Abstract
Enterotoxigenic Escherichia coli (ETEC) produces different virulence factors allowing the bacterium to colonize and develop watery diarrhea. Proteomics studies have also introduced new protein belonging to the secretion pathways, antigen 43 (Ag43), which plays important role in E. coli pathogenesis. The objective of this study was to investigate O-types and virulence factors of E. coli isolates from neonatal calves diarrhea. Total of 120 isolates from diarrheic calves were genotyped for their O groups and the presence of virulence genes K99, F41 and STa as well as Ag43. The predominant O-type was O101 (51.00%) and the prevalence of K99, F41 and STa was 7 (5.80%). The Ag43 was detected in all samples with three different allelic patterns. Our results indicated that K99 positive isolates certainly have one of each 2200 bp or 1800 bp or both copies of Ag43 passenger domain, while negative K99 isolates lack the Ag43. The results reported here provide informative data regarding the prevalence of E. coli O-types and their virulence factors in enteric colibacillosis. The Ag43 that was more found in K99 positive isolates might be associated with diarrhea-causing E. coli strains in neonatal calves.
Key Words: Antigen 43, Calf, Diarrhea, Entrotoxigenic Escherichia coli, O-type
Introduction
Neonatal calf diarrhea is an important cause of morbidity and mortality in newborn calves and still a major cause of productivity and economic loss to cattle producers worldwide. 1,2 According to the report of the National Animal Health Monitoring System for United States, about 57.00% of weaning calf mortality was due to diarrhea.3 Among diarrheagenic strains of Escherichia coli, the most common cause of neonatal diarrhea is Enterotoxigenic E. coli (ETEC).4 The ETEC strains produce different virulence factors associating with colonization in the small intestine, avoiding the immune response and stimulating the deleterious inflammatory response.5,6 Susceptible age of calves to ETEC infections and colibacillosis is first four days after birth.7,8
One of the main fimbrial antigens in ETEC is the plasmid-encoded K99 (F5)9 mediating adherence to the ileum and establishing the first steps of colonization in the bovine intestine.4 The F5 positive strains are usually isolated from calves between one and five days of age with diarrhea and it is believed that this observation is due to the decline of the F5 receptors expression in the bovine epithelia within days.10 Other virulence factors such as heat-stable enterotoxin (STa)11 and F41 may also be associated with diarrhea in newborn calves.9,12 Although fimbria and enterotoxins are two known groups of the classic virulence factors, proteomics studies have recently introduced several new proteins such as autotransporters involving in E. coli pathogenesis.13-15
Autotransporter proteins are belonging to the largest secretion pathways in gram-negative bacteria. They all have a same three-part structure including a signal peptide directing the protein secretion into the inner membrane, a passenger domain which is exported to the extracellular space and performed the function of protein and a translocator domain that is placed in the outer membrane and formed a beta-barrel pore in which passenger domain transmission occurs.16,17 Some of the autotransporters are specified to the E. coli pathovars and some of them are conserved in most E. coli strains. Antigen 43 (Ag43) is one of these proteins with close homologues in many of E. coli pathovars, especially uropathogenic and diarrheagenic strains. The gene encoding Ag43 (agn43) exists as multiplex alleles with different copy numbers in different strains.18-20 The Ag43 as a non fimbrial adhesion is involved in auto-aggregation and biofilm formation. Its passenger domain is presented on the bacterial cell surface by bounding to the translocator domain via non-covalent interaction and mediates colony aggregation through the self-recognizing mechanism. However, this capacity varied between Ag43 allelic variants.21-23
Surveys on the new antigens and identification of fimbriae and enterotoxins are important in the epidemiology and pathogenesis of ETEC strains.24-26
This study was performed to investigate the prevalence of O-types and virulence genes of E. coli strains isolated from diarrheic calves. We further examined the possible relatedness of the classic virulence factors (K99, F41, and STa) to the presence of Ag43.
Materials and Methods
Feces samples of diarrheic calves (n = 120) under five days of age were obtained from industrial farms in two provinces of Alborz and Qazvin, Iran. The farm owners gave informed consent for using the samples for this study. The criteria for sampling included animals with symptoms of diarrhea. Samples were plated on selective MacConkey (Merck, Darmstadt, Germany) agar plates within 12 hr following collection. After 24 hr of incubation at 37 ˚C, at least three lactose-positive colonies of each sample were selected and incubated in Luria-Bertani broth (Merck) at 37 ˚C for one night. The bacterial suspension was centrifuged at 13,000 rpm for 30 sec. The pellet was suspended in 50 μL of sterile water, incubated at 100 ˚C for 10 min and centrifuged. The supernatant was used in the polymerase chain reaction (PCR) analysis. All of isolates were examined for O-type. A pair of 16S rDNA specific primer was used as an E. coli positive control. The PCR primer pairs were designed based on the sequences of O15, O35, O26, O78, O115, O119, O86, O9, O101, and O8. The primers used for O-typing are listed in Table 1. For multiplex PCR, 50 to 100 ng DNA as a template was used in final volume of 30 μL containing 1X PCR buffer (50 mM KCl, 10 m M Tris-HCl [pH = 8.30]), 2.50 mM MgCl2, 167 mM each of dATP, dCTP, dGTP and dTTP, 0.05 to 0.15 mM of the respective primers and 2.50 U Taq DNA polymerase (Cinaclone, Tehran, Iran). The PCR was performed with a thermocycler (Eppendorf, Montesson, Franc) with microtubes in the following conditions: initial denaturation stage of 95 ˚C for 10 min followed by denaturation in 30 cycles at 95 ˚C for 30 sec and then on annealing at 50 ˚C for 45 sec and extension stage at 72 ˚C for 70 sec which ends by a final extension at 72 ˚C for 5 min. The PCR products were loaded on 2.00% agarose gel with ethidium bromide. After electrophoresis, gels were photographed under ultraviolet (UV) light. Molecular identification was done by PCR for the detection of the K99, F41, STa and Ag43. The primers used in this part of the study are also listed in Table 1.
Table 1.
List of the forward (F) and reverse (R) primers used in the study
| Types | Primers |
|---|---|
| O8 | F: CAATCGCCAGAGGCATAA |
| R: TCTGGCTGCCCTTGTGAG | |
| O9 | F: TGGGTGTTAAAAGACATCAA |
| R: CCCAGAAATCCATGCTC | |
| O15 | F: ATTTTCACGAGGCATAGC |
| R: AAGACTCACAATCGCACC | |
| O26 | F: GCTAAAATTCAATGGGCG |
| R: ACATAAGCAATTGCAGCG | |
| O35 | F: GTTTCCCAGATAATCTCCTC |
| R: AAATACCCTGTCACTACCG | |
| O78 | F: GGTATGGGTTTGGTGGTA |
| R: AGAATCACAACTCTCGGCA | |
| O86 | F: GAGTTATTTTGGTTCACCCTT |
| R: TAGCCCACCTATGAATAGAGC | |
| O101 | F: GTGTTACTTTCATATCGTCCAG |
| R: ATGCAATGCGGTTTCTAC | |
| O115 | F: TCTAGATGGTGTTCTGAGGT |
| R: GTCCATGACGATAAACCTG | |
| O119 | F: GTTAACAATCAGCTCGATAAAC |
| R: TTTGCAAGTAAACACCCTAAAC | |
| 16S | F: AGAGTTTGATCC/ATGGCTCAG |
| R: CCGTCAATTCCTTTGAGTTT | |
| K99 | F: TATTATCTTAGGTGGTATGG |
| R: GGTATCCTTTAGCAGCAGTATTTC | |
| F41 | F: GCATCAGCGGCAGTATCT |
| R: GTCCCTAGCTCAGTATTATCACCT | |
| STa | F: GCTAATGTTGGCAATTTTTATTTCTGTA |
| R: AGGATTACAACAAAGTTCACAGCAGTAA | |
| Ag43 | F: GTGGCGATTGCGCTGTCT |
| R: TACACCGGTCTGATGGCT |
The PCR was performed in the Eppendorf termocycler (Eppendorf) by using 0.20 mM of each dNTP, 0.50 μM of each primer, 2 μL of DNA, 0.75 mM of MgCl2, 1X of buffer and 1 U of DNA Taq polymerase for Ag43 and 2.50 U of it for others. The cycling program for all virulence factors except Ag43 was comprised of an initial activation step at 94 ˚C for 4 min, 25 cycles of amplification (denaturation at 94 ˚C for 30 sec, annealing for 45 sec at 50 ˚C followed by an extension at 70 ˚C for 90 sec) and a final extension step at 70 ˚C for 10 min and chilling at 4 ˚C. For amplification of Ag43, the steps were followed by 29 cycles of denaturing (95 ˚C for 30 sec), annealing (57.30 ˚C for 30 sec), extension (72 ˚C for 3.5 min) and a final extension (72 ˚C for 5 min), respectively. The PCR products were analyzed by electro-phoresis in agarose gels stained with ethidium bromide and photographed under UV light. A 1 Kb DNA ladder was used to determine the molecular size of the PCR products.
Results
All of the isolates were identified as E. coli on the basis of staining property, colony characteristics, and standard biochemical reaction. The isolated E. coli strains belonged to O101: 51 (42.50%), O8: 18 (15.00%), O9: 10 (8.30%), O115: 6 (5.00%) and 35 (29.10%) were unknown (Table 2).
Table 2.
O-types, virulence factors genes of E. coli isolates from calves with neonatal diarrhea
| O-type | Number of isolates (%) |
Number of isolates expressing the target genes
|
||||||
|---|---|---|---|---|---|---|---|---|
| F5(K99) | F41 | STa | Ag43(1800bp) | Ag43(2200bp) | Ag43 (1800&2200bp) | Ag43(NO) | ||
| O101 | 51 (42.50%) | 6 (5.00%) | 6 (5.00%) | 6 (5.00%) | 20 (16.66%) | 11 (9.16%) | 9 (7.50%) | 11 (9.16%) |
| O8 | 18 (15.00%) | 0 | 0 | 0 | 4 (3.33%) | 4 (3.33%) | 4 (3.33%) | 6 (5.00%) |
| O9 | 10 (8.33%) | 0 | 0 | 0 | 5 (4.16%) | 0 | 4 (3.33%) | 1 (0.83%) |
| O115 | 6 (5.00%) | 0 | 0 | 0 | 3 (2.50%) | 0 | 2 (1.66%) | 1 (0.83%) |
| Non typable | 35 (29.16%) | 1 (0.83%) | 1 (0.83%) | 1 (0.83%) | 21 (17.50%) | 7 (5.83%) | 7 (5.83%) | 0 |
| Total | 120 (100%) | 7 (5.83%) | 7 (5.83%) | 7 (5.83%) | 53 (44.16%) | 22 (18.33%) | 26 (21.66%) | 19 (15.83%) |
All samples were examined by multiplex PCR method for the main ETEC three virulence genes including F5, F41, and STa with the sizes of 314 bp, 380 bp and 190 bp, respectively (Table 2). It was found that among all cases, only 7 (5.80%) of them were positive for F5, F41 and STa. The agn43 was detected in 101 samples. The region, placed between the end of the putative signal peptide and the beginning of the beta barrel domain containing the complete passenger domain were amplified and three different allelic patterns of gene were observed (Fig. 1); isolates having one copy of 1800 bp with the frequency of 44.16% (53/120), isolates having one copy of 2200 bp with the frequency of 18.33% (22/120) and isolates having the both copies with the frequency of 21.66% (26/120). Isolates coding no similar region were 19 (15.83%; Table 2).
Fig. 1.
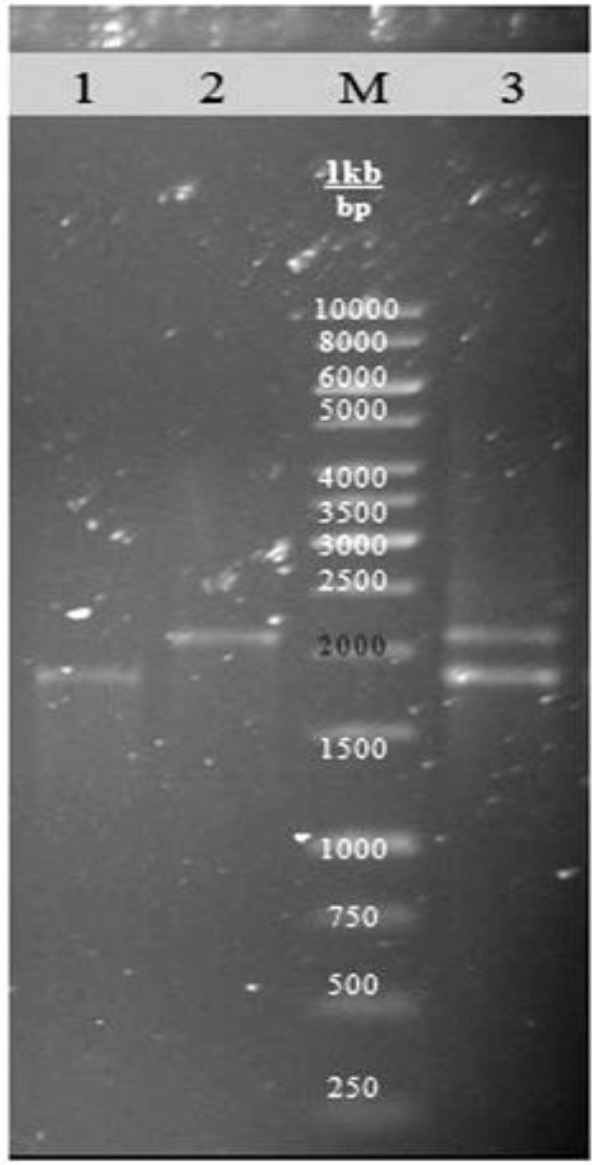
Three different allelic patterns of Ag43 in enterotoxigenic Escherichia coli strains. Lane 1: Colony having a copy of passenger domain 1800 bp; Lane 2: Colony having a copy of passenger domain 2200 bp; Lane 3: Colony having two copies of passenger domain 1800 bp and 2200 bp; Lane M: DNA ladder
The frequency of different Ag43 profiles along with O-types and K99, F41 and STa virulence factors presented together to investigate their relationship (Table 3). Different patterns of agn43 as well as isolates having no Ag43 were observed in all O-types. Allele encoding one copy of 2200 bp was absent in O9 and O115 groups.
Table 3.
The relationship between O-Type, K99, F41, STa and Ag43 adhesion protein
| Types |
Ag43
|
|||
|---|---|---|---|---|
| 1800 bp | 2200 bp | Both 1800 and 2200 bp | Neither | |
| O101 | 20 | 11 | 9 | 11 |
| O8 | 4 | 4 | 4 | 6 |
| O9 | 5 | 0 | 4 | 1 |
| O115 | 3 | 0 | 2 | 1 |
| K99 | 2 | 2 | 3 | 0 |
| F41 | 2 | 2 | 3 | 0 |
| STa | 2 | 2 | 3 | 0 |
All isolates having K99, F41 and STa virulence factors were also positive for Ag43. The frequency of isolates having only one copy of alleles, 1800 bp or 2200 bp, was the same (28.57%), while the isolates encoding both of the alleles were predominant (42.85%).
Discussion
The E. coli is a versatile bacterial species encompassing both commensal and pathogenic strains. Pathogenic strains are commonly identified by their virulence factors. Diarrheagenic E. coli has specific virulence factors for intra-intestinal colonization and propagation.
In the first stages of colonization, the most essential relevant adhesion is F5.5,10,27 This fimbria was found to be the most common virulence factor of E. coli strains isolated from colibacillosis in dairy calves.28 In comparing the virulence factors found in this survey to those isolated in other countries, strains carrying the F5 fimbrial adhesion gene also possessed the STa and F41 genes.29 Although, data indicated a positive relationship between fimbrial adhesins and enterotoxins, at least among detected O-types, about 62.20% were negative for known ETEC virulence factors. It has been indicated that enterotoxin STa was not detected in any E. coli K99 isolates from the diarrheic calves.30 Based on the other study, only 3.00 to 10.00% of strains isolated from calves diarrhea had F5 factor that is approximately in agreement with our findings.31 However, in some investigations, the prevalence rate of ETEC K99+ was about 30.00-100%.32-34 A complication for fimbrial identification arises from the fact that an isolate may produce a subclone which may or may not produce a fimbrial antigen, whereas another one may produce multiple fimbriae of different types.35 The similar frequency for K99 and F41 (5.30%) and a lower percentage of STa (4.02%) have been reported previously.7 A similar results was reported by Younis and El-Naker8 and lower prevalence (0.57%, 2.30%, and 7.30%) was recorded by Zhang et al.36 All of ETEC pathotypes reported in Shahrani et al. study were K99+, while the sta gene was detected in only 4.46% of them.33 Although, 25.00% of E. coli diarrheic samples of their study hadn’t any current virulence factors. Besides these findings, Acha et al. work has shown that K99 adhesion is found in both healthy and diarrheal calves.30 According to the results of present study, low percentage of K99, B41 and STa (5.80%), common factors in neonatal calves colibacillosis, also confirms the results of other studies that these factors may not play a role in the pathogenesis of this disease. In the case of O-types found in this survey, even, Kauffman has published a diagnostic scheme based on the distribution of H, O and K antigens as detected from septicemic and enteric colibacillosis in calves including O8, O9, O15, O20, O26, O35, O78, O86, O101, O115, O117 and O119.37 Although a variety of O-types has been associated with diarrhea, only a limited number of them has been reported in enteric infections of newborn calves. There is an evidence that reveal certain factors such as O-type is important in key steps for survival in the blood and resistance to bacterial killing by the host.38 Based on the results of this work, K99 positive isolates were O101. According to literature data, the majority of K99 isolates belonged to the O groups of O8, O9, O20 and O101.9 The scarce in knowledge about relationships between the presence of virulence factors and disease progression is more obvious and it is necessary to evaluate more virulence gene content of E. coli strains. Several studies were performed to determine the frequency of agn43 in different pathotypes of E. coli. Restieri et al. have illustrated that agn43 is the most common sequence of atuotransporters identified in clinical isolates of E. coli.18 It exists in uropathogenic E. coli (UPEC), diarrheagenic E. coli and avian pathogenic E. coli isolates with a high frequency of 94.00%, 91.00%, and 66.00%, respectively, whereas 56.00% of the commensal fecal isolates contained agn43 sequences.18 Wells et al. have identified 215 auto-transporter proteins in 28 genomic sequences of E. coli including Ag43 with 54.30% identity and 84.50% similarity in all 28 ones.39 Sahl et al. have displayed a common genomic core in ETEC isolates by comparing seven sequenced human ETEC genomes and 42 non-ETEC strains of E. coli.20 Virulence factors and conserved proteins were identified as potential targets that could be included in vaccine development against a diversity of ETEC strains. Among them, agn43 existed 62.50% in ETEC genomes and 35.10% in non-ETEC genomes, respectively.20 In the present study, we found the gene encoding Ag43 in 101 of 120 (84.17%) fecal ETEC isolates verifying the high frequency of this protein. Furthermore, we identified strains containing more than one copy of agn43. It is believed that a gene duplication is a vital event for organisms to overcome the host immune response and to adapt to different microenvironments.19 Several copies of agn43 exist in E. coli strains belonging to different pathotypes. For example, ETEC strain H10407 possesses two copies, while EAEC strain 042 and EPEC strain B171 possess three and four copies, respectively.39,40 Here, we obtained that E. coli strains isolated from neonatal calf diarrhea possessing two copies of agn43 and interestingly the others seemed to have each one of those copies. Evidently, our data demonstrated the gene encoding 1800 bp as a predominant allelic variant of Ag43, particularly in O101 versus 2200 bp. These results indicated the diverse distribution of agn43 in bovine ETEC strains and verified the conservation of this gene in E. coli pathovars.
The important point is the conservation of Ag43 protein among isolated strains that make it difficult to establish a potent relationship between this antigen and others. However, the presence of Ag43 in K99, F41 and STa positive strains has an emphasis on investigating for other novel antigenic factors plying pathogenic roles in ETEC strains.
In the end, our results provide data regarding the prevalence and relation of virulence factors that may be applicable to design widespread control and prevention strategies against calves enteric colibacillosis such as vaccines. Also, passenger domains of Ag43 gene were indicated as possible candidates for ETEC profiling.
Acknowledgments
This study was funded by the University of Tehran under Grant No. 27/6/7502015.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Constable PD. Antimicrobial use in the treatment of calf diarrhea. J Vet Int Med. 2004;18:8–17. doi: 10.1111/j.1939-1676.2004.tb00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorenz I, Fagan J, More SJ. Calf health from birth to weaning Management of diarrhea in pre-weaned calves. Irish Vet J. 2011;64(1) doi: 10.1186/2046-0481-64-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho YI, Yoon J. An overview of calf diarrhea - infectious etiology, diagnosis, and intervention. J Vet Sci. 2014;15(1):1–17. doi: 10.4142/jvs.2014.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franck SM, Bosworth BT, Moon HW. Multiplex PCR for enterotoxigenic, attaching and effacing, and Shiga toxin-producing Escherichia coli strains from calves. J Clin Mic. 1998;36(6):1795–1797. doi: 10.1128/jcm.36.6.1795-1797.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster D, Smith GW. Pathophysiology of diarrhea in calves. Vet Clin North Am Food Anim Pract. 2009;25(1):13–36. doi: 10.1016/j.cvfa.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagy B, Fekete , PZ Enterotoxigenic Escherichia coli in veterinary medicine. Int J Med Microbiol. 2005;295(6-7):443–454. doi: 10.1016/j.ijmm.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Shams Z, Tahamtan Y, Pourbakhsh A, et al. Detection of enterotoxigenic K99 (F5) and F41 from fecal sample of calves by molecular and serological methods. Comp Clin Path. 2012;21(4):475–478. doi: 10.1007/s00580-010-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Younis EE, Ahmed AM, El-Khodery SA, et al. Molecular screening and risk factors of enterotoxigenic Escherichia coli and Salmonella spp in diarrheic neonatal calves in Egypt. Res Vet Sci. 2009;87(3):373–379. doi: 10.1016/j.rvsc.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy B, Fekete , PZ Enterotoxigenic Escherichia coli (ETEC) in farm animals. Vet Res. 1999;30(2-3):259–284. [PubMed] [Google Scholar]
- 10.Gyles CL, Fairbrother JM. Escherichia coli. In: Gyles CL, Prescott JF, Songer JG, et al., editors. Pathogenesis of bacterial infections in animals. Ames, USA: Blackwell ; 2004. pp. 193–223. [Google Scholar]
- 11.Blanco J, Blanco M, Garabal JL, et al. Enterotoxins, colonization factors and serotypes of enterotoxigenic Escherichia coli humans and animals. Microbiologia. 1991;7(2):57–73. [PubMed] [Google Scholar]
- 12.Güler L, Gündüz K, Ok Ü. Virulence factors and antimicrobial susceptibility of Escherichia coli isolated from calves in Turkey. Zoonoses Public Health. 2008;55(5):249–257. doi: 10.1111/j.1863-2378.2008.01121.x. [DOI] [PubMed] [Google Scholar]
- 13.Harris JA, Roy K, Woo-Rasberry V, et al. Directed evaluation of enterotoxigenic Escherichia coli auto-transporter proteins as putative vaccine candidates. PLoS Negl Trop Dis. 2011;5(12):e1428. doi: 10.1371/journal.pntd.0001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo Q, Qadri F, Kansal R, et al. Conservation and immunogenicity of novel antigens in diverse isolates of enterotoxigenic Escherichia coli. PLoS Negl Trop Dis. 2015;9(1):e0003446. doi: 10.1371/journal.pntd.0003446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Bloois E, Winter RT, Kolmar H, et al. Decorating microbes: Surface display of proteins on Escherichia coli. Trends Biotechnol. 2011;29(2):79–86. doi: 10.1016/j.tibtech.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Leo JC, Grin I, Linke D. Type V secretion: Mechanism(s) of autotransport through the bacterial outer membrane. Philos Trans R Soc B Biol Sci. 2012;367(1592):1088–1101. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramesh B, Sendra VG, Cirino PC, et al. Single-cell characterization of autotransporter-mediated Escherichia coli surface display of disulfide bond-containing proteins. J Bio Chem. 2012;287(46):38580–38589. doi: 10.1074/jbc.M112.388199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Restieri C, Garriss G, Locas MC, et al. Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Appl Environ Microbiol. 2007;73(5):1553–1562. doi: 10.1128/AEM.01542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roche AJ, McFadden JP, Owen P. Antigen 43, the major phase-variable protein of the Escherichia coli outer membrane, can exist as a family of proteins encoded by multiple alleles. Microbiology. 2001;147(Pt 1):161–169. doi: 10.1099/00221287-147-1-161. [DOI] [PubMed] [Google Scholar]
- 20.Sahl JW, Steinsland H, Redman JC, et al. A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect Immun. 2011;79(2):950–960. doi: 10.1128/IAI.00932-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caffrey P, Owen P. Purification and N-terminal sequence of the alpha subunit of antigen 43, a unique protein complex associated with the outer membrane of Escherichia coli. J Bacteriol. 1989;171(7):3634–3640. doi: 10.1128/jb.171.7.3634-3640.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heras B, Totsika M, Peters KM, et al. The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping. Proc Nat Acad Sci USA. 2014;111(1):457–462. doi: 10.1073/pnas.1311592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallecha A, Oreh H, Marjan W, et al. Control of gene expression at a bacterial leader RNA, the agn43 gene encoding outer membrane protein Ag43 of Escherichia coli. J Bacteriol. 2014;196(15):2728–2735. doi: 10.1128/JB.01680-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray C. Salmonella and Escherichia coli from veterinary and human sources in Australia during 1985 and 1986. Aust Vet J. 1987;64(8):256–257. doi: 10.1111/j.1751-0813.1987.tb09698.x. [DOI] [PubMed] [Google Scholar]
- 25.Ojeniyi B, Ahrens P, Meyling A. Detection of fimbrial and toxin genes in Escherichia coli and their prevalence in piglets with diarrhea The application of colony hybridization assay, polymerase chain reaction and phenotypic assays. Zentralbl Veterinarmed B. 1994;41(1):49–59. doi: 10.1111/j.1439-0450.1994.tb00205.x. [DOI] [PubMed] [Google Scholar]
- 26.Wray C, McLaren I, Carroll P. Escherichia coli isolated from farm animals in England and Wales between 1986 and 1991. Vet Rec. 1993;133(18):439–442. doi: 10.1136/vr.133.18.439. [DOI] [PubMed] [Google Scholar]
- 27.Mainil J. Escherichia coli virulence factors. Vet Immunol Immunopathol. 2013;152(1):2–12. doi: 10.1016/j.vetimm.2012.09.032. [DOI] [PubMed] [Google Scholar]
- 28.Picco NY, Alustiza FE, Bellingeri RV, et al. Molecular screening of pathogenic Escherichiacoli strains isolated from dairy neonatal calves in Cordoba province, Argentina. Rev Argent Microbiol. 2015;47(2):95–102. doi: 10.1016/j.ram.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pourtaghi H, Dahpahlavan V, Momtaz H. Virulence genes in Escherichia coli isolated from calves with diarrhea in Iran. Comp Clin Patho. 2013;(22):513–515. [Google Scholar]
- 30.Acha S, Kühn I, Jonsson P, et al. Studies on calf diarrhea in Mozambique: Prevalence of bacterial pathogens. Acta Vet Scand. 2004;45(1-2):27–36. doi: 10.1186/1751-0147-45-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherwood D, Snodgrass D, Lawson G. Prevalence of enterotoxigenic Escherichia coli in calves in Scotland and northern England. Vet Rec. 1983;113(10):208–212. doi: 10.1136/vr.113.10.208. [DOI] [PubMed] [Google Scholar]
- 32.Quinn PJ, Carter ME, Markey B, Carter GR. Clinical veterinary microbiology. Madrid, Spain: Wolfe Publishing. 1994:209–236. [Google Scholar]
- 33.Shahrani M, Dehkordi FS, Momtaz H. Characterization of Escherichia coli virulence genes, pathotypes and antibiotic resistance properties in diarrheic calves in Iran. Biol Res. 2014;47(1) doi: 10.1186/0717-6287-47-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzipori S. The relative importance of enteric pathogens affecting neonates of domestic animals. Adv Vet Sci Comp Med. 1985;29:103–206. [PubMed] [Google Scholar]
- 35.Thorns C, Sojka M, Roeder P. Detection of fimbrial adhesins of ETEC using monoclonal antibody-based latex reagents. Vet Rec. 1989;125(4):91–92. doi: 10.1136/vr.125.4.91. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Zhao M, Ruesch L, et al. Prevalence of virulence genes in Escherichia coli strains recently isolated from young pigs with diarrhea in the US. Vet Microbiol. 2007;123(1-3):145–152. doi: 10.1016/j.vetmic.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 37.Kauffmann F. The serology of the coli group. J Immunol. 1947;57(1):71–100. [PubMed] [Google Scholar]
- 38.Dezfulian H, Batisson I, Fairbrother JM, et al. Presence and characterization of extraintestinal pathogenic Escherichia coli virulence genes in F165-positive E coli strains isolated from diseased calves and pigs. J Clin Microbiol. 2003;41(4):1375–1385. doi: 10.1128/JCM.41.4.1375-1385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wells TJ, Totsika M, Schembri MA. Autotransporters of Escherichia coli: A sequence-based characterization. Microiology. 2010;156(Pt 8):2459–2469. doi: 10.1099/mic.0.039024-0. [DOI] [PubMed] [Google Scholar]
- 40.Van der Woude MW, Henderson IR. Regulation and function of Ag43 (flu) Annu Rev Microbiol. 2008;(62):153–169. doi: 10.1146/annurev.micro.62.081307.162938. [DOI] [PubMed] [Google Scholar]


