Abstract
Deriving from logical and mechanical interactions between DNA strands and complexes, DNA-based artificial reaction networks (RNs) are attractive for their high programmability, as well as cascading and fan-out ability, which are similar to the basic principles of electronic logic gates. Arising from the dream of creating novel computing mechanisms, researchers have placed high hopes on the development of DNA-based dynamic RNs and have strived to establish the basic theories and operative strategies of these networks. This review starts by looking back on the evolution of DNA dynamic RNs; in particular’ the most significant applications in biochemistry occurring in recent years. Finally, we discuss the perspectives of DNA dynamic RNs and give a possible direction for the development of DNA circuits.
Nucleic Acids and Molecular Computation
The great advances of silicon-based semiconductors in the past century have been key to the development of high-performance processors [1]. Moore’s law, which states that the number of transistors in a dense integrated circuit doubles approximately every 2 years, has guaranteed a continuous evolution of both the efficiency and energy-cost of silicon-based processors. Both of these consequences play core roles in building sophisticated computers in various realms.
Artificial logic networks propose the use of molecules to realize basic computations, with the ultimate goal of constructing submicroscopic computers [1]. DNAs, as naturally occurring programmable molecules, were first adopted to build such algorithms in 1994, when Adleman used a DNA strand as a parallel computing tool to solve a simplified Hamiltonian path problem (see Glossary), with the help of some biotechnologies like ligation and the polymerase chain reaction [2]. A number of subsequent efforts [3–5] seemed to indicate a bright future for DNA-based molecular computation, because the parallel computation ability of DNA molecules is more efficient than single-thread computation on a chip. However, subsequent progress in constructing molecular computing devices has been frustratingly slow [6]. It seems that the computing power of DNA-based circuits cannot rival that of silicon computers in the execution of any algorithms. But when microviewed environments, especially biological environments, are considered, there are many tasks beyond the abilities of traditional silicon-based processors [7]. For instance, it is inconceivable to use silicon chips and sensors to build nanodevices that can directly interact with biomolecules or even cells in biological environments. Compared with nanomaterials, silicon chips are large and do not function normally in electrolyte solutions like those found in cell microenvironments and cell cytoplasm [6]. An effective alternative is use of DNA-based dynamic RNs, or DNA logic circuits that are composed of a series of cascading hybridization or strand-displacement reactions, benefiting from the rigid principles of Watson-Crick base pairing (A-T and C-G) [8]. The advantages of DNA molecules as the basic building blocks for network construction are overwhelming when compared with existing artificial materials, due to the following reasons. (i) DNA is a naturally biocompatible and water-soluble molecule. Since nearly all biological functions occur in aqueous electrolyte solution, soluble DNA molecules can be easily accommodated [9]. (ii) Due to the directionality and programmability of nucleic acid molecules, there is an exponentially large number of different possible combinations of nucleotides, leading to high information density [2]. With rational design, DNA strands can be prepared with specific sequences to meet the demands of circuit design. (iii) A large number of functional DNAs have been reported, such as aptamers and DNAzymes, which can be used for effective target recognition and signal conversion [10,11]. In this way, biomolecules can be directly involved in DNA-based logic operations.
All these properties make DNA an excellent candidate for construction of functional dynamic circuits. Such DNA-based RNs are materially different from traditional solid silicon chips and exist and work in buffered solutions like the cellular environment. In this review, we first describe the progress of DNA dynamic RN development and some classic strategies realized by strand displacements in vitro. Then, we focus on and summarize the most recent advances in biochemical applications of DNA-based logic circuits. Finally, we propose a possible direction of DNA-based artificial RNs in the future.
Evolution of DNA Dynamic RNs
With increasing thorough understanding of DNA chemistry, including formation of phosphodiester bonds between adjacent nucleotides, as well as Watson-Crick base pairing and the double-helical structure, researchers found that the hybridization and strand displacement of DNA can be a controllable process for implementation as logic elements [12,13]. This process, also known as dynamic DNA nanotechnology, usually focuses on nonequilibrium dynamics rather than the equilibrium end states of the DNA hybridization process. As noted above, one of the first attempts to use DNA hybridization to implement an algorithm was reported in 1994 by Adleman et al. [2]. Their work indicated the unique advantages of DNA circuits for parallel computing. In 1997, Ouyang et al. used DNA computation to solve the maximal clique problem, which further proved the high parallelism of DNA computing [4]. However, limitations caused by hybridization efficiency issues and the relationship between library size and computational ability created a bottleneck that inhibited promotion of DNA computing ability. Displacement of one strand from a double-helical nucleic acid by concomitant replacement with an equivalent nucleotide sequence is a familiar and integral aspect of DNA or RNA replication and genetic recombination in vivo [14]. The phenomenon of branch migration, which can be regarded as a result of base-pair breathing [15], was first recognized in vitro by Lee et al. in 1970 when studying the renatured molecules of terminally repetitious, circularly permuted bacteriophage DNA [16]. A simple strand displacement is shown in Figure 1A.
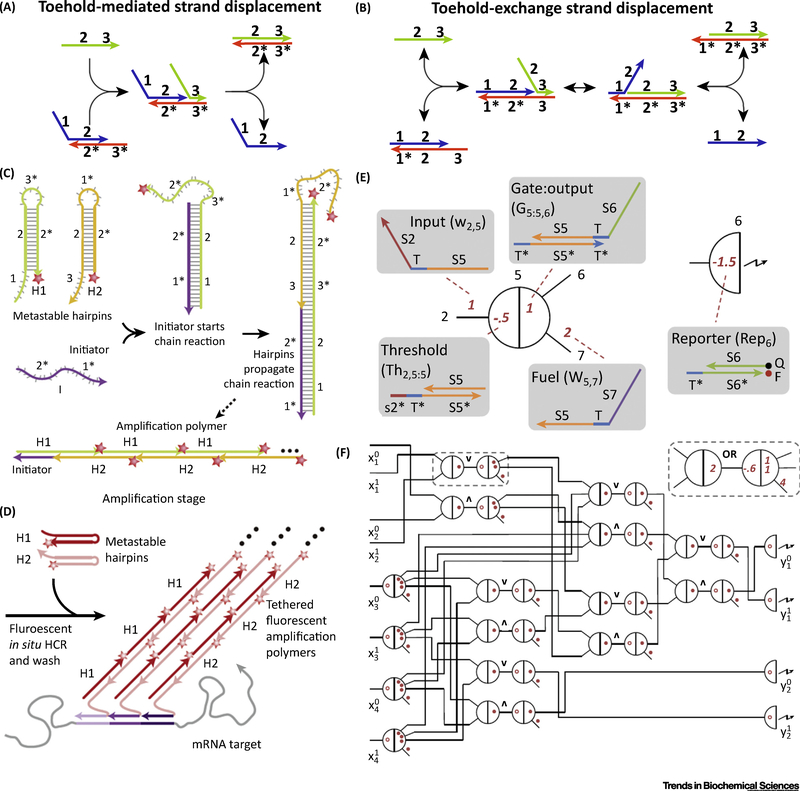
Figure 1. Basic Designs of DNA Dynamic Reaction Networks.
(A) Scheme of toehold-mediated strand displacement reaction. Double-stranded complex (blue and orange) has an overhang domain (3*), known as a toehold. The competitor strand (green) also has a domain 3 which is complementary to 3*. The hybridization of complementary toehold domains acts as the initiation of the following strand migration process, in domain 2/2*. As a result, the competitor strand completely hybridizes with the orange strand while blue strand is released. This process is driven forward by the decrease in free energy on formation of new base pairs. (B) Scheme of toehold exchange strand displacement reaction. The competitor strand (green) hybridizes with the double-stranded complex (blue and orange) via domain 3/3* and a strand migration occurs in domain 2/2*. After the blue strand is released, a new toehold is formed as domain 1, so that the entire process is reversible. (C) Scheme of HCR reaction. In the presence of a single-stranded catalyst, two hairpin structures, H1 and H2, can be opened one by one, forming a long and periodic double stranded DNA chain. (D) Use of HCR as a multiplex amplification strategy for mRNA mapping. (E) See-saw DNA node. (F) Abstract diagram of the seesaw circuit that is equivalent to the square-root digital logic function.
It has been demonstrated that the free energy released by the formation of 10 new base pairs is comparable with that released by the hydrolysis of ATP in standard conditions (ΔG°ATP ≈ – 7.7 kcal/mol) and in cellular conditions (AGatp ≈ –14 kcal/mol). By contrast, the free energy released by the formation of a new base is ΔG°hYbridization ≈ –1.4 kcal/mol/base pair [13,17,18]. Thus, unpaired DNA can be used as a resource of free energy. Yurke et al. used single-stranded DNA as the fuel strand of a molecular machine [12]. Their DNA molecular machine, named molecular tweezers, can be controlled by an added fuel DNA strand and operates between the closed and open states of the tweezers. This work was the first systematic use of a toehold [19,20] domain to design a DNA machine, and proved that a DNA machine is sufficiently reliable to undergo a series of operations. Subsequent reports by Yan et al. showed that the force generated by formation of new base pairs is so strong that it can even change the orientation of DNA origami [21]. Seelig et al. reported the first logic gates totally constructed of DNA strands [22]. They demonstrated AND, OR, and NOT gates, signal restoration, amplification, feedback, and cascading using rational design of DNA molecules (Box 1).
Box 1. Toehold Protection.
The thermodynamics and kinetics of DNA strand displacement reactions were studied in detail by Zhang et al. [23]. According to their results, the length of the toehold domain is related to the rate constant of displacement reaction over six orders of magnitude, from 1 M−1 s−1 to6 × 106 M−1 s−1.These results indicated that the occurrence of strand displacement depends on the existence of a toehold domain, while the rate of strand displacement is determined by the length of the toehold domain. Given the key role of the toehold domain in DNAstrand displacement reactions, when constructing seriesor parallel computing circuits, it is essential to protect the toehold domain before initiation or between layers.
Turberfield et al. designed a DNA-catalyzed nanomachine using a toehold-mediated strand displacement reaction to catalytically open many DNA loop-stem structures [63]. The loop domain also served as a toehold but was locked by a stable stem domain so that no reaction could happen before initiation. Inspired by this strategy, Dirks et al. reported the famous hybridization chain reaction (HCR) using two kinds of DNA hairpin structures [64] (Figure 1C). Such a catalysis is so effective that it has been widely adopted in numerous reports [65–67]. Choi et al. designed a series of HCR probes and mapped five target mRNAs simultaneously in fixed whole-mount and sectioned zebrafish embryos [68] (Figure 1D). A strategy similar to HCR is catalytic hairpin amplification reported by Yin et al. [69–71]. Another type of toehold protection widely used in the past decade is formation of double-stranded DNA by hybridizing the toehold with its complementary strand [8,72].
In addition, with the help of chemically modified artificial bases, physical signals like photons can be used to release an active toehold domain. Prokup et al. used 6-nitropiperonyloxymethylene as a photoresponsive caging group to protect the thymidine nucleotides in the toehold. In the presence of 365 nm light as input signal, the caging group is deprotected and the toehold domain is activated, initiating the process of strand displacement reaction [73]. Similarly, Huang et al. used photochemical excitation to release the toehold domain protected in the loop domain to initiate a branch-migration reaction [74].
Although effective, the traditional toehold-mediated strand displacement reaction suffers from some obvious weaknesses. For example, it is difficult to construct multilayered DNA circuits, because the competitor strand must be longer than the released strand [23]. In order to eliminate this problem, Winfree et al. reported a strategy called toehold-exchange displacement reaction [24]. The incoming strand hybridizes with the bottom strand and displaces the top strand (Figure 1B). Different from the traditional strand displacement reaction, the resulting double-stranded DNA still has an active toehold that can be used to initiate the reverse displacement reaction. As such, toehold-exchange has two obvious advantages. First, the generated toehold after strand displacement allows the construction of downstream layers. Second, toehold exchange weakens the coupling between the kinetics of strand displacement and the thermodynamics of the reaction, so that a strand displacement reaction based on toehold exchange can be rapid despite being only weakly thermodynamically favorable or even thermodynamically unfavorable [23]. Based on this strategy, Winfree’s group designed a catalytic reaction driven forward exclusively by the entropy gain of the entire system [24]. Since then, the toehold-exchange displacement reaction has proved to be an effective strategy for artificial RN construction because of the tunable kinetics of the DNA reaction. By rational design of some auxiliary DNA complexes, Soloveichika et al. demonstrated that the DNA RN can be used to compile formal chemical RNs. This means that DNA RN can somewhat be regarded as a useful language, sufficient for implementation of arbitrarily complex chemical reaction kinetics; just as a few basic electronic elements, such as transistors and wires, are sufficient for the construction of arbitrarily complex logic circuits [25]. Such potential was further adopted by Qian et al., who proposed several powerful DNA circuits with awesome computing abilities, like square root calculation [26], as well as neural network mimicry [27] (Figure 1E,F). The neural network mimicry system consists of 110 DNA strands and demonstrates associative memory capable of answering 81 possible questions. Chen et al. further applied these strategies to mathematically express chemical RNs as DNA circuits [28].
DNAzymes and RNAzymes, which can cleave or ligate the substrate strands in the presence of cofactors, have also been used to design logic circuits [29]. By using an input strand to initiate the activity of a blocked DNAzyme, Stojanovic et al. first reported a set of DNAzyme-based logic gates capable of generating any Boolean function, including the AND, OR, and NOT gates, and the first serial molecular logic gate circuits depending on gate-to-gate communication [30,31]. Kolpash-chikov et al. then demonstrated that the output signal of DNAzyme-based logic gates can be connected to a downstream event, such as release of a small molecule or control of enzyme activity, other than cleavage or formation of oligonucleotides [32]. Pei etal. even designed DNAzyme-based automation to play a game with a human by covering all possible responses, so that the automation won the game nearly each time [33]. Elbaz et al. used DNAzyme-based circuits and demonstrated multilayered gate cascades, fan-out gates and parallel logic gate operations [34].
Initially, the reaction direction of DNA circuits was exclusively determined by the specificity of DNA hybridization. However, recently, Chatterjee et al. discovered that the spatial organization of DNA strands can also act as a control of circuit direction [35]. A DNA origami chip is used as the substrate on which DNA hairpins are immobilized. Only if the distance between two immobilized hairpins is short enough can a strand migration occur. Otherwise, the reaction is inhibited because the distance is longer than the intended DNA bridge. They named this strategy the DNA domino architecture, meaning that spatial organization is used to realize rapid arbitrary logic at the molecular scale.
Applications of DNA-Based RNs
A DNA network can act as a controllable platform for the construction and simulation of chemical reactions with different orders, based on the well-established kinetics model of DNA hybridization and strand displacement. Franco et al. coupled an artificial biochemical oscillator with a variety of load processes, such as the operation of a DNA-based nanomechanical device (‘DNA tweezers’) or the production of a functional RNA molecule (an aptamer for malachite green) [36]. Such an oscillator system under load is like an in vitro molecular clock, whose frequency and amplitude are affected by the loaded reaction. The oscillator may therefore serve as a model system for the study of modularity, coupling of subcircuits, and ensuring robustness in biochemical networks [37].
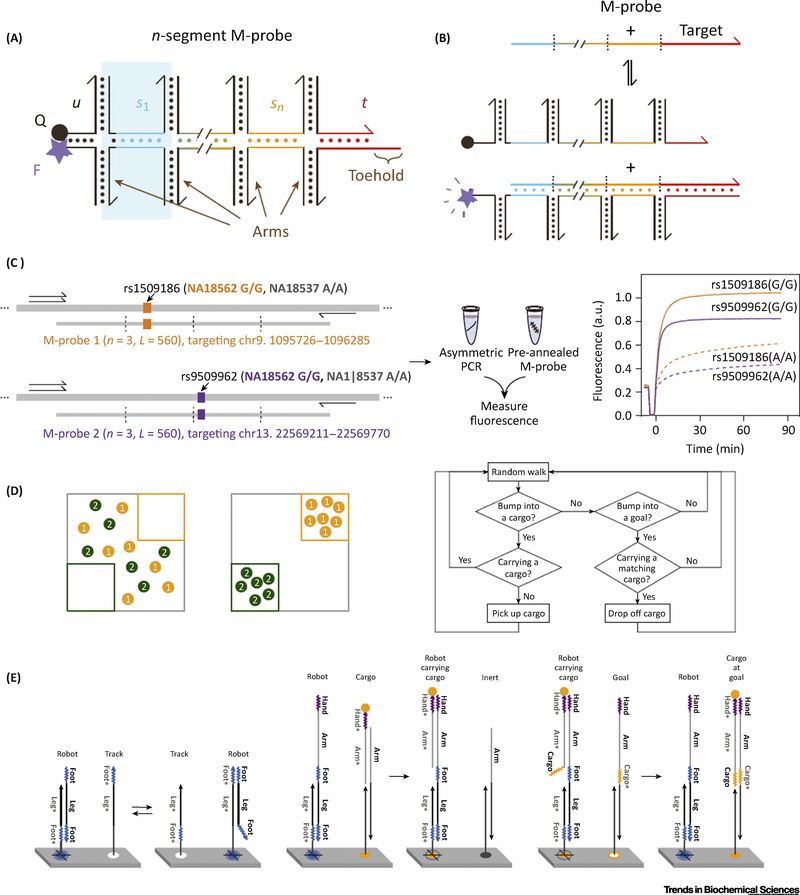
The theories of DNA dynamic networks offer effective strategies for nucleic acid detection. An impressive design of DNA RN in bioanalysis is called the M probe, which is based on the previously reported X probe [38]. The M probe is composed of n segments consisting of two oligonucleotides hybridized to each other via a horizontal region as the recognition domain and a reporter section at the end of the probe (Figure 2A) [39]. With this multistranded equivalent of the toehold probe, hypervariable, long, or repetitive sequences can be selectively bound and enriched with toleration of sequence variations up to 7 nucleotides at prescribed positions, while maintaining single nucleotide sensitivity at other positions (Figure 2B,C).
Figure 2. In Vitro Applications of DNA Dynamic Networks.
(A) A conditionally fluorescent M-Probe bearing internal segments. The lower oligonucleotides have a sequence complementary to sub-sequences of the target, and the upper oligonucleotides have a sequence identical to sub-sequences of the target. (B) Hybridization of the M-Probe to the target results in displacement of the upper oligos as a multistranded complex. Fluorescence increases through this process due to delocalization of the fluorophore and quencher. The hybridization reaction is designed to be both reversible and sequence-specific. (C) The M-Probes are selective for even single nucleotide variants across a 560 nucleotide target sequence. (D, E) The cargo-sorting algorithm. (D) Left: schematic diagram of sorting arbitrarily distributed molecules into distinct piles at specified destinations. Right: flow chart of a simple cargo-sorting algorithm. In the molecular implementation, choices for picking up and dropping off cargos are not always taken as designed – the robot may instead return to random walking with a small probability. (E) Mechanism of the three building blocks for the random walk, cargo pickup, and cargo drop-off.
In the field of mechanical engineering, by programming a concise algorithm, Thubagere et al. built DNA circuits on a chip to sort cargos at the molecular level [40]. They designed a cargo strand and a robot strand and anchored some track strands onto a DNA origami. After several steps of strand displacement-based random walk (~300 steps), the robot strand can capture the cargo strand and drop it upon reaching the destination site (Figure 2D,E). The cargo sorting process is so reliable that the robot can distinguish different cargos without any energy supply.
Applications of Tool Molecule-Integrated DNA RNs
The design of DNA dynamic RNs can be so powerful that more and more problems and algorithms in math or graph theory can be solved and programmed. However, as molecular interaction-based artificial networks, DNA circuits have obvious drawbacks, like system leakage caused by interactions or impurities in the DNA strands, which have limited the reliability and further application of DNA logic circuits in pure computations, as usually performed with silicon chips. Although leakage can be eliminated by introducing a mismatch site when designing the strands [41], this ingenious strategy does not solve the root problem. Otherwise, the introduction of a mismatch site may lead to an unwanted side reaction when the circuit is scaled up. Also, in spite of the abundant theory and strategy of DNA RNs, the input signal or molecules are always DNA strands [42]. However, DNA, as a type of naturally occurring programmable molecules, can be easily used to interact with naturally existing targets. It is assumed that, if biomolecules can be used as the input signal of a DNA RN, some biofunctions will be logically controlled or adopted by the network. Fortunately, this assumption benefits from the explosive growth in the number of functional nucleic acids, such as aptamers and DNAzymes, which further make nucleic acids effective as both recognition and computational elements in logic circuits. Thus, a wide range of biomolecules can be easily involved in predetermined logic computations defined by DNA-based logic circuits.
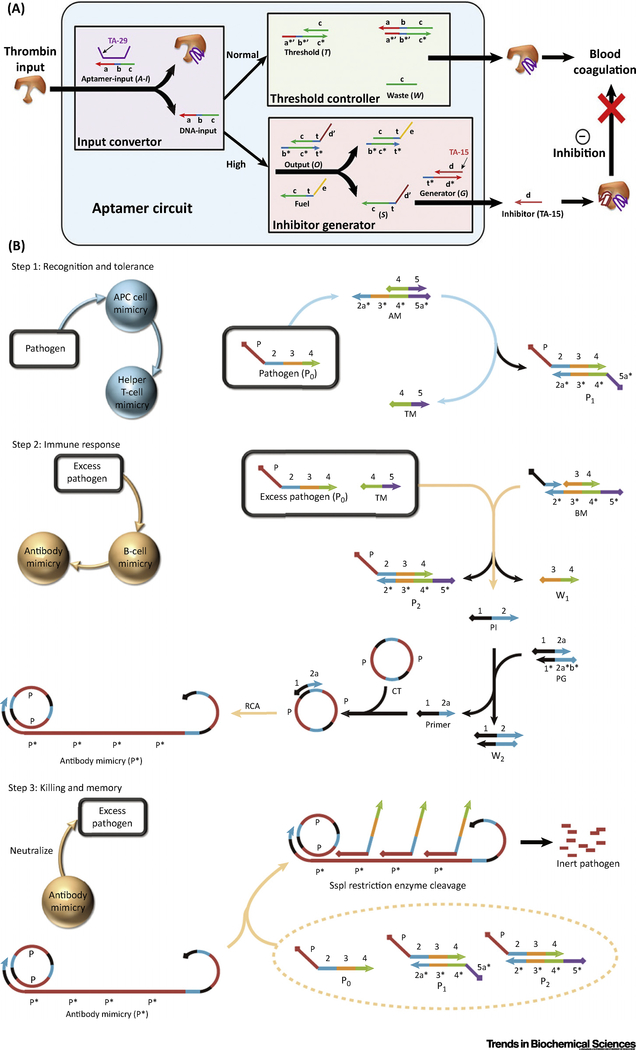
The first example of using aptamers in DNA circuits was reported by Kolpashchikov et al. [32]. They used complementary strands to change the recognition ability of aptamers, so that the target of the aptamer (e.g., malachite green or Taq DNA polymerase) could be released or captured according to a Boolean calculation result. However, in Kolpashchikov’s work, the binding of targets by aptamers is somewhat more like a signal output strategy instead of a signal input. In order to demonstrate the potential of using biomolecules as input, Han et al. developed a modular composition of DNA logic circuits with accurate threshold control, enabling autonomous, self-sustained, and programmable manipulation of enzyme activity in vitro (Figure 3A) [43]. When the enzyme (e.g., thrombin) concentration is higher than the given threshold, the circuit generates an inhibitor to suppress the activity, keeping the activity of thrombin under a certain level. In this DNA circuit, two antithrombin aptamers are used to smartly control the function of thrombin: a 29mer (TA-29) that binds to the heparin exosite without inhibitory function and a 15mer (TA-15) that binds to the fibrinogen exosite with strong inhibitory function. The input signal is the concentration of enzyme, and the output signal is the enzyme activity. Such accurate control of enzyme activity clarified the strong potential of DNA circuits in biomedicine, especially personalized medicine. In the field of bionic engineering, a DNA dynamic RN can be designed to mimic the basic functions of certain organismic pathways (e.g., the mammalian adaptive immune responsive system; AIRS) in a concise manner [44] (Figure 3B), including three steps: (i) recognition and tolerance, (ii) immune response, and (iii) killing and memory. This is an inspiring application of DNA circuits in bionics, indicating that an artificial RN can, to a certain degree, achieve some basic functions of a natural pathway.
Figure 3. Applications of DNA Logical Networks in Enzyme Activity Control and Biomimicry.
(A) DNA logic circuits with accurate threshold control, enabling autonomous, self-sustained, and programmable manipulation of protein activity in vitro. Such a circuit consists of three modules, including: (i) an input convertor that converts the protein input to DNA input for downstream cascade reactions; (ii) a threshold controller that sets the threshold concentration for the system to maintain regular protein activity; and (iii) an inhibitor generator that inhibits excessively high protein activity once it surpasses the threshold. (B) Three steps in AIRS. (Step 1)When there is a pathogenic challenge with a small amount of pathogen DNA, the DNA is first recognized by AM in AIRS and TM is released. (Step2) When the concentration of pathogen DNA increases sufficiently, the threshold defined by Step 1 is overwhelmed. With help of released TM, pathogen DNA is transferred to BM. Activated BM generates a single-stranded antibody initiator that triggers the subsequent rolling circle reaction to generate a long and repeated antibody-mimicry strand. (Step 3) Pathogen DNA is captured by the generated antibody-mimicry strands via hybridization, then hydrolyzed by endonuclease SspI and finally cut into inert pieces. The memory part of the AIRS is realized by released TM. When pathogen DNA invades the next time, the first step is skipped because existing TM can directly trigger the second and third steps of AIRS and finally remove the pathogen DNA more efficiently. Abbreviations: AIRS, adaptive immune responsive system; AM, antigen-presenting cell mimicry; BM, B cell mimicry; TM, T cell mimicry.
Building DNA Networks on Cell Membranes
The cellular environment is an electrolyte-rich buffered solution containing various ions and biomolecules necessary for the sophisticated pathways in living cells. It is both exciting and meaningful to build DNA networks on the cell membrane or inside a cell so that cell states or metabolism can be controlled logically by exogenous stimuli. Inspired by the signal amplification strategy of nucleic acid probes, Han et al. used an aptamer as an anchor to target cancer cells and a prolonged initiator domain to trigger a circuit-based enzyme-free amplification on the cell membrane to generate numerous photosensitizer-labeled strands in the cellular environment [45]. The enriched local photosensitizer concentration showed obvious cytotoxicity toward target cells compared with nontarget cells.
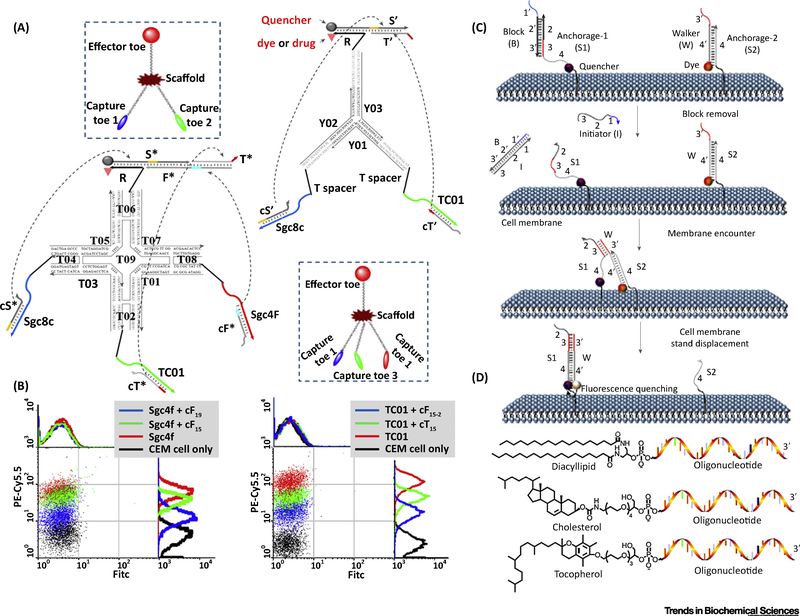
Locating a DNA logic network on the cell surface is important because some cells express membrane proteins with only a limited difference in abundance. To uniquely target cells that have no distinctive markers on their surfaces, a set of multiple markers is needed for each subpopulation in a Boolean manner. DNA logic networks offer a solution to distinguish different cancer cells by analyzing multiple markers on cell surfaces. The first example of using DNA circuits to analyze and selectively label target cells was reported by Rudchenko [46]. Only CD45 and CD20 were simultaneously expressed on the membrane, and the cell was labeled by a fluorophore as a result of DNA computation. This is an always-on binding strategy, because all probe modified-antibodies must be preincubated with cells. In order to develop a smarter and more independent theranostic nanoplatform, You et al. further constructed a DNA ‘nanoclaw’ for logic-based autonomous cancer targeting and therapy (Figure 4A,B) [47]. They used an aptamer as the recognition unit and designed the input-binding/logic-analysis/output-generation process for single-step operation. This strategy can be extended to a larger general platform to facilitate high-order and programmable multiple cell-surface marker identification, including 12 three-input conditions and two four-input conditions [48].
Figure 4. Cellular Applications of DNA Logic Networks.
(A) DNA circuit-based ‘nanoclaw’ for logic-based autonomous cancer targeting and therapy. For cells overexpressing three kinds of target proteins, the three corresponding aptamers (Sgc8c for tyrosine-protein kinase-like 7; targets of TC01 and Sgc4f are unknown, according to the principle of cell Systematic Evolution of Ligands by Exponential Enrichment) act as a three-input AND gate to give a TRUE result, identifying the cell as the target, which is labeled with a fluorophore or treated by photodynamic therapy. (B) Only when multiple biomarkers exist on the cell membrane is the cell labeled with a fluorophore. (C) DNA probe for monitoring dynamic and transient molecular encounters on live cell membranes. (D) Three kinds of anchor molecules on cell membranes. Abbreviations: CEM, human T lymphoblastic leukemic cell line; FITC, fluorescein isothiocyanate.
Apart from membrane proteins, lipid molecules can also be used as anchors for DNA logic circuits. You et al. reported a circuit-based DNA probe to monitor dynamic and transient molecular encounters on live cell membranes (Figure 4C) [49]. Because the encounters of membrane molecules are so fast and the signals are so weak, the detection of membrane events has been a significant challenge. The design overcame this challenge by using a DNA circuit to accumulate each molecular encounter and make the enhanced signal strong enough to be detected by a standard confocal microscope or flow cytometer. By adopting kinetics and 2D fusion-based data analysis, the encounter rates of three kinds of membrane lipid molecules were calculated and the encounter preference derived (Figure 4D).
Building DNA Networks Inside of Cells
DNA dynamic networks inside cells offer the possibility to logically detect various targets in a Boolean manner instead of determining one target simply in a quantitative or qualitative manner. For intracellular applications, Pei et al. used DNA tetrahedra as a platform to construct DNA logic gates inside cells [50,51]. Hemphill et al. [52] published an early report of intracellular logic circuit construction. They successfully used two kinds of miRNA, miR-21 [53] and miR-122 [54], which are usually overexpressed in some cancer cells, as input to construct an AND gate in a living cell. If the cell expresses both mRNAs, the circuit gives a fluorescence signal as the TRUE output. Wu et al. realized a circuit-based amplified detection of mRNA in living cells, and further expanded the application fields of enzyme-free strategies [55]. Differences in molecular levels between normal and diseased cells are always adopted as the activation clue of smart drug delivery strategies. However, since the correlation between biomarker and disease is not always a one-to-one relationship, logical initiation of a DNA network by more than one biomarker is suitable for construction of multi-target-triggered smart nanotheranostics. Douglas et al. developed a DNA drug-loaded origami-based nanobox that is locked by two kinds of aptamers [56]. Only in the presence of both targets (e.g., tyrosine-protein kinase-like 7 for sgc8c and platelet-derived growth factor for 41t) can the box be opened and the encapsulated drugs exposed to take action. This device can be loaded with a variety of materials in a highly organized fashion and is controlled by an aptamer-encoded logic gate, enabling response to a wide array of cues.
siRNA is a convenient signal output due to its biological functions, which can be easily determined by tracking the translated proteins or monitoring cell metabolism [57]. Bindewald et al. designed an RNA switch that can be triggered by a mRNA and converted into an RNA-induced silencing complex for RNAi of another mRNA [58]. Groves et al. systematically tested the effects of chemical modifications and transfection methods on the performance of four-way strand exchange reactions inside living cells and further established that functional siRNA could be activated via strand exchange [59]. siRNA can also be used for gene-edition based logic circuits. Rinaudo et al. designed the first example of a universal RNAi-based logic evaluator that operates in mammalian cells [60]. They used siRNA to silence the 3′ untranslated regions of mRNA, so that the encoded protein (e.g., Zs Yellow) would not be translated. Based on this strategy, Xie et al. further constructed a logic circuit for identification of specific cancer cells [61]. They first identified a list of mRNA markers for a certain kind of cancer cells. If all the markers are confirmed in the circuit, then the cell is identified as the target cancer cell and apoptosis is triggered without affecting non target cell types.
Concluding Remarks and Future Perspectives
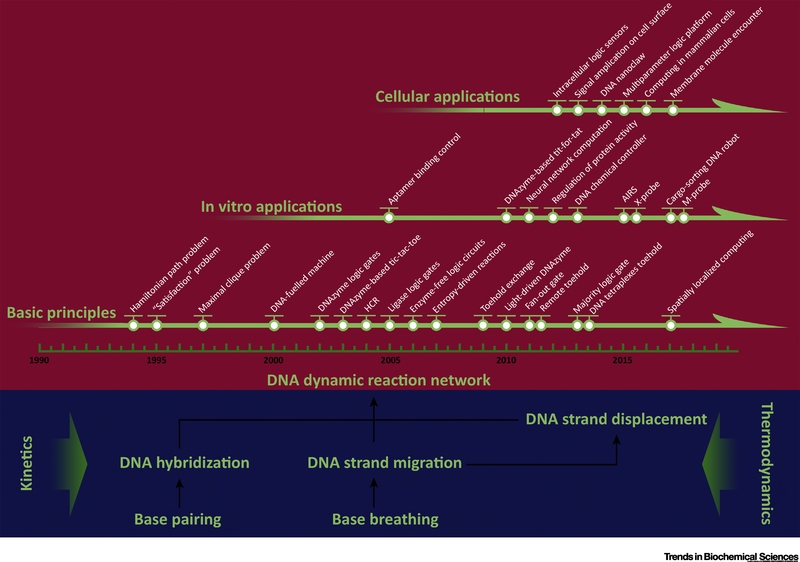
After decades of development, DNA dynamic reactions have grown from fundamental Boolean gates to entire functional logic networks. Studies of the thermodynamics and kinetics of DNA hybridization and strand displacement have made DNA hybridization a programmable tool to construct algorithms at the molecular level with predictable results. Although some researchers have tried to encapsulate artificial chemical circuits into a solid chip, DNA circuits seem to be more suitable for operation in aqueous solution [1]. However, because they function in an aqueous environment, DNA circuits suffer limitations related to chemical equilibria and reaction rates. For example, slow reaction kinetics lead to slow computing rates; unintended hybridization and impurities in DNA strands cause systematic errors; and lack of robustness to interactions between DNA and DNA and between other molecules and DNA make DNA uncompetitive in purely computational tasks in contrast to mature solid chips. Compared with silicon-based chips which perform all the computations on a solid platform, one of the unique features of DNA logic circuits is the biological molecular properties, which allow DNA circuits to function seamlessly with biological inputs and outputs, like other nucleic acids, proteins, small molecules, and even cellular states. As such, DNA circuits can be regarded as ‘aqueous chips’ that have great potential for insertion into naturally occurring systems to perform customized tasks. Although the natural function of DNA is as carriers of genetic information, not as building blocks, DNA seems to be unparalleled at least for biological applications of artificial RNs. Applications of DNA dynamic RNs are summarized in Figure 5, Key Figure.
Figure 5. Key Figure Applications of DNA Dynamic Reaction Network.
Initiated from the DNA computing algorithm realized by DNA parallel hybridization, DNA dynamic reactions have evolved from basic logic gates to entire functional networks, with applications ranging from mechanical operations to biological regulation. Abbreviations: AIRS, adaptive immune responsive system.
For the future perspective part of this review, foreseeable advances may lie in the emergence of more sophisticated computational circuits in vitro and more powerful biofunctional circuits for intracellular applications (see Outstanding Questions) [7]. More effective strategies with reduced leakage can be expected by using modified domains with slower base breathing on the terminus, just like researchers are trying to do right now. Besides, some rationally programmed algorithms can also be used to eliminate mistakes caused by defects of DNA hybridization and strand displacements, which may improve the computational reliability of DNA RNs [62]. Considering the biological application potentials of DNA RNs, the stability of oligonucleotides should be studied in a comprehensive way to further clarify the mechanism of DNA RNs in cellular environments. Alternative to normal nucleic acids, enzyme resistant analogs of natural nucleic acids with modifications on pentose (e.g., 2-methoxy-modified nucleic acids) and backbones (e.g., phosphorothioate oligonucleotide and peptide nucleic acids), or the mirror-image enantiomer (e.g. l-nucleic acids) that cannot be recognized by naturally existing proteins, have obvious advantages in the construction of biostable DNA RNs. However, studies about thermodynamics and kinetics of these non-natural DNA-based hybridizations and strand displacements are still limited. Another limitation of applying DNA RNs in living systems is the diluted concentration of DNA species after being delivered into a complex humoral environment. A possible solution toward this problem is to encapsulate DNA RN into an artificial vesicle to allow all the molecular computations to be performed in an isolated environment. All in all, the merits of DNA RNs are still obvious, and implementations that focus on the molecular or biological properties of DNA should be the direction of DNA circuit growth.
Outstanding Questions.
One of the most primary problems that limit the efficiency of DNA circuits is leakage caused by the high rate of base breathing. Is there a way to eliminate the leakage problem without affecting the performance of DNA circuits?
How is the stability of circuits made by natural oligonucleotides when used inside of cells or in cellular microenvironments?
For intracellular applications of DNA circuits, the enzyme resistant analogs of natural nucleic acids like phosphor-othioate oligonucleotide, 2-methoxy-modified nucleic acids, peptide nucleic acids or l-nucleic acids are preferred. However, the kinetics or thermodynamic properties of these analogs are unclear compared with natural oligonucleotides. Can the principles of building DNA circuits by natural nucleic acids be used for those of modified nucleic acids?
Is there an efficient way to read the output signal produced by living cells?
How can the concentrations of different DNA species be controlled and monitored when delivering into cells?
Is there a possibility that a totally DNA reaction network-based bio computing core with special functions can be built inside a cell to regulate a certain pathway?
How can DNA RNs be made more deliverable and resistant to dilution, so that the entire computation can run in complex humoral environments?
Supplementary Material
Highlights.
If only the disassociation constant is considered, a strand migration reaction should be extremely slow (with a half-life of even thousands of year). However, it is a super-fast process with a half-life of seconds, and the reason is base breathing.
Toehold exchange reactions are more flexible compared with traditional strand displacement reactions. Thus, toehold exchange can be used, at least in theory, to construct higher scaled circuits.
DNA circuits are like logically programmed mechanical controllers that allow temporally and spatially programmable regulation of biological systems.
Considering the electrolyte-rich cellular microenvironment and abundant membrane molecules which play significant roles in metabolism, the cell surface has been found to be an effective and essential platform to build a DNA RN.
Acknowledgments
This work is supported by NSFC grants (NSFC 21521063), and by NIH GM R35 127130 and by NSF 1645215.
Glossary
- Aptamer
a single-stranded oligonucleotide (DNA or RNA) that can specifically bind with its target, which can be an ion, small molecule, protein, or even a biomarker on whole cells with Kd in the pM to μM range. Aptamers are screened using a method called SELEX (Systematic Evolution of Ligands by Exponential Enrichment). A library pool is incubated with the target. After removing the unbound strands, bound strands are purified and enriched by pCR to generate a new pool, which is incubated with target again. After several rounds of screening, when the current pool has sufficient affinity and specificity toward target, the candidate strands are sequenced and synthesized as aptamers.
- Base-pair breathing
nucleic acid hybridization is stabilized by a series of weak interactions (stacking and hydrogen bonding) between individual base pairs. For doublestranded DNA in buffer solution, each base pair is always undergoing an equilibrium between bound and unbound states so that some bases are temporarily unbound, especially the base pairs at the ends of helices. This breathing-like process in DNA helices is called base-pair breathing.
- DNAzyme
also known as a deoxyribozyme, or catalytic DNA, is a short oligonucleotide with high catalytic activity toward specific substrates with the help of certain cofactors like metal ions and small molecules. DNAzymes are usually isolated using an in vitro selection process from an initial library pool.
- Domain
subsequence tract section. Several continuous nucleotides in a strand that act as a unit in hybridization, branch migration, dissociation, structure, or (deoxy) ribozyme function.
- Hamiltonian path problem
problem in the mathematical field of graph theory about whether a Hamiltonian path (a path in an undirected or directed graph that visits each vertex exactly once) or a Hamiltonian cycle exists in a given graph (whether directed or undirected)
- Leakage
refers to the part of reactions in a DNA dynamic RN that happen without initiation, usually caused by reversible DNA hybridization equilibrium, defects of synthesized DNA strands, or imperfect DNA hybridization.
- Parallel computation
(including temporal parallel and spatial parallel computation) different calculations or executions are performed simultaneously in time or in space. For DNA computation, since the initial pool can maintain a large number of different strands, hybridizations can happen simultaneously. In such a way, the computation ability can be promoted with a larger initial pool.
- RNAi
biological process of inhibition of gene expression or mRNA translation by two types of small RNA molecules –miRNA and siRNA.
- Toehold
particular type of domain that serves to colocalize nucleic acid strands and complexes. Toeholds are typically short (4–10 nucleotides).
Footnotes
Supplemental Information
Supplemental information associated with this article can be found, in the online version, at https://doi.org/10.10167j.tibs.2018.04.010.
References
- 1.Ball P (2000) Chemistry meets computing. Nature 406,118–120 [DOI] [PubMed] [Google Scholar]
- 2.Adleman L (1994) Molecular computation of solutions to combinatorial problems. Science 266, 1021–1024 [DOI] [PubMed] [Google Scholar]
- 3.Sakamoto K et al. (2000) Molecular computation by DNA hairpin formation. Science 288, 1223–1226 [DOI] [PubMed] [Google Scholar]
- 4.Ouyang Q et al. (1997) DNA solution of the maximal clique problem. Science 278, 446–449 [DOI] [PubMed] [Google Scholar]
- 5.Lipton R (1995) DNA solution of hard computational problems. Science 268, 542–545 [DOI] [PubMed] [Google Scholar]
- 6.Benenson Y (2009) Biocomputers: from test tubes to live cells. Mol. BioSyst 5, 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J et al. (2017) Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation. Nat. Chem 9, 1056–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang DY and Seelig G (2011) Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem 3, 103–113 [DOI] [PubMed] [Google Scholar]
- 9.Jones MR et al. (2015) Programmable materials and the nature of the DNA bond. Science 347, 1260901. [DOI] [PubMed] [Google Scholar]
- 10.Lyu Y et al. (2016) Generating cell targeting aptamers for nanotheranostics using cell-SELEX. Theranostics 6, 1440–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X-B et al. (2011) Metal ion sensors based on DNAzymes and related DNA molecules. Ann. Rev. Anal. Chem 4, 105–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yurke B et al. (2000)ADNA-fuelled molecular machine made of DNA. Nature 406, 605–608 [DOI] [PubMed] [Google Scholar]
- 13.SantaLucia J (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Nati. Acad. Sci. U. S. A 95, 1460–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapique N and Benenson Y (2017) Genetic programs can be compressed and autonomously decompressed in live cells. Nat. Nanotechnoi 13, 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cubero E et al. (1999) Observation of spontaneous base pair breathing events in the molecular dynamics simulation of adifluorotoluene-containing DNA oligonucleotide. J. Am. Chem. Soc 121, 8653–8654 [Google Scholar]
- 16.Lee CS et al. (1970) A physical study by electron microscopy of the terminally repetitious, circularly permuted DNA from the coli-phage particles of Escherichia coli 15. J. Moi. Biol 48, 1IN19–8IN322 [DOI] [PubMed] [Google Scholar]
- 17.Zhao B et al. (2017) Visualizing intercellular tensile forces by DNA-based membrane molecular probes. J. Am. Chem. Soc 139, 18182–18185 [DOI] [PubMed] [Google Scholar]
- 18.Alberty RA and Goldberg RN (1992) Standard thermodynamic formation properties for the adenosine 5′-triphosphate series. Biochemistry 31, 10610–10615 [DOI] [PubMed] [Google Scholar]
- 19.Genot AJ et al. (2011) Remote toehold: a mechanism for flexible control of DNA hybridization kinetics. J. Am. Chem. Soc 133, 2177–2182 [DOI] [PubMed] [Google Scholar]
- 20.Tang W et al. (2013) DNA tetraplexes-based toehold activation for controllable DNA strand displacement reactions. J. Am. Chem. Soc 135, 13628–13631 [DOI] [PubMed] [Google Scholar]
- 21.Yan H et al. (2002) A robust DNA mechanical device controlled by hybridization topology. Nature 415, 62–65 [DOI] [PubMed] [Google Scholar]
- 22.Seelig G et al. (2006) Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 [DOI] [PubMed] [Google Scholar]
- 23.Zhang DY and Winfree E (2009) Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc 131,17303–17314 [DOI] [PubMed] [Google Scholar]
- 24.Zhang DY et al. (2007) Engineering entropy-driven reactions and networks catalyzed by DNA. Science 318, 1121–1125 [DOI] [PubMed] [Google Scholar]
- 25.Soloveichik D et al. (2010) DNA as a universal substrate for chemical kinetics. Proc. Natl. Acad. Sci. U. S. A 107, 5393–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian L and Winfree E (2011) Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 [DOI] [PubMed] [Google Scholar]
- 27.Qian L et al. (2011) Neural network computation with DNA strand displacement cascades. Nature 475, 368–372 [DOI] [PubMed] [Google Scholar]
- 28.Chen Y-J et al. (2013) Programmable chemical controllers made from DNA. Nat. Nanotechnoi 8, 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu L et al. (2015) Switchable catalytic DNA catenanes. Nano Lett 15, 2099–2103 [DOI] [PubMed] [Google Scholar]
- 30.Stojanovic MN et al. (2002) Deoxyribozyme-based logic gates. J. Am. Chem. Soc 124, 3555–3561 [DOI] [PubMed] [Google Scholar]
- 31.Stojanovic MN and Stefanovic D (2003) A deoxyribozyme-based molecular automaton. Nat. Biotechnol 21 (9), 1069–1074 [DOI] [PubMed] [Google Scholar]
- 32.Kolpashchikov DM and Stojanovic MN (2005) Boolean control of aptamer binding states. J. Am. Chem. Soc 127 (32), 11348–11351 [DOI] [PubMed] [Google Scholar]
- 33.Pei R et al. (2010) Training a molecular automaton to play a game. Nat. Nanotechol 5 (11), 773–777 [DOI] [PubMed] [Google Scholar]
- 34.Elbaz J et al. (2010) DNA computing circuits using libraries of DNAzyme subunits. Nat. Nanotechol 5 (6), 417–422 [DOI] [PubMed] [Google Scholar]
- 35.Chatterjee G et al. (2017) A spatially localized architecture for fast and modular DNA computing. Nat. Nanotechol 12 (9), 920. [DOI] [PubMed] [Google Scholar]
- 36.Franco E et al. (2011) Timing molecular motion and production with a synthetic transcriptional clock. Proc. Natl. Acad. Sci 108 (40), E784–E793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genot A et al. (2016) High-resolution mapping of bifurcations in nonlinear biochemical circuits. Nat. Chem. 8 (8), 760–767 [DOI] [PubMed] [Google Scholar]
- 38.Wang JS and Zhang DY (2015) Simulation-guided DNA probe design for consistently ultra specific hybridization. Nat. Chem 7 (7), 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang JS et al. (2017) Modular probes for enriching and detecting complex nucleic acid sequences. Nat. Chem 9 (12), 1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thubagere AJ et al. (2017) A cargo-sorting DNA robot. Science 357 (6356), eaan6558. [DOI] [PubMed] [Google Scholar]
- 41.Jiang YS et al. (2014) Mismatches Improve the performance of strand-displacement nucleic acid circuits. Angew. Chem 126(7), 1876–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han D et al. (2014) Nucleic acid based logical systems. Chem. Eur. J 20 (20), 5866–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han D et al. (2012) A logical molecular circuit for programmable and autonomous regulation of protein activity using DNA aptamer-protein interactions. J. Am. Chem. Soc 134 (51), 20797–20804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han D et al. (2015) A cascade reaction network mimicking the basic functional steps of adaptive immune response. Nat. Chem 7 (10), 835–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han D et al. (2013) Engineering a cell-surface aptamer circuit for targeted and amplified photodynamic cancer therapy. ACS Nano 7 (3), 2312–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudchenko M et al. (2013) Autonomous molecular cascades for evaluation of cell surfaces. Nat. Nanotechol 8 (8), 580–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.You M et al. (2014) DNA “nano-claw”: logic-based autonomous cancer targeting and therapy. J. Am. Chem. Soc 136 (4), 1256–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.You M et al. (2014) Programmable and multiparameter DNA-based logic platform for cancer recognition and targeted therapy. J. Am. Chem. Soc 137 (2), 667–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.You M et al. (2017) DNA probes for monitoring dynamic and transient molecular encounters on live cell membranes. Nat. Nanotechol 12, 453–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang L et al. (2014) Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew. Chem. Int Ed 53 (30), 7745–7750 [DOI] [PubMed] [Google Scholar]
- 51.Pei H et al. (2012) Reconfigurable three-dimensional DNA nanostructures for the construction of intracellular logic sensors. Angew. Chem 124 (36), 9154–9158 [DOI] [PubMed] [Google Scholar]
- 52.Hemphill J and Deiters A (2013) DNA computation in mammalian cells: microRNA logic operations. J. Am. Chem. Soc 135 (28), 10512–10518 [DOI] [PubMed] [Google Scholar]
- 53.Medina PP et al. (2010) OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467 (7311), 86–90 [DOI] [PubMed] [Google Scholar]
- 54.Negrini M et al. (2011) microRNA involvement in hepatocellular carcinoma. Anticancer Agents Med. Chem 11 (6), 500–521 [DOI] [PubMed] [Google Scholar]
- 55.Wu C et al. (2015) A nonenzymatic hairpin DNA Cascade reaction provides high signal gain of mrna imaging inside live cells. J. Am. Chem. Soc 137 (15), 4900–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Douglas SM et al. (2012) A logic-gated nanorobot for targeted transport of molecular payloads. Science 335 (6070), 831–834 [DOI] [PubMed] [Google Scholar]
- 57.Ren K et al. (2016) A DNA dual lock-and-key strategy for cell-subtype-specific siRNA delivery. Nat. Commun 7, 13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bindewald E et al. (2016) Multistrand structure prediction of nucleic acid assemblies and design of RNA switches. Nano Lett. 16 (3), 1726–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Groves B et al. (2016) Computing in mammalian cells with nucleic acid strand exchange. Nat. Nanotechol 11 (3), 287–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rinaudo K et al. (2007) A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol 25 (7), 795–801 [DOI] [PubMed] [Google Scholar]
- 61.Xie Z et al. (2011) Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333 (6047), 1307–1311 [DOI] [PubMed] [Google Scholar]
- 62.Heath JR et al. (1998)Adefect-tolerant computer architecture: opportunities for nanotechnology. Science 280 (5370), 1716–1721 [Google Scholar]
- 63.Turberfield AJ et al. (2003) DNA fuel for free-running nanomachines. Phys. Rev. Lett 90 (11), 118102. [DOI] [PubMed] [Google Scholar]
- 64.Dirks RM and Pierce NA (2004) Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. U. S. A 101 (43), 15275–15278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu G et al. (2013) Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc. Natl. Acad. Sci 110 (20), 7998–8003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu G et al. (2013) Building fluorescent DNA nanodevices on target living cell surfaces. Angew. Chem. Int. Ed 52 (21), 5490–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang J et al. (2011) Pyrene-excimer probes based on the hybridization chain reaction for the detection of nucleic acids in complex biological fluids. Angew. Chem. Int. Ed 50 (2), 401–404 [DOI] [PubMed] [Google Scholar]
- 68.Choi HM et al. (2010) Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol 28 (11), 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yin P et al. (2008) Programming biomolecular self-assembly pathways. Nature 451 (7176), 318–322 [DOI] [PubMed] [Google Scholar]
- 70.Li B et al. (2011) Rational, modular adaptation of enzyme-free DNA circuits to multiple detection methods. Nucleic Acids Res. 39(16), e110–e110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X et al. (2013) Stacking nonenzymatic circuits for high signal gain. Proc. Natl. Acad. Sci 110 (14), 5386–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xing Y et al. (2011) A responsive hidden toehold to enable controllable DNA strand displacement reactions. Angew. Chem. Int. Ed 50 (50), 11934–11936 [DOI] [PubMed] [Google Scholar]
- 73.Prokup A et al. (2012) DNA computation: a photochemically controlled AND gate. J. Am. Chem. Soc 134 (8), 3810–3815 [DOI] [PubMed] [Google Scholar]
- 74.Huang F et al. (2013) DNA branch migration reactions through photocontrollable toehold formation. J. Am. Chem. Soc 135(21), 7967–7973 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.