Abstract
Poly (ADP-ribose) polymerases (PARP), particularly PARP1, play an essential role in the detection and repair of DNA single strand breaks (SSB) and double strand breaks (DSB). PARP1 accumulates at DNA damage sites within seconds after DNA damage to catalyze the massive induction of substrate protein poly ADP-ribosylation (PARylation). However, the molecular mechanisms underlying the recruitment and activation of PARP1 in DNA repair are not fully understood. Here we show that PNUTS is a robust binding partner of PARP1. Inhibition of PNUTS led to strong accumulation of endogenous DNA damage and sensitized the cellular response to a wide range of DNA-damaging agents, implicating PNUTS as an essential and multifaceted regulator of DNA repair. Recruitment of PNUTS to laser-induced DNA damage was similar to that of PARP1, and depletion or inhibition of PARP1 abrogated recruitment of PNUTS to sites of DNA damage. Conversely, PNUTS was required for efficient induction of substrate PARylation after DNA damage. PNUTS bound the BRCA1 C-terminal (BRCT) domain of PARP1 and was required for the recruitment of PARP1 to sites of DNA damage. Finally, depletion of PNUTS rendered cancer cells hypersensitive to PARP inhibition. Taken together, our study characterizes PNUTS as an essential partner of PARP1 in DNA repair and a potential drug target in cancer therapy.
Keywords: PNUTS, PARP1, DNA damage, DNA repair, PARP inhibition
INTRODUCTION
DNA damage is frequently induced by various exogenous and endogenous DNA damaging agents. It was estimated that over 50,000 SSBs and 10 DSBs occur in a single cell of our body every day (1,2). These DNA lesions, if not dealt with properly, can lead to genomic instability and the progression of cancer, aging, neurodegeneration, and other diseases (3-5). It is therefore crucial for cells to promptly sense the occurrence of DNA damage and mount a sophisticated DNA damage response (DDR) system to repair DNA damage, halt cell proliferation, and potentially trigger cell death (3,6,7).
PARPs catalyze the attachment of poly (ADP-ribose) chains to substrate proteins, a process termed PARylation (8-11). In particular, PARP1 plays an important role in the cellular response to DNA damage by acting as an early and upstream sensor for a variety of DNA damage. As perhaps the first wave of the DDR, PARP1 is recruited to DNA damage sites within seconds to catalyze PARylation at or near DNA damage sites (12). The rapid and massive induction of PAR chains then mediates the subsequent recruitment of various DNA repair factors, many of which exhibit PAR-binding activities. In turn, PARP1 promotes the repair of various types of DNA damage, controls chromatin dynamics, regulates gene transcription, and influences cell fate determination (8-12).
Consistent with the important function of PARP1 in DNA repair, its inhibition has been considered as a promising approach to enhance the cytotoxic effect of radiation and chemotherapeutics, as well as to exploit synthetic lethality in tumors with defective homologous recombination (HR) (10,13). Olaparib, a PARP inhibitor (PARPi), was approved by the Food and Drug Administration (FDA) and the European Medicines Agency (EMA) in 2014 for the treatment of ovarian cancer with BRCA1 and BRCA2 mutations. Following the success of olaparib, additional PARPi gained approval for BRCA-deficient ovarian cancer treatment. In Jan 2018, FDA expanded the approved use of olaparib to include the treatment of breast cancer patients with BRCA gene mutations. Furthermore, a large number of ongoing clinical trials are further evaluating the clinical potential of PARPi in ovarian, breast, pancreatic, prostate, head and neck, and other types of cancer, either as stand-alone regimens or in combination with radiation, cytotoxic drugs, cell cycle checkpoint inhibitors, anti-angiogenesis, and immunotherapy. Given the enormous interests of PARPi in cancer treatment, it is important and urgent to better understand the mechanisms underlying the function and regulation of PARP1 in DNA repair.
Emerging biochemical and structural studies revealed significant insights into the molecular mechanism of PARP1 activation and function. It has been shown that the initial recruitment of PARP1 to DNA damage occurs via direct DNA-binding mediated by the zinc-finger domains at the N-terminus of PARP1. In addition, a BRCT domain localized in the central region of PARP1 is also required for the DNA damage recruitment of PARP1 (14). Upon binding to damaged DNA, PARP1 becomes enzymatically active via conformational reorganization, and subsequently, mediates the PARylation of various proteins, including itself (autoPARylation) at the DNA damage sites (11,12,14). Building upon these recent advances, further uncovering the detailed mechanism of PARP1 recruitment and activation is likely a key step toward a better understanding of the early and initiating events of the DDR. For example, it is intriguing if additional factors are involved in the recruitment and activation of PARP1.
Phosphatase 1 nuclear targeting subunit 1 (PNUTS), also known as PPP1R10, was described as a nuclear regulator of PP1 (15). PNUTS was implicated in the DNA damage response and maintenance of telomere stability (16-19). More recently, we reported that PNUTS and PP1 mediate NHEJ by fine-tuning the phosphorylation of DNA-dependent protein kinase catalytic subunit (DNA-PKcs) (20). In this study, we identified PARP1 as a robust binding partner of PNUTS. PNUTS is recruited to DNA damage in a PARP1-dependent manner, and is required for the induction of PARylation after DNA damage. Although PNUTS is known to act by modulating PP1, the role of PNUTS in promoting PARP1 function and suppressing endogenous DNA damage is largely independent of its PP1-binding, indicating a new mode of PNUTS function. Further analysis revealed that the PNUTS-PARP1 association is mediated by multiple motifs of PNUTS and the BRCT domain of PARP1, and that PNUTS is required for the recruitment of PARP1 to DNA damage.
MATERIALS AND METHODS
Cell culture and treatment
Human cervix carcinoma (HeLa) and bone osteosarcoma epithelial (U2OS) lines, authenticated by ATCC, were maintained in Dulbecco’s modified Eagle medium (DMEM, Hyclone) with 10% fetal bovine serum (FBS, Hyclone). Human head and neck squamous cell carcinoma UM-SCC-38 cells were authenticated and maintained as in previous studies (21,22). Mycoplasma contamination was examined by DAPI staining/fluorescence microscopy. Cell viability and death assays were performed as in our previous study (23). Briefly, cells were incubated for 1-4 days. The numbers of viable cells were counted using a hemocytometer. To measure cell death, trypan blue staining was performed by mixing 0.4% trypan blue in PBS with cell suspension at a 1:10 ratio. Ionized radiation was performed using an X-ray cabinet (RS-2000 Biological irradiator). Clonogenic assay was performed as described in a previous study (24). Briefly, cells were transfected with or without PNUTS siRNA, and seeded into 6-well plates at a density of 1,000 cells per well. After 24 hours, cells were treated with or without drugs. After incubation for 10 days, cells were then fixed in 1% glutaraldehyde for 30 minutes, stained with 5% crystal violet, and counted for colony numbers. Multiple siRNA targeting PNUTS (target sequence #1 UCUGACAAGUACAACCUU or #2 GGCGGCUACAAACUUCUU), and PARP1 (target sequence UGACUUGGAAGUGAUCGA) were purchased from Integrated DNA Technologies (IDT), and transfected into cells using a protocol recommended by the manufacturer. A non-targeting control, or scramble siRNA was used as a control. Olaparib and bleomycin were purchased from Selleckchem; H2O2 and camptothecin were obtained from Sigma.
Immunoblotting
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting were carried out as previously described (25), using the following antibodies:, γH2AX, PNUTS, PARP1, H2B, BRCA1 phospho-S1524, CHK1 phospho-S317 antibodies from Bethyl Laboratories; β-actin, GFP, Poly (ADP-Ribose) Polymer antibodies from Abcam; α-tubulin, BRCA1 phospho-S1524, active caspase-3 antibodies from Cell Signaling Technology.
Immunofluorescence and imaging
Immunofluorescence was performed as described in a previous study (26). Briefly, cells were fixed in 3% formaldehyde with 0.1% Triton X-100, washed, and then blocked in 10% goat serum in PBS. Primary antibodies were diluted in blocking buffer, and incubated with the cells for 2 h. The cells were then washed and incubated with the Alexa Fluor 488/555 conjugated secondary antibody (Invitrogen, 1: 2,000) for 1 h. Imaging was performed using a Zeiss Axiovert 200M fluorescence microscope at the UNMC Advanced Microscopy Core Facility. Laser microirradiation was performed using 405nm laser under the Zeiss Axiovert 200M Microscope with Marianas Software (Intelligent Imaging Innovations, Inc. Denver, CO).
Protein expression and pull-down
GFP-PNUTS was constructed by inserting human PNUTS to pEGFP-C1 (Clontech). W401A PNUTS was generated by site-directed mutagenesis. ΔC (aa 1-904) and ΔN (aa 288-940) PNUTS were obtained by PCR amplification. GFP-PARP1 was characterized as in a previous study (27). PARP1-BRCT (aa 345-540) was obtained by PCR amplification and inserted into the pEGFP-C1 vector for expression. Four segments of PARP1 (N: aa 1-372; BRCT: aa 368-524; WGR: aa 525-660; and C: aa 661-1014) were tagged with maltose-binding protein (MBP), expressed in BL21 cells, and purified. Five segments of Xenopus PNUTS (N: aa 1-305; M: aa 301-544; C: 540-819; ΔC: aa 1-544; ΔN: aa 301-819) were tagged with glutathione S-transferases (GST) or MBP, expressed in BL21, and purified. For pull-down assays, these recombinant proteins were incubated in HeLa cell lysates or Xenopus egg extracts, and re-isolated. As described in our previous study (20), PNUTS pull-down was performed in Xenopus egg extracts, and samples were then resolved by SDS-PAGE. The gel sections were dissected and subjected to mass spectrometric analysis (Taplin mass spectrometry facility, Harvard).
Single cell gel electrophoresis (comet assay)
The comet assay was performed as in our previous study (22). Briefly, cells were washed with PBS, trypsinized, washed with PBS again, and plated in 0.65% low melting agarose. After solidification, slides were incubated in lysis solution (1 mol/L NaCl, 3.5 mM N-laurylsarcosine, 50 mM NaOH) for 2 h. Slides were then washed, and incubated in alkaline electrophoresis buffer (50 mM NaOH, 2 mM EDTA) for 30 min. Electrophoresis was carried out for 10 min at 20 V. The slides were washed and stained with propidum iodide (25 μg/mL).
RESULTS
PNUTS depletion leads to accumulation of endogenous DNA damage.
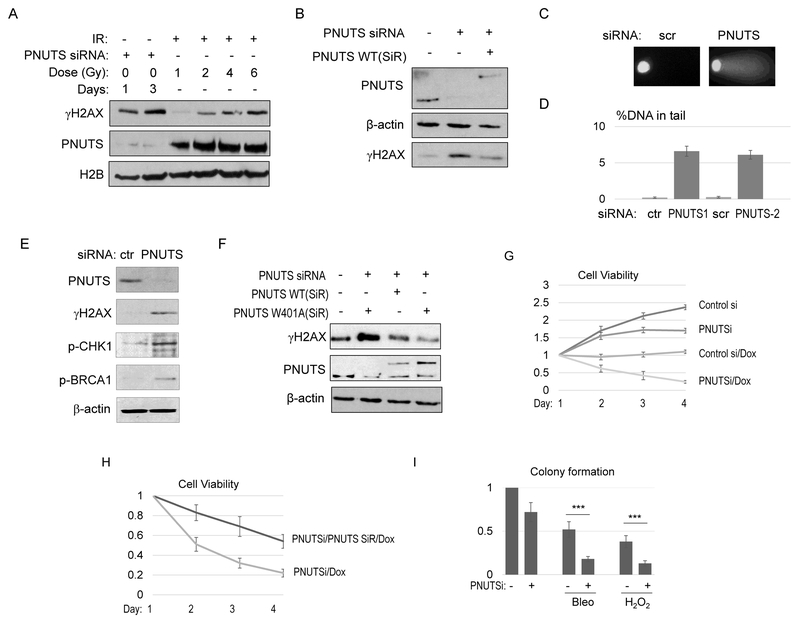
PNUTS has been implicated in the DDR by a number of previous studies (16,17,19,20). In better defining the role of PNUTS in the DDR, we observed that depletion of PNUTS by siRNA induced accumulation of endogenous DNA damage in HeLa cells, as labeled by γH2AX (Fig. 1A, S1A & S1B), a phosphorylated form of histone H2AX that is commonly used as a marker of DNA damage, particularly DSBs (28). The levels of overall γH2AX (Fig. 1A), and γH2AX foci formation (Fig. S1A & S1B) resulted from PNUTS suppression were comparable to that induced by 2-4 Gy ionized radiation (IR). A similar induction of DNA damage was confirmed in SCC38 cells using a different PNUTS-targeting siRNA (Fig. S1C). The re-expression of RNAi-resistant PNUTS suppressed the induction of γH2AX (Fig. 1B), confirming the specific effect of PNUTS knockdown. In addition to the induction of γH2AX, DNA damage caused by PNUTS depletion was detected in the comet assay (Fig. 1C & 1D). Consistently, DNA damage signaling, as evidenced by the phosphorylation of CHK1 and BRCA1, was activated in cells treated with PNUTS siRNA (Fig. 1E).
Figure 1. PNUTS functions as a multifaceted regulator of DNA repair.
(A) HeLa cells were transfected with control, non-targeting, or PNUTS siRNA (#1) for 1 day. The cells were then treated with IR at the indicated doses, incubated for 30 min, harvested and analyzed by immunoblotting for γH2AX, PNUTS, and H2B. (B) HeLa cells were transfected with PNUTS siRNA (#1), and siRNA-resistant (SiR) WT PNUTS, as indicated. The cell lysates were analyzed by immunoblotting for γH2AX, PNUTS, and β-actin. (C, D) HeLa cells were treated with PNUTS siRNA (#1 and #2), as indicated. The comet assay was performed as described in Materials and Methods. Representative images are shown in panel C. The percentage of DNA in the tail section was quantified, the mean values and standard derivations are shown in panel D (N>20). (E) HeLa cells were treated with control or PNUTS siRNA (#1) for 1 day. The cell lysates were analyzed by immunoblotting for γH2AX, phospho-CHK1, phospho-BRCA1, PNUTS, and β-actin. (F) SCC38 cells were transfected with PNUTS siRNA (#1), and siRNA-resistant (SiR) WT or W401A PNUTS, as indicated. The cell lysates were analyzed by immunoblotting for γH2AX, PNUTS, and β-actin. (G) SCC38 cells were treated with control or PNUTS siRNA (#1) at day 0, incubated with doxorubicin (Dox, 2 μg/ml) at day 1, and incubated for 3 days. Cell viability was determined and normalized to that of day 1. The mean value and standard deviation were calculated from 3 independent experiments. (H) SCC38 cells were treated with PNUTS siRNA (#1) and siRNA-resistant (SiR) PNUTS, as in panel G. These cells were then treated with Dox (2 μg/ml), and incubated for 1-4 days. Cell viability was determined and normalized to that of the first day. The mean value and standard deviation were calculated from 3 independent experiments. (I) The colonogenic assay was performed as described in Materials and Methods. The numbers of colonies formed were normalized to the untreated control. The mean value and standard deviation were calculated from 3 independent experiments. Statistical significance was analyzed using an unpaired 2-tailed Student’s t-test. A p-value<0.001 was considered highly significant (***).
As PNUTS has been characterized as a regulator of PP1, we asked if the same function of PNUTS is required to suppress the induction of DNA damage. Surprisingly, expression of W401A PNUTS, a mutant that does not bind or inhibit PP1 (20,25), efficiently rescued the accumulation of DNA damage (Fig. 1F). Thus, our findings suggested a new, and PP1-independent role of PNUTS in DNA repair.
PNUTS suppression rendered cells hypersensitive to a wide range of DNA damaging agents.
Although we showed previously that PNUTS promotes DNA DSB repair via DNA-PKcs-mediated NHEJ (20), the striking level of DNA damage induced by PNUTS suppression may suggest additional roles of PNUTS in multiple DNA repair pathways, given that endogenous DNA DSBs are relatively rare, and the vast majority of endogenous DNA damage are breaks and base damage on one strand. To this end, we discovered that PNUTS knockdown substantially sensitized cells to various DNA damaging drugs, including topoisomerase II inhibitor (doxorubicin, Fig. 1G), ionized radiation (IR, Fig. S2A), DNA alkylating and oxidative drug (bleomycin, Fig. S2B), topoisomerase I inhibitor (camptothecin, Fig. S2C), and oxidative agent (H2O2) (Fig. S2D), as evaluated by cell viability. Confirming the specificity of PNUTS knockdown, an independent PNUTS siRNA also enhanced the cell response to various doses of doxorubicin (Fig. S2E). Moreover, re-expression of siRNA-resistant PNUTS partially rescued the cell hypersensitivity to doxorubicin (Fig. 1H). The colony formation assay was carried out to further assess the impact of PNUTS depletion on the cell response to DNA damaging drugs. Colony formation was significantly reduced with the combined treatment of PNUTS suppression with bleomycin or H2O2 (Fig. 1I). In particular, PNUTS knockdown alone reduced colony formation by approximately 25%, but caused more than 60% decrease in cells that were treated with Bleo or H2O2 (compared to Bleo or H2O2 alone, Fig. 1I).
PARP1 is a major binding partner of PNUTS.
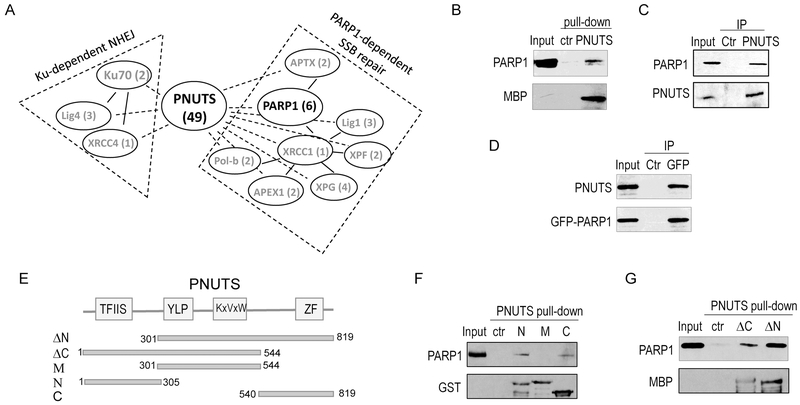
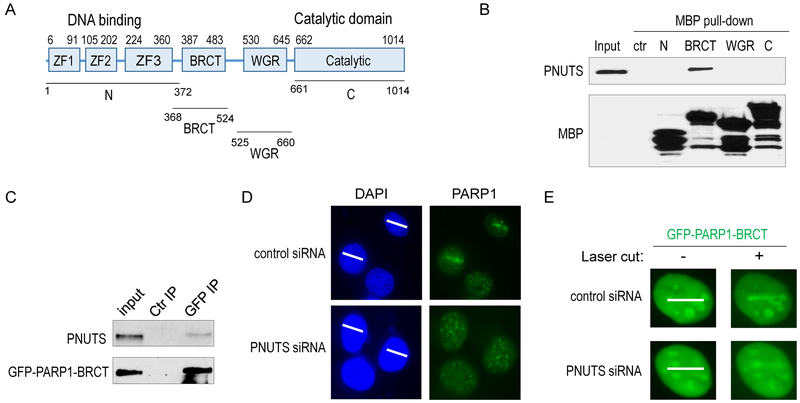
Our findings described above suggested that PNUTS functions as an essential and multifaceted regulator of DNA repair. Notably, our proteomic analysis revealed a number of factors involved in both Ku-dependent NHEJ and PARP1-related SSB repair as associated proteins of PNUTS (Fig. 2A). Among these factors, PARP1 appeared as a major binding partner of PNUTS (Fig. 2A & 2B). We subsequently confirmed the PARP1 and PNUTS association by reciprocal co-immunoprecipitation (Fig. 2C & 2D). Next, we sought to delineate the motif(s) of PNUTS that mediates the PARP1 association (Fig. 2E). We observed that both the N-terminus and C-terminus segments of PNUTS bound PARP1, whereas the middle segment containing PP1 binding and inhibiting elements exhibited no appreciable PARP1 association (Fig. 2F). Truncated forms of PNUTS which lack either N or C segment retained the capability to pull down PARP1 (Fig. 2G). Therefore, PNUTS associates with PARP1 via multiple and distinct binding motifs, potentially reflecting an intimate functional relationship between PARP1 and PNUTS. As a result, our efforts to further identify smaller, and minimal binding cassettes turned out unsuccessful.
Figure 2. PNUTS associates with PARP1.
(A) PARP1 is a major binding partner of PNUTS. MBP-PNUTS pull-down was performed in Xenopus egg extracts and analyzed by mass spectrometry, as described in Materials and Methods. A number of DNA repair proteins were identified as binding patterns of PNUTS, as shown here with the numbers of peptides. (B) MBP-PNUTS pull-down was performed as in panel A. 20% extract input, control beads pull-down and MBP-PNUTS pull-down samples were analyzed by immunoblotting for PARP1 and MBP. (C) PNUTS IP was performed in HeLa cell lysates. The lysate input at 20%, control IP with blank beads, and PNUTS IP products were analyzed by immunoblotting for PARP1 and PNUTS. (D) GFP-PARP1 was expressed in HeLa cells. GFP IP was performed cell lysates. The lysate input at 20%, control IP, and GFP IP products were analyzed by immunoblotting for PARP1 and PNUTS. (E) The schematic diagram of Xenopus PNUTS mutants generated in this study. (F) Three segments of Xenopus PNUTS (N: aa 1-305; M: aa 301-544; C: 540-819) were tagged with GST, expressed, and purified. These recombinant proteins were incubated in Xenopus egg extracts, re-isolated by pull-down. The extract input at 20%, control pull-down, PNUTS N, M, and C pull-down samples were analyzed by immunoblotting for PARP1 and GST. (G) Two segments of Xenopus PNUTS (ΔC: aa 1-544; ΔN: aa 301-819) were tagged with MBP, expressed, and purified. These recombinant proteins were incubated in Xenopus egg extracts, re-isolated by pull-down. The extract input at 20%, control pull-down, PNUTS ΔC and ΔN pull-down samples were analyzed by immunoblotting for PARP1 and MBP.
PNUTS is recruited to laser-induced DNA damage sites in a PARP1-dependent manner.
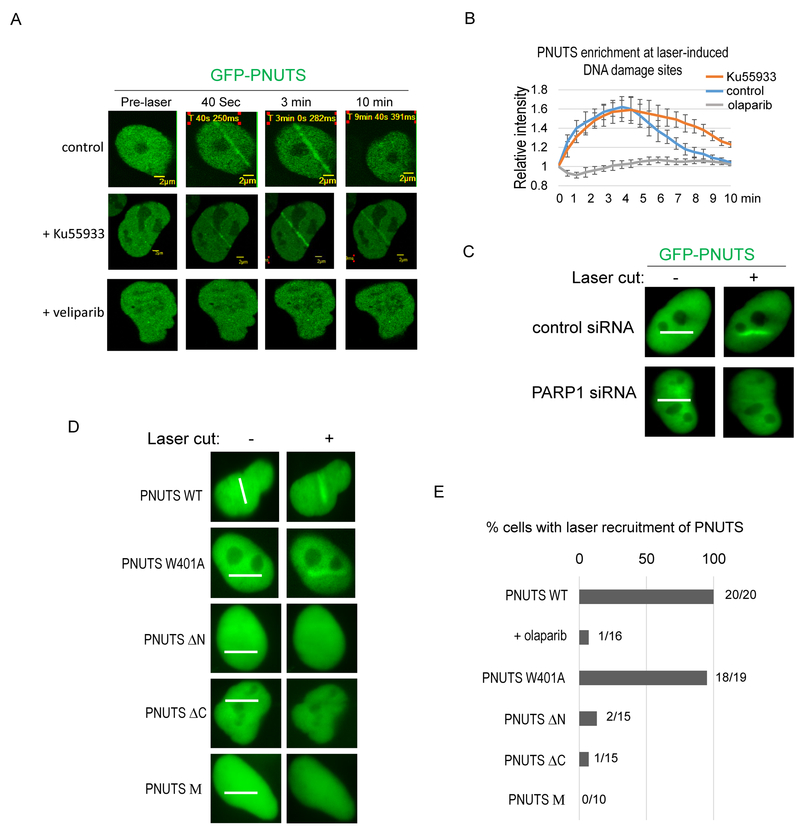
The recruitment of PNUTS to laser-induced DNA damage sites was reported in previous studies (17,20). However, the mechanism responsible for PNUTS recruitment is yet to be uncovered. As shown in Fig. 3A, PNUTS underwent fast and transient recruitment to DNA damage induced by laser microirradiation. The enrichment of PNUTS to DNA damage sites occurred within seconds, and was diminished after approximately 10 min (Fig. 3A, 3B & movie S1). This pattern of PNUTS recruitment is very similar to that of PARP1. Interestingly, the recruitment of PNUTS to DNA damage sites was abolished by PARP1 inhibition, using either veliparib or olaparib (Fig. 3A, 3B, S3, & movie S2). Inhibition of ATM, an upstream sensor kinase of DNA repair and checkpoint signaling (29), did not disrupt PNUTS recruitment (Fig. 3A & 3B). Furthermore, suppression of PARP1 expression using a siRNA impaired the recruitment of PNUTS to laser-induced DNA damage (Fig. 3C). As we discovered that the N and C-termini of PNUTS mediate PARP1 association, we asked if these motifs are required for the DNA damage recruitment of PNUTS. Interestingly, deletion of either the PNUTS N-terminus or the C-terminus Zinc-finger domain substantially attenuated the recruitment of PNUTS (Fig. 3D & 3E). By comparison, the PP1-binding deficient mutant form of PNUTS was efficiently localized to the laser-irradiated sites (Fig. 3D & 3E).
Figure 3. The recruitment of PNUTS to DNA damage sites is dependent on PARP1.
(A) GFP-PNUTS expressing U2OS cells were microirradiated with laser (405 nm). The recruitment of GFP-PNUTS to laser induced DNA damage is shown. PARP inhibition with veliparib (ABT-888,10 μM), but not ATM inhibition with Ku55933 (10 μM), prevented the recruitment of PNUTS. (B) The intensity of the GFP-PNUTS signal at laser-cut sites was normalized to that outside of the laser-cut sites for the relative enrichment of GFP-PNUTS. The mean values and standard deviations are shown (N=5). (C) HeLa cells treated with control or PARP1 siRNA were microirradiated by laser. The recruitment of GFP-PNUTS to laser-induced DNA damage site 3 min after the laser treatment is shown. The path of laser microirradiation is marked with white lines. The recruitment of GFP-PNUTS is largely absent in cells treated with PARP1 siRNA. Consistent findings were observed in more than 20 cells. (D, E) HeLa cells expressing GFP-tagged WT, W401A, ΔN (aa 288-940), ΔC (1-904), and M (320-539) PNUTS were microirradiated by laser. The recruitment of these proteins 3 min after laser treatment is shown in panel D. The path of laser microirradiation is marked with white lines. Quantification of cells with laser recruitment is shown in panel E.
PNUTS is required for the induction of PARylation after DNA damage.
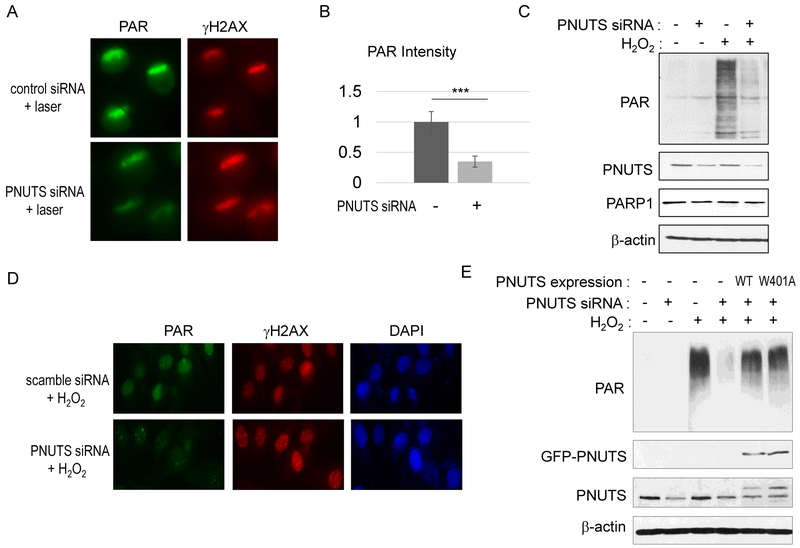
Upon sensing DNA damage, PARP1 mediates the PARylation of substrate proteins at the DNA damage sites to facilitate DNA repair (8,9). The binding of PNUTS to PARP1 suggested a potential role of PNUTS as a new functional partner of PARP1. Strikingly, we observed that PNUTS knockdown reduced the induction of PARylation after DNA damage. Our immunofluorescent analysis showed that PNUTS knockdown reduced the induction of PARylation at sites of laser microirradiation (Fig. 4A & 4B). A similar effect was confirmed using an independent PNUTS siRNA (Fig. S4). Moreover, PARylation upon the treatment of H2O2 was suppressed by PNUTS knockdown, as measured by immunoblotting (Fig. 4C) and immunofluorescence (Fig. 4D). Moreover, the expression of both wild-type (WT) and the PP1 binding-deficient PNUTS restored PARylation, indicating that PNUTS functions in the PARP1 pathway independent of PP1 (Fig. 4E).
Figure 4. PNUTS is required for the induction of poly (ADP-ribosylation) after DNA damage.
(A) HeLa cells with control or PNUTS siRNA (#1) were treated with laser microirradiation. Cells were analyzed 5 min after laser treatment by immunofluorescence for the induction of Poly (ADP-ribosylation) and γH2AX. (B) The intensity of PARylation induction, as in panel A, was quantified, and compared between control and PNUTS siRNA-treated cells. Mean values and standard deviations are shown (N>10). Statistical significance was analyzed using an unpaired 2-tailed Student’s t-test. A p-value<0.001 was considered highly significant (***). (C) HeLa cells were treated with control or PNUTS siRNA (#1, for 1 day) and with H2O2 (for 5 min), as indicated. The induction of PARylation, and the levels of PNUTS, PARP1 and β-actin are shown by immunoblotting. (D) HeLa cells were treated with scramble or PNUTS siRNA (#2, for 1 day) and with H2O2 (for 5 min). Cells were analyzed by immunofluorescence for PAR and γH2AX. (E) HeLa cells were treated with H2O2 and PNUTS siRNA (#1) as indicated. To rescue PNUTS expression, RNAi-resistant WT or W401A PNUTS was expressed. Immunoblotting of PAR, GFP, PNUTS, and β-actin is shown.
PNUTS binds the BRCT domain of PARP1 and mediates the recruitment of PARP1 to laser-induced DNA damage sites.
PARP1 contains multiple functional motifs that have been well characterized in previous studies (10,11). In particular, the N-terminal Zinc finger motifs yield direct binding affinity to DNA SSB and DSB; the BRCT domain is also required for the DNA damage recruitment of PARP1, potentially via PAR-binding and/or auto-modification; the tryptophan-glycine-arginine (WGR) domain partially mediates the DNA binding and allosteric activation of PARP1 (14,30) (Fig. 5A). Interestingly, the BRCT domain, but not the other domains, of PARP1 associated with PNUTS (Fig. 5B). Consistently, immunoprecipitation of the BRCT domain, and the BRCT-containing, middle segment of PARP1 recovered a portion of PNUTS (Fig. 5C & S5A).
Figure 5. PNUTS knockdown impairs the recruitment of PARP1 to laser-induced DNA damage sites.
(A) The schematic diagram of PARP1 motifs and mutants. (B) As described in Materials and Methods, four segments of PARP1 (N: aa 1-372; BRCT: aa 368-524; WGR: aa 525-660; and C: aa 661-1014) were tagged with MBP, expressed in BL21 cells, and purified. The recombinant proteins were incubated in HeLa cell lysates. 20% input, control pull-down, and PARP1 pull-down samples were analyzed by immunoblotting for PNUTS and MBP. (C) As described in Materials and Methods, the BRCT domain of PARP1 was tagged with GFP and expressed in HeLa cells. GFP IP was performed. 20% input, control IP, and GFP IP samples were analyzed by immunoblotting for PNUTS and GFP. (D) HeLa cells with control or PNUTS siRNA (#1) treatment were microirradiated with 405nm laser. The path of laser microirradiation is marked by white lines (panels on the left). The localization of PARP1 3 min after laser treatment is shown by immunofluorescence (panels on the right). More than 10 cells were examined. (E) The BRCT domain of PARP1, tagged with GFP, was expressed in HeLa cells with control or PNUTS knockdown (#1). Cells were microirradiated with 405nm laser, and examined for the localization of GFP 3 min after laser treatment. More than 20 cells were examined.
Given that the BRCT domain is required for the efficient recruitment of PARP1 to DNA damage, we speculated that PNUTS may play a role in the DNA damage recruitment of PARP1. Indeed, immunofluorescent and direct fluorescent analyses revealed that PNUTS knockdown reduced the recruitment of endogenous PARP1 (Fig. 5D), and GFP-PARP1 (Fig. S6), to the site of laser microirradiation. A similar effect was confirmed using a distinct PNUTS siRNA (Fig. S7). Next, as PNUTS binds the BRCT domain of PARP1, we evaluated the impact of PNUTS depletion on the recruitment of BRCT domain. Interestingly, the BRCT domain, or the middle segment of PARP1, which enriched in the area of laser microirradiation in control cells, was not efficiently recruited in cells treated with PNUTS siRNA (Fig. 5E & S5B).
PNUTS targeting enhances the effectiveness of PARP inhibition.
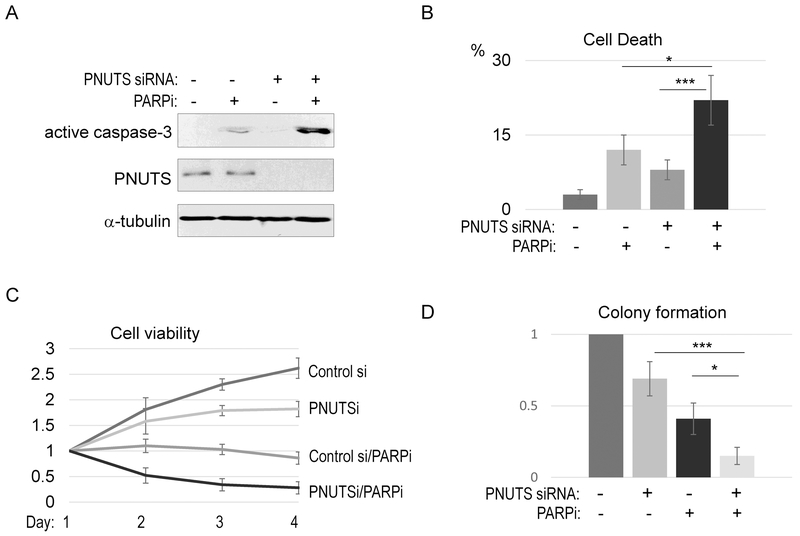
As we characterized PNUTS as a new functional partner of PARP1, we hypothesized that PNUTS is targetable to sensitize the PARPi response in cancer cells. In support of this notion, we showed that PNUTS depletion synergized with PARPi in inducing cell death, as measured by either caspase-3 activation (Fig. 6A) or trypan blue exclusion (Fig. 6B). In turn, PNUTS depletion profoundly sensitized SCC38 cells to olaparib, as judged by cell viability (Fig. 6C) and colony formation (Fig. 6D). Using an independent PNUTS siRNA, we further confirmed that PNUTS depletion enhanced the cell response to various doses of PARPi (Fig. S8). Similar results were observed also in HeLa cells (Fig. S9A & S9B), demonstrating that the functional impact of PNUTS depletion is not cell line specific.
Figure 6. PNUTS targeting sensitizes the tumor cell response to PARP inhibition.
(A) SCC38 cells were treated with PNUTS siRNA (#1) and PARPi (olaparib, 10 μM), as indicated. One day after the treatment, cells were harvested and analyzed by immunoblotting for active caspase-3, PNUTS, and α-tubulin. (B) SCC38 cells were treated with as in panel A, and measured by the trypan blue exclusion assay for cell death. The mean value and standard deviation were calculated from 3 independent experiments. Statistical significance was analyzed using an unpaired 2-tailed Student’s t-test. A p-value<0.05 was considered statistically significant (*), and p<0.001 was highly significant (***). (C) SCC38 cells were treated with PNUTS siRNA and olaparib as in panel A. The relative cell viability is shown. (D) The colonogenic assay was performed as described in Materials and Methods. The numbers of colonies were normalized to the untreated control. The mean value and standard deviation were calculated from 3 independent experiments. Statistical significance was analyzed using an unpaired 2-tailed Student’s t-test.
DISCUSSION
PNUTS is a new functional partner of PARP1.
PARP1 plays an essential role in the detection and repair of various types of DNA damage (8-11). The activation and recruitment of PARP1 occur within seconds after DNA damage, resulting in massive induction of PARylation at the DNA damage sites (8,12). It has been shown the recruitment and activation of PARP1 following DNA damage is achieved in a multi-step and feedback-regulated manner (11,14). The N-terminus of PARP1 possesses multiple zinc finger domains that bind a variety of DNA lesions, including DNA SSBs and DSBs. Moreover, the BRCT domain resided at the C-terminus of the zinc finger domains is also involved in the recruitment of PARP1 to DNA damage sites. Functional investigations indicated that the BRCT mediates the PAR-binding activity of PARP1, and modulates the molecular conformation of PARP1. The initial binding of PARP1 to DNA damage, in turn, leads to PARP1 activation and substrate PARylation at the DNA damage sites, which serves as a platform to recruit various DNA repair proteins, including additional PARP1. This positive feedback enables efficient PARP1 recruitment and PARylation within seconds after DNA damage. Ultimately, extensive auto-PARylation of PARP1 reduces its DNA binding, whereas de-PARylation occurs in a timely manner to allow for the access of downstream repair factors (11,12,14).
The current understanding of PARP1 function, as summarized above, highlights complex and well-coordinated molecular actions of PARP1, and at the same time, leads to an intriguing question about the potential involvement of additional regulatory factors in the process. Although PARP1 is regarded as one of the earliest and most upstream sensors of DNA damage, it is plausible the initial recruitment and activation of PARP1 is achieved in the context of a multi-factor complex. For example, a recent study showed that Src-associated substrate during mitosis of 68 kDa (Sam68) bound the C-terminal catalytic domain of PARP1 to promote its activation after DNA damage (31). Also interact with the catalytic domain of PARP1 and facilitate the function of PARP1 are histone PARylation factor 1 (HPF1) and TIMELESS (32-34). In this project we identified PNUTS as a robust binding partner of PARP1. PNUTS plays an essential role in the induction of PARylation at the laser-induced DNA damage sites or in cells treated with H2O2. Conversely, the recruitment of PNUTS to DNA damage sites is dependent on PARP1, suggesting a mode of reciprocal regulation. The detailed mechanism underlying PARP1-mediated recruitment of PNUTS remains to be revealed in future studies. Potentially, PNUTS may exhibit PAR-binding activity, or is itself a substrate of PARP1. Taken together, our study supports a role of PNUTS as an essential partner of PARP1 in mediating the early DDR.
PNUTS is a multifaceted regulator of DNA repair with PP1-independent functions.
PNUTS was originally described as a nuclear regulator of PP1 that retains a portion of PP1 in the nucleus (15). PNUTS has been implicated in RNA processing (35), transcription (36), DDR and maintenance of telomere stability (16-19), and modulation of retinoblastoma (Rb) and Phosphatase and tensin homolog (Pten) (37-39). The function of PNUTS as an inhibitory regulator of PP1 was best characterized in the case of Rb (37,40). PNUTS-conferred PP1 modulation is mediated by a central domain containing the PP1-binding motif (KSVTW) and several additional motifs that make contact with PP1 (40). Furthermore, PNUTS encompasses a number of additional motifs, including an N-terminal Transcription factor II S-like (TFIIS-Like) domain, and a C-terminal Zinc finger domain. These domains are well conserved evolutionarily, but yet to be functionally defined.
Landsverk et al. showed for the first time that PNUTS is enriched at DNA damage sites, and that PNUTS knockdown led to prolonged DNA damage foci and checkpoint activation (17). A potential role of PNUTS in DDR was supported by another study showing that PNUTS upregulation attenuated DNA damage-induced cell death in cardiomyocytes (19). We recently revealed a detailed mechanism by which PNUTS promotes DNA DSB repair via nonhomologous end joining (NHEJ) (20). We showed that PP1 binds multiple motifs of DNA-PKcs to regulate DNA-PKcs phosphorylation and activation, whereas PNUTS associates with the DNA-PK complex and is required for DNA-PKcs phosphorylation at Ser-2056 and Thr-2609. Thus, PNUTS and PP1 together fine-tune the dynamic phosphorylation of DNA-PKcs after DNA damage to mediate NHEJ (20). In the current study, we showed that PNUTS depletion led to substantial accumulation of endogenous DNA damage, and rendered cell hypersensitive to a wide range of agents that induce DNA SSB, DSB, alkylation, and oxidation. Thus, PNUTS is likely to function beyond DNA-PK-mediated NHEJ. Indeed, in this study we delineated the role of PNUTS in mediating the recruitment and function of PARP1.
Surprisingly, a PP1-binding deficient mutant form of PNUTS efficiently rescued DNA damage-induced PARylation from PNUTS depletion, indicating that the role of PNUTS in the PARP1 pathway is independent of PP1. The PP1-binding activity of PNUTS was also largely dispensable for the suppression of endogenous DNA damage. Thus, our study suggested a new mode of PNUTS function that does not involve its PP1 binding or inhibition. We further reported that both the N-terminal and C-terminal domains of PNUTS bind PARP1, and are required for the recruitment of PNUTS to DNA damage sites. In addition, we mapped PNUTS binding to the BRCT domain of PARP1, a region that was shown to mediate the recruitment of PARP1 to DNA damage sites (14). We subsequently confirmed that PNUTS depletion disrupted the DNA damage recruitment of PARP1. We reason that PNUTS may govern the function of the PARP1 BRCT domain in PAR-binding, auto-modification, and inter-/intra-molecular conformational changes. Future investigations using biochemical and structural techniques are necessary to provide clear insights into this process.
PNUTS is an emerging anti-cancer drug target.
The gene locus of PNUTS, 6p21.3, is frequently amplified in cancer (41-43). In particular, several previous studies discovered PNUTS upregulation in cancer, and implicated PNUTS as a potential oncogene (39,44-47). For example, studies showed PNUTS upregulation in gliomas (44), breast cancer (48), gastric cancer (45), melanomas (46), sarcomas (47), and other cancers. Building on our mechanistic characterization of PNUTS in DNA repair, we asked if the expression level of PNUTS influences the cell sensitivity to DNA damaging drugs and PARPi, and if targeting PNUTS, in conjunction with existing DNA damage drugs and PARPi, leads to enhanced cytotoxic effects. Interestingly, we observed that PNUTS knockdown sensitized cells to a wide range of cytotoxic drugs that are commonly used in cancer treatment. Moreover, PNUTS depletion potentiated cancer cells to PARP inhibition. On one hand, the synergy between PNUTS and PARP1 suppression can be caused by the PARP1-independent function of PNUTS, particularly in DSB repair (20). On the other hand, PNUTS depletion disrupts the function of PARP1, via a mechanism that differs from olaparib or other PARP1 inhibitors, such that the combination treatment can more robustly target PARP to confer higher cytotoxicity. Thus, future studies shall uncover how PNUTS depletion influences the outcome of PARPi. For example, while we showed here that PNUTS facilitates the initial (~5 min) recruitment and function of PARP1 after DNA damage, the effects of PNUTS depletion on the sustained chromatin-binding (trapping) of PARP1 (49,50), and on the cell fate determination after PARPi (13,51), remain to be examined. Taken together, our findings shed new light on the role of PNUTS in PARP1-mediated DNA repair, and suggest PNUTS as a potential drug target for cancer therapy.
Supplementary Material
SIGNIFICANCE.
Findings reveal PNUTS as an essential functional partner of PARP1 in DNA repair and suggest its inhibition as a potential therapeutic strategy in conjunction with DNA-damaging agents or PARP inhibitors.
ACKNOWLEDGEMENT
We thanks Dr. Jay Reddy (University of Nebraska-Lincoln) and Gregory Oakley for technical support and stimulating discussions. The UNMC Advanced Microscopy Core Facility was supported by the Nebraska Research Initiative, the Fred and Pamela Buffett Cancer Center Support Grant (P30CA036727), and an Institutional Development Award (IDeA) from the NIGMS of the NIH (P30GM106397). This work was partially supported by NIH grant CA172574 to A.P.
FINANCIAL SUPPORT: This work was partially supported by NIH grant CA172574 to A.P.
Footnotes
CONFLICT OF INTEREST: The authors declare no conflict of interest.
REFERENCES
- 1.Tubbs A, and Nussenzweig A (2017) Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 168, 644–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vilenchik MM, and Knudson AG (2003) Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci U S A 100, 12871–12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson SP, and Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jalal S, Earley JN, and Turchi JJ (2011) DNA repair: from genome maintenance to biomarker and therapeutic target. Clinic cancer res 17, 6973–6984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huen MS, and Chen J (2010) Assembly of checkpoint and repair machineries at DNA damage sites. Trends biochem sci 35, 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, and Linn S (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annual rev biochem 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 7.Lou Z, and Chen J (2005) Mammalian DNA damage response pathway. Adv exp med biol 570, 425–455 [DOI] [PubMed] [Google Scholar]
- 8.Ray Chaudhuri A, and Nussenzweig A (2017) The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nature reviews. Mol cell biol 18, 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin-Hernandez K, Rodriguez-Vargas JM, Schreiber V, and Dantzer F (2017) Expanding functions of ADP-ribosylation in the maintenance of genome integrity. Semin cell dev biol 63, 92–101 [DOI] [PubMed] [Google Scholar]
- 10.Dulaney C, Marcrom S, Stanley J, and Yang ES (2017) Poly(ADP-ribose) polymerase activity and inhibition in cancer. Semin cell dev biol 63, 144–153 [DOI] [PubMed] [Google Scholar]
- 11.Luo X, and Kraus WL (2012) On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes dev 26, 417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu C, Vyas A, Kassab MA, Singh AK, and Yu X (2017) The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic acids res 45, 8129–8141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lord CJ, and Ashworth A (2017) PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mortusewicz O, Ame JC, Schreiber V, and Leonhardt H (2007) Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic acids res 35, 7665–7675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen PB, Kwon YG, Nairn AC, and Greengard P (1998) Isolation and characterization of PNUTS, a putative protein phosphatase 1 nuclear targeting subunit. J Biol Chem 273, 4089–4095 [DOI] [PubMed] [Google Scholar]
- 16.Bounaix Morand du Puch C, Barbier E, Kraut A, Coute Y, Fuchs J, Buhot A, Livache T, Seve M, Favier A, Douki T, Gasparutto D, Sauvaigo S, and Breton J (2011) TOX4 and its binding partners recognize DNA adducts generated by platinum anticancer drugs. Arch Biochem Biophys 507, 296–303 [DOI] [PubMed] [Google Scholar]
- 17.Landsverk HB, Mora-Bermudez F, Landsverk OJ, Hasvold G, Naderi S, Bakke O, Ellenberg J, Collas P, Syljuasen RG, and Kuntziger T (2010) The protein phosphatase 1 regulator PNUTS is a new component of the DNA damage response. EMBO rep 11, 868–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H, Lee OH, Xin H, Chen LY, Qin J, Chae HK, Lin SY, Safari A, Liu D, and Songyang Z (2009) TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat struct mol biol 16, 372–379 [DOI] [PubMed] [Google Scholar]
- 19.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, Muller OJ, Potente M, Zeiher AM, Hermeking H, and Dimmeler S (2013) MicroRNA-34a regulates cardiac ageing and function. Nature 495, 107–110 [DOI] [PubMed] [Google Scholar]
- 20.Zhu S, Fisher LA, Bessho T, and Peng A (2017) Protein phosphatase 1 and phosphatase 1 nuclear targeting subunit-dependent regulation of DNA-dependent protein kinase and non-homologous end joining. Nucleic acids res 45, 10583–10594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner JC, Graham MP, Kumar B, Saunders LM, Kupfer R, Lyons RH, Bradford CR, and Carey TE (2010) Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head neck 32, 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Mosel AJ, Oakley GG, and Peng A (2012) Deficient DNA damage signaling leads to chemoresistance to Cisplatin in oral cancer. Mol Cancer Ther 11, 2401–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Luong VQ, Giannini PJ, and Peng A (2014) Mastl kinase, a promising therapeutic target, promotes cancer recurrence. Oncotarget 5, 11479–11489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manthey KC, Glanzer JG, Dimitrova DD, and Oakley GG (2010) Hyperphosphorylation of Replication Protein a in Cisplatin-Resistant and -Sensitive Head and Neck Squamous Cell Carcinoma Cell Lines. Head Neck 32, 636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher LA, Wang L, Wu L, and Peng A (2014) Phosphatase 1 nuclear targeting subunit is an essential regulator of M-phase entry, maintenance, and exit. J Biol Chem 289, 23745–23752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng AM, Lewellyn AL, Schiemann WP, and Maller JL (2010) Repo-Man Controls a Protein Phosphatase 1-Dependent Threshold for DNA Damage Checkpoint Activation. Curr Biol 20, 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y, Li C, Wei L, Teng Y, Nakajima S, Chen X, Xu J, Leger B, Ma H, Spagnol ST, Wan Y, Dahl KN, Liu Y, Levine AS, and Lan L (2017) SSRP1 Cooperates with PARP and XRCC1 to Facilitate Single-Strand DNA Break Repair by Chromatin Priming. Cancer Res 77, 2674–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, and Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273, 5858–5868 [DOI] [PubMed] [Google Scholar]
- 29.Shiloh Y (2003) ATM and related protein kinases: Safeguarding genome integrity. Nat Rev Cancer 3, 155–168 [DOI] [PubMed] [Google Scholar]
- 30.Langelier MF, Planck JL, Roy S, and Pascal JM (2012) Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun X, Fu K, Hodgson A, Wier EM, Wen MG, Kamenyeva O, Xia X, Koo LY, and Wan F (2016) Sam68 Is Required for DNA Damage Responses via Regulating Poly(ADP-ribosyl)ation. PLoS biol 14, e1002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibbs-Seymour I, Fontana P, Rack JGM, and Ahel I (2016) HPF1/C4orf27 Is a PARP-1-Interacting Protein that Regulates PARP-1 ADP-Ribosylation Activity. Mol cell 62, 432–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie S, Mortusewicz O, Ma HT, Herr P, Poon RY, Helleday T, and Qian C (2015) Timeless Interacts with PARP-1 to Promote Homologous Recombination Repair. Mol cell 60, 163–176 [DOI] [PubMed] [Google Scholar]
- 34.Young LM, Marzio A, Perez-Duran P, Reid DA, Meredith DN, Roberti D, Star A, Rothenberg E, Ueberheide B, and Pagano M (2015) TIMELESS Forms a Complex with PARP1 Distinct from Its Complex with TIPIN and Plays a Role in the DNA Damage Response. Cell rep 13, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim YM, Watanabe T, Allen PB, Lee SJ, Greengard P, Nairn AC, and Kwon YG (2003) PNUTS, a protein phosphatase 1 (PP1) nuclear targeting subunit. Characterization of its PP1- and RNA-binding domains and regulation by phosphorylation. J Biol Chem 278, 13819–13828 [DOI] [PubMed] [Google Scholar]
- 36.Lee JH, You J, Dobrota E, and Skalnik DG (2010) Identification and characterization of a novel human PP1 phosphatase complex. J Biol Chem 285, 24466–24476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Udho E, Tedesco VC, Zygmunt A, and Krucher NA (2002) PNUTS (phosphatase nuclear targeting subunit) inhibits retinoblastoma-directed PP1 activity. Biochem biophys res commun 297, 463–467 [DOI] [PubMed] [Google Scholar]
- 38.Krucher NA, Rubin E, Tedesco VC, Roberts MH, Sherry TC, and De Leon G (2006) Dephosphorylation of Rb (Thr-821) in response to cell stress. Exp cell res 312, 2757–2763 [DOI] [PubMed] [Google Scholar]
- 39.Kavela S, Shinde SR, Ratheesh R, Viswakalyan K, Bashyam MD, Gowrishankar S, Vamsy M, Pattnaik S, Rao S, Sastry RA, Srinivasulu M, Chen J, and Maddika S (2013) PNUTS functions as a proto-oncogene by sequestering PTEN. Cancer Res 73, 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choy MS, Hieke M, Kumar GS, Lewis GR, Gonzalez-DeWhitt KR, Kessler RP, Stein BJ, Hessenberger M, Nairn AC, Peti W, and Page R (2014) Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code. Proc Natl Acad Sci U S A 111, 4097–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seute A, Sinn HP, Schlenk RF, Emig R, Wallwiener D, Grischke EM, Hohaus S, Dohner H, Haas R, and Bentz M (2001) Clinical relevance of genomic aberrations in homogeneously treated high-risk stage II/III breast cancer patients. Int J Cancer 93, 80–84 [DOI] [PubMed] [Google Scholar]
- 42.Richard F, Pacyna-Gengelbach M, Schluns K, Fleige B, Winzer KJ, Szymas J, Dietel M, Petersen I, and Schwendel A (2000) Patterns of chromosomal imbalances in invasive breast cancer. Int J Cancer 89, 305–310 [PubMed] [Google Scholar]
- 43.Santos GC, Zielenska M, Prasad M, and Squire JA (2007) Chromosome 6p amplification and cancer progression. J clinic path 60, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, Rosenblum M, Mikkelsen T, and Fine HA (2006) Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 9, 287–300 [DOI] [PubMed] [Google Scholar]
- 45.D’Errico M, de Rinaldis E, Blasi MF, Viti V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D, Palombo F, Giuliani A, and Dogliotti E (2009) Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer 45, 461–469 [DOI] [PubMed] [Google Scholar]
- 46.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, and Wang Y (2005) Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clinic cancer res 11, 7234–7242 [DOI] [PubMed] [Google Scholar]
- 47.Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, Shah K, Socci ND, Weir BA, Ho A, Chiang DY, Reva B, Mermel CH, Getz G, Antipin Y, Beroukhim R, Major JE, Hatton C, Nicoletti R, Hanna M, Sharpe T, Fennell TJ, Cibulskis K, Onofrio RC, Saito T, Shukla N, Lau C, Nelander S, Silver SJ, Sougnez C, Viale A, Winckler W, Maki RG, Garraway LA, Lash A, Greulich H, Root DE, Sellers WR, Schwartz GK, Antonescu CR, Lander ES, Varmus HE, Ladanyi M, Sander C, Meyerson M, and Singer S (2010) Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat genet 42, 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, Gelmon K, Chia S, Mar C, Wan A, Laks E, Biele J, Shumansky K, Rosner J, McPherson A, Nielsen C, Roth AJ, Lefebvre C, Bashashati A, de Souza C, Siu C, Aniba R, Brimhall J, Oloumi A, Osako T, Bruna A, Sandoval JL, Algara T, Greenwood W, Leung K, Cheng H, Xue H, Wang Y, Lin D, Mungall AJ, Moore R, Zhao Y, Lorette J, Nguyen L, Huntsman D, Eaves CJ, Hansen C, Marra MA, Caldas C, Shah SP, and Aparicio S (2015) Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518, 422–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, and Pommier Y (2012) Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res 72, 5588–5599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen Y, Aoyagi-Scharber M, and Wang B (2015) Trapping Poly(ADP-Ribose) Polymerase. J Pharm Exp Ther 353, 446–457 [DOI] [PubMed] [Google Scholar]
- 51.Bouchard VJ, Rouleau M, and Poirier GG (2003) PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol 31, 446–454 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.