Abstract
Histone modifications, largely regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), have been recognized as major regulatory mechanisms governing human diseases including cancer. Despite significant effort and recent advances, the mechanism by which the HAT and transcriptional coactivator p300 mediates tumorigenesis remains unclear. Here, we use a genetic and chemical approach to identify the Microphthalmia-associated transcription factor (MITF) as a critical downstream target of p300 driving human melanoma growth. Direct transcriptional control of MITF by p300-dependent histone acetylation within proximal gene regulatory regions was coupled to cellular proliferation, suggesting a significant growth regulatory axis. Further analysis revealed Forkhead Box M1 (FOXM1) as a key effector of the p300-MITF axis driving cell growth that is selectively activated in human melanomas. Targeted chemical inhibition of p300 acetyltransferase activity using a potent and selective catalytic p300/CBP inhibitor demonstrated significant growth inhibitory effects in melanoma cells expressing high levels of MITF. Collectively, these data confirm the critical role of the p300-MITF-FOXM1 axis in melanoma and support p300 as a promising novel epigenetic therapeutic target in human melanoma.
Keywords: Melanoma, MITF, FOXM1, p300, small molecule inhibitor, epigenetic regulation, cell cycle, senescence
Introduction
Dynamic changes in histone acetylation are major regulatory mechanisms governing gene transcription in human diseases including cancers (1,2). Large-scale analyses of chromatin modifications in human cancers have prompted the development of new epigenetic therapies including histone deacetylase (HDAC) and DNA-methytransferase (DNMT) inhibitors. Among newly identified epigenetic targets, histone acetyltransferases (HATs) and their therapeutic efficacies remain unclear. The transcriptional coactivator p300 possesses both lysine acetyltransferase (KAT) enzymatic activity as well as scaffolding abilities, which regulate the transcriptional activity of target genes, and have demonstrated complex roles in determining cell fate in both normal and diseased tissues(3). p300 has notably been found to be amplified in subsets of human melanomas and has been implicated as an oncogene in this and other malignancies (4). Our group previously reported the development of a small-molecule inhibitor of p300/CBP HAT, C646, (5–7) and its potential therapeutic efficacy in cancer, including myeloid leukemia and melanoma (5); however, its modest potency and electrophilic functionality have limited its pharmacological applications, necessitating the search for more potent and specific reagents targeting p300. More recently, investigators have used a virtual ligand screen to identify A-485, a potent, selective, and drug-like catalytic p300/CBP inhibitor that targets lineage-specific tumours including hematologic malignancies and prostate cancers (8).
Previously, p300 was found to serve as a coactivator for the MITF transcription factor (9,10), regulating the expression of a subset of downstream target genes through consensus DNA binding E-box and M-box motifs (11). Additionally, varying levels of MITF expression have been associated with melanoma development and progression, and have been found to contribute to BRAF-inhibitor therapeutic resistance (12–15). Transcriptional regulators such as SOX10, PAX3, CREB, LEF-1, and ATF2 have been shown to control MITF expression, although the precise mechanisms remain to be elucidated (10,16–23). Given the importance of MITF in melanoma biology, and the significance of p300 acetyltransferase activity in regulating melanoma cell growth, we sought to determine the role of p300 in melanoma development and progression and its potential relevance to the master melanocyte differentiation gene, MITF. Here we explore the functional role of p300 in human melanoma using both a genetic and chemical approach using the potent and specific inhibitor of p300/CBP HAT, A-485*, to further dissect the specific functional contributions of p300 HAT activity to melanoma development and progression. These studies have allowed us to identify MITF as a critical downstream effector of p300 HAT activity and important stimulus for melanoma growth. Additionally, bioinformatic analysis of MITF target genes allowed us to identify Forkhead Box M1 (FOXM1) as a specific target of the p300-MITF signaling axis. Analysis of primary human melanoma genetic data from the TCGA database identified specific and exclusive alterations in this signaling axis in primary human melanomas, suggesting a critical growth regulatory pathway. Moreover, chemical inhibition of p300/CBP histone acetyltransferase (HAT) activity by A-485* was found to significantly inhibit proliferation of multiple melanoma lines in an MITF-dependent fashion, supporting the role of p300 as a promising therapeutic target in human melanoma and promoting a therapeutic strategy for p300 HAT inhibitor therapies in tumors expressing high levels of MITF.
Materials and Methods
Cell culture
Melanoma cell lines were kindly provided by Dr. Meenhard Herlyn at the Wistar Institute (Philadelphia, PA) and Dr. Levi Garraway at the Broad Institute (Cambridge, MA). Melanoma cells were maintained in Dulbecco’s modified Eagle medium. The medium was supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin (pen-strep), and 1% L-glutamine. The media, pen-strep, and L-glutamine, FBS were purchased from Invitrogen. All cell lines were grown at 37 °C in an atmosphere containing 5% CO2.
Plasmids and Transfection
Lentiviral expression vectors pCW45-GFP and pCW45-MITF were kindly provided by Dr. David Fisher (Massachusetts General Hospital, Boston). pGL2-MITF and control vector were generous gifts from Dr. Widlund (Brigham and Women’s Hospital, Boston). The Lentiviral pLKO1-based shRNA vectors targeting p300 and MITF were purchased from Sigma Aldrich (St. Louis, MO). Packaging and envelope expressing plasmids (psPAX2 and pMD2.G) were gifts from Didier Trono (Addgene plasmid #12260).
Lentivirus production
Selected p300 and MITF shRNA plasmids were co-transfected into HEK293T cells along with expression vectors containing lentiviral envelope and packaging plasmids via Lipofectamine 2000 according to the manufacturer’s protocol. Lentiviruses were harvested 48 hours following transfection. 250,000 human melanoma cells were transduced with each harvested lentivirus (500μL) in the presence of 8 μg/mL of polybrene. Subsequently, melanoma cells transduced with pLKO1-based shRNAs were selected in 1.5 μg/mL of puromycin after 48 hour following transduction in their respective culture medium. Information regarding p300 shRNAs used in this study is described in a previous publication (7).
Cell treatment with a small molecule inhibitor
To treat cells, compound A-485* was dissolved in anhydrous DMSO to make a 10 mM solution and added to culture medium to the desired concentration. An equal amount of DMSO was used as the vehicle control.
Cell cycle analysis
Cells were stained with propidium iodide according to a published protocol (24). Data acquisition and analysis were performed on a FACSCalibur flow cytometer with the CellQuest software (BD Biosciences, San Jose, CA).
Microarray studies
RNA from melanoma cells transduced with either shp300 or scrambled lentiviruses was purified using the Qiagen RNAeasy Plus Kit. Samples were submitted to Boston University Microarray and Sequencing Resource Core Facility for analysis on the Affymetrix GeneChip Human Gene 2.0 ST. The initial data processing and normalizations were performed by the core facility. The gene ontology analysis was performed with Ingenuity Pathway Analysis (Qiagen, Redwood City, CA).
qRT-PCR
Complementary DNA was synthesized using the Superscript III First Strand Synthesis System (Invitrogen, Grand Island, NY). qRT-PCR was performed using the SYBR Green PCR Master Mix (ABI/Invitrogen) as described previously (5). The primer pairs were designed using the NCBI PrimerBlast tool and individually optimized. Gene expression values were determined with the ΔΔCt method. GAPDH was used as an internal control. The absolute copy number of MITF was determined by using a standard curve generated from a 500bp MITF amplicon with varying concentrations. The list of primers used in this study is provided in the supporting information.
ChIP assay followed by qPCR
ChIP was performed based on a previously described protocol (5). The antibodies used in the study include normal rabbit and mouse IgG (sc-2027x and sc-2025, Santa Cruz), or antibodies against p300 (sc-585, Santa Cruz), histone H3K18 (13998, Cell Signaling), histone H3K27 (8173, Cell Signaling), or RNA polymerase II (2629, Cell Signaling). Primers for qPCR were designed to amplify a region near transcription start (Supplementary Table S1). qPCR was performed using SYBR Green PCR Master Mix (ABI/Invitrogen) on StepOnePlus real time PCR system (Applied Biosystems).
Histone acetyltransferase assays
Histone acetyltransferase assays were performed as described previously (25). Briefly, reactions measured the p300-catalyzed incorporation of 14C from the acetyl-CoA substrate (60mCi/mmol) into purified histone H3. A-485* was dissolved in 100% DMSO and diluted in 10% DMSO for a final reaction concentration of 1%. Reactions were performed in a buffer composed of 50 mM HEPES (pH 7.9), 50 mM NaCl, 1 mM TCEP, and 25 ug/ml BSA at 30°C and initiated by the addition of 14C-acetyl-CoA to a final concentration of 200 nM. After 5 minutes, the reaction was quenched and acetylated histone product was separated on a 16% tris-tricine gel and visualized by autoradiography. A 14C-BSA standard was run in parallel and used to quantify product formation.
Western blotting
Whole-cell lysates were prepared as described in the supplement. Western blots were performed as previously described (26). A complete list of primary and secondary antibodies used in this study is included in the supplemental information.
Senescence detection
Senescent cells were detected by staining for lysosomal senescence-activated beta-galactosidase activity with a commercial kit from Cell Signaling Technology (#9860) and by immunofluorescent staining of promyelocytic leukemia protein nuclear bodies using an established protocol (27).
Luciferase Assay
For reporter assays, cells were plated in a 6-well culture plate (Corning) to a 500,000 cells/well density and treated with either DMSO (control) and A485 (10μM) 2 days. The following day, cells were transfected with 5μg of pCW45-MITF vector (kind gift from Dr. Widlund and Dr. Fischer), 0.5μg of pRL.null (Promega) using Lipofectamine 3000 transfection reagent (ThermoFisher). Cells were allowed to incubate with the transfection mixture for 24 hours. Cells were washed once with phosphate-buffered saline and lysed with 200ml of 1 lysis buffer/well (Promega). The assay samples were then analyzed on a luminometer using the Dual-Luciferase kit (Promega). Luciferase signals were normalized to corresponding Renilla signals. Results are expressed as fold activation over DMSO-treated control and are plotted as the means from at least three independent experimental data points with error bars representing standard deviation.
Data Availability
Structural data that support the findings of this study have been deposited in PDB with the accession code 5KJ2, and microarray data have been deposited in GEO with the accession code
Results
p300 is an essential mediator of human melanoma cell growth
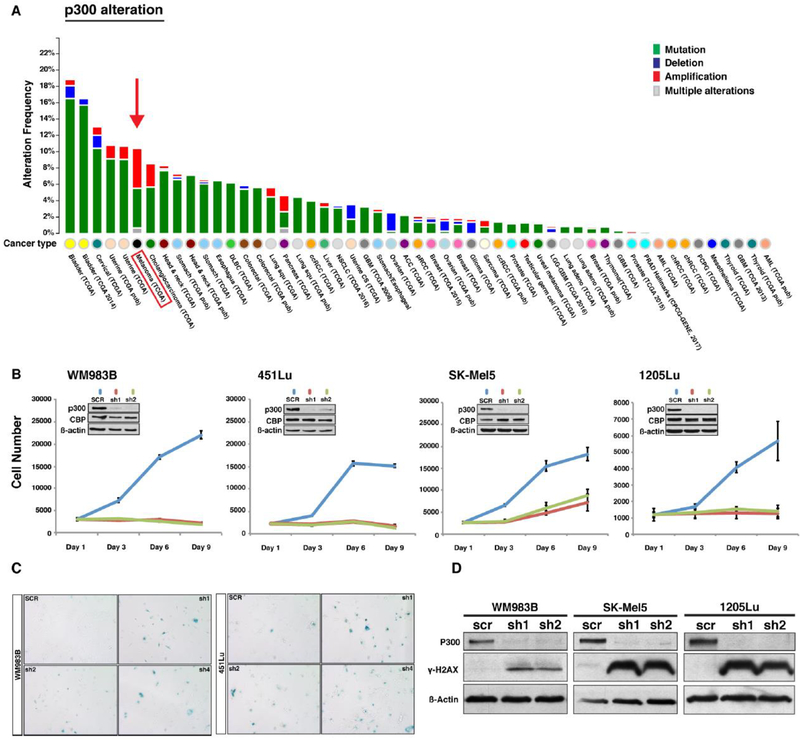
In order to analyze connections between p300 function and melanoma development, we evaluated the frequency of EP300 gene alterations present in human cancers using publicly available data sets from The Cancer Genome Atlas (TCGA) (28,29). Fifty-four different types of human cancers were evaluated through TCGA where melanomas displayed an approximately 10% incidence of EP300 genetic alterations, half of which were classified as amplifications (Figure 1A). In addition, the EP300 genetic locus was noted to be the site of frequently gained copy numbers in melanomas identified via a comprehensive genomic analysis of 101 melanoma short-term cultures and cell lines (4). Together, these findings suggest that p300 may play an important role in melanomagenesis. In order to investigate the functional role of p300 in human melanoma, we first evaluated basal levels of EP300 gene expression in a panel of melanoma cell lines and found that EP300 expressed uniformly throughout all cell lines examined (Figure S1A). We subsequently used targeted shRNA to knock down p300 expression in four human metastatic melanoma cell lines (WM893B, 451Lu, SK-Mel5, 1205Lu) (Figure 1B). Forty-eight hours following the depletion of p300, melanoma cells were evaluated phenotypically and found to display appreciable changes in morphology including an enlarged and flattened appearance (Figure S1B). Melanoma cell growth was also found to be significantly inhibited in all melanoma cell lines evaluated (Figure 1B) in association with a G0/G1 growth arrest (Figure S1C). In addition, p300 knockdown was found to induce increased expression of senescence-associated ß-galactosidase (Figure 1C) and associated cellular senescence, which was similar to previously reported changes seen following p300 knockdown in primary human melanocytes (30). There was no appreciable increase in apoptosis following p300 knockdown as measured by annexin V staining and FACS analysis (Figure S1D); however, melanoma cells were found to exhibit increased expression of the DNA-damage response and cellular scenescence marker, γ-H2AX. (Figure 1D).
Figure 1. Genetic alteration frequency of EP300 gene in human cancers and functional assessment of the p300 in melanoma cell lines.
(A) EP300 gene alteration frequency in various human cancers available in the TCGA data set. Melanoma is the cancer type displaying the highest EP300 gene alteration frequency. (B) Functional role of p300 in melanoma cell growth. EP300 expression was silenced by shRNAs in four melanoma cell lines (WM983B, 451Lu, SK-Mel5, and 1205Lu) and subjected to a cell proliferation assay. Insert demonstrates p300 knockdown efficiency via western blot. (C) Senescence-associated ß-galactosidase staining assay of p300 silenced melanoma cell lines. ß-galactosidase staining positive cell numbers were increased after p300 depletion in 451Lu and WM983B cells when compared to those of control. (D) Western blot analysis of cellular senescence marker γ-H2AX. p300-depleted melanoma cells were assayed for activation of the cellular senescence response using antibody against phosphorylated H2AX. (E) Cell cycle profile analysis of p300-depleted melanoma cells (1205Lu and 451Lu). Error bars, SD; *p<0.05.
Expression Profiling Reveals p300 Downstream Effector Genes and Targeted Pathways in Human Melanoma Cells
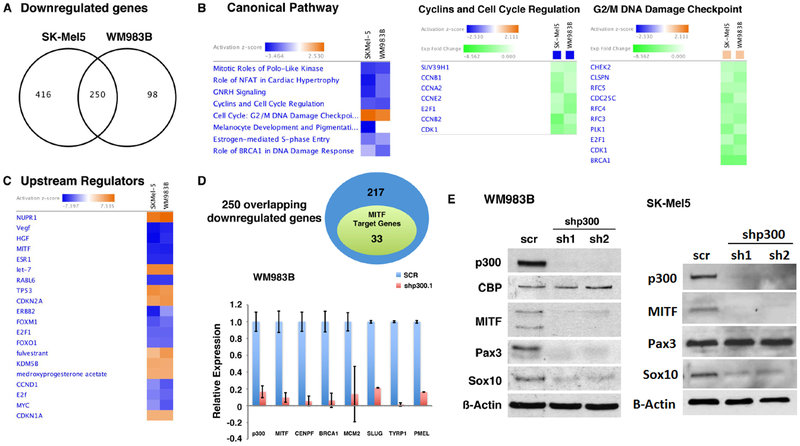
In order to obtain a comprehensive assessment of p300 downstream effector genes in human melanoma, we performed genome-wide expression profiling analysis of WM983B and SK-Mel5 melanoma cells following p300 depletion (Figure 2). We identified 666 and 348 down-regulated genes (>2-fold) in SK-Mel5 and WM983B, respectively, following silencing of p300. 250 genes were found to be significantly downregulated in both melanoma cell lines following silencing of p300 and were considered as potential bona fide p300 downstream effectors which were selected for further evaluation (Figure 2A). Ingenuity Pathway Analysis (IPA) of the common p300 effector genes found them to be statistically (z-score) enriched for downregulation of cell cycle control genes and upregulation of genes associated with the DNA damage response (Figure 2B). The two highest ranked genes identified in gene set enrichment analysis (GSEA) were associated with cell cycle control and DNA replication and showed similar negative enrichment scores (Figure S2A). Expression profiling data were confirmed via qRT-PCR analysis which was notable for significantly upregulated expression of the p21 cyclin-dependent kinase inhibitor CDKN1A following p300 knockdown, while expression of Cyclin A2 (CCNA2) was significantly reduced (Figure S2B), consistent with a cellular growth arrest phenotype.
Figure 2. Identification of MITF as a transcriptional target of p300 via global gene expression profile analysis.
(A) Genes differentially expressed following EP300 gene silencing in human melanoma cells. Two hundred and fifty overlapping genes in both p300-depleted WM983B and SK-Mel5 cells were identified as p300 downstream targets. (B) Canonical pathways regulated by p300 in melanoma cells. Ingenuity Pathway Analysis (IPA) of the p300 downstream target genes revealed that pathways associated with cell proliferation are altered in p300 depleted melanoma cells. (C) Identification of potential upstream regulators of p300 target genes by IPA. MITF was found as one of the upstream regulators of p300 target gene expression. (D) Number of known MITF-target genes that are also included in the p300-downstream effector genes. IPA identified 33 genes from the p300 target gene set as known MITF target genes. Decreased transcript levels of MITF and its target genes were further verified in p300-depleted melanoma cells via qRT-PCR. (E) Western blot analysis evaluated PAX3 and SOX10 levels in p300-silenced melanoma WM983B and SK-Mel 5 cells.
p300 Regulates MITF Expression in Human Melanoma Cells
Upstream Regulator Analysis (URA) by IPA was used to further identify global regulatory mechanisms associated with p300 knockdown in human melanoma cells. These studies allowed us to identify the MITF transcription factor as a significant upstream regulator affecting p300 downstream target gene expression (Figure 2C). Among the 250 overlapping p300 target genes identified, 33 genes were listed as MITF transcriptional target genes (Figure 2D and S2C), which was confirmed through qRT-PCR analysis (Figure 2D) and previously published ChIP-seq analysis (31). As p300 is a known transcriptional cofactor for MITF (10), we were not surprised to discover that loss of p300 was associated with downregulation of MITF target gene expression; however, our microarray analysis further demonstrated that depletion of p300 was also associated with reduced expression of MITF itself, suggesting a direct effect of p300 on MITF transcription (Figure 2D). Additionally, several known upstream regulators of MITF, such as Pax3 and Sox10, were found to be downregulated in the setting of p300 knockdown (Figure 2D). These data were confirmed by qRT-PCR and western blot analysis of knockdown cell lysates in melanoma WM983B and SK-Mel 5 cells; however, downregulation of Pax3 appeared to be cell line-dependent (Figure 2E).
MITF Controls Human Melanoma Cell Proliferation
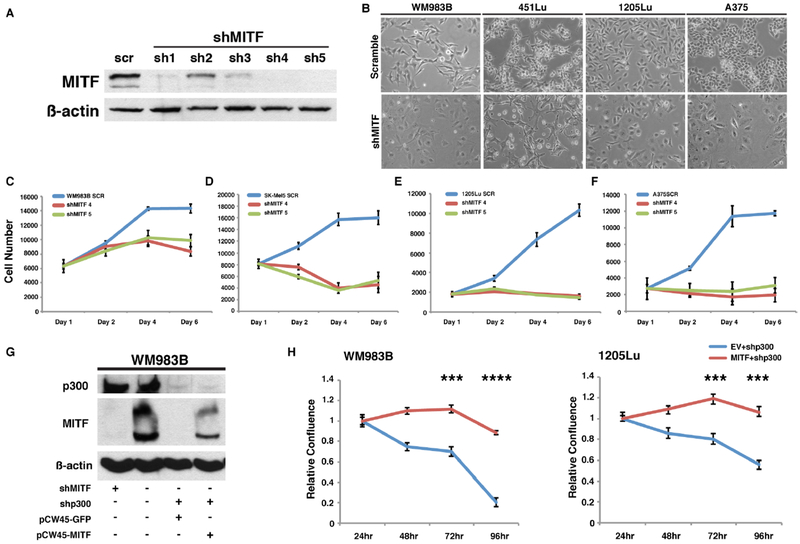
In order to further confirm the functional impact of p300 and MITF expression in human melanoma cells, we used shRNA to knockdown MITF expression in four melanoma lines (WM983B, 451Lu, 1205Lu, and A375) (Figure 3A). Morphologically, all melanoma cell lines evaluated showed an enlarged and flattened cell phenotype following MITF depletion, which was similar to the morphologic phenotype of p300 knockdown (Figure 3B and S1B) and suggestive of cellular senescence as has been previously observed (31,32). Interestingly, knockdown of MITF was associated with inhibition of cell growth in all melanoma cells tested, similar to that seen in p300-depleted melanoma cells (Figure 3C–F, S3A). In order to further define the significance of MITF as a mediator of p300-controlled cell growth, we ectopically expressed MITF in p300-knockdown melanoma cell lines (Figure 3G). Strikingly, ectopic expression of MITF in p300-silenced WM983B and 1205Lu human melanoma cells was found to rescue the p300 knockdown growth inhibitory phenotype (Figure 3H, S3B) suggesting that MITF is an important mediator of p300-associated cell growth in human melanoma.
Figure 3. MITF rescues the p300-dependent growth phenotype in p300 knockdown melanoma cells.
(A) Illustrative western blot analysis validating stable knockdown of MITF by shRNAs in melanoma cells. WM983B cell line is shown. (B) Morphological changes present in melanoma cell lines after MITF depletion. (C to F) Cell proliferation assay demonstrating decreased cell growth rates in the melanoma cell lines following stable depletion of MITF. (G) Representative western blot analysis validating ectopic re-expression of MITF in p300 stable knockdown cells. p300 knockdown demonstrates inhibition of MITF expression (also shown in the Figure 2E). Plasmid vectors expressing MITF (pCW45-MITF) and control GFP (pCW45-GFP) were transiently transfected in p300-depleted melanoma cells (WM983B). (H) Relative cell growth assay of p300-depleted WM983B and 1205Lu melanoma cells following transient re-expression of MITF versus GFP control. Error bars, SD; ***p<0.001.
p300 Controls MITF Expression Through Direct Chromatin Modifications at upstream promoter sites
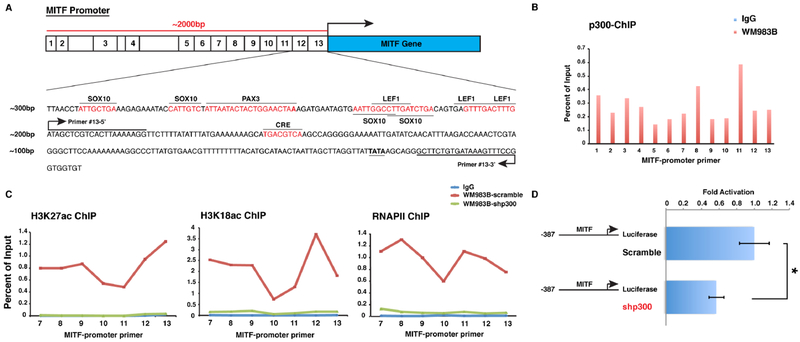
We evaluated MITF promoter accessibility in the setting of p300 knockdown using chromatin immunoprecipitation (ChIP) analysis spanning 2000bp of the promoter region proximal to the transcription start site (TSS) of MITF (Figure 4A). p300 pulldown ChIP analysis identified significant occupancy of p300 within the promoter region proximal to the TSS of the MITF gene in human melanoma cells (Figure 4B). ChIP analysis of the signature p300 histone marks, acetylated H3K18 and H3K27, has previously identified ‘active’ regions of the MITF promoter (33), which demonstrate significant overlap with p300-associated regulatory regions. Acetylated H3K18 and H3K27 were readily detected within the promoter region of MITF, which was completely abolished following p300-depletion (Figure 4C). Additionally, recruitment of RNA polymerase II (RNPII) to the proximal TSS was also eliminated following gene silencing of p300. Further evaluation of p300 regulation of MITF transcription was performed using an MITF promoter reporter assay (17), where MITF promoter activity was found to be significantly reduced following p300-depletion in human melanoma cells (Figure 4D). Together, these data indicate that p300 mediates core histone acetylation within the promoter region of the MITF gene and critically regulates MITF expression in human melanoma cells.
Figure 4. p300 transcriptionally regulates MITF expression via histone acetylation at proximal upstream regulatory regions of the MITF gene.
(A) Illustrative map of ChIP-PCR amplicons in the MITF upstream locus is depicted. (B) p300 protein is highly enriched at MITF upstream promoter sites. Normal rabbit immunoglobulin G (IgG) is used as antibody control. (C) ChIP-PCR analysis of H3K27-Ac, H3K18-Ac, and RNAPII within the MITF gene promoter locus in p300-wildtype (scramble) and p300-knockdown melanoma cells. IgG serves as antibody control. (D) MITF gene promoter luciferase assay in WM983B melanoma cells with p300-wildtype (scramble) and p300-knockdown. Error bars, SD; *p<0.05.
The p300-MITF signaling axis transcriptionally regulates FOXM1
Further evaluation of our microarray data was pursued in order to identify potential downstream effectors of the p300-MITF regulatory axis. Notably, expression of Forkhead Box M1 (FOXM1), a pro-proliferative and pro-survival MEK-target gene that has been shown to be an important regulator of cell cycle in many human cancers (34–36), was found to be markedly suppressed following p300 depletion (Figures 5A, 5B). Additionally, target genes of FOXM1 were consistently downregulated in both SK Mel-5 and WM983B cells following EP300 gene silencing (Figure 2C, 5A and 5C), while FOXM1 protein levels were also found to be reduced following p300 depletion (Figure 5B). This is particularly noteworthy since human melanomas have recently been found to express elevated levels of FOXM1, which has been shown to be a potential therapeutic target for this disease (37,38).
Figure 5. The p300-MITF signaling axis transcriptionally regulates FOXM1 expression and determines clinical outcomes.
(A) List of transcriptional targets of FOXM1 which are down-regulated by p300 depletion in melanoma cell lines as shown Figure 2A. (B) Western blot analysis showing decreased FOXM1 and MITF protein levels in p300-depleted melanoma cell lines. (C and D) Evaluation of mRNA and protein levels of FOXM1 and cell cycle regulatory proteins in MITF knockdown melanoma cells. (E) RT-PCR validation analysis of the known FOXM1 target genes selected from (A). (F) Reverse Phase Protein Analysis (RPPA) of FOXM1 expression in human melanoma patient samples from the TCGA dataset. FOXM1 protein expression levels were significantly higher in melanoma patients with either EP300 or MITF gene amplification (altered), (N = 205). (G) Alterations in gene expression of EP300, MITF, CCNA2, CDK2, and FOXM1 display patterns of mutual exclusion in the TCGA dataset. Tumor samples are shown in columns; genes in rows. Only samples with at least one alteration are shown. (H) Kaplan-Meier plot of overall survival of patients stratified by altered transcription levels of the EP300/MITF/CCNA2/CDK2/FOXM1 genes as shown in (F).
Since our work identified the p300-MITF signaling axis as a critical pathway for melanoma survival and proliferation, we hypothesized that a key function of MITF in melanoma may be to regulate the expression of FOXM1 in these tumors. In order to test this hypothesis, we performed MITF knockdown in three melanoma cell lines and investigated changes in FOXM1 expression. Notably, we found that both transcript and protein levels of FOXM1 were sharply reduced following MITF depletion in human melanoma cells, suggesting an important regulatory role in these cells (Figures 5D, 5E). Since Strub and colleagues (31) had previously used ChIP-seq to explore the MITF transcriptome, we reviewed their dataset to evaluate MITF occupancy at the FOXM1 promoter, and remarkably identified a peak within the proximal promoter region of FOXM1. Of note, reverse phase protein array analysis from the TCGA cutaneous melanoma data set (28,29) revealed that tumors from patients with elevated expression of p300 or MITF (denoted by gene amplifications in this dataset) also displayed significantly higher levels of FOXM1 protein expression (Figure 5F), further suggesting that the p300-MITF-FOXM1 signaling axis is critical for the proliferative phenotype of human melanomas.
While direct transcriptional regulation of FOXM1 by MITF may be a principal mechanism governing FOXM1 expression and melanoma cell growth, alternative regulation of FOXM1 protein expression and function may be achieved through direct phosphorylation of FOXM1, which contributes to its protein stability (39). Since FOXM1 phosphorylation has been shown to be catalyzed by cyclin-dependent kinases (38), and CDK2 expression is transcriptionally regulated by MITF (40), we hypothesized that down-regulated expression of CDK2/CyclinA complexes following MITF depletion would result in reduced phosphorylation of FOXM1, leading to decreased FOXM1 stability and subsequent degradation (41). Indeed, we found that both mRNA and protein expression levels of CDK2 and cyclin A2 were significantly reduced following MITF-depletion which could contribute to loss of FOXM1 stability and function (Figure 5C and 5D).
Following the observation that the p300-MITF axis controls the expression of FOXM1, we explored the significance of these findings in human melanoma specimens by mining genetic data from 471 patient samples available through the TCGA database, using cBioPortal (28,29). Our analysis of gene alteration patterns for p300, MITF, cyclin A2, CDK2, and FOXM1 demonstrated a strong pattern of mutual exclusivity, suggesting that genes within this set contribute to a common downstream pathway (42) (Figure 5F). Furthermore, patients with gene alterations within the p300/MITF/cyclinA2/CDK2/FOXM1 pathway showed significantly reduced overall survival compared to patients without such gene alterations (Figure 5G, 5H). We therefore conclude that the p300-MITF-FOXM1 axis is important for tumor cell survival and may be predictive of patient outcomes.
Characterization of a selective p300/CBP HAT inhibitor, A-485*, in human melanomas
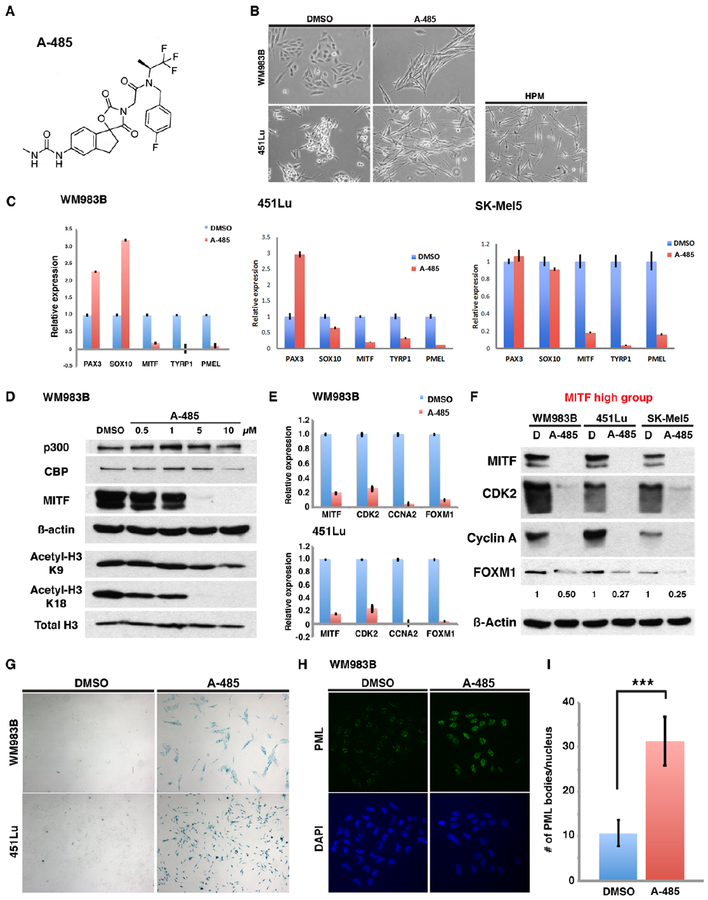
Since p300-mediated histone acetylation appears to play a major role in regulating MITF transcription (Figure 4), we sought to evaluate whether targeted inhibition of p300-mediated lysine acetylation would be sufficient to block MITF transcription and inhibit proliferation in human melanomas. The small molecule inhibitor of p300 HAT activity, A-485*, (43), was synthesized as a mixture of diastereomers following the methods described in the supplementary materials (Figure 6A). To confirm that A-485* is a potent inhibitor of p300 acetyltransferase activity, we performed in vitro enzymatic assays using purified recombinant full-length p300 with purified histone H3 protein as substrate. Acetyltransferase reactions were performed with 14C-acetyl-CoA and 14C incorporation into target histone H3 was evaluated using phosphorimage analysis of SDS-PAGE (Figure S4). Compound A-485* was found to inhibit p300 acetyltransferase activity on purified H3 with an IC50 of ~0.5μM (Figure S5) under the conditions of our assays, which was significantly more potent than that reported for C646 (IC50 ~20 μM) determined under similar conditions (7). Upon treatment with A-485*, melanoma cells displayed altered morphologic features with an elongated, refractile, and spindled appearance reminiscent of that seen in primary human melanocytes (Figure 6B). We therefore sought to determine whether inhibition of p300 HAT activity by A-485* might directly promote melanocyte differentiation. We found that the melanocyte differentiation genes, PAX3 and SOX10 were variably altered in expression following A-485* treatment of WM983B, 451Lu and SK-Mel5 cells, while expression of MITF, and its target differentiation genes TYRP1, and PMEL was markedly reduced in all cell lines (Figure 6C). Compound A-485* treatment of WM983B melanoma cells demonstrated effective inhibition of histone H3 acetylation at lysine 18 while sparing lysine 9, a histone site that is not preferentially acetylated by p300/CBP (Figure 6D). Moreover, MITF protein expression was dramatically suppressed following A-485* treatment while protein levels of p300 and CBP remained unchanged (Figure 6D).
Figure 6. Chemical inhibition of p300 HAT activity by a potent and selective inhibitor promotes melanoma senescence through suppression of the p300/MITF/FOXM1 transcriptional axis.
(A) Chemical structure of A-485*. (B) Cell morphology changes following A-485* treatment of melanoma cells (5 μM) for 72 hours. (C) qRT-PCR analysis of mRNA levels of MITF and melanocyte differentiation-associated genes following treatment of melanoma cells with A-485*. (D) Western blot of p300-dependent histone acetylation and MITF expression in melanoma cells following treatment A-485*. (E and F) Relative mRNA and protein levels of MITF, CDK2, CCNA2 and FOXM1 in melanoma cells following treatment with A-485* (10 μM) for 5 days. (G) Senescence-associated ß-galactosidase staining of melanoma cells treated with A-485* (10 μM) for 7 days. (H) Immunofluorescence staining of the cellular senescence protein, PML, in melanoma cells treated with A-485* (10 μM) for 7 days. (I) Quantification of PML bodies in melanoma cells following A-485* treatment. Error bars =1 SD; ***p<0.001.
Since we determined that FOXM1 gene expression is regulated by the p300-MITF signaling axis in human melanoma cells, we sought to evaluate whether A-485* treatment of melanoma cells would also suppress FOXM1 expression in a similar fashion to p300 knockdown. Consistent with our previous results, A-485* treatment of WM983B and 451Lu melanoma cells led to dramatically reduced transcription of MITF, CDK2, cyclin A2, and FOXM1 (Figure 5B, 5CD and Figure 6E). In addition, protein levels of FOXM1 were reduced following A-485* treatment of three metastatic melanoma cell lines along with reduced expression of MITF, CDK2 and cyclin A (Figure 6F). These results suggest that p300 HAT inhibition potently inhibits expression of the pro-proliferative FOXM1 gene in human melanomas which may be mediated by down-regulated MITF expression. Since FOXM1 serves as a major regulator of cell proliferation and survival, we sought to determine whether A-485* treatment of human melanoma cells would induce a cellular senescence phenotype. We found that both WM983B and 451Lu melanoma cells demonstrated marked increases in senescence-associated ß-galactosidase staining following A-485* treatment, consistent with induction of cellular senescence (Figure 6G). Additionally, melanoma cells treated with A-485* displayed increased intensity of senescence-associated promyelocytic leukemia (PML) and a rise in PML nuclear bodies following treatment, supporting the induction of cellular senescence by this compound (Figures 6H and 6I).
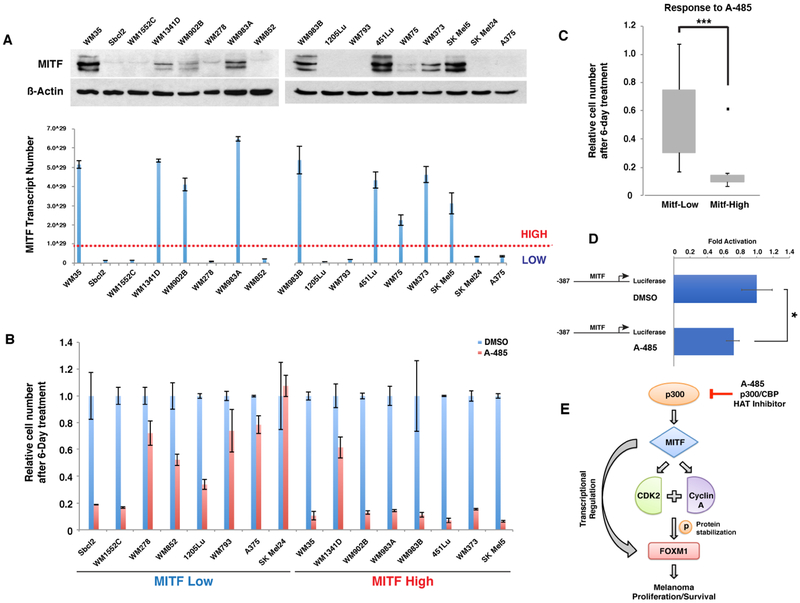
A-485* treatment of human melanoma cells promotes targeted growth inhibition in melanoma cell lines expressing high levels of MITF
Melanoma cells have been noted to express varying levels of MITF, with MITF-high expressing tumors exhibiting a more proliferative phenotype and MITF-low expressing tumors demonstrating a more invasive/metastatic phenotype (44). Our library of human melanoma cell lines consists of melanoma cells possessing both high and low levels of MITF baseline expression (Figure 7A and S1A). We therefore sought to evaluate the effect of p300 inhibitor A-485* in human melanoma cells expressing varying levels of MITF, in order to determine whether the specific inhibition of MITF expression through p300 HAT allowed for targeted cell growth inhibition. Remarkably, we found that melanoma cells possessing high-MITF expression levels displayed significantly greater growth inhibition by A-485* (10μM) compared to low-MITF expressing melanoma cells (Figure 7B and 7C). MITF promoter reporter assay was performed to evaluate the regulation of MITF transcription by A-485*, where MITF promoter activity was found to be significantly reduced following A-485* treatment in human melanoma cells (Figure 7D). Together, these data suggest a model for p300 as a critical regulator of MITF expression in human melanoma cells and the specific therapeutic efficacy of targeting p300 HAT activity in human melanomas possessing increased expression of MITF (Figure 7E).
Figure 7. Selective targeting of melanoma cells with high endogenous MITF expression by the p300 histone acetyltransferase inhibitor, A-485*.
(A) Panel of melanoma cell lines evaluated for MITF expression by immunoblot blot and qRT-PCR. (B) Relative growth of human melanoma cell lines following A-485* treatment. Cells were subjected to 6-days of treatment with A-485* (10 μM). (C) Box-plot analysis of cytotoxic effects of A-485* in two groups of melanoma cell lines expressing high and low levels of MITF. Error bars, SD; ***p<0.001. (D) MITF gene promoter luciferase assay in WM983B melanoma cells treated with A-485* and DMSO. Error bars, SD; *p<0.05. (E) Schematic model illustrating the p300-MITF-FOXM1 transcriptional axis regulating melanoma cell growth.
Discussion
While genetic mutations in human melanomas have been explored extensively over the past decade, the role of epigenetic alterations in melanoma development and progression has been less clearly defined. We have undertaken a series of experiments to determine the functional significance of p300-mediated chromatin remodeling in human melanomas using both a genetic and chemical approach. Our studies have allowed us to identify a highly conserved molecular pathway driving melanoma cell growth which is mediated by p300-associated epigenetic modifications of core histones at the MITF promoter. We have further determined that restoration of MITF expression in p300 knockdown melanoma cells reverses the growth-arrest phenotype seen with depletion of p300, suggesting that MITF is a key regulator of p300-associated proliferation in these cells. MITF has been identified as a critical mediator of “phenotype switching” in melanoma whereby cells are able to convert from a proliferative (high-MITF) phenotype to an invasive/migratory (low-MITF) phenotype (45); however, the precise mechanism governing this switch remains to be determined. In addition, single-cell RNA-seq of metastatic melanoma tissues identified subsets of cells within a tumor expressing high and low levels of MITF, supporting the potential interchangeability of this phenotype switch (46). Furthermore, MITF levels have been shown to be critically regulated in the context of tumor development, with up to 20% of advanced melanomas demonstrating MITF gene amplifications (12), (Cancer Genome Atlas Network, 2015, http://cancergenome.nih.gov/), supporting its role as a bona fide oncogene in melanoma. Moreover, MITF has been identified as a key driver of acquired resistance to MAPK-pathway inhibition through a variety of pro-survival mechanisms, as well as innate resistance to such treatments (47–50). Taken together, these data implicate MITF as a central mediator of melanoma cell fate and an important therapeutic target for melanoma; however, previous attempts to target MITF in human melanoma cells have been largely unsuccessful (51). Recently, Smith and colleagues undertook a unique approach to understanding acquired MAPK-targeted therapeutic resistance by exploring mediators of non-mutational drug tolerance in human melanoma cell lines, which allowed them to identify MITF as a key mediator of early drug tolerance (50). A large-scale drug repurposing screen allowed for identification of the HIV1 protease inhibitor, nelfinavir, as an inhibitor of MITF expression which promoted resensitization of tolerant cell lines to inhibitors of BRAF and MEK through indirect transcriptional repression of MITF (50). While this work was intriguing, the lack of understanding of nelfinavir’s molecular mechanism(s) and its narrow application to cases of non-mutational drug tolerance may limit nelfinavir’s clinical development. In contrast, we have identified direct activation of the MITF promoter through p300-mediated acetylation of core histones at critical promoter sites, and identified a potent and specific inhibitor, A-485*, of this process and melanoma cell growth. This clear mechanism of A-485* effects on MITF, and the potent growth inhibitory effects in tumors possessing increased expression of MITF, raise the possibility of development of this and other novel epigenetic therapies for melanoma. Additionally, as A-485* treatment of WM983B melanoma cells demonstrated effective inhibition of histone H3 acetylation at lysine 18 while sparing lysine 9, we suggest that H3K18 acetylation may be used as a surrogate marker for therapeutic targeting of p300 HAT activity in vivo. Indeed, as effective agents targeting the MAPK pathway have been developed for tumors with activating BRAF gene mutations, their sustained therapeutic utility has been stymied by the near-universal acquired resistance to these drugs over time, necessitating the need for a multi-pronged approach (52–54) and targeting of the numerous and varied resistance mechanisms (14,15,49,55–57). Development of novel strategies targeting epigenetic pathways may promote more durable therapeutic responses in melanoma, as bypass mechanisms of resistance would be less probable.
The recent investigation of A-485, the pure stereoisomer form of A-485* across >100 cancer cell lines showed a wide range of sensitivities of the anti-proliferative effects mediated by A-485 across these lines, including in human melanomas. In the case of prostate cancer, expression of the androgen receptor or a splice variant conferred sensitivity to A-485 and AML1-ETO appeared to predict sensitivity to p300 inhibition in acute leukemia. For the other cancer types, no clear biomarker that correlated with A-485 sensitivity had been established; however, the studies here now add MITF in melanoma as such a biomarker.
Our studies of downstream effectors of p300 in human melanomas allowed us to identify a specific growth regulatory axis involving MITF, cyclin A and CDK2, and the pro-survival transcription factor, FOXM1. Notably, these genes were found to be altered in a mutually exclusive way in primary human tumors (Figure 5G) and associated with a poor prognosis (Figure 5H). Indeed, increased FOXM1 expression in melanoma has previously been associated with accelerated tumor progression and poor prognosis (58) as well as suppression of the senescence phenotype (38). Our studies also support a cell survival phenotype associated with the p300-MITF axis and FOXM1 expression in human melanoma cells, which is abrogated following treatment with the p300 HAT inhibitor, A-485*, and associated with subsequent cellular senescence (Figure 6). Interestingly, recent data suggests that the novel anticancer agents Honokiol (51) and Imipramine Blue (52) exhibit anti-tumor effects through inhibition of FOXM1. Thus, combination treatment of melanomas with these agents and A-485 may demonstrate synergistic activity and should be further investigated.
Recent significant advances in melanoma immunotherapies have been the cause for great excitement in the cancer treatment field, but thus far have proven highly effective for only subsets of patients with advanced disease (59) suggesting a significant need for the development of additional therapeutic strategies for this disease. Moreover, while epigenetic modifying agents such as DNA methyltransferase inhibitors (DNMTis), have been demonstrated to enhance the host response to immunotherapies in mouse models for melanoma (60), such studies have had limited impact with regard to disease treatment to date. Additionally, while several epigenetic modifying therapies have been approved by the US Food and Drug Administration (FDA) for use in patients, including DNA methyltransferase inhibitors (DNMTi), and histone deacetylase inhibitors (HDACi), such agents have not demonstrated widespread benefits in human cancers. Our data suggest that the time may be right to pursue new therapeutic strategies targeting epigenetic alterations associated with histone acetyltransferase activity in human melanoma. Our data further suggest that such therapies may prove beneficial in directly targeting tumors expressing high levels of MITF, but perhaps also by augmenting immune responsiveness (61). Finally, we expect that A-485* should prove useful, not only as a chemical probe to specifically dissect out the functional significance of MITF in human melanomas, but more broadly in understanding the role of protein acetylation in tumor biology and immunomodulation.
Supplementary Material
Statement of Significance:
These results show that MITF is a major downstream target of p300 in human melanoma whose expression is predictive of melanoma response to small molecule inhibition of p300 HAT activity.
Acknowledgments
We thank members of the Alani, Ryu, and Cole labs for helpful discussions. DJM is supported by CTSA grant UL1TR001079 and FAMRI (Flight Attendant Medical Research Institute). BEZ is supported by a Jane Coffin Childs Fund postdoctoral fellowship. EK is supported by an ASA Medical Student Grant. PAC is supported by NIH grant GM62437. PAC is a cofounder, shareholder, and paid consultant for Acylin Therapeutics Inc. which is developing p300/CBP HAT inhibitors. He is also a paid consultant for Abbvie Inc. which is pursuing the development of p300/CBP HAT inhibitors. RMA is a cofounder and shareholder for Acylin Therapeutics Inc. which is developing p300/CBP HAT inhibitors. The results shown here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Footnotes
Conflict of Interest Statement: PAC is a cofounder, shareholder, and paid consultant for Acylin Therapeutics Inc. which is developing p300/CBP HAT inhibitors. He is also a paid consultant for Abbvie Inc. which is pursuing the development of p300/CBP HAT inhibitors. RMA is a cofounder and shareholder for Acylin Therapeutics Inc. which is developing p300/CBP HAT inhibitors.
References
- 1.Feinberg AP, Koldobskiy MA, Gondor A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 2016;17:284–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nature reviews Molecular cell biology 2014;15:703–8 [DOI] [PubMed] [Google Scholar]
- 3.Dancy BM, Cole PA. Protein lysine acetylation by p300/CBP. Chemical reviews 2015;115:2419–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer research 2008;68:664–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan G, Eller MS, Elm C, Larocca CA, Ryu B, Panova IP, et al. Selective inhibition of p300 HAT blocks cell cycle progression, induces cellular senescence, and inhibits the DNA damage response in melanoma cells. The Journal of investigative dermatology 2013;133:2444–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chemistry & biology 2010;17:471–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dancy BM, Crump NT, Peterson DJ, Mukherjee C, Bowers EM, Ahn YH, et al. Live-cell studies of p300/CBP histone acetyltransferase activity and inhibition. Chembiochem : a European journal of chemical biology 2012;13:2113–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 2017;550:128–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vachtenheim J, Sestakova B, Tuhackova Z. Inhibition of MITF transcriptional activity independent of targeting p300/CBP coactivators. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society 2007;20:41–51 [DOI] [PubMed] [Google Scholar]
- 10.Sato S, Roberts K, Gambino G, Cook A, Kouzarides T, Goding CR. CBP/p300 as a co-factor for the Microphthalmia transcription factor. Oncogene 1997;14:3083–92 [DOI] [PubMed] [Google Scholar]
- 11.Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes & development 2000;14:1712–28 [PubMed] [Google Scholar]
- 12.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005;436:117–22 [DOI] [PubMed] [Google Scholar]
- 13.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends in molecular medicine 2006;12:406–14 [DOI] [PubMed] [Google Scholar]
- 14.Muller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C, et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nature communications 2014;5:5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature 2013;504:138–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, Le Caignec C, et al. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet 2000;9:1907–17 [DOI] [PubMed] [Google Scholar]
- 17.Huber WE, Price ER, Widlund HR, Du J, Davis IJ, Wegner M, et al. A tissue-restricted cAMP transcriptional response: SOX10 modulates alpha-melanocyte-stimulating hormone-triggered expression of microphthalmia-associated transcription factor in melanocytes. The Journal of biological chemistry 2003;278:45224–30 [DOI] [PubMed] [Google Scholar]
- 18.Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ. Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Human genetics 2000;107:1–6 [DOI] [PubMed] [Google Scholar]
- 19.Price ER, Ding HF, Badalian T, Bhattacharya S, Takemoto C, Yao TP, et al. Lineage-specific signaling in melanocytes. C-kit stimulation recruits p300/CBP to microphthalmia. The Journal of biological chemistry 1998;273:17983–6 [DOI] [PubMed] [Google Scholar]
- 20.Takeda K, Yasumoto K, Takada R, Takada S, Watanabe K, Udono T, et al. Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. The Journal of biological chemistry 2000;275:14013–6 [DOI] [PubMed] [Google Scholar]
- 21.Verastegui C, Bille K, Ortonne JP, Ballotti R. Regulation of the microphthalmia-associated transcription factor gene by the Waardenburg syndrome type 4 gene, SOX10. The Journal of biological chemistry 2000;275:30757–60 [DOI] [PubMed] [Google Scholar]
- 22.Watanabe A, Takeda K, Ploplis B, Tachibana M. Epistatic relationship between Waardenburg syndrome genes MITF and PAX3. Nature genetics 1998;18:283–6 [DOI] [PubMed] [Google Scholar]
- 23.Yasumoto K, Takeda K, Saito H, Watanabe K, Takahashi K, Shibahara S. Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling. The EMBO journal 2002;21:2703–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darzynkiewicz Z, Juan G. DNA content measurement for DNA ploidy and cell cycle analysis. Curr Protoc Cytom 2001;Chapter 7:Unit 7 5 [DOI] [PubMed] [Google Scholar]
- 25.Zucconi BE, Luef B, Xu W, Henry RA, Nodelman IM, Bowman GD, et al. Modulation of p300/CBP Acetylation of Nucleosomes by Bromodomain Ligand I-CBP112. Biochemistry 2016;55:3727–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cummings SD, Ryu B, Samuels MA, Yu X, Meeker AK, Healey MA, et al. Id1 delays senescence of primary human melanocytes. Mol Carcinog 2008;47:653–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vernier M, Bourdeau V, Gaumont-Leclerc MF, Moiseeva O, Begin V, Saad F, et al. Regulation of E2Fs and senescence by PML nuclear bodies. Genes & development 2011;25:41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling 2013;6:pl1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2012;2:401–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bandyopadhyay D, Okan NA, Bales E, Nascimento L, Cole PA, Medrano EE. Down-regulation of p300/CBP histone acetyltransferase activates a senescence checkpoint in human melanocytes. Cancer research 2002;62:6231–9 [PubMed] [Google Scholar]
- 31.Strub T, Giuliano S, Ye T, Bonet C, Keime C, Kobi D, et al. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene 2011;30:2319–32 [DOI] [PubMed] [Google Scholar]
- 32.Giuliano S, Cheli Y, Ohanna M, Bonet C, Beuret L, Bille K, et al. Microphthalmia-associated transcription factor controls the DNA damage response and a lineage-specific senescence program in melanomas. Cancer research 2010;70:3813–22 [DOI] [PubMed] [Google Scholar]
- 33.Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. The EMBO journal 2011;30:249–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madureira PA, Varshochi R, Constantinidou D, Francis RE, Coombes RC, Yao KM, et al. The Forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. The Journal of biological chemistry 2006;281:25167–76 [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Banerjee S, Kong D, Li Y, Sarkar FH. Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer research 2007;67:8293–300 [DOI] [PubMed] [Google Scholar]
- 36.Gemenetzidis E, Bose A, Riaz AM, Chaplin T, Young BD, Ali M, et al. FOXM1 upregulation is an early event in human squamous cell carcinoma and it is enhanced by nicotine during malignant transformation. PloS one 2009;4:e4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kruiswijk F, Hasenfuss SC, Sivapatham R, Baar MP, Putavet D, Naipal KA, et al. Targeted inhibition of metastatic melanoma through interference with Pin1-FOXM1 signaling. Oncogene 2016;35:2166–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders L, Ke N, Hydbring P, Choi YJ, Widlund HR, Chick JM, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer cell 2011;20:620–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Molecular and cellular biology 2007;27:1007–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, et al. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer cell 2004;6:565–76 [DOI] [PubMed] [Google Scholar]
- 41.Laoukili J, Alvarez M, Meijer LA, Stahl M, Mohammed S, Kleij L, et al. Activation of FoxM1 during G2 requires cyclin A/Cdk-dependent relief of autorepression by the FoxM1 N-terminal domain. Molecular and cellular biology 2008;28:3076–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, et al. High-throughput oncogene mutation profiling in human cancer. Nature genetics 2007;39:347–51 [DOI] [PubMed] [Google Scholar]
- 43.Michaelides MU, Hansen TU, Dai YU, Zhu GU, Frey RU, Gong JU, et al. ; Abbvie, Inc., assignee, assignee. Spirocyclic HAT inhibitors and methods for their use2016. [Google Scholar]
- 44.Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society 2006;19:290–302 [DOI] [PubMed] [Google Scholar]
- 45.Hoek KS, Goding CR. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res 2010;23:746–59 [DOI] [PubMed] [Google Scholar]
- 46.Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352:189–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gopal YN, Rizos H, Chen G, Deng W, Frederick DT, Cooper ZA, et al. Inhibition of mTORC½ overcomes resistance to MAPK pathway inhibitors mediated by PGC1alpha and oxidative phosphorylation in melanoma. Cancer research 2014;74:7037–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer cell 2013;23:302–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konieczkowski DJ, Johannessen CM, Abudayyeh O, Kim JW, Cooper ZA, Piris A, et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer discovery 2014;4:816–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith MP, Brunton H, Rowling EJ, Ferguson J, Arozarena I, Miskolczi Z, et al. Inhibiting Drivers of Non-mutational Drug Tolerance Is a Salvage Strategy for Targeted Melanoma Therapy. Cancer cell 2016;29:270–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawakami A, Fisher DE. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab Invest 2017;97:649–56 [DOI] [PubMed] [Google Scholar]
- 52.Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med 2014;371:1867–76 [DOI] [PubMed] [Google Scholar]
- 53.Long GV, Fung C, Menzies AM, Pupo GM, Carlino MS, Hyman J, et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nature communications 2014;5:5694. [DOI] [PubMed] [Google Scholar]
- 54.Grob JJ, Amonkar MM, Karaszewska B, Schachter J, Dummer R, Mackiewicz A, et al. Comparison of dabrafenib and trametinib combination therapy with vemurafenib monotherapy on health-related quality of life in patients with unresectable or metastatic cutaneous BRAF Val600-mutation-positive melanoma (COMBI-v): results of a phase 3, open-label, randomised trial. The Lancet Oncology 2015;16:1389–98 [DOI] [PubMed] [Google Scholar]
- 55.Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell 2015;162:1271–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roesch A Tumor heterogeneity and plasticity as elusive drivers for resistance to MAPK pathway inhibition in melanoma. Oncogene 2015;34:2951–7 [DOI] [PubMed] [Google Scholar]
- 57.Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 2014;508:118–22 [DOI] [PubMed] [Google Scholar]
- 58.Miyashita A, Fukushima S, Nakahara S, Yamashita J, Tokuzumi A, Aoi J, et al. Investigation of FOXM1 as a Potential New Target for Melanoma. PloS one 2015;10:e0144241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nature reviews Clinical oncology 2017;14:463–82 [DOI] [PubMed] [Google Scholar]
- 60.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015;162:974–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu H, Sui X, Li X, Li Y. Lyoniresinol inhibits melanogenic activity through the induction of microphthalmia-associated transcription factor and extracellular receptor kinase activation. Mol Cell Biochem 2013;373:211–6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural data that support the findings of this study have been deposited in PDB with the accession code 5KJ2, and microarray data have been deposited in GEO with the accession code