Abstract
BUB-related 1 (BubR1) encoded by Budding Uninhibited by Benzimidazole 1B (BUB1B) is a crucial mitotic checkpoint protein ensuring proper segregation of chromosomes during mitosis. Mutations of BUB1B are responsible for mosaic variegated aneuploidy (MVA), a human congenital disorder characterized by extensive abnormalities in chromosome number. Although microcephaly is a prominent feature of MVA carrying the BUB1B mutation, how BubR1 deficiency disturbs neural progenitor proliferation and neuronal output and leads to microcephaly is unknown. Here we show that conditional loss of BubR1 in mouse cerebral cortex recapitulates microcephaly. BubR1-deficient cortex includes a strikingly reduced number of late-born, but not of early-born, neurons, although BubR1 expression is substantially reduced from an early stage. Importantly, absence of BubR1 decreases the proportion of neural progenitors in mitosis, specifically in metaphase, suggesting shortened mitosis owing to premature chromosome segregation. In the BubR1 mutant, massive apoptotic cell death, which is likely due to the compromised genomic integrity that results from aberrant mitosis, depletes progenitors and neurons during neurogenesis. There is no apparent alteration in centrosome number, spindle formation or primary cilia, suggesting that the major effect of BubR1 deficiency on neural progenitors is to impair the mitotic checkpoint. This finding highlights the importance of the mitotic checkpoint in the pathogenesis of microcephaly. Furthermore, the ependymal cell layer does not form in the conditional knockout, revealing an unrecognized role of BubR1 in assuring the integrity of the ventricular system, which may account for the presence of hydrocephalus in some patients.
Introduction
Mosaic variegated aneuploidy (MVA) is a congenital disorder characterized by widespread abnormalities in chromosome number (aneuploidy). Individuals with this autosomal recessive syndrome show growth retardation, microcephaly, intellectual disabilities, developmental delays, mild dysmorphia, structural central nervous system abnormalities and an increased predisposition to cancer (1–6). MVA has also been classified as a ciliopathy due to overlap with the features of cilia dysfunction, such as polycystic kidneys and, in some cases, Dandy–Walker malformation and hydrocephalus (7). In agreement with this classification, fibroblasts cultured from patients with MVA have shown impaired ciliogenesis (7,8). MVA has been linked to mutations in the Budding Uninhibited by Benzimidazole 1B (BUB1B) gene, which encodes BUB-related 1 (BubR1) protein (2,9). While some reported cases of MVA do not carry genetic mutations in BUB1B, these cases are comparatively mild and do not present with microcephaly, suggesting that BubR1 loss is one of the important causes of MVA-related microcephaly (10).
To model this disorder, a hypomorphic allele of BubR1, the mouse ortholog of BUB1B, has been generated to circumvent early embryonic lethality resulting from complete loss of the gene. BubR1H/H (BubR1 hypomorphic allele) reduces protein production; its embryonic fibroblasts express ~11% of the proteins found with normal endogenous BubR1 (11). Similar to the human phenotype, these mice exhibit small stature, cancer predisposition and reduced lifespan, but whether the cortical BubR1 protein level is correspondingly reduced was not examined (11). Recent studies demonstrated that the hypomorphic allele does not significantly alter cortical progenitor cell division and that cortical size does not significantly differ from wild type (WT) controls up until the young adult stage (12). Thus, although it is likely that BubR1 is directly involved in cortical development, perhaps through directing faithful segregation of chromosomes, there is no information about its function in cortical progenitor cell division and ultimate cortical size. This is partly due to the lack of an animal model that features a substantial reduction in BubR1 expression in cortical progenitors without affecting viability.
Abnormal regulation of mitosis plays a critical role in the pathogenesis of microcephaly as proteins encoded by microcephaly-causing genes are associated with the mitotic apparatus and their deficiency causes mitotic defects (13–24). For instance, MCPH6 (CENPJ), whose deletion results in the absence of centrioles (25), and MCPH2 (Wdr62) were shown to regulate mitotic progression through activation of the SAC (15). Recent studies have also identified a link between mitotic delay and microcephaly: a delay in mitosis leads to both cell death and to premature differentiation, which ultimately reduce progenitor cells in number (15,26,27). Interestingly, substantial chromosomal aneuploidy has been found in normal neural progenitor cells (28). However, when aneuploidy becomes extensive (>5 chromosomal variations), cells are subject to apoptosis, which provides an additional mechanism through which aberrant mitosis can deplete the progenitor pool (29). It is therefore necessary to investigate the role of the mitotic checkpoint in assuring faithful chromosome segregation and genomic integrity and to evaluate its contribution to the pathogenesis of microcephaly.
BubR1, a core component of the mitotic checkpoint complex, is required to prevent anaphase until all chromosomes are properly aligned and attached to microtubule spindles at the metaphase plate (30). BubR1 works directly at the kinetochore, the link between the centromere and the spindle, to ensure stable kinetochore-microtubule attachments, and as a member of the spindle assembly checkpoint to help maintain inhibition of the anaphase-promoting complex (31–35). In addition to its functions in preventing premature progression of mitosis, BubR1 is implicated in suppression of centrosome amplification by inhibiting Polo-like kinase activity (36). Evidence also supports a critical function in ciliogenesis: primary cilia are malformed in the fibroblasts of MVA patients and in medaka fish with morpholino knockdown (7). Primary cilia are antenna-like, microtubule-based cellular protrusions important for diverse cellular processes, including mitogenic signaling such as Sonic Hedgehog (37). Defects in ciliogenesis may contribute to the pathogenesis of Dandy–Walker syndrome, hydrocephalus and microcephaly (7). A recent study of knockdown of BubR1 and of several other ciliopathy genes demonstrated its role in maintaining progenitor populations and neuronal migration during cortical development (38). Thus BubR1 is implicated in multiple critical cellular processes, but which defective function alters the neural progenitor population and whether this alteration is sufficient to generate microcephaly remain to be determined.
Here we show that conditional knockout (CKO) of BubR1 in the developing cortex can mimic the microcephaly found in human MVA. We demonstrate that nearly complete reduction of BubR1 is required to generate microcephaly and that drastic reduction of stem cell–like apical neural progenitors (aNPCs) affects the output of intermediate progenitors and postmitotic neurons, and ultimately cortical size. Furthermore, our progenitor analyses, including flow cytometry data, reveal that, in the absence of BubR1, the proportion of cells in mitosis, especially in metaphase, is reduced. Defective mitosis is therefore likely to cause faulty chromosome segregation, which may result in subsequent cell death due to compromised genomic integrity. Remarkably, although primary cilia did not appear to be affected by loss of BubR1, we observed complete absence of the ependymal layer in the BubR1 CKO. Thus, our findings reveal that BubR1 plays a novel role in the ventricular system by properly generating and/or maintaining the ependymal layer.
Results
BubR1 is highly expressed along ventricular neuroepithelium in the developing cortex
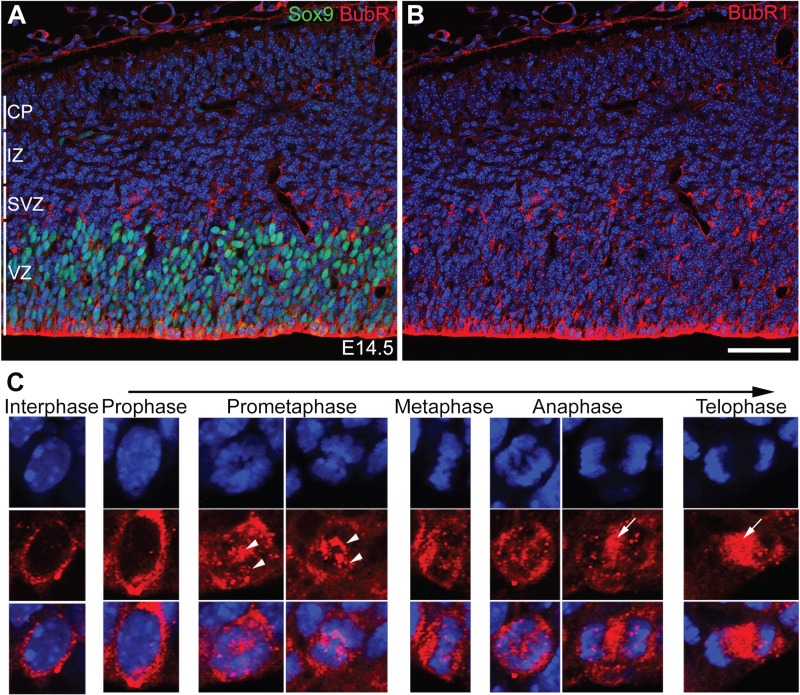
To determine the role of BubR1 in progenitor cell division and cortical development, we first assessed BubR1 expression and subcellular localization during neurogenesis. As expected, at embryonic day (E) 14.5, BubR1 expression was limited to proliferating cells in a distinct ventricular zone (VZ) enriched with Sox9-expressing aNPCs and in Sox9-negative proliferating subventricular zones (SVZ) comprising intermediate progenitors. Conversely, BubR1 was lacking in intermediate zone (IZ) and cortical plate (CP) where postmitotic neurons were migrating and forming layers, respectively. BubR1 was expressed throughout the VZ/SVZ (Fig. 1A and B) and enriched along the ventricular lining of the developing cortex where cells were undergoing mitosis (Fig. 1A). In interphase, BubR1 showed a granular, diffuse cytoplasmic distribution with minimal nuclear localization. The intensity of its signal varied among cells in interphase, likely representing cell cycle status and whether the cell was close to either entering or leaving mitosis. During mitosis, localization of BubR1 was dynamic, depending on the mitotic phases, as previously demonstrated in cell culture (35). As its distribution has been shown to be concentrated on kinetochores, very small amounts of BubR1 began to appear on chromosomes in prophase, but expression on chromosomes was robust during prometaphase and metaphase. BubR1 proteins moved to opposite poles of the chromosomes as they segregated in anaphase (Fig. 1C). In late anaphase and telophase, they were largely decreased and less punctate in chromosomes, but highly concentrated in spindles (Fig. 1C). This dynamic expression pattern is consistent with its roles in stabilizing microtubule attachment to the kinetochore and spindle checkpoint. In summary, our studies of BubR1 expression in NPCs demonstrated that it is localized on chromosomes and spindles during NPC mitosis and that it persists in the cytoplasm of cycling cells during interphase.
Figure 1.
BubR1 is dynamically localized in NPCs. (A and B) At E14.5, BubR1 expression is confined to actively proliferating cells, such as Sox9+ apical NPCs in the VZ and Sox9− intermediate NPCs in the SVZ, and is absent from IZ and CP, where postmitotic cells are situated. (C) In the VZ/SVZ, BubR1 is present in the cytoplasm of cells in interphase. It is abundant in chromosomes from prometaphase until early anaphase and is concentrated in the kinetochore during mitosis (arrowheads); its association with the spindle is evident at the end of mitosis, including in late anaphase and telophase (arrows). Scale bar, 50 μm.
Significant loss, but not reduction, of BubR1 generates microcephaly
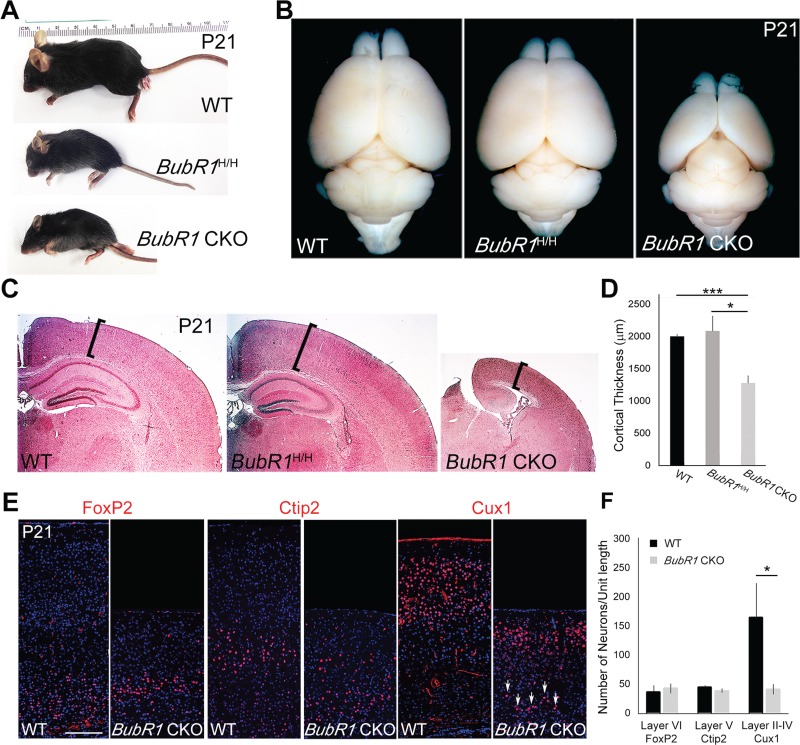
Because microcephaly is a hallmark of BubR1-associated MVA, we first determined whether BubR1H/H mice show any alterations in brain size; these mice demonstrate a significant reduction of BubR1 expression in various organs and embryonic fibroblast culture (11). Consistent with previous reports, BubR1H/H mice were markedly smaller than WT littermates (Fig. 2A) (11). At postnatal day (P) 21, the size of their cortex appeared normal (Fig. 2B and C). We also analyzed the cerebellum for structural abnormalities such as Dandy–Walker malformation, which also occurs in MVA. We observed no obvious alterations in the size or shape of the cerebellum (Supplementary Material, Fig. S1). These results provide evidence that reduced BubR1 expression in a hypomorphic allele is compatible with normal histogenesis of the brain. Our study of BubR1H/H therefore corroborates previous reports that BubR1 reduction does not produce significant changes in embryonic brain development despite its importance in age-related decline of myelination and in proliferation and differentiation of adult neural stem cells (12,39).
Figure 2.
Loss of BubR1 induces microcephaly. (A) BubR1H/H and BubR1 CKO mice exhibit dramatic reduction in body size. (B) BubR1 CKO, but not BubR1H/H, has smaller cortex compared to WT. (C and D) H&E staining shows significantly reduced cortical thickness in BubR1 CKO, but not in BubR1H/H compared to WT. (E and F) Production of early-born neurons (FoxP2+ cells in layer VI, Ctip2+ cells in layer V) is not significantly affected, but late-born neurons (Cux1+ cells in layers II–IV) are severely depleted in BubR1 CKO cortex. Some Cux1+ late-born neurons are present in deeper cortical layers (arrows). *P < 0.05, ***P < 0.001. Scale bar, 200 μm.
To further investigate the impact of complete BubR1 loss in the developing cortex, without affecting viability of the animals, we took advantage of CKO technology using Emx1-Cre, which ablates the gene of interest in the dorsal telencephalon, beginning around E9.5, on BubR1H/H background (designated as BubR1 CKO; Supplementary Material, Fig. S2). BubR1 CKOs, like BubR1H/H mice, were much smaller than their age-matched littermates (Fig. 2A). However, in contrast to the BubR1H/H mice, BubR1 CKO had a grossly obvious decrease in cortical size (Fig. 2B). Histological analyses demonstrated that cortical thickness of BubR1 CKO was significantly reduced compared to both BubR1H/H and WT and that the overall number of neurons was globally reduced (Fig. 2C). The hippocampus of BubR1 CKO showed some lamination but a striking reduction in size (Fig. 2C).
Next, we used layer-specific markers to characterize lamination and neuronal composition in the cortex of BubR1 CKO. Interestingly, the number of FoxP2+ (layer VI) and Ctip2+ (layer V) early-born neurons was comparable to WT, but Cux1+ late-born neurons (layer II–IV) were significantly decreased (Fig. 2E and F). It is therefore possible that progenitors of late-born neurons have a specific requirement for BubR1 or that they are mainly affected because defects accumulate in progenitors through multiple cell divisions. However, we cannot rule out the possibility that the number of early-born neurons is affected to a smaller extent because incomplete Cre recombination or prolonged protein stability caused insufficient removal of BubR1 from the progenitors of early-born neurons.
A striking reduction of late-born neuron numbers is therefore a major contributor to decreased cortical thickness in BubR1 CKO. Cortical size was also reduced and late-born neurons specifically reduced at early postnatal stages (P8), which demonstrate a consistent congenital phenotype (Supplementary Material, Fig. S3) rather than gradual loss due to neuronal degeneration over time. Cux1+ late-born neurons were also located ectopically in deeper cortical layers, suggesting deficits in neuronal migration (Fig. 2E, arrows). This result is consistent with a previous report showing that BubR1 knockdown impairs neuronal migration in developing cortex (38).
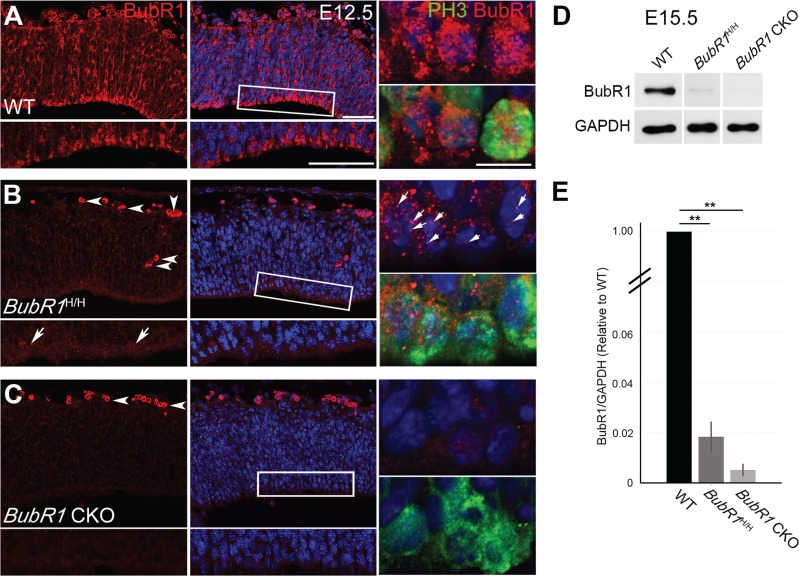
Due to the striking difference in cortical phenotype, we then investigated whether the BubR1H/H and BubR1 CKO mice express distinctly different levels of BubR1. BubR1 immunostaining showed that BubR1 expression was strikingly diminished in both BubR1H/H and BubR1 CKO mice at E12.5 (Fig. 3A–C). However, loss of BubR1 expression was more pronounced in BubR1 CKO than BubR1H/H as immunohistochemistry detected residual expression in BubR1H/H at E12.5 (Fig. 3A, arrows). Of note, the residual BubR1 protein was localized to the chromosomes in BubR1H/H, but largely absent in BubR1 CKO (Fig. 3B and C). A previous study using overexpression of BubR1 domain demonstrated that expression in the kinetochore correlates with improved phenotypic outcomes (40). Thus, it is possible that the difference in kinetochore expression in BubR1H/H and BubR1CKO accounts for why only BubR1 CKO demonstrates a severe cortical phenotype. To quantify the reduction of BubR1 expression in the neuroepithelium of both BubR1H/H and BubR1 CKO, we used western blot to analyze cortical lysates at E15.5. We normalized the level of BubR1 to Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and then compared the relative amount to WT. BubR1 expression in the BubR1H/H was about 1.8% of that of WT, in the BubR1 CKO, about 0.5% of that of WT (Fig. 3D and E, N = 3 for each genotype). Taken together, this data supports the idea that substantial loss, approaching complete absence, of BubR1 in cortical progenitors is required to induce microcephaly in mice.
Figure 3.
Depletion of BubR1 during neurogenesis is nearly complete in BubR1 CKO. (A–C) At E12.5, minimal BubR1 is present at the ventricular surface in BubR1H/H neuroepithelium (B, inset, arrows), where it is located on the chromosomes of PH3+ cells undergoing mitosis (arrows). It is almost completely absent in BubR1 CKO (C) but highly enriched in WT (A). Arrowheads indicate nonspecific or auto-fluorescence signals from blood cells (B and C). (D) Western blot of E15.5 cortices confirms significant reduction of BubR1 expression in BubR1H/H and BubR1 CKO compared to WT. (E) The graph compares the relative levels of BubR1 protein in the BubR1H/H and BubR1 CKO to the WT level after normalization with GAPDH. **P < 0.01. Scale bar, 50 μm; enlarge image, 10 μm.
Cortical progenitors are reduced in BubR1 CKO embryos
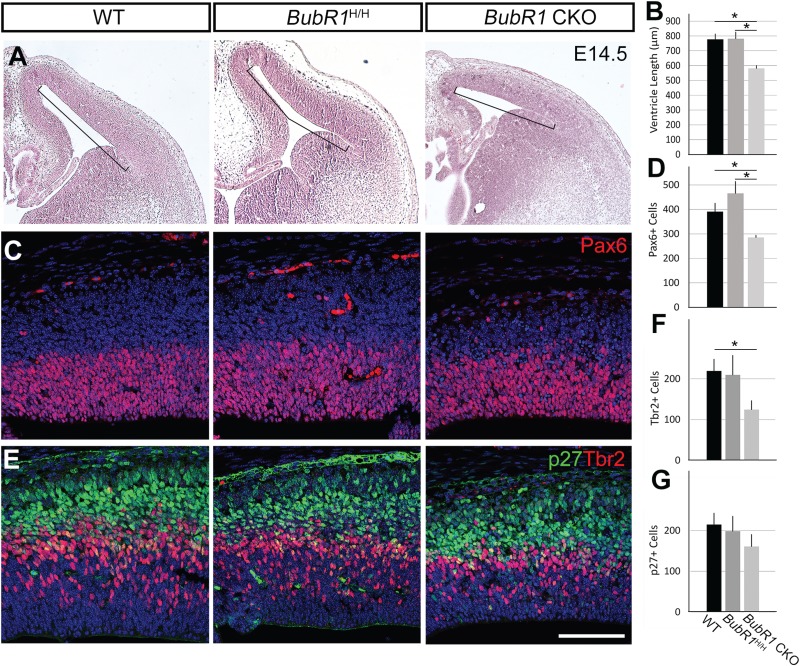
To determine the pathogenic mechanism underlying reduced brain size and decreased number of late-born neurons, we investigated anatomical alterations that occurred during neurogenesis. Interestingly, despite the significant reduction in cortical thickness in the BubR1 CKO mice seen postnatally, cortical thickness was variably affected during early to mid-neurogenesis (Fig. 4A). Importantly, there was a significant reduction in the length of the ventricular surface beginning at E14.5 compared to both WT and BubR1H/H cortices (Fig. 4A and B). As ventricular surface comprises the endfeet of aNPCs and aNPCs undergoing mitosis, reduction of ventricular length suggests a reduction in aNPC population and potentially cells undergoing mitosis. Accordingly, we detected reduced Pax6+ aNPC number in CKO, but not in WT or BubR1H/H cortices at E14.5 (Fig. 4C and D). Since aNPCs generate intermediate progenitors and neurons through asymmetric cell division, we also examined the changes in population of Tbr2+ intermediate progenitors. As expected, we found a significant reduction in Tbr2+ intermediate progenitors (Fig. 4E and F). Furthermore, cell cycle inhibitor P27+ postmitotic cells were also decreased in the BubR1 CKO, as compared to WT or BubR1H/H though the difference did not reach statistical significance (Fig. 4E and G). Because progenitors and neurons were reduced in number in the BubR1 CKO compared to both BubR1H/H and WT, it is unlikely that BubR1 loss either causes premature cell cycle exit leading to precocious differentiation or promotes asymmetric cell division that increases intermediated progenitors or neurons at the expense of apical progenitors. Together, the loss of BubR1 reduces cortical ventricular surface and cortical progenitors, including apical and intermediate progenitors, which subsequently decreases neuronal production.
Figure 4.
Cortical progenitors are reduced in developing cortex in the absence of BubR1. (A and B) At E14.5, ventricular length is significantly reduced in BubR1 CKO compared to WT and BubR1H/H. (C–F) Compared to WT, numbers of cortical progenitors are significantly reduced in BubR1 CKO, including apical NPCs (Pax6+ C, D) and basal intermediate progenitors (Tbr2+ E, F). (E and G) Post-mitotic cells (P27+ E, G) are also decreased in BubR1 CKO to a lesser degree. WT and BubR1 CKO cells counted from images of three sections from each animal are compared (n = 3 for each genotype) *P < 0.05. Scale bar, 100 μm.
BubR1 deficiency causes massive cell death in progenitors and postmitotic cells
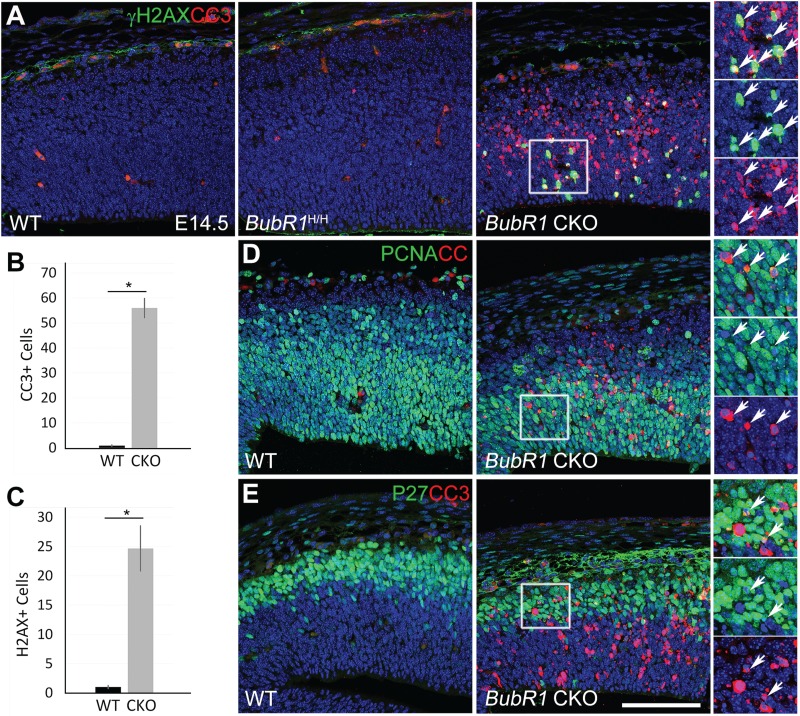
Because pyknotic nuclei were common in the BubR1 CKO cortex but not in WT or BubR1H/H, a decrease in overall cell viability likely accounts for the reduction across all cell types. To test this, we first used cleaved caspase-3 (CC3) immunostaining to detect cells undergoing apoptotic cell death at E14.5. As expected, we found significantly more CC3+ cells in BubR1 CKO than in WT (Fig. 5A and B). Next, to determine if the DNA damage caused cell death, we examined the prevalence of γH2AX (S139), a marker of double-strand DNA breaks, in BubR1 CKO (Fig. 5A). γH2AX+ cells in the BubR1 CKO were significantly increased compared to WT, but CC3+ cells were twice as abundant as γH2AX+ cells, suggesting that the DNA damage may not be the main cause of apoptosis (Fig. 5A–C). As expected, double immunostaining of CC3 and γH2AX revealed that γH2AX+ cells partially overlapped with CC3+ apoptotic cells (Fig. 5A, inset).
Figure 5.
Absence of BubR1 severely compromises cell survival. (A–C) Numbers of dying cells stained for apoptosis (CC3+) or cells exhibiting double-strand DNA damage (γH2AX+) are drastically increased in the developing cortex of BubR1 CKO but not BubR1H/H or WT at E14.5. Arrows indicate examples of cells expressing both markers (inset). (D and E) Subsets of progenitors (PCNA+) and post-mitotic cells (P27+) undergo apoptosis in BubR1 CKO (arrows in inset).*P < 0.05. Scale bars, 100 μm.
To identify cells undergoing apoptotic cell death, double immunostaining using CC3 and markers for progenitor (PCNA) and post-mitotic (P27) cells was performed at E14.5. As shown in Figure 5D and E, apoptosis occurred in the subsets of progenitor (PCNA+) and post-mitotic (P27+) cell populations (insets). Cell death in these cell populations was observed as early as E12.5 (Supplementary Material, Fig. S4). Although a few cells were found in which CC3 overlapped with P27, it rarely overlapped with Ctip2+ layer 5 neurons (Supplementary Material, Fig. S5). This result suggests that the dying cells are mainly progenitors or cells that have just become postmitotic but not yet differentiated into neurons. Taken together, our results provide clear evidence that massive cell death reduces progenitors and cortical neurons in BubR1 CKO and that this reduction is likely to be a major contributor to the pathogenesis of microcephaly.
BubR1 is required for proper progression of mitosis
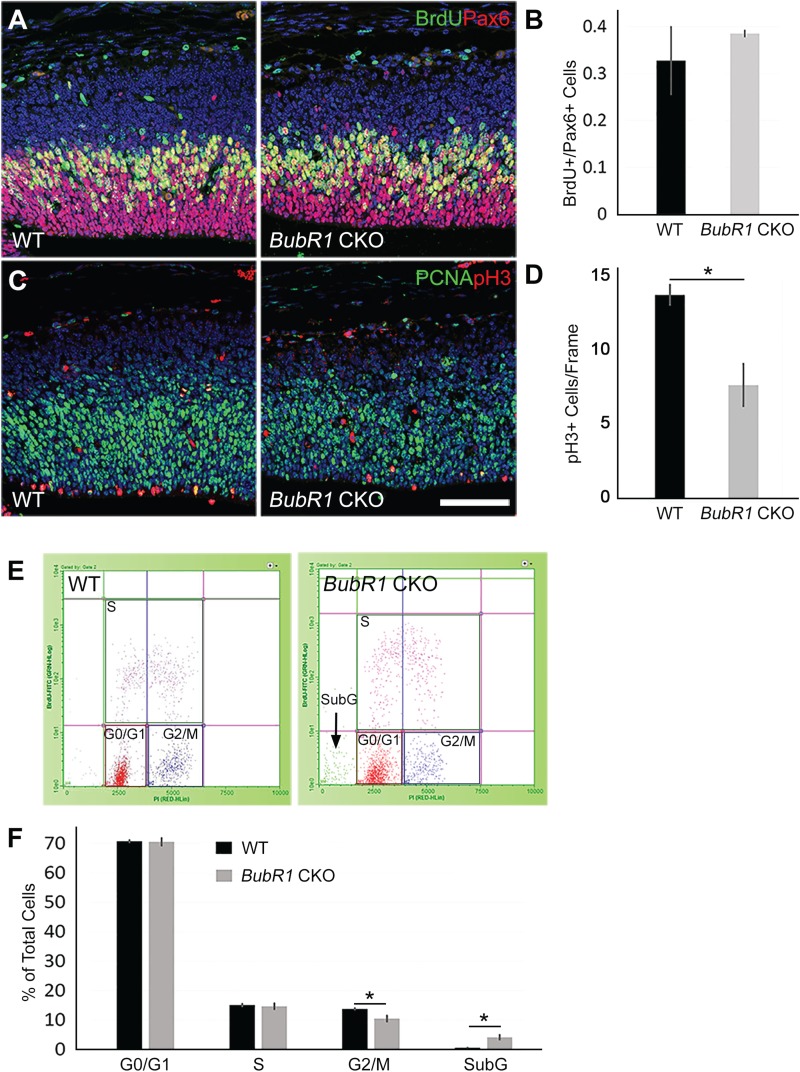
To determine the BubR1 functions required for the viability of neural progenitors, we compared several features of neural progenitors in BubR1 CKO and WT. First, we looked for changes in cell cycle progression by determining mitotic index. The fraction of S-phase cells among total aNPCs was calculated from BrdU+ Pax6+/total Pax6+ cells at E14.5. Although the number of Pax6+ aNPCs was reduced, there was no significant change in the fraction of S-phase cells in BubR1 CKO (Fig. 6A and B). Next, because BubR1 acts predominantly during M-phase, we used PH3 immunostaining to determine the number of mitotic cells (Fig. 6C). In contrast to the majority of microcephaly models, in which the number of mitotic cells increases due to mitotic delay, BubR1 CKO mice showed a significant decrease in the number of M-phase cells compared to WT (Fig. 6C and D). Given the known role of BubR1 in controlling progression through the mitotic checkpoint until all conditions are satisfied, it is plausible that M-phase cells are reduced because loss of BubR1 allows cells to progress through mitosis prematurely and without being properly checked.
Figure 6.
BubR1 loss reduces the number of cells in mitosis. (A and B) Loss of BubR1 does not affect the proportion of S-phase cells marked by 30-min BrdU pulse labeling, among total proliferating aNPCs (BrdU+/Pax6+), in BubR1 CKO at E14.5. (C and D) The total number of mitotic cells across the ventricular lining is significantly reduced in BubR1 CKO. PH3+ cells counted from three sections from each animal are compared between WT and BubR1 CKO (n = 3 for each genotype). (E and F) Cell cycle analyses using flow cytometry after 30-min BrdU incorporation and propidium iodide staining show a reduction of mitotic cells (G2+M) without affecting other cell cycle phases in BubR1 CKO. BubR1 CKO cortices also include a significant proportion of SubG cells.*P < 0.05. Scale bar, 100 μm.
We confirmed reduction of cells in mitosis by using flow cytometry to compare the proportion of cells in each phase of the cell cycle at E14.5 in the BubR1 CKO and WT cortices. Timed pregnant females were injected with BrdU to label S-phase cells 30 min prior to embryonic cortical tissue harvest. All nuclei were labeled with propidium iodide and plots of propidium iodide to BrdU labeling were analyzed to determine the fraction of cells in cell cycle phases. Flow cytometry analysis revealed no change in the proportion of S-phase cells in the BubR1 CKO mice compared to WT, confirming our immunofluorescent assay results using BrdU (Fig. 6B, E and F). Consistent with our observation of abundant apoptotic cells, subG cells were significantly increased, corroborating the presence of substantial cell death at this stage (Fig. 6E and F and Supplementary Material, Fig. S6). Importantly, G2/M-phase cells were significantly reduced, which corroborates the results of our PH3+ mitotic cell counting and suggests that cells were more likely to exit M-phase prematurely than to be arrested in mitosis (Fig. 6C, D and F).
We hypothesized that the changes in BubR1 CKO mitotic cells resulted from cell autonomous alterations caused by BubR1 loss. To evaluate the effect of BubR1 loss at a single cell level, we used in utero electroporation of BubR1 shRNA. E13.5 embryos of timed pregnant females were electroporated with either shBubR1 or empty vector control. Plasmids were co-electroporated with pCAG-GFP to detect electroporated cells. Embryos were harvested 2 days later (E15.5). Decreased BubR1 expression confirmed BubR1 knockdown in the majority of GFP+ electroporated cells (Supplementary Material, Fig. S7A). As we expected, BubR1 shRNA electroporation significantly decreased the proportion of M-phase cells among total electroporated cells compared to control shRNA (Supplementary Material, Fig. S7B and C). Overall, our data suggests that loss of BubR1 allows NPCs to prematurely bypass the mitotic checkpoint, which accelerates the progression of mitosis and decreases the proportion of mitotic cells among cycling NPCs.
BubR1 is crucial to maintain cells in metaphase
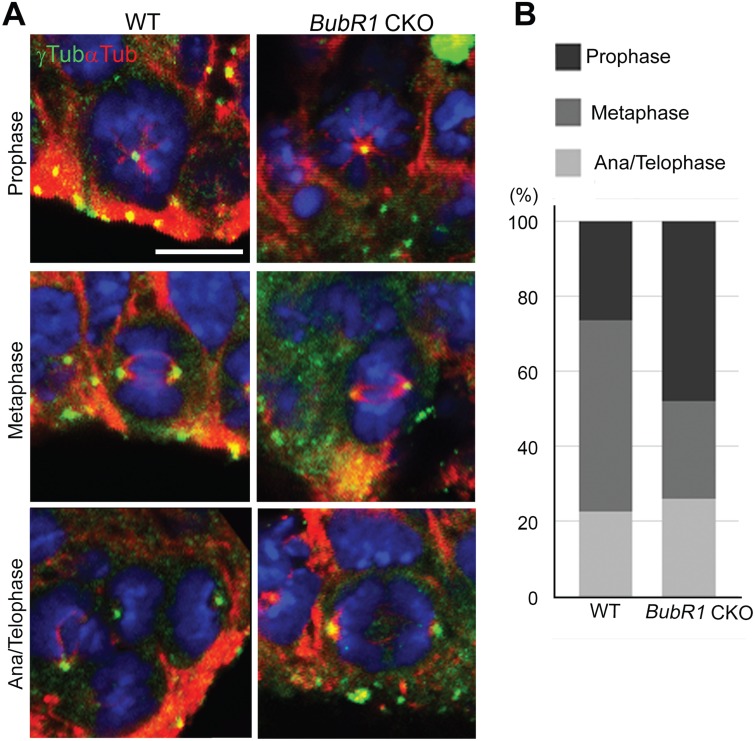
Since BubR1 has a role both in regulating microtubule- kinetochore attachments and in preventing premature anaphase, it is possible that the loss of BubR1 affects metaphase plate formation. For instance, the delay in establishing stable microtubule-kinetochore attachments or failure to inhibit initiation of anaphase can reduce the mitotic cells in metaphase. To analyze the potential mitotic progression defects in BubR1 CKO mice, we determined the proportion of cells in each mitotic phase. We subdivided neural progenitors in mitosis into four phases: 1) prophase—chromosomes begin to condense; 2) metaphase—microtubules attach to chromosomes, which align at midplate; 3) anaphase—mitotic spindle starts to shorten and chromosomes begin to separate; and 4) telophase—chromosomes have fully separated. We labeled both centrosomes (γ-tubulin or pericentrin) and the mitotic spindle (α-tubulin) to classify cells in each phase. Our analyses revealed that the proportion of total mitotic cells in metaphase was significantly decreased in BubR1 CKO compared to WT (Fig. 7A and B). This finding supports the importance of the mitotic checkpoint function of BubR1; when it is absent, metaphase cells are decreased, probably due to premature transition to anaphase. Interestingly, while the proportion of cells in anaphase/telophase remained similar to WT, prophase cells were increased. One possible explanation for these findings is that chromosome congression to midplate is delayed due to the defects in establishing a stable kinetochore-spindle attachment. However, since the total length of mitosis is likely to be reduced in BubR1-deficient progenitors, prophase length may not differ from that of WT.
Figure 7.
BubR1 is required for normal mitotic progression. (A) Mitotic cells were analyzed and classified according to stage of mitosis: prophase, metaphase or ana/telophase. Representative images from each phase are shown for WT and BubR1 CKO. BubR1-deficient mitotic cells show the normal number and appearance of centrosomes and bipolar spindles. (B) In BubR1 CKO, as compared to WT, the fraction of cells in metaphase among total mitotic cells is reduced but the proportion of cells in prophase is increased (n > 50, from three to four animals for each genotype). Scale bar, 5 μm.
Importantly, our analyses of mitotic cells did not detect multipolar cell divisions that have been reported in in vitro studies following BubR1 loss (36). Failure to inhibit Plk1-mediated centrosome amplification has previously been shown to generate multiple centrosomes, resulting in multipolar cell division in lymphocytes carrying mutations in BUB1B gene (36). We did not observe an abnormal number of centrosomes, suggesting that centrosome biogenesis is not disrupted in the absence of BubR1 in cortical progenitors (Fig. 7A). Bipolar spindle formation also appeared to be unaffected (Fig. 7A). Collectively, our study suggests that the major effect of the absence of BubR1 is that metaphase is shortened because inhibition of the onset of premature anaphase is impaired.
BubR1 affects generation of ependymal cell layer
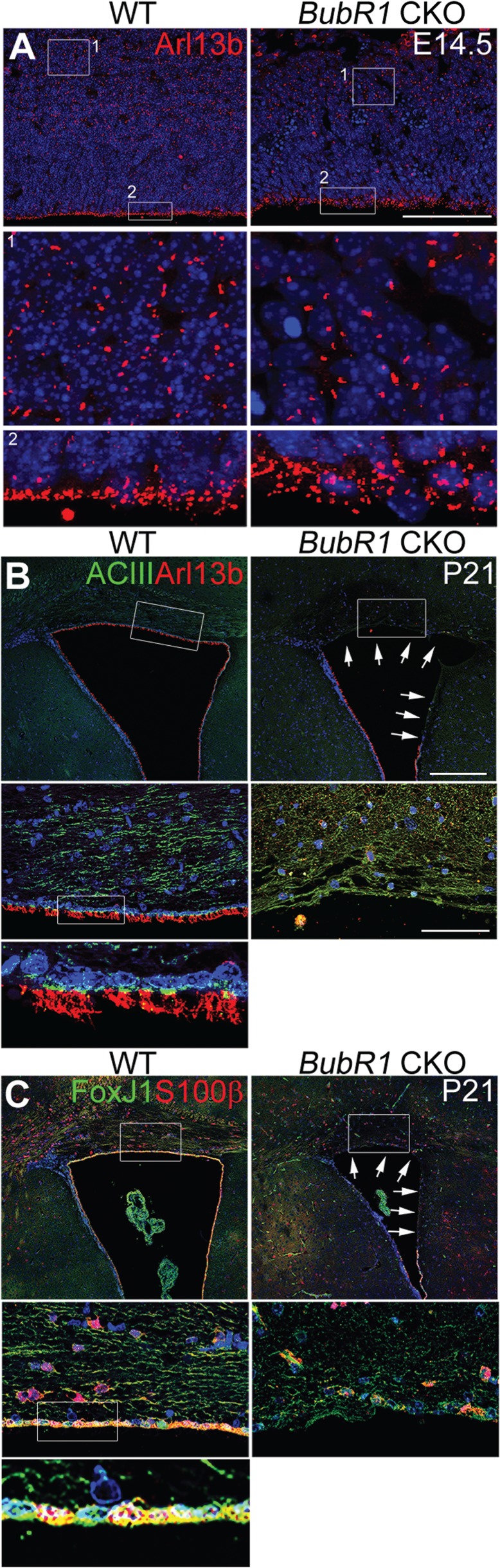
Previous reports have established BubR1 as a ciliopathy gene, but whether ciliogenesis defects arise in developing or mature cortices when BubR1 is decreased has not been explored (7). Since primary cilia play a critical role in transmitting mitogenic signaling such as Shh, the reduction of cortical progenitor cells may have resulted from primary cilia defects. To examine whether primary cilia were present and their morphology, we used cilia markers such as adenylate cyclase type III (AC3) and Arl13b. First, we investigated the cilia phenotype in embryonic cortex of the BubR1 CKO. At E14.5, there were no apparent abnormalities in the number, size or localization of primary cilia (Fig. 8A). We then examined postnatal cerebellar granule neuron precursors in BubR1H/H mice at P8 (Supplementary Material, Fig. S8). Since Emx1-Cre is not expressed in the cerebellum, we did not examine primary cilia of those cells in BubR1 CKO mice. Shh emanating from Purkinje cells is essential for proliferation of cerebellar granule cell precursors (41), and the effects of primary cilia on the regulation of their proliferation are well established (37). In BubR1H/H mice, we failed to detect changes in the primary cilium of cerebellar granule cell precursors, consistent with the proper cerebellar development observed (Supplementary Material, Figs S1 and S8).
Figure 8.
BubR1 is required to generate the ependymal cell layer. (A) BubR1 loss does not produce defects in presence or gross morphology of Arl13b+ primary cilia in either post-mitotic cells (Box #1) or progenitor cells (Box #2) as compared to WT at E14.5. (B, C) At P21, markers for cilia, AC3+ and Arl13b+ (B), and ependymal cells, S100β and FoxJ1 (C), line the entire lateral ventricle in WT, but no ependymal cells line the dorsal or medial ventricle in BubR1 CKO (arrows). Scale bars, B (low magnification): 200 μm, A and B (high magnification): 50 μm.
We next determined whether there were defects in the motile cilia of the ependymal cell layer lining lateral cortex. Surprisingly, in the BubR1 CKO, multiple motile cilia, which can be visualized with cilia markers such as Arl13b and AC3, were completely absent in the ventricular lining of dorsal cortex where Emx1-Cre is expressed (Fig. 8B). Staining for ependymal cell markers, S100β and FoxJ1, confirmed the absence of ependymal cells in the ventricular lining of BubR1 CKO (Fig. 8C). Because motile cilia of ependymal cells propel cerebrospinal fluid (CSF), disruption of the ependymal layer and dysfunction of cilia are associated with hydrocephalus. Despite absence of the ependymal layer of dorsal cortex, BubR1 CKO showed no evidence of hydrocephalus. This suggests that an intact ependymal cell layer along the ventral telencephalon, where Emx1-Cre is not expressed, may be sufficient for normal flow of CSF. Together, these findings demonstrate a previously unrecognized function of BubR1 in ependymal cell generation and/or maintenance.
Discussion
Our results reveal that BubR1 is a microcephaly-causing gene that acts primarily by affecting the mitotic progression of cortical progenitors. As many known microcephaly genes are associated with components of the mitotic apparatus, such as the centrosome, spindle and kinetochore, mutations in these genes have been observed to cause mitotic defects in animal models and/or patient fibroblasts. These defects result from delay of mitosis due to abnormal alignment of spindle orientation, disorganization of microtubules and anomalous centrosome duplication or maturation, which lead to premature differentiation and/or cell death (42–44). Importantly, mitotic delay induced by pharmacological agents causes P53-independent premature differentiation or P53-dependent cell death, suggesting a pathogenic mechanism related to delayed mitosis (27). However, our results demonstrate that the absence of BubR1 differs from previously described microcephaly-inducing genes because it does not cause an increase in cells undergoing mitosis due to mitotic delay. Instead, it induces a 2-fold decrease in mitotic cells, probably due to unchecked, and thus rapid, progression through mitosis. How does rapid mitosis cause such massive cell death? We propose that, because cells with minor aneuploidy will undergo additional rounds of mitosis, the mitotic checkpoint defects can amplify chromosomal defects through subsequent mitoses. That whole chromosome aneuploidy, as in MVA, is insufficient to trigger apoptosis or even prevent proliferation in retinal pigment epithelium (RPE) cell culture (45) supports this proposal. Eventually, aneuploidy cells with significant loss or gain of chromosomes will undergo cell death due to their severely compromised genomic content; NPCs have been shown to tolerate aneuploidy of up to five anomalies before cell death is triggered (29). The mechanism responsible for triggering cell death in aneuploid cells is not fully elucidated and may occur through both P53 dependent and independent mechanisms (46,47). It will be important in future studies to determine precisely how the mitotic defects found in BubR1 CKO compromise genomic integrity through accumulation of chromosomal anomalies and lead to cell death in conjunction with the P53 pathway.
Our study reveals that BubR1 acts by a distinctive pathogenic mechanism despite having massive cell death and small brain size in common with other microcephaly mutants. Interestingly, the kinetochore-associated protein, Blinkin, which interacts directly with BubR1, is encoded by a microcephaly-causing gene (MCPH4/CASC5) (18). The N-terminus of BubR1 contains a tetratricopeptide repeat, which relays a signal to prevent anaphase by forming a connection with unattached kinetochores through Blinkin (48,49). It is therefore plausible that similar mitotic defects, such as failure to inhibit premature onset of anaphase, may be responsible for the pathogenesis of MCPH4-microcephaly. However, patient fibroblast analyses have shown that the number of PH3+ mitotic cells is increased compared to WT, consistent with the mitotic delay (50). Furthermore, because patients with Blinkin mutations differ from those with MVA and do not show predisposition to cancer, it is possible that mitotic defects due to Blinkin might differ from those due to BubR1 (18,50). It would be interesting to determine whether shortened mitosis is unique to the loss of BubR1 and not caused by other microcephaly-causing genes, including kinetochore component gene mutations.
Our finding that the number of mitotic cells in metaphase is reduced in BubR1 CKO can be explained by the loss of BubR1’s roles in mitosis. Such roles include facilitating chromosome congression by ensuring proper KT-MT stabilization and preventing premature initiation of anaphase as a member of the checkpoint complex. Although it is difficult to segregate these two functions, we observed that BubR1 mutant progenitors, albeit reduced in number, demonstrated typical chromosome congression at the metaplate with a seemingly normal spindle organization. This observation suggests that BubR1’s function in KT-MT stabilization is not predominant in the pathogenesis of microcephaly. As BubR1 includes distinct domains that mediate specific functions, including kinetochore attachment regulatory domain (KARD), manipulations of KARD could provide direct support for this argument. It has been shown that phosphorylation of the KARD phospho sites (S676, S670 and T680) at the onset of metaphase allows BubR1 to recruit PP2A-B56α to unattached kinetochores (51). PP2A-B56α then promotes KT-MT connections by counteracting Aurora B kinase activity that phosphorylates the kinetochore complex proteins and decreases their affinity for microtubules (52,53). Accordingly, an Aurora B inhibitor partially rescued the chromosome congression defects caused by BubR1 knockdown (51). Rescue experiments in BubR1 CKO mice using KARD domain deletion, phospho site mutants or an Aurora B inhibitor could therefore clarify whether BubR1 function in KT-MT stabilization has a substantial role in cortical progenitor cell division. Alternatively, rescue experiments based on manipulation of the KEN domain that includes binding site of CDC20, an activator of the anaphase promoting complex (54–56), could be used to identify which loss of function is critical for the pathogenesis of microcephaly. Now that we have established an animal model for microcephaly with novel features, future experiments could use it to identify pathogenic mechanisms in vivo by dissecting functional interactions with distinct binding partners. Furthermore, this model may serve as a platform for developing therapeutic approaches, including gene therapy and pharmacological treatments.
Our work demonstrates that only a minimal level of BubR1 is required for proper cortical development and that nearly complete loss of BubR1 is necessary to induce microcephaly. It is puzzling that there is very little difference in BubR1protein between BubR1H/H and CKO but only the CKO shows the severe defects. It is possible that E15 is not the optimal time to detect differences in BubR1 protein levels responsible for progenitor cell division because many cells are postmitotic at this late point in neurogenesis. Nonetheless, the quantitative data provide evidence that BubR1 protein level is profoundly reduced in both the hypomorphic allele and CKO. Importantly, we have found that there is some residual protein in the kinetochore of BubR1H/H but not of CKO. A previous study using overexpression of the BubR1 domain demonstrated that BubR1 localization to the kinetochore is beneficial because it correlates with improvement of phenotypes such as aneuploidy and cancer (40). Therefore, the residual protein remaining in the kinetochore in the BubR1H/H may suffice to avoid the mitosis defects found in BubR1 CKO. The lack of a severe phenotype in the hypomorphic allele despite a substantial loss of BubR1 also suggests functional compensation by other proteins. Because the BubR1 CKO mice used in the current study are generated on a hypomorphic background, it is likely that the same functional compensation would occur in the BubR1H/H and CKO mice. Of note, BubR1 knockdown experiments consistently show the reduced number of mitotic cells observed in our experiments and in a previous study (38). These results are consistent with functional compensation in the hypomorphic background, which lowers the amount of BubR1 necessary for proper progression of mitosis. Since there is no such functional compensation in the WT background, knockdown mediated by BubR1 shRNA can cause mitotic defects although knockdown is unlikely to achieve nearly complete removal of BubR1 protein. Interestingly, overexpression of BubR1 in mice is implicated in extension of life expectancy through precise and faithful chromosome segregation (57), suggesting that the function of BubR1 in preventing faulty mitosis is dependent on a critical dose. Consistent with this notion, regardless of allelic mutation, BubR1 expression levels appear to be most representative of phenotypic presentation. While BubR1 expression varies widely among people who harbor BUB1B mutations regardless of whether or not they are symptomatic, a downward trend of BubR1 expression seems to correspond to more severe disease (58). It is also interesting that BubR1 levels vary significantly in different cell types within the same individual (59). Variations in BubR1 tissue expression may account for the phenotypic disparity and significant heterogeneity observed in MVA. Although levels of BubR1 expression in patients’ brains have not been determined, our findings may explain why rare individuals with BUB1B mutations do not develop microcephaly.
Our analyses of BubR1 mutants show distinct defects in ependymal cell layer generation but no apparent defects in primary cilia. MVA has been proposed to be a ciliopathy due to overlap with the features of other ciliopathies, such as cerebellar hypoplasia and polycystic kidneys (7). Ciliogenesis is impaired in the fibroblasts of MVA patients because of compromised apical docking of the basal body (7). Similarly, knockdown of BubR1 in medaka fish impairs ciliogenesis and cerebellar development, but whether cortical development is also impaired has not been explored. We found that primary cilia in the developing cortex are unperturbed by loss of BubR1, though we studied only their number and gross appearance. Interestingly, absence of primary cilia in IFT88 mutant mice does not alter the cortical progenitor population or cause neuronal loss (25), suggesting that cortical progenitor cell division does not require intact primary cilia structure. Importantly, consistent with a previous report that knockdown of BubR1 and other ciliopathy genes disrupts cortical lamination, we observed that a subset of superficial neurons remained in the deep cortical layers (38). Also of considerable interest was our observation that the ependymal layer, which extends motile cilia along the ventricle, is absent in the BubR1 CKO mice. This was an unexpected finding because BubR1 has not previously been implicated in ependymal cell generation/maintenance. We did not observe hydrocephalus in BubR1 CKO mice although it is a common feature of animal models with defective cilia or absent ependymal cells (60–64). Hydrocephalus may not have developed because Emx1-Cre expression was restricted, which induced loss of the ependymal layer in only a limited area, including the dorsal cortex. Hydrocephalus is not considered a characteristic feature of MVA, but it is present in about 30% of BubR1-associated MVA patients (1,10). Although the contribution of the lack of ependymal cells to the hydrocephalus in MVA patients remains to be addressed, our results suggest new possible mechanisms.
In summary, we describe the first animal model of BubR1 insufficiency that demonstrates early onset microcephaly and therefore represents the human disease MVA more accurately than those that have been described previously. Our work shows that substantial reduction of BubR1 is compatible with normal cortex development and that nearly complete loss of BubR1 is required for the pathogenesis of microcephaly. We also demonstrate, for the first time, that microcephaly can be caused by rapid progression through mitosis due to a failure of the mitotic checkpoint, rather than mitotic delay. Finally, our study demonstrates the crucial role that BubR1 plays in cellular functions that affect mitotic progression, cell survival and formation of ependymal cell layer in the cortex.
Materials and Methods
Mice
All animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of Temple University Lewis Katz School of Medicine. The BubR1H/H mice were genotyped as previously described (11). Emx1-Cre mice were obtained from Jackson Lab and genotyped accordingly (65).
Histology
Embryos (E15 and older) and postnatal animals were perfused, and their brains isolated and fixed overnight as described previously (60). Brain tissues were embedded in paraffin and sectioned at 7 μm. Hematoxylin and eosin staining was done using standard procedures.
Immunohistochemistry
Paraffin-embedded tissue was cut into 7-μm sections and rehydrated with a xylene-ethanol series into distilled water. Antigen retrieval was achieved through a sodium citrate boil cycle and rinsed in phosphate-buffered saline (PBS). Samples were then incubated overnight at 4°C with primary antibody and 5% normal goat or donkey serum in PBS. Samples were then rinsed in PBS. Secondary antibody (Alexa Flour 488 anti-mouse, Alexa Flour 488 anti-chicken, Alexa Flour 488 anti-rat, Cy3 anti-rabbit, Alexa Flour 647 anti-mouse) in PBS with 5% normal goat was added to each sample and incubated at room temperature for 3 h. Samples were again washed in PBS and stained with Hoechst 33258. Samples were mounted using Fluoromount G. Images were acquired using Axioplan 2 (Carl Zeiss) and confocal microscopes (SP8, Leica) and analyzed with LAS AF (Leica) and Photoshop (Adobe).
Primary antibodies
Immunostaining: Sox9 (Millipore, AB5535), BubR1 (BD, 612503), FoxP2 (Abcam, ab16046), Ctip2 (Abcam, ab18465), Cux1 (Santa Cruz, sc-13024), Pax6 (Covance, PRB-278P), Tbr2 (Abcam, ab183991), P27 (BD, 610241), γH2AX (Millipore, 05-636), CC3 (Cell signaling, 9661), PCNA (Cell signaling, 2586), BrdU (Abcam, ab6326), PH3 (Millipore, 06-570), γ-Tubulin (Proteintech, 15176-1-AP), α-Tubulin (Proteintech, 66031-1-Ig), Arl13b (Abcam, ab136648), AC3 (Santa Cruz, sc-588), S100b (Abcam, ab52642) and FoxJ1(Invitrogen, 14-9965-82).
Western blot: BubR1 (BD, 612503) and GAPDH (Proteintech, 60004-1-Ig)
In utero electroporation
Timed pregnant females at E13.5 were anesthetized using isoflurane gas anesthesia. The uterine horn was then exposed to allow the injection of 2-μl plasmid DNA (4 μg/μl) and 0.05% fast green dye (Sigma) in PBS into the lateral ventricles of the embryos. Injections were performed manually using a pulled glass micropipette. Electroporation was induced using an electroporation generator (ECM 830, BTX, Harvard Apparatus). Each injected embryo received five 50-ms pulses at 40 V with a 950-ms interval.
Flow cytometry analyses
Timed pregnant females at E14.5 were injected intraperitoneally (IP) with 50-mg/kg BrdU. Embryos were harvested after 30 min and the dorsal cortex isolated. Cortical tissue was manually dissociated, washed in 0.1% bovine serum albumin (BSA) in PBS and fixed in 100% ethanol. Cells were treated with 2N HCl/0.5% Triton X-100 and washed. Primary antibody anti-BrdU was added at 1:200 in PBS with 1% BSA and 0.5% Tween20 and left overnight at 4°C. Cells were washed in PBS with 1% BSA. Secondary antibody Alexa Flour anti-rat 488 was added at 1:200 in PBS with 1% BSA and 0.5% Tween20 and left 1 h at room temperature. Cells were pelleted and resuspended in PBS with 10-μg/ml RNAse A and 20-μg/ml propidium iodide in PBS. Following incubation for 30 min at room temperature, cells were analyzed using Guava EasyCyte (Millipore). At least 3000 cells were counted per sample (N = 3 for each genotype) per run and each sample was run in duplicate. If width to area ratio was inconsistent with cellular shape, cells were excluded. Cells were stratified based on green (BrdU) and red (PI) staining to segregate cells in G0/G1, S and G2/M phase. The proportion of cells in each phase was compared across genotype and significance determined with Student’s t-test.
Mitotic phase analyses
Embryonic samples were dissected out, embedded in OCT media and cryosectioned at 16 μm. Antigen retrieval, immunohistochemistry and confocal imaging were completed as described above. Z-stacks were taken at 1-μm intervals spanning up to 20 μm to cover entire mitotic cells. Individual z-stacks for each sample were examined to determine the phase of mitosis. Mitotic phase was assigned by intensity and nuclear shape of Hoechst staining and presence, location and quantity of centrosomes labeled by γ-tubulin or pericentrin. A minimum of 50 cells per genotype from at least three animals was analyzed (E12.5–E14.5).
Quantification and statistical analyses
All cell counting and quantification were completed manually on two to four non-consecutive sections from a minimum of three animals per test group. Results were tested for statistical significance using Student’s t-test.
Supplementary Material
Acknowledgements
The authors thank Dr Alan Tessler for editing the manuscript and Kim lab members for helpful discussions.
Conflict of Interest statement. None declared.
Funding
National Institute of Neurological Disorders and Stroke (R56NS104038-01 to S.K.); Shriners Hospitals for Children (86100, 85109 to S.K.); National Institutes of Health (grants CA191956, CA06927 to T.J.Y.); Department of Defense (W81XWH-17-1-0136 to T.J.Y.); Appropriations from the Commonwealth of Pennsylvania (to T.J.Y.); PA Dept. of Health CURE (to T.J.Y.); Greenberg Foundation (to T.J.Y.).
References
- 1. Kajii T.,Ikeuchi T., Yang Z.Q., Nakamura Y., Tsuji Y., Yokomori K., Kawamura M., Fukuda S., Horita S. andAsamoto A. (2001) Cancer-prone syndrome of mosaic variegated aneuploidy and total premature chromatid separation: report of five infants .Am. J. Med. Genet., 104, 57–64. [DOI] [PubMed] [Google Scholar]
- 2. Suijkerbuijk S.J., Osch M.H., Bos F.L., Hanks S., Rahman N. and Kops G.J. (2010) Molecular causes for BUBR1 dysfunction in the human cancer predisposition syndrome mosaic variegated aneuploidy .Cancer Res., 70, 4891–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wijshake T., Malureanu L.A., Baker D.J., Jeganathan K.B., Sluis B. and Deursen J.M. (2012) Reduced life- and healthspan in mice carrying a mono-allelic BubR1 MVA mutation .PLoS Genet., 8, 1–0.e1003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kawame H., Sugio Y., Fuyama Y., Hayashi Y., Suzuki H.,Kurosawa K. and Maekawa K. (1999), Syndrome of microcephaly, Dandy–Walker malformation, and Wilms tumor caused by mosaic variegated aneuploidy with premature centromere division (PCD): report of a new case and review of the literature .J. Hum. Genet., 44, 219–224. [DOI] [PubMed] [Google Scholar]
- 5. Kajii T., Kawai T., Takumi T., Misu H., Mabuchi O., Takahashi Y., Tachino M., Nihei F. and Ikeuchi T. (1998) Mosaic variegated aneuploidy with multiple congenital abnormalities: homozygosity for total premature chromatid separation trait .Am. J. Med. Genet., 78, 245–249. [DOI] [PubMed] [Google Scholar]
- 6. Warburton D., Anyane-Yeboa K., Taterka P., Yu C.Y. and Olsen D. (1991) Mosaic variegated aneuploidy with microcephaly: a new human mitotic mutant? Ann. Genet., 34, 287–292. [PubMed] [Google Scholar]
- 7. Miyamoto T., Porazinski S., Wang H., Borovina A., Ciruna B., Shimizu A., Kajii T., Kikuchi A., Furutani-Seiki M. and Matsuura S. (2011) Insufficiency of BUBR1, a mitotic spindle checkpoint regulator, causes impaired ciliogenesis in vertebrates .Hum. Mol. Genet., 20, 2058–2070. [DOI] [PubMed] [Google Scholar]
- 8. Miyamoto T., Hosoba K., Ochiai H., Royba E., Izumi H., Sakuma T., Yamamoto T., Dynlacht B.D. and Matsuura S. (2015) The microtubule-depolymerizing activity of a mitotic kinesin protein KIF2A drives primary cilia disassembly coupled with cell proliferation. Cell Rep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hanks S., Coleman K., Reid S., Plaja A., Firth H., Fitzpatrick D., Kidd A., Mehes K., Nash R., Robin N. et al. (2004) Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B .Nat. Genet., 36, 1159–1161. [DOI] [PubMed] [Google Scholar]
- 10. Garcia-Castillo H., Vasquez-Velasquez A.I., Rivera H. and Barros-Nunez P. (2008) Clinical and genetic heterogeneity in patients with mosaic variegated aneuploidy: delineation of clinical subtypes .Am. J. Med. Genet. A, 146A, 1687–1695. [DOI] [PubMed] [Google Scholar]
- 11. Baker D.J., Jeganathan K.B., Cameron J.D., Thompson M., Juneja S., Kopecka A., Kumar R., Jenkins R.B., Groen P.C., Roche P. et al. (2004) BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice .Nat. Genet., 36, 744–749. [DOI] [PubMed] [Google Scholar]
- 12. Yang Z., Jun H., Choi C.I., Yoo K.H., Cho C.H., Hussaini S.M.Q., Simmons A.J., Kim S., Deursen J.M., Baker D.J. et al. (2017) Age-related decline in BubR1 impairs adult hippocampal neurogenesis .Aging Cell, 16, 598–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Faheem M., Naseer M.I., Rasool M., Chaudhary A.G., Kumosani T.A., Ilyas A.M., Pushparaj P., Ahmed F., Algahtani H.A., Al-Qahtani M.H. et al. (2015) Molecular genetics of human primary microcephaly: an overview .BMC Med. Genomics, 8(Suppl 1): S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jackson A.P., Eastwood H., Bell S.M., Adu J., Toomes C., Carr I.M., Roberts E., Hampshire D.J., Crow Y.J., Mighell A.J. et al. (2002) Identification of microcephalin, a protein implicated in determining the size of the human brain .Am. J. Hum. Genet., 71, 136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J.F., Zhang Y., Wilde J., Hansen K.C., Lai F. and Niswander L. (2014) Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat. Commun., 5, 3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu T.W., Mochida G.H., Tischfield D.J., Sgaier S.K., Flores-Sarnat L., Sergi C.M., Topcu M., McDonald M.T., Barry B.J., Felie J.M. et al. (2010) Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture .Nat. Genet., 42, 1015–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fong K.W., Choi Y.K., Rattner J.B. and Qi R.Z. (2008) CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex .Mol. Biol. Cell, 19, 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Genin A., Desir J., Lambert N., Biervliet M., Van Der Aa N., Pierquin G., Killian A., Tosi M., Urbina M., Lefort A. et al. (2012) Kinetochore KMN network gene CASC5 mutated in primary microcephaly .Hum. Mol. Genet., 21, 5306–5317. [DOI] [PubMed] [Google Scholar]
- 19. Carmo Avides M. and Glover D.M. (1999) Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centers .Science, 283, 1733–1735. [DOI] [PubMed] [Google Scholar]
- 20. Bond J., Roberts E., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O. et al. (2005) A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size .Nat. Genet., 37, 353–355. [DOI] [PubMed] [Google Scholar]
- 21. Woods C.G., Bond J. and Enard W. (2005) Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings .Am. J. Hum. Genet., 76, 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar A., Girimaji S.C., Duvvari M.R. and Blanton S.H. (2009) Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly .Am. J. Hum. Genet., 84, 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hussain M.S., Baig S.M., Neumann S., Nurnberg G., Farooq M., Ahmad I., Alef T., Hennies H.C., Technau M., Altmuller J. et al. (2012) A truncating mutation of CEP135 causes primary microcephaly and disturbed centrosomal function .Am. J. Hum. Genet., 90, 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guernsey D.L., Jiang H., Hussin J., Arnold M., Bouyakdan K., Perry S., Babineau-Sturk T., Beis J., Dumas N., Evans S.C. et al. (2010) Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4 .Am. J. Hum. Genet., 87, 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Insolera R., Bazzi H., Shao W., Anderson K.V. and Shi S.H. (2014) Cortical neurogenesis in the absence of centrioles .Nat. Neurosci., 17, 1528–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marthiens V., Rujano M.A., Pennetier C., Tessier S., Paul-Gilloteaux P. and Basto R. (2013) Centrosome amplification causes microcephaly .Nat. Cell Biol., 15, 731–740. [DOI] [PubMed] [Google Scholar]
- 27. Pilaz L.J., McMahon J.J., Miller E.E., Lennox A.L., Suzuki A., Salmon E. and Silver D.L. (2016) Prolonged mitosis of neural progenitors alters cell fate in the developing brain .Neuron, 89, 83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang A.H., Kaushal D., Rehen S.K., Kriedt K., Kingsbury M.A., McConnell M.J. and Chun J. (2003) Chromosome segregation defects contribute to aneuploidy in normal neural progenitor cells. J. Neurosci., 23, 10454–10462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peterson S.E., Yang A.H., Bushman D.M., Westra J.W., Yung Y.C., Barral S., Mutoh T., Rehen S.K. and Chun J. (2012) Aneuploid cells are differentially susceptible to caspase-mediated death during embryonic cerebral cortical development. J. Neurosci., 32, 16213–16222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bolanos-Garcia V.M. and Blundell T.L. (2011) BUB1 and BUBR1: multifaceted kinases of the cell cycle .Trends Biochem. Sci., 36, 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mao Y., Abrieu A. and Cleveland D.W. (2003) Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1 .Cell, 114, 87–98. [DOI] [PubMed] [Google Scholar]
- 32. Burke D.J. and Stukenberg P.T. (2008) Linking kinetochore-microtubule binding to the spindle checkpoint .Dev. Cell, 14, 474–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shirayama M., Toth A., Galova M. and Nasmyth K. (1999) APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5 .Nature, 402, 203–207. [DOI] [PubMed] [Google Scholar]
- 34. Michaelis C., Ciosk R. and Nasmyth K. (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids .Cell, 91, 35–45. [DOI] [PubMed] [Google Scholar]
- 35. Johnson V.L., Scott M.I., Holt S.V., Hussein D. and Taylor S.S. (2004) Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression .J. Cell Sci., 117, 1577–1589. [DOI] [PubMed] [Google Scholar]
- 36. Izumi H., Matsumoto Y., Ikeuchi T., Saya H., Kajii T. and Matsuura S. (2009) BubR1 localizes to centrosomes and suppresses centrosome amplification via regulating Plk1 activity in interphase cells .Oncogene, 28, 2806–2820. [DOI] [PubMed] [Google Scholar]
- 37. Spassky N., Han Y.G., Aguilar A., Strehl L., Besse L., Laclef C., Ros M.R., Garcia-Verdugo J.M. and Alvarez-Buylla A. (2008) Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool .Dev. Biol., 317, 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo J., Higginbotham H., Li J., Nichols J., Hirt J., Ghukasyan V. and Anton E.S. (2015) Developmental disruptions underlying brain abnormalities in ciliopathies. Nat. Commun., 6, 7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Choi C.I., Yoo K.H., Hussaini S.M., Jeon B.T., Welby J., Gan H., Scarisbrick I.A., Zhang Z., Baker D.J., Deursen J.M. et al. (2016) The progeroid gene BubR1 regulates axon myelination and motor function .Aging (Albany NY), 8, 2667–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weaver R.L., Limzerwala J.F., Naylor R.M., Jeganathan K.B., Baker D.J. and Deursen J.M. (2016) BubR1 alterations that reinforce mitotic surveillance act against aneuploidy and cancer .Elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wechsler-Reya R.J. and Scott M.P. (1999) Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog .Neuron, 22, 103–114. [DOI] [PubMed] [Google Scholar]
- 42. Bianchi F.T., Berto G.E. and Di Cunto F. (2018) Impact of DNA repair and stability defects on cortical development. Cell Mol. Life Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Wolf B. and Kops G. (2017) Kinetochore malfunction in human pathologies .Adv. Exp. Med. Biol., 1002, 69–91. [DOI] [PubMed] [Google Scholar]
- 44. Gilmore E.C. and Walsh C.A. (2013) Genetic causes of microcephaly and lessons for neuronal development .Wiley Interdiscip. Rev. Dev. Biol., 2, 461–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soto M., Raaijmakers J.A., Bakker B., Spierings D.C.J., Lansdorp P.M., Foijer F. and Medema R.H. (2017) p53 prohibits propagation of chromosome segregation errors that produce structural aneuploidies .Cell Rep., 19, 2423–2431. [DOI] [PubMed] [Google Scholar]
- 46. Ohashi A., Ohori M., Iwai K., Nakayama Y., Nambu T., Morishita D., Kawamoto T., Miyamoto M., Hirayama T., Okaniwa M. et al. (2015) Aneuploidy generates proteotoxic stress and DNA damage concurrently with p53-mediated post-mitotic apoptosis in SAC-impaired cells. Nat. Commun., 6, 7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yasuhira S., Shibazaki M., Nishiya M. and Maesawa C. (2016) Paclitaxel-induced aberrant mitosis and mitotic slippage efficiently lead to proliferative death irrespective of canonical apoptosis and p53. Cell Cycle, 15, 3268–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kiyomitsu T., Murakami H. and Yanagida M. (2011) Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1 .Mol. Cell. Biol., 31, 998–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kiyomitsu T., Obuse C. and Yanagida M. (2007) Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1 .Dev. Cell, 13, 663–676. [DOI] [PubMed] [Google Scholar]
- 50. Szczepanski S., Hussain M.S., Sur I., Altmuller J., Thiele H., Abdullah U., Waseem S.S., Moawia A., Nurnberg G., Noegel A.A. et al. (2016) A novel homozygous splicing mutation of CASC5 causes primary microcephaly in a large Pakistani family .Hum. Genet., 135, 157–170. [DOI] [PubMed] [Google Scholar]
- 51. Suijkerbuijk S.J., Vleugel M., Teixeira A. and Kops G.J. (2012) Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments .Dev. Cell, 23, 745–755. [DOI] [PubMed] [Google Scholar]
- 52. Welburn J.P., Vleugel M., Liu D., Yates J.R. 3rd, Lampson M.A., Fukagawa T. and Cheeseman I.M. (2010) Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface .Mol. Cell, 38, 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Foley E.A., Maldonado M. and Kapoor T.M. (2011) Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase .Nat. Cell Biol., 13, 1265–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Elowe S., Dulla K., Uldschmid A., Li X., Dou Z. and Nigg E.A. (2010) Uncoupling of the spindle-checkpoint and chromosome-congression functions of BubR1 .J. Cell Sci., 123, 84–94. [DOI] [PubMed] [Google Scholar]
- 55. Lara-Gonzalez P., Scott M.I., Diez M., Sen O. and Taylor S.S. (2011) BubR1 blocks substrate recruitment to the APC/C in a KEN-box-dependent manner. J. Cell Sci., 124, 4332–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Malureanu L.A., Jeganathan K.B., Hamada M., Wasilewski L., Davenport J. and Deursen J.M. (2009) BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase .Dev. Cell, 16, 118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baker D.J., Dawlaty M.M., Wijshake T., Jeganathan K.B., Malureanu L., Ree J.H., Crespo-Diaz R., Reyes S., Seaburg L., Shapiro V. et al. (2013) Increased expression of BubR1 protects against aneuploidy and cancer and extends healthy lifespan .Nat. Cell Biol., 15, 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Matsuura S., Matsumoto Y., Morishima K., Izumi H., Matsumoto H., Ito E., Tsutsui K., Kobayashi J., Tauchi H., Kajiwara Y. et al. (2006) Monoallelic BUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome .Am. J. Med. Genet. A, 140, 358–367. [DOI] [PubMed] [Google Scholar]
- 59. Rio Frio T., Lavoie J., Hamel N., Geyer F.C., Kushner Y.B., Novak D.J., Wark L., Capelli C., Reis-Filho J.S., Mai S. et al. (2010) Homozygous BUB1B mutation and susceptibility to gastrointestinal neoplasia .N. Engl. J. Med., 363, 2628–2637. [DOI] [PubMed] [Google Scholar]
- 60. Park R., Moon U.Y., Park J.Y., Hughes L.J., Johnson R.L., Cho S.H. and Kim S. (2016) Yap is required for ependymal integrity and is suppressed in LPA-induced hydrocephalus. Nat. Commun., 7, 10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Del Bigio M.R., Ependymal cells: biology and pathology .Acta Neuropathol., 2010. 119, 55–73. [DOI] [PubMed] [Google Scholar]
- 62. Abouhamed M., Grobe K., San I.V., Thelen S., Honnert U., Balda M.S., Matter K. and Bahler M. (2009) Myosin IXa regulates epithelial differentiation and its deficiency results in hydrocephalus .Mol. Biol. Cell, 20, 5074–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tissir F., Qu Y., Montcouquiol M., Zhou L., Komatsu K., Shi D., Fujimori T., Labeau J., Tyteca D., Courtoy P. et al. (2010) Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus .Nat. Neurosci., 13, 700–707. [DOI] [PubMed] [Google Scholar]
- 64. Wilson G.R., Wang H.X., Egan G.F., Robinson P.J., Delatycki M.B., O'Bryan M.K. and Lockhart P.J. (2010) Deletion of the Parkin co-regulated gene causes defects in ependymal ciliary motility and hydrocephalus in the quakingviable mutant mouse .Hum. Mol. Genet., 19, 1593–1602. [DOI] [PubMed] [Google Scholar]
- 65. Gorski J.A., Talley T., Qiu M., Puelles L., Rubenstein J.L. and Jones K.R. (2002) Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage .J. Neurosci., 22, 6309–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.