Abstract
Purpose of review
Migraine is a primary headache disorder and one of the most common and disabling neurological diseases worldwide. Genome-wide association studies have identified ≈40 genetic loci associated with migraine. How these and other genetic findings are used to expand our knowledge on the pathophysiological mechanism of common migraine and rare migraine variants will be discussed.
Recent findings
The genetic load, based on common polygenic variation, is higher in familial migraine cases than in nonfamilial cases, and higher for migraine with aura and hemiplegic migraine. Migraine shares common genetic variant risks with depression. Specific clinical features of common migraine seem to be determined by genetic factors. A stronger family history of migraine is associated with lower age-at-onset, higher frequency and number of medication days and the migraine with aura subtype. Mild hemiplegic migraine is likely caused by complex polygenic interaction of multiple gene variants and environmental factors, like in common migraine subtypes. Phenotypical features in hemiplegic migraine patients may guide physicians in providing adequate genetic counseling.
Summary
Integration of genetic, phenotypic and epigenetic data will help to identify the biological mechanisms by which genetic factors contribute to migraine pathogenesis. Recent studies show the impact of genetics on clinical features and comorbidities in migraine and may guide clinicians to an adequate genetic advice for patients.
Keywords: depression, genetics, genome-wide association study, hemiplegic migraine, migraine
INTRODUCTION
Migraine is a disabling episodic brain disorder characterized by headache attacks associated with photophobia and phonophobia and/or nausea or vomiting [1]. In approximately one-third of patients, headaches are preceded by transient focal neurological symptoms, so-called auras. Hemiplegic migraine is a rare subtype of migraine with aura in which the aura phase includes motor weakness [2]. Migraine is rated as the second most disabling disorder [3] and is often associated with other neurological or psychiatric disorders, such as stroke, epilepsy and depressive disorder [4–6]. A study in a genetic isolate suggested that the bidirectional association between depression and migraine, in particular, migraine with aura, can be partly explained by shared genetic factors [7].
Over the past decade, genome-wide association studies (GWAS) have made important contributions to identifying genetic components involved in migraine. These studies provided further evidence that common types of migraine are genetically complex in the sense that multiple genetic variants, with small effect sizes, together with environmental factors confer migraine susceptibility. Epidemiological studies suggested that familial occurrence is most profound with an early disease onset and the aura subtype, indicating a possibly higher genetic susceptibility in migraine with aura [8,9]. Therefore, it seemed paradoxal that for migraine without aura, more robust associations with genetic loci were discovered in the migraine GWAS [10]. It was hypothesized that this suggests that medium/high-risk alleles with rare(r) prevalence, not captured in GWAS, confer susceptibility to migraine with aura instead. This was further supported by the fact that hemiplegic migraine, a rare monogenic syndrome at the end of the spectrum of the aura subtype, is caused by missense mutations in the CACNA1A (FHM1), ATP1A2 (FHM2) and SCN1A (FHM3) genes [1,11]. Recent reports suggested that the PRRT2 gene might be the fourth hemiplegic migraine gene. There is, however, reasonable doubt, already because PRRT2 was typically shown to be associated with paroxysmal kinesigenic dyskinesia, benign familial infantile seizures, or infantile convulsion choreoathetosis syndrome in hundreds of patients [12]. Only in a few typical PRRT2 families, hemiplegic migraine was reported in some mutation carriers, most of which also had the typical phenotype. Vice versa, PRRT2 mutations were found in less than 5% of index cases with hemiplegic migraine who often also diplayed the typical phenotypes [13]. The most logical conclusion is that a PRRT2 mutation is not sufficient to cause hemiplegic migraine. Likely, in those few PRRT2 mutation carriers who displayed hemiplegic migraine, additional gene variants interacting with the PRRT2 mutation may be involved. This would, thus, imply that in part of the hemiplegic migraine families a complex genetic rather than a monogenic mechanism causes the disorder [13].
The present review aims to highlight the most recent advances in migraine genetics, focusing on studies that have used GWAS data to explore pathophysiological mechanisms, and on studies that investigated to what extent (endo)phenotypes, individual features of migraine, familial occurrence of migraine and comorbid neuropsychiatric conditions are determined by genetic susceptibility. Additionally, the clinical spectrum of hemiplegic migraine and chances of finding a pathogenic mutation will be discussed.
Box 1.

no caption available
FROM GENOME-WIDE ASSOCIATION STUDIES TO INCREASING INSIGHT IN MIGRAINE PATHOGENESIS
Several GWAS have been performed in migraine [10,14–18]. In a GWAS several million common DNA variants, so called single-nucleotide polymorphisms (SNPs), that are dispersed over the genome are tested in large cohorts for association with a trait. By identifying different allele frequencies between patients and controls that survive rigorous multiple testing correction genomic regions that associate with a disease are identified. GWAS have been undertaken that focus on migraine without aura [15], migraine with aura [17], clinic-based samples [15,17] and population-based samples [16]. The most recent meta-analysis GWAS included 59 674 cases and 316 078 controls and identified 38 distinct genomic regions associated with migraine (Table 1) [14]. The likely implicated genes related to these SNPs are involved in neuronal, vascular, metalloproteinases, pain and metal-iron-related pathways [10,14,19▪]. This GWAS also employed pathway analysis, which indicated typical enrichment of genes from the vascular pathway, highlighting a possible vascular cause of migraine [14]. However, the analysis was limited as only the genes closest to the respective significant index SNP were considered and the gene nearest to the associated SNP may not always be the causal gene [14]. Hence, further analyses are needed for each SNP to clarify the exact relationship to migraine. A study by Gupta et al. [20] nicely illustrates how integration of genetic, phenotypic and epigenetic data led to the identification of the biologic mechanism. The common, noncoding, GWAS hit implied the PHACTR1 as the locus for no less than five vascular diseases, including migraine. Through genetic fine-mapping and epigenomic analysis, the disease-associated SNP rs9349379 in the third intron of the PHACTR1 gene, was demonstrated to regulate expression of endothelin 1 (EDN1), a gene with a clear vascular function, that is located 600 kb upstream of PHACTR1. The known physiologic effects of EDN1 on the vasculature may explain the pattern of risk for the five associated diseases including migraine. Furthermore, current evidence indicates that endothelin 1 may have a role in migraine attack induction [21].
Table 1.
Loci associated with common migraine and headache and their association with clinic-based samples and migraine with and without aura
| Index SNPa | Chromosome | Gene nearest index SNP | Gene prioritized with DEPICTb | Clinic-based migraine samplec | Migrained | Migraine without aurae | Migraine with auraf | Headacheg |
| Migraine loci found in Gormley et al., 2016. | ||||||||
| rs10218452 | 1 | PRDM16 | PRDM16 | X | X | X | ||
| rs1572668 | 1 | LRRIQ3 | X | |||||
| rs2078371 | 1 | TSPAN2 | NGF | X | X | |||
| rs6693567 | 1 | ADAMTSL4 | ECM1 | X | X | |||
| rs1925950 | 1 | MEF2D | X | X | X | X | ||
| rs138556413 | 2 | CARF | NBEAL1 | X | ||||
| rs10166942 | 2 | TRPM8 | X | X | X | Xh | X | |
| rs6791480 | 3 | TGFBR2 | TGFBR2 | X | X | X | ||
| rs13078967 | 3 | GPR149 | ARHGEF26 | X | X | |||
| rs7684253 | 4 | SPINK2 | REST | X | ||||
| rs9349379 | 6 | PHACTR1 | X | X | X | X | ||
| rs140002913 | 6 | NOTCH4 | X | |||||
| rs10456100 | 6 | KCNK5 | X | |||||
| rs67338227 | 6 | FHL5/UFL1 | X | X | X | Xh | X | |
| rs28455731 | 6 | GJA1 | GJA1 | X | X | |||
| rs1268083 | 6 | HEY2 | HEY2 | X | ||||
| rs186166891 | 7 | SUGCT | X | X | ||||
| rs10155855 | 7 | DOCK4 | X | |||||
| rs6478241 | 9 | ASTN2 | X | X | X | X | ||
| rs2506142 | 10 | NRP1 | NRP1 | X | ||||
| rs10786156 | 10 | PLCE1 | PLCE1 | X | ||||
| rs12260159 | 10 | HPSE2 | HPSE2 | X | ||||
| rs2223089 | 10 | ARMS2 | HTRA1 | X | X | |||
| rs4910165 | 11 | MRVI1 | MRVI1 | X | X | |||
| rs11031122 | 11 | MPPED2 | X | |||||
| rs10895275 | 11 | YAP1 | YAP1 | X | ||||
| rs561561 | 11 | IGSF9B | X | |||||
| rs1024905 | 12 | FGF6 | FGF6 | X | Xi | |||
| rs11172113 | 12 | LRP1 | LRP1 | X | X | X | Xh | X |
| rs11624776 | 14 | ITPK1 | X | |||||
| rs77505915 | 16 | CFDP1 | X | X | ||||
| rs4081947 | 16 | ZCCHC14 | ZCCHC14 | X | ||||
| rs75213074 | 17 | WSCD1 | X | |||||
| rs17857135 | 17 | RNF213 | X | |||||
| rs111404218 | 20 | JAG1 | JAG1 | X | ||||
| rs4814864 | 20 | SLC24A3 | X | |||||
| rs144017103 | 20 | CCM2L | CCM2L | X | ||||
| rs12845494 | X | MED14 | X | |||||
| Additional migraine loci identified in previous GWAS | ||||||||
| rs10915437 | 1 | AJAP1 | - | X | ||||
| rs1835740 | 8 | MTDH | - | X | X | |||
| rs10504861 | 8 | MMP16 | - | X | ||||
| rs11172113 | 12 | TARBP2/NPFF | - | Xh | Xh | |||
| Additional headache loci identified by Meng et al., 2018 | ||||||||
| rs2036465 | 1 | MACF1 | - | X | ||||
| rs7555006 | 1 | CDKN2C | - | X | ||||
| rs3748784 | 1 | PTBP2 | - | X | ||||
| rs2072806 | 6 | BTN2A2 | - | X | ||||
| rs4596713 | 9 | FXN | - | X | ||||
| rs2895526 | 10 | CAMK1D | - | X | ||||
| rs56349329 | 11 | ATG13 | - | X | ||||
| rs6606710 | 12 | MYO1H | - | X | ||||
| rs7300001 | 12 | IFT81 | - | X | ||||
| rs8614 | 17 | NUFIP2 | - | X | ||||
| rs77804065 | 17 | LINC02210-CRHR1-MAPT | - | X | ||||
| rs4941139 | 18 | ZCCHC2 | - | X | ||||
| rs1555132 | 20 | NOL4L | - | X | ||||
GWAS, genome-wide association studies; SNPs, single-nucleotide polymorphisms.
bGene-prioritization function used to identify likely causal gene at each GWAS locus with DEPICT, data-driven expression-prioritized integration for complex traits.
dData from previous GWAS [14].
eData from previous GWAS [15].
fData from previous GWAS [17].
gData from previous GWAS [29].
hIdentified by using a gene-based pleiotropy approach across migraine with and without aura patient groups [25].
iLoci not found in top 28 SNPs, however, SNP present in list of 3343 significant SNPs.
Shared genetic architecture for migraine with and without aura
There has been continuous debate whether migraine with and without aura are distinct disorders or whether these subtypes are genetically related. Previous observations supported that the migraine subtypes are a continuum of severity, with migraine with aura being more severe but not etiologically distinct from migraine without aura [11,22–24]. Clinical and epidemiological studies showed that in many patients, the prevailing form of migraine changes over time [24]. Moreover, the same prophylactic and acute migraine drugs may be effective in both migraine subtypes. Family studies demonstrated that subtypes of migraine co-occur within families [11,24]. Initially, GWAS focused on migraine with aura or without aura and found different genomic loci for both migraine subtypes [15,17]. These studies, however, had relative small sample sizes and only focused on the top SNP hits, so only on SNPs with P values below the genome-wide significance threshold (5 × 10−8). Recently, significant overlap was found in the portion of shared genes associated with migraine with and without aura, which led to the conclusion that genetically the two migraine subtypes are more alike than different [25].
GENETIC LOAD OF MIGRAINE AND ITS ENDOPHENOTYPES
Family and twin studies estimated the heritability for migraine to be ∼42% (95%CI: 36–47%) [26]. This is in line with the observation that migraine aggregates in families. A recent study, using a novel approach, used results of the most recent migraine GWAS [14] to create polygenic risk scores to see if the genetic load of these common variants contribute to the aggregation of migraine in families (1589 Finnish families were included) [27▪]. Indeed, the aggregation of migraine in families can at least partly be explained by the contribution of common polygenic variations. Familial cases carried higher common variant burden compared with nonfamilial cases.
Migraine features determined by genetic predisposition
In this polygenic risk score study, the genetic load was higher in migraine with aura and hemiplegic migraine cases compared with migraine without aura (Fig. 1) [27▪]. This could partially explain the lower prevalence of migraine with aura and hemiplegic migraine, as conceivably a higher genetic burden is required for developing these subtypes. There were no differences found between genetic common variant burden in migraine with aura and hemiplegic migraine (sporadic or familial) [27▪]. This may be because of a lack of power as there were fewer hemiplegic migraine patients included. Another likely explanation is that rare genetic variants (not captured in GWAS), in combination with common variants, contribute to hemiplegic migraine. Importantly, the involvement of common variants in hemiplegic migraine is further indicated by the observation that 41% of the familial cases were in the highest quartile of polygenic risk (Fig. 1) [27▪]. These findings provide compelling evidence that not all hemiplegic migraine cases seem to be caused by single-gene mutations (see also paragraph predictors of finding a pathological mutation in hemiplegic migraine).
FIGURE 1.
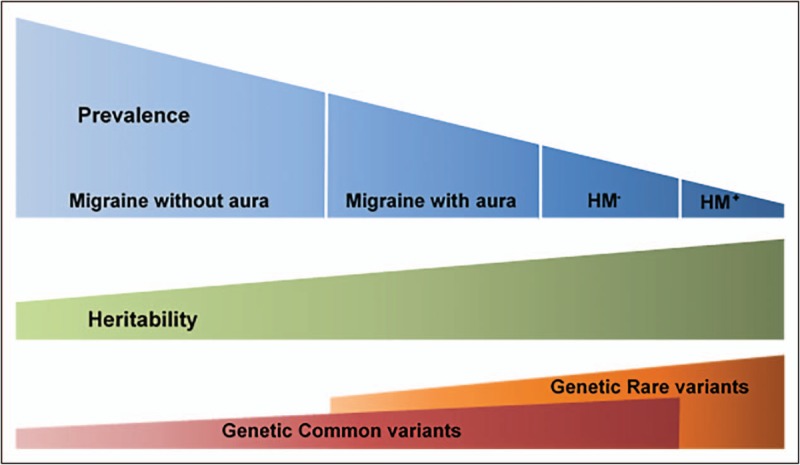
Migraine spectrum. As the prevalence of migraine decreases, common variant load appears to increase. Data from [27▪]. HM− hemiplegic migraine without mutation in one of the hemiplegic genes; HM+, hemiplegic migraine with a mutation in one of the hemiplegic migraine genes.
The Finnish study also showed that higher polygenetic risk scores were associated with stronger family history of migraine, lower age-at-onset and the migraine with aura subtype [27▪]. This is in accordance with a large Dutch clinical study in which a stronger family history of migraine, defined as one or both parents being affected, was associated with lower age-at-onset, higher frequency and higher number of medication days and the migraine with aura subtype [28▪].
Overlap between genetic loci causing migraine and headache
A recent study from UK Biobank that had 74 461 cases and 149 312 controls identified 28 genomic loci associated with a broadly defined ‘headache’ phenotype (Table 1) [29]. Of note, ‘only’ 14 of the found loci were previously identified in a migraine GWAS (Table 1) [14]. There are three possible explanations for this finding. Firstly, these loci are involved in a broader headache pathway, as part of the cases may in fact be patients with tension type headache. Epidemiological studies showed co-occurrence of migraine and tension type headache previously [30]. Secondly, not all migraine patients in the migraine GWAS may be true migraine patients and may have been misdiagnosed patients with another headache phenotype. Given the robustness of the associated SNPs in the migraine GWAS and the fact that six of the loci identified in the headache GWAS were also identified in clinical-based migraine GWAS, this scenario seems unlikely [10,14]. Thirdly, part of the headache cases suffered from migraine.
GENETIC RELATION BETWEEN MIGRAINE AND OTHER BRAIN DISORDERS
Substantial epidemiological comorbidity is found between neuropsychiatric disorders, including migraine [31,32▪]. To quantify the degree of genetic overlap between different brain disorders, the Brainstorm Consortium was formed. This consortium used GWAS summary statistics of a staggering 265 218 cases and 784 643 controls to assess the genetic risk factor overlap of 25 common brain disorders [32▪].
Genetic relation between migraine and psychiatric disorders, especially depression
Of note, the Brainstorm Consortium study indicated that migraine was correlated to several psychiatric disorders, including attention deficit hyperactivity disorder and major depressive disorder (Fig. 2) [32▪]. This is in agreement with a recent Australian study specifically searching for a shared genetic background for depression and migraine. In that meta-analysis in the combined 8 045 569 SNPs of the migraine GWAS and the top 10 000 SNPs from a major depressive disorder GWAS, three novel risk loci were identified that were associated with both disorders [33]. Of the putative candidate genes, CDCA2 is especially interesting, as it has been shown to be involved in a pathway linked to both migraine with and without aura [25]. Furthermore, gene-based association analyses revealed significant enrichment of genes associated with both migraine and major depressive disorder. Pathway analyses suggested several important pathways, especially neural-related pathways of signaling and ion channel regulation to be involved in this shared etiology [33]. This study shows that valuable genetic information can be found when considering all SNPs, more than only the significant (top) SNPs.
FIGURE 2.
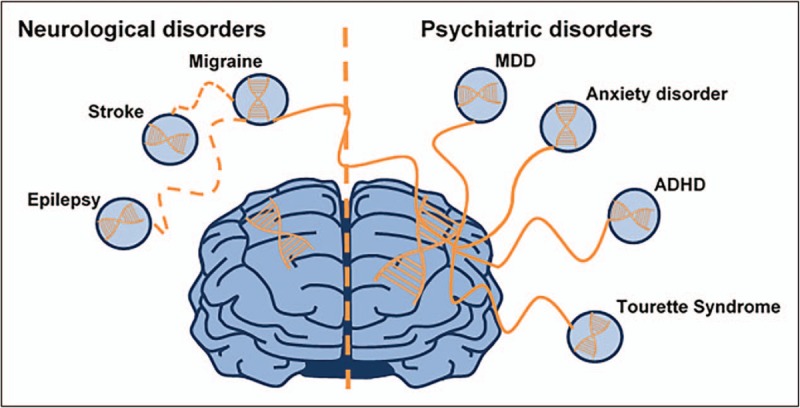
Shared genetic heritability between migraine and common brain disorders. Adapted from Anttila et al.[32▪]. MDD, major depressive disorder.
Genetic correlation between migraine and neurological disorders
The Brainstorm Consortium found no significant genetic risk overlap across neurological disorders. However, their results may indicate a possible genetic sharing between epilepsy and migraine, albeit not significant (Fig. 2) [32▪]. Not reaching significance may be explained by several factors including the relative small epilepsy cohort that was used, the heterogeneity of the epilepsy phenotype, and the fact that only summary statistics were used for the analyses [32▪]. The correlation between migraine and epilepsy is not surprising. For instance, in patients with hemiplegic migraine, there is often co-occurrence of epileptic and (hemiplegic) migraine attacks [34], and comorbidity between common migraine and epilepsy was also seen in epidemiological studies [31]. The Brainstorm Consortium also failed to find a significant genetic risk between ischemic stroke and migraine, despite the fact that an earlier GWAS had revealed substantial genetic overlap between both disorders [35] and epidemiological studies demonstrated increased risk of ischemic stroke in migraine patients [5,36]. Future studies with larger cohorts and more detailed data may be able to show genetic correlation for migraine and comorbid neurological disorders, such as epilepsy and stroke.
PREDICTORS OF FINDING A PATHOLOGICAL MUTATION IN HEMIPLEGIC MIGRAINE
Genetic and neurological counseling of patients with hemiplegic migraine is not only of value for themselves, but also for their offspring. Pelzer and coworkers investigated whether the clinical characteristics of patients with hemiplegic migraine with and without autosomal dominant mutations in CACNA1A, ATP1A2 or SCN1A differ, and whether disease in the latter may be caused by mutations in other genes. The clinical characteristics of 208 patients with familial (n = 199) or sporadic (n = 9) hemiplegic migraine because of a mutation in CACNA1A, ATP1A2, or SCN1A were compared with those of 73 patients with familial (n = 49) or sporadic (n = 24) hemiplegic migraine without a mutation in these genes [37▪]. In addition, 47 patients (familial: n = 33; sporadic: n = 14) without mutations in CACNA1A, ATP1A2, or SCN1A were scanned for mutations in novel genes by using whole exome sequencing (WES). The study showed that the chance of finding a mutation in one of the three FHM genes in a hemiplegic migraine patient is higher if attacks are characterized by more extensive motor weakness, associated with brainstem manifestations, confusion, or brain edema, or are triggered by mild head trauma (Fig. 3). Moreover, only mutation carriers have mental retardation, progressive ataxia or both [37▪]. Additionally, patients with mutations often have a younger age of disease onset and more affected family members [37▪].
FIGURE 3.
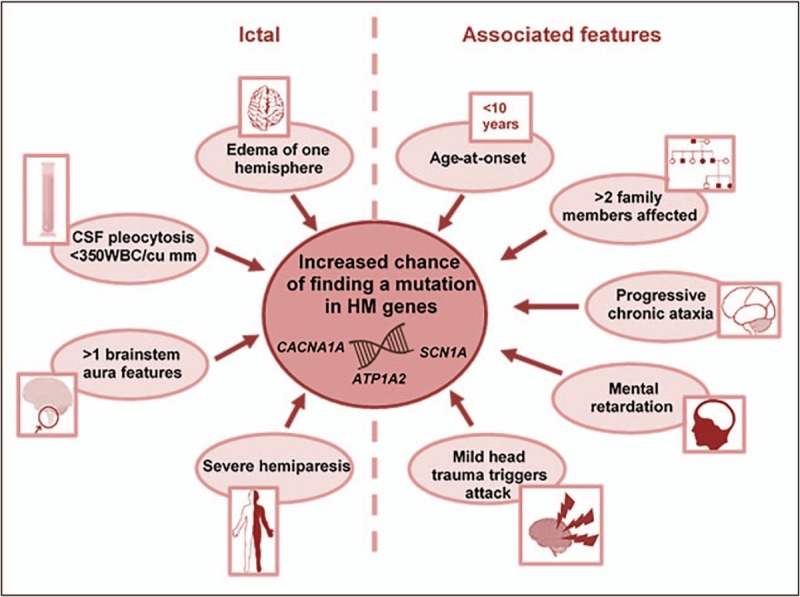
Predictors of finding a mutation in hemiplegic migraine. HM, hemiplegic migraine; WBC, white blood cell count. Adapted from Pelzer et al.[37▪].
Most patients with hemiplegic migraine without a mutation in CACNA1A, ATP1A2 or SCN1A display a mild phenotype that is more akin to that of common (nonhemiplegic) migraine. This seems reflected by the fact that WES failed to identify pathogenic mutations in new genes that fit a Mendelian inheritance. A major fourth autosomal dominant gene for hemiplegic migraine may be unlikely, although this cannot be fully excluded. As a consequence, not all offspring of hemiplegic migraine cases have an a priori 50% chance to inherit hemiplegic migraine. Mild hemiplegic migraine is more likely caused by complex polygenic interaction of multiple gene variants with small effect, like in common migraine subtypes [27▪,37▪]. Phenotypical features in hemiplegic migraine patients may guide physicians in selecting patients for mutation screening and in providing adequate genetic counseling [37▪].
CONCLUSION
A new era in the genetics of migraine has begun with the identification of an increasing number of common genetic variants using the GWAS approach. Recent studies showed that the genetic load, based on common polygenic variation, is higher in familial migraine cases than in nonfamilial cases, and higher for migraine with aura and hemiplegic migraine. Mild hemiplegic migraine is likely caused by complex polygenic interaction of multiple gene variants, like in common migraine subtypes. Phenotypical features in hemiplegic migraine patients may guide physicians in providing adequate genetic counseling. Specific clinical features of common migraine seem to be determined by genetic factors. A stronger family history of migraine is associated with lower age-at-onset, higher frequency and number of medication days and the migraine with aura subtype. Migraine shares common genetic variant risks with depression.
Integration of genetic, phenotypic and epigenetic data will help to identify the biological mechanisms by which genetic factors contribute to migraine pathogenesis. It also shows the impact of genetics on clinical features and comorbidities in migraine and may guide clinicians to an adequate genetic advice for patients.
Acknowledgements
None.
Financial support and sponsorship
None.
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
Footnotes
Arn M.J.M. van den Maagdenberg and Gisela M. Terwindt contributed equally to this manuscript.
REFERENCES
- 1.Headache Classification Committee of the International Headache Society (IHS). The International Classification of Headache Disorders, 3rd edition. Cephalalgia 2018; 38:1–211. [DOI] [PubMed] [Google Scholar]
- 2.Pelzer N, Stam AH, Haan J, et al. Familial and sporadic hemiplegic migraine: diagnosis and treatment. Curr Treat Options Neurol 2013; 15:13–27. [DOI] [PubMed] [Google Scholar]
- 3.Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017; 390:1211–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louter MA, Wardenaar KJ, Veen G, et al. Allodynia is associated with a higher prevalence of depression in migraine patients. Cephalalgia 2014; 34:1187–1192. [DOI] [PubMed] [Google Scholar]
- 5.Schurks M, Rist PM, Bigal ME, et al. Migraine and cardiovascular disease: systematic review and meta-analysis. BMJ 2009; 339:b3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ottman R, Lipton RB. Comorbidity of migraine and epilepsy. Neurology 1994; 44:2105–2110. [DOI] [PubMed] [Google Scholar]
- 7.Stam AH, de Vries B, Janssens AC, et al. Shared genetic factors in migraine and depression: evidence from a genetic isolate. Neurology 2010; 74:288–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart WF, Bigal ME, Kolodner K, et al. Familial risk of migraine: variation by proband age at onset and headache severity. Neurology 2006; 66:344–348. [DOI] [PubMed] [Google Scholar]
- 9.Russell MB, Iselius L, Olesen J. Migraine without aura and migraine with aura are inherited disorders. Cephalalgia 1996; 16:305–309. [DOI] [PubMed] [Google Scholar]
- 10.Anttila V, Winsvold BS, Gormley P, et al. Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat Genet 2013; 45:912–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrari MD, Klever RR, Terwindt GM, et al. Migraine pathophysiology: lessons from mouse models and human genetics. Lancet Neurol 2015; 14:65–80. [DOI] [PubMed] [Google Scholar]
- 12.Ebrahimi-Fakhari D, Saffari A, Westenberger A, Klein C. The evolving spectrum of PRRT2-associated paroxysmal diseases. Brain 2015; 138:3476–3495. [DOI] [PubMed] [Google Scholar]
- 13.Pelzer N, de Vries B, Kamphorst JT, et al. PRRT2 and hemiplegic migraine: a complex association. Neurology 2014; 83:288–290. [DOI] [PubMed] [Google Scholar]
- 14.Gormley P, Anttila V, Winsvold BS, et al. Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. Nat Genet 2016; 48:856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freilinger T, Anttila V, de Vries B, et al. Genome-wide association analysis identifies susceptibility loci for migraine without aura. Nat Genet 2012; 44:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chasman DI, Schurks M, Anttila V, et al. Genome-wide association study reveals three susceptibility loci for common migraine in the general population. Nat Genet 2011; 43:695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anttila V, Stefansson H, Kallela M, et al. International Headache Genetics Consortium. Genome-wide association study of migraine implicates a common susceptibility variant on 8q22.1. Nat Genet 2010; 42:869–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ligthart L, de Vries B, Smith AV, et al. Meta-analysis of genome-wide association for migraine in six population-based European cohorts. Eur J Hum Genet 2011; 19:901–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19▪.Nyholt DR, Borsook D, Griffiths LR. Migrainomics - identifying brain and genetic markers of migraine. Nat Rev Neurol 2017; 13:725–741. [DOI] [PubMed] [Google Scholar]; Comprehensive review on results regarding current and potential neuroimaging and genetic markers of migraine.
- 20.Gupta RM, Hadaya J, Trehan A, et al. A genetic variant associated with five vascular diseases is a distal regulator of endothelin-1 gene expression. Cell 2017; 170:522.e515–533.e515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iljazi A, Ayata C, Ashina M, Hougaard A. The role of endothelin in the pathophysiology of migraine—a systematic review. Curr Pain Headache Rep 2018; 22:27. [DOI] [PubMed] [Google Scholar]
- 22.Nyholt DR, Gillespie NG, Heath AC, et al. Latent class and genetic analysis does not support migraine with aura and migraine without aura as separate entities. Genet Epidemiol 2004; 26:231–244. [DOI] [PubMed] [Google Scholar]
- 23.Ligthart L, Boomsma DI, Martin NG, et al. Migraine with aura and migraine without aura are not distinct entities: further evidence from a large Dutch population study. Twin Res Hum Genet 2006; 9:54–63. [DOI] [PubMed] [Google Scholar]
- 24.Launer LJ, Terwindt GM, Ferrari MD. The prevalence and characteristics of migraine in a population-based cohort: the GEM study. Neurology 1999; 53:537–542. [DOI] [PubMed] [Google Scholar]
- 25.Zhao H, Eising E, de Vries B, et al. Gene-based pleiotropy across migraine with aura and migraine without aura patient groups. Cephalalgia 2016; 36:648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polderman TJ, Benyamin B, de Leeuw CA, et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 2015; 47:702–709. [DOI] [PubMed] [Google Scholar]
- 27▪.Gormley P, Kurki MI, Hiekkala ME, et al. Common variant burden contributes to the familial aggregation of migraine in 1,589 families. Neuron 2018; 98:743.e744–753.e744. [DOI] [PMC free article] [PubMed] [Google Scholar]; With the use of a polygenetic risk score, the authors demonstrated that genetic load is higher in familial migraine cases than in nonfamilial cases, and higher for migraine with aura and hemiplegic migraine.
- 28▪.Pelzer N, Louter MA, van Zwet EW, et al. Linking migraine frequency with family history of migraine. Cephalalgia 2018; 39:229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]; A stronger family history of migraine is associated with lower age-at-onset, higher frequency and number of medication days, and the migraine with aura subtype.
- 29.Meng W, Adams MJ, Hebert HL, et al. A genome-wide association study finds genetic associations with broadly-defined headache in UK biobank (N = 223,773). EBioMedicine 2018; 28:180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagen K, Asberg AN, Uhlig BL, et al. The epidemiology of headache disorders: a face-to-face interview of participants in HUNT4. J Headache Pain 2018; 19:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipton RB, Fanning KM, Buse DC, et al. Identifying natural subgroups of migraine based on comorbidity and concomitant condition profiles: results of the Chronic Migraine Epidemiology and Outcomes (CaMEO) Study. Headache 2018; 58:933–947. [DOI] [PubMed] [Google Scholar]
- 32▪.Anttila V, Bulik-Sullivan B, Finucane HK, et al. Analysis of shared heritability in common disorders of the brain. Science 2018; 360. [DOI] [PMC free article] [PubMed] [Google Scholar]; Migraine shares common genetic variant risks with psychiatric diseases among which depression. These results highlight the importance of common genetic variation as a risk factor for brain disorders and the value of heritability based methods in understanding their cause.
- 33.Yang Y, Zhao H, Boomsma DI, et al. Molecular genetic overlap between migraine and major depressive disorder. Eur J Hum Genet 2018; 26:1202–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prontera P, Sarchielli P, Caproni S, et al. Epilepsy in hemiplegic migraine: genetic mutations and clinical implications. Cephalalgia 2018; 38:361–373. [DOI] [PubMed] [Google Scholar]
- 35.Malik R, Freilinger T, Winsvold BS, et al. Shared genetic basis for migraine and ischemic stroke: a genome-wide analysis of common variants. Neurology 2015; 84:2132–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurth T, Chabriat H, Bousser MG. Migraine and stroke: a complex association with clinical implications. Lancet Neurol 2012; 11:92–100. [DOI] [PubMed] [Google Scholar]
- 37▪.Pelzer N, Haan J, Stam AH, et al. Clinical spectrum of hemiplegic migraine and chances of finding a pathogenic mutation. Neurology 2018; 90:e575–e582. [DOI] [PubMed] [Google Scholar]; This article will help guide physicians in selecting hemiplegic migraine patients for mutation screening and will also help with providing adequate genetic counseling.


