogers MB, Downing T, Smith BA, Imamura H, Sanders M, Svobodova M, Volf P, Berriman M, Cotton JA, Smith DF. Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet 2014; 10:e1004092 10.1371/journal.pgen.1004092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toews DP, Brelsford A. The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol. 2012; 21:3907–3930. 10.1111/j.1365-294X.2012.05664.x [DOI] [PubMed] [Google Scholar]
- 53.Lawton SP, Bowen LI, Emery AM, Majoros G. Signatures of mito-nuclear discordance in Schistosoma turkestanicum indicate a complex evolutionary history of emergence in Europe. Parasitology. 2017; 144:1752–1762. 10.1017/S0031182017000920 [DOI] [PubMed] [Google Scholar]
- 54.Yanagida T, Carod JF, Sako Y, Nakao M, Hoberg EP, Ito A. Genetics of the pig tapeworm in Madagascar reveal a history of human dispersal and colonization. PLoS One. 2014; 9:e109002 10.1371/journal.pone.0109002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamane K, Suzuki Y, Tachi E, Li T, Chen X, Nakao M, Nkouawa A, Yanagida T, Sako Y, Ito A, Sato H, Okamoto M. Recent hybridization between Taenia asiatica and Taenia saginata. Parasitol Int. 2012; 61:351–355. 10.1016/j.parint.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 56.Yamane K, Yanagida T, Li T, Chen X, Dekumyoy P, Waikagul J, Nkouawa A, Nakao M, Sako Y, Ito A, Sato H, Okamoto M. Genotypic relationships between Taenia saginata, Taenia asiatica and their hybrids. Parasitology. 2013; 140:1595–1601. 10.1017/S0031182013001273 [DOI] [PubMed] [Google Scholar]
- 57.Sato MO, Sato M, Yanagida T, Waikagul J, Pongvongsa T, Sako Y, Sanguankiat S, Yoonuan T, Kounnavang S, Kawai S, Ito A, Okamoto M, Moji K. Taenia solium, Taenia saginata, Taenia asiatica, their hybrids and other helminthic infections occurring in a neglected tropical diseases' highly endemic area in Lao PDR. PLoS Negl Trop Dis. 2018; 12:e0006260 10.1371/journal.pntd.0006260 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The dark gray areas show the Andean plateau (>1,000 m altitude), and the light gray areas show highland jungle or Andean slopes (400–1,000 m elevation). 1. San Lorenzo, 2. Esmeraldas, and 3. Atacames, Province of Esmeraldas; 4. Pedernales, 5. Montalvo, and 6. Pedro Pablo Gomez, Province of Manabi; 7. Cielo Verde, Province of Imbabura; 8. Puerto Quito, 9. Pedro Vicente Maldonado, 10. Los Bancos, 11. Nanegalito, 12. Pachijal, and 13. Quinche, Province of Pichincha; 14. Valle Hermoso, Province of Santo Domingo; 15. Balsapamba, Province of Bolivar; 16. Chanchan, Province of Chimborazo; 17. La Troncal, Province of Cañar; 18. El Triunfo, 19. Naranjal, and 20. Balao, Province of Guayas; 21. Santa Rosa, Province of El Oro; 22. Cascales, 23. Lago Agrio, and 24. Palma Roja, Province of Scumbios; 25. Coca, 26. Shangrila, 27. La Joya de los Sachas, 28. Pompeya, 29. Union Milagrena, and 30. Loreto, Province of Orellana; 31. Puyo, Province of Pastaza; 32. Palanda, and 33. Zumba, Province of Zamora-Chinchipe. (Adapted from a map available at http://english.freemap.jp/)
(TIF)
1. L. (V.) guyanensis, 2. L. (V.) panamensis, 3. L. (V.) lainsoni.
(TIF)
(TIF)
Direct sequence analysis showing a species-specific polymorphic site of Leishmania mpi gene (A) or 6pgd gene (B) fragments.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.


