Abstract
Bacteriophage, also called phage, is a human-safe bacteria-specific virus. It is a monodisperse biological nanostructure made of proteins (forming the outside surface) and nucleic acids (encased in the protein capsid). Among different types of phages, filamentous phages have received great attention in tissue regeneration research due to their unique nanofiber-like morphology. They can be produced in an error-free format, self-assemble into ordered scaffolds, display multiple signaling peptides site-specifically, and serve as a platform for identifying novel signaling or homing peptides. They can direct stem cell differentiation into specific cell types when they are organized into proper patterns or display suitable peptides. These unusual features have allowed scientists to employ them to regenerate a variety of tissues, including bone, nerves, cartilage, skin, and heart. This review will summarize the progress in the field of phage-based tissue regeneration and the future directions in this field.
Keywords: phage, tissue regeneration, nanofibers, stem cells
1. Introduction
1.1. Tissue regeneration
Tissue regeneration is the restoration of injured tissue to normal state with full functions [1–7]. Most tissues are composed of specialized (differentiated) cells and their supporting matrix, called extra cellular matrix (ECM) [8, 9]. In daily tissue maintenance activities or small injury repairs, the cells in the lost/damaged tissue can be replaced by the proliferation of surrounding undamaged counterparts, including adult stem cells (ASCs) and/or differentiated normal cells [10, 11]. However, for non-self-healable extensive damages and large defects, special regeneration therapies are usually required, including the use of supporting scaffolds (e.g. artificial polymers, decellularized tissues, and naturally occurring biomacromolecules) with or without bioactive molecules (e.g. growth factors, peptides, cytokines, and genes), tissue-specific therapeutic cells (e.g. implanted stem cells, and endogenous stem cells), and their combinations [1, 10]. Scaffolds are mostly made of biodegradable materials to mimic the native ECM. They provide 3D environments to support the proliferation and differentiation of therapeutic cells in vitro and in vivo, go through gradual biodegradation after implantation, and finally, are replaced by the newly formed connective tissue framework or ECM [10–13]. Recent findings have even demonstrated that scaffolds with special patterns could direct some of the cellular behaviors, such as seeding and differentiation. Bioactive molecules are small biomolecules capable of regulating some crucial processes to promote the regeneration, such as cell differentiation, cell proliferation, cell migration, vascularization, and so on [14, 15]. They could be introduced into the scaffolds by various methods, including direct loading, chemical conjugation, cellular expression, and are expected to function over an extended period of time [14]. Therapeutic cells play a central role in the tissue regeneration. Differentiated cells with proliferative capacity have been approved by FDA, such as autologous fibroblasts, and used for the regeneration of labile and quiescent tissues (e.g. skin and liver) which are composed of cells that can proliferate in response to injuries [11]. However, most tissue cells are incapable of proliferation, especially in non-dividing tissues (e.g. cardiac). In addition, the source of autologous differentiated cells with proliferative capacity is very limited. With the expanding of our knowledge of ASCs, these cells have been considered as a better choice for the tissue regeneration purpose for several reasons: (1) ASCs can repair the nearby damaged tissue and some small tissue injuries can be self-healed by the endogenous ASCs; (2) ASCs are self-renewable; (3) Autologous ASCs can be isolated from patients and culture-expanded in vitro; (4) ASCs are multipotent and can be differentiated into many tissue-specific cell types [16, 17].
Although current tissue regeneration therapies have shown promising results, there are still several challenges in developing them into efficient regenerative medicines. The first challenge is how to produce bioactive molecules, particularly the growth factors, in large scale with lower costs. Most growth factors used for tissue regeneration are recombinant polypeptides expressed by E. Coli or mammalian cells from commercial sources [18]. However, the high production costs (~$40/µg on the market) and short half-lives limit their broad clinical applications for tissue regeneration. The second challenge is how to improve the integration of growth factors into the scaffolds to ensure their long-term functionalities. Current strategies for incorporating growth factors into the scaffolds include direct loading, chemical conjugation, and cellular expression. Growth factors can be directly loaded into the scaffolds and released by diffusion, but the releasing period is too short for the long term regeneration process [14, 15]. Growth factors can also be chemically conjugated onto the scaffolds and released as the scaffolds are being degraded, but the chemical reactions may deactivate their regular bio-functions [14, 15]. Alternatively, growth factors can be expressed by genetically modified therapeutic stem cells, but the efficiency of the gene transfection into stem cells by non-viral vectors is low [14, 15]. The third challenge is how to regulate ASC function and control their fate in the scaffolds. Stem cell survival, self-renewal, and differentiation in vivo are regulated by their local microenvironments [12], also called a niche, including cell-cell interactions, growth factors, and cell-matrix adhesions [13]. For the stem cells seeded inside the scaffolds, they are controlled by the formed specialized niches from the scaffolds and/or bioactive molecules [13]. However, a systematic understanding of the regulation of ASC fate has not been achieved. ASCs without proper control may lead to serious safety issues [19]. For example, implanted neural stem cells induced brain tumor to an ataxia telangiectasia patient four years after implantation [20]. Ideally, the implanted scaffolds carrying bioactive molecules mimic the natural extracellular matrix (ECM) of tissues and thus regulate the therapeutic stem cells to promote tissue regeneration process [1, 10]. This review is focused on the use of genetically modifiable filamentous bacteriophage (phage) bio-nanofibers and phage biopanning identified functional peptides to overcome these challenges in the field of tissue regeneration.
1.2. Phage and phage display
1.2.1. Classification
Phages are a type of virus that infects and replicates within their host bacteria. They are categorized into three major branches including lytic phage, temperate phage, and non-lytic phage. Specifically, a wide variety of phage types exists, such as T4 phage (lytic phage), T7 phage (lytic phage), P22 phage (lytic phage), λ phage (temperate phage), M13 phage (non-lytic phage), and fd phage (non-lytic phage) [21]. Lytic phages lyse the bacteria to release harmful endotoxins and may cause safety issues [22]. Temperate phages integrate their DNA into the bacterial chromosome and may increase pathogenic risk [23]. Non-lytic phages pose minimum safety concerns and have been mostly used for biomedical applications [21, 24]. To the best of our knowledge, non-lytic phages, especially the M13 and fd filamentous phages [25], are almost the only reported phages for tissue regeneration purpose [24, 26]. We will focus on the use of bioengineered filamentous phage for tissue regeneration in this review.
1.2.2. Phage structure
Filamentous phages are composed of a circular single-stranded DNA (ssDNA) packed inside a coat protein shell [25, 26]. With a unique fiber-like shape, they can be pictured as a semi-flexible bio-nanofiber with ~900 nm in length and ~6.5 nm in width [21, 24]. The circular ssDNA encodes 10 phage genes including the 5 coat proteins (named pIII, pVI, pVII, pVIII, and pIX) [25]. The coat protein shell is composed of a sidewall assembled from several thousand copies (~2700 for wild-type M13 phage [27, 28], and ~4000 for wild type fd phage [29]) of highly ordered major coat protein (pVIII) capped with 5 copies of two minor coat proteins (pVII and pIX) at one end and two other minor proteins (pIII and pVI) at the other end.[25] Since the length of a phage depends on the length of the ssDNA within, bioengineered phages with genetically modified ssDNA usually show different lengths [26].
1.2.3. Phage display
Phage display was first reported by George Smith in 1985 [30]. This technique is the genetic modification of phage coat proteins to display foreign peptides by inserting DNA sequences encoding the peptides into the genes of the coat proteins. Peptides have been successfully fused to the N-terminus of pIII (called pIII display) [31], the N-terminus of pVI (called pVI display) [32], and the N-terminus of pVIII (called pVIII display) [29, 33]. Besides these single displays, two or even three different peptides have been displayed on single phages to form double-displayed (pIII & pVIII) [34] and triple-displayed (pIII, pVI, and pVIII) phages [35]. By displaying various functional peptides at two tips or along the side-wall, the surface chemistry of phage nanofibers could be precisely controlled to form multi-functional phage bio-nanofibers.
1.2.4. Phage biopanning
By displaying peptides on coat proteins, phage-displayed random peptide libraries that contain billions of phage nanofibers with each phage displaying a unique peptide sequence could be constructed [29]. There are two types of reported phage libraries: pIII (available from NEB Inc.) [36] and pVIII phage library [37]. These phage libraries have been successfully used to select the peptides that specifically bind to the selection targets, including inorganic crystals [36], polymers [38], proteins [29], cells [39], and tissues [40, 41]. The process of the selections is known as phage biopanning. During a typical phage biopanning, a phage library is allowed to interact with the targets. The non-bound phage particles are washed away with a detergent added buffer. The target-bound phage particles are eluted with an acidic buffer (pH = ~2.2) and amplified by infecting host bacteria. The amplified phages serve as a new enriched pool (or sub-library) with significantly reduced diversity to interact with targets again. This selection process is repeated for several rounds until a few peptide sequences are dominant in the output and they are regarded as target specific peptides. The phage biopanning identified target-binding functional peptides/phages have been extensively used for tissue regeneration purpose (Table 1).
Table 1:
Summary of phage display derived functional peptides and bioengineered phages for tissue engineering.
| Functional peptide/protein identified from phage biopanning |
Functions and/or applications | References |
|---|---|---|
| NPYHPTIPQSVH | HAP-binding & HAP-nucleating | [42] |
| SVSVGMKPSPRP | HAP-binding | [43, 44] |
| MLPHHGA | HAP-nucleating | [45, 46] |
| VTKHLNQISQSY | Apatite-binding | [47, 48] |
| DPIYALSWSGMA | human bone marrow stromal cell-binding | [49] |
| TRIP-1 protein | TRAP activation and inducing osteoblast differentiation | [50] |
| GGGAWEAFSSLSGSRVG | BMP-2 binding and inducing osteogenesis | [51] |
| TSPHVPYGGGS | BMP-2 binding and inducing osteogenesis | [52, 53] |
| GCPFSSTKTE | bone marrow-homing, hematopoietic stem cell-binding, and inducing osteogenic differentiation |
[54, 55] |
| SGHQLLLNKMPN | MSC-binding for enhanced gene delivery into MSCs | [56, 57] |
| VTAMEPGQ | MSC-binding for enhanced gene delivery into MSCs | [39, 58, 59] |
| RRQTLSHQMRRP | bind to the 1–35 residues of the Nogo-66 enhance neuronal neurite outgrowth on CNS myelin substrates, and promote CNS regeneration after spinal cord injury (SCI) |
[60, 61] |
| HIYTALV | NgR1-binding promote axonal regeneration |
[62] |
| CGLPYSSVC | murine NSC-targeting | [63] |
| FAQRVPP | NSC-derived NPC-targeting promote the proliferation and neural differentiation of human and mouse NSCs in vitro and improve neural tissue regeneration in acute contusive spinal cord injury in vivo |
[64] |
| KLPGWSG | murine NSC-targeting enhance NSC neuronal differentiation |
[65] |
| HGEVPRFHAVHL | specific to the NSCs derived from rhesus monkey NSC labeling |
[66] |
| PFSSTKT | bone marrow-homing and primitive hematopoietic stem cell- binding induce NSC differentiation and promote neural regeneration |
[67–71] |
| QTRFLLH & VPTQSSG | neural precursor cell-targeting enhance the gene delivery into neural precursor cells in vitro and in vivo |
[72] |
| CLEVSRKNC | ischemic stroke tissue-homing | [73] |
| HSNGLPL | TGF-β1-binding retain TGF-β1 to induce the chondrogenic differentiation of human MSCs in vitro and promote the regeneration of articular cartilage in a chondral defect in a rabbit model with or without additional TGF-β1 growth factor |
[74–76] |
| WYRGRL | collagen II α 1-binding articular cartilage-homing |
[76–78] |
| EPLQLKM | bone marraw derived MSC-targeting promote the proliferation and chondrogenic differentiation of MSCs in vitro and in vivo |
[79–83] |
| GAHWQFNALTVR | Hyaluronan (HA)-binding retain HA and promote the proliferation and chondrogenic differentiation of MSCs |
[78, 84, 85] |
| DWRVIIPPRPSA | chondrocyte-homing serve as a chondrocyte targeting probe for cartilage regeneration gene therapies |
[86, 87] |
| STMMSRSHKTRSHHV | hyaluronan-binding block the RHAMM/HA signaling to reduce the wound macrophage number, fibroblast number, and blood vessel density and thus promote the scarless healing of skin |
[88] |
| CFKHSSHQC & SQIDYATGPRQA |
wound-healing matrix (Integra®)-binding introduce drugs into the matrix for improved performance |
[89] |
| CARSKNKDC | target skin and tendon wound-homing favor early stages of wound healing |
[90, 91] |
| ACHSALTKHCG | skin penetrating and cell entering penetrate across stratum corneum into epidermis and dermis layers and into cells in skin tissue |
[92–96] |
| CGLLPVGRPDRNVWRWLC & CKGQCDRFKGLPEWC |
bind to the TGF-β type II receptor block the interaction between TGF-β1 and TGF-β type II receptor induce the VSMC migration and affect the expression of TGF- β1 related proteins |
[97] |
| QPFTTSLTPPAR | mouse ESC-derived cardiomyocyte-targeting | [98] |
| APWHLSSQYSRT | cardiac-homing | [99] |
| CSTSMLKAC | home to ischemic heart tissue | [100] |
| KLWVLPK | vascular wall-binding home to injured carotid arteries |
[101, 102] |
| TVKHRPDALHPQ & LTTAPKLPKVTR |
human ESC-binding | [103] |
| LTGKNFPMFHRN & MHRMPSFLPTTL |
bind to the extracellular domain of TGF-β receptor I and II concentrate endogenous TGF-β and regulate TGF-β responsive genes to induce the epithelial to mesenchymal transition, which has potential applications in tissue regeneration |
[104, 105] |
|
Bioengineered phage as the substrate/scaffold component for bone regeneration |
Applications | |
| Wild type phage | Form the HAP-phage hybrid bundle structure | [106] |
| Phage displaying EEEEEEEE | Form the HAP-phage hybrid bundle structure | [107] |
| Phage displaying ESQES Phage displaying QESQSEQDS |
Form the HAP-phage hybrid bundle structure | [28] |
| Phage displaying RGD | Enhance cell adhesion | [108–110] |
| Phage displaying GRGDS Phage displaying DDYD |
A mixture of both phages and alginate forms hydrogels in the presence of calcium ion. Enhance cell (MC3T3-E1) viability, proliferation, and osteogenesis |
[111] |
| Phage displaying DGEA | Support the adhesion, proliferation, spreading, and induce the early osteogenic differentiation of the bone progenitor cells (MC3T3) |
[112] |
| Phage displaying an OCN- derived peptide (PDPLEPRREVCE) Phage displaying an OGP- derived peptide (YGFGG) |
Phage ridge/groove films enhance the MSC elongation. Phage ridge/groove films accelerate and enhance the osteogenic differentiation of MSCs in osteogenic medium. |
[27] |
| Phage displaying a fibronectin-derived adhesive peptide (PHSRN) Phage displaying a |
Phage ridge/groove films greatly favored the proliferation of MSCs. Phage ridge/groove films assembled from a mixture of RGD- phage and PHSRN-phage promote the osteogenic |
[113] |
| fibronectin-derived adhesive peptide (RGD) |
differentiation of MSCs in the primary media without osteogenic supplements. |
|
| Phage displaying the OGP (ALKRQGRTLYGFGG) Phage displaying a BMP-2- derived peptide (KIPKASSVPTELSAISTLYL) |
Phage ridge/groove films control the morphology of iPSCs Phage ridge/groove films induce the osteoblastic differentiation of iPSCs |
[114] |
| Phage displaying RGD | Phage functionalized 3D scaffolds could induce both osteogenesis and angiogenesis for vascularized bone regeneration in vivo. |
[115] |
| Phages displaying RGD, IKVAV, and HPQ |
RGD-phage formed films could direct the elongation of cell growth. Films made of a mixture of HPQ-phage and RGD-phage could facilitate the immobilization of growth factors/ligands and direct orientation of the growth of NPCs. Films made of a mixture of IKVAV-phage and RGD-phage could support the proliferation and differentiation of NPCs and control their morphology and growth. |
[116–118] |
| Phage displaying RGD | promote the proliferation and myogenic differentiation of CAC12 myoblasts in the presence of graphene oxide |
[119] |
| Wild type phage Phage displaying RGD |
Phages form films to induce the directional growth of human NIH-3T3 fibroblasts |
[120–125] |
1.2.5. Phage properties
Filamentous phages are bio-nanofibers with the modifiable surface [24]. They can be considered as an excellent biomaterial for biomedical applications for several reasons: (1) More than one foreign peptides can be genetically displayed on the surface of a single phage through phage display technique to construct multi-functional phage bio-nanofibers. (2) The phage-based peptide library is an ideal tool to select target specific peptides through phage biopanning process. (3) Now it is known that phages are naturally present in human body, in particular, in human microbiota and bladder [126, 127], indicating that in vivo use of phages is safe to the human body. Several groups have injected phage libraries (up to 1014 phage particles into one patient) into patients and conducted in vivo biopanning to identify human tissue-specific peptides without obvious side effects, suggesting its good tolerability in human beings [103, 128–130]. (4) Phages are stable under physiological conditions for hours so that they could even retain their infectivity during intravenous (IV) injection and circulation in the human body for in vivo biopanning or biodistribution studies [103, 128–130]. (5) Due to the specificity of infection, error-free nature of replication, and easy procedure of production, phage nanofibers could be manufactured in large scale to form the white macroscopic pellets with high cost effectiveness [24, 131] and easily quantified by counting the macroscopic phage plaques which are the clear zones formed in a lawn of bacteria due to bacterial infection by the phages. (6) The capability of forming liquid crystalline structures at certain concentrations makes phages a great biomaterial to self-assemble into highly ordered 3D scaffolds.
2. Roles of phages in tissue regeneration
2.1. Advantages
Recent research has demonstrated that filamentous phages could be genetically engineered to promote tissue regeneration [21, 24, 132–135] in four different ways or their combinations (Figure 1): (i) Bioengineered phages displaying functional peptides serve as growth factor alternatives to improve tissue regeneration; (ii) Bioengineered phages self-assemble into 3D scaffolds to regulate the ASCs by providing both physical and chemical cues; (iii) ASC-binding peptides selected from a phage-displayed random peptide library are used as targeting molecules to enhance ASC gene/drug delivery so that the transfected ASCs could continually express growth factors over a long period of time after implantation; (iv) Phage biopanning identified functional peptides could be conjugated with biomaterials to form scaffolds for tissue regeneration. In addition, filamentous phage nanofibers could be manufactured in large scale using fermentation systems [131] with low costs to support their increasing applications for tissue regeneration.
Figure 1.
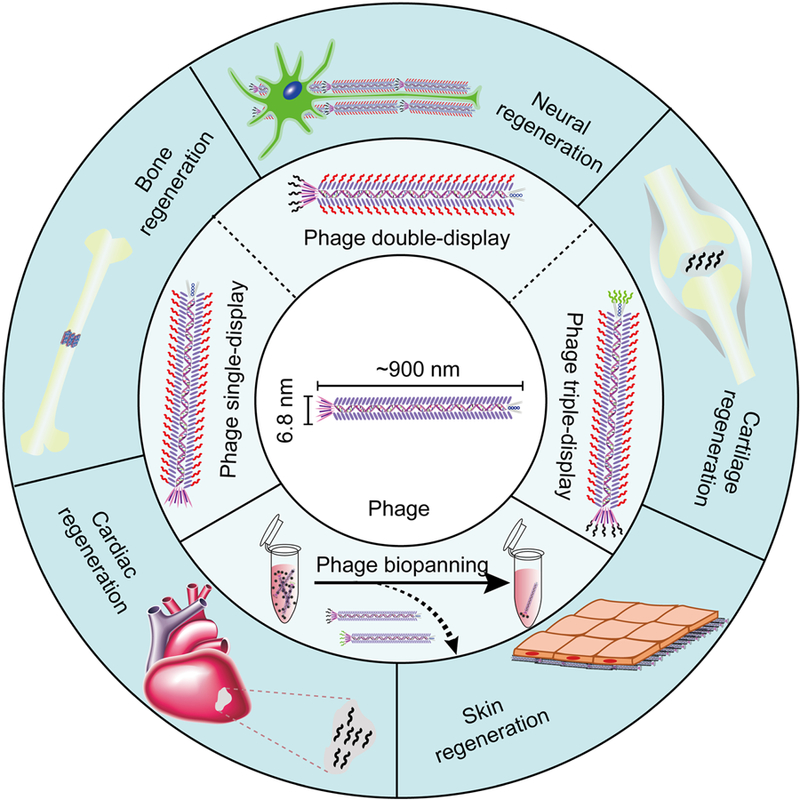
Overview of phage- and phage display-based tissue regeneration strategies. Phage biopanning enables the identification of regeneration associated functional phages/peptides. The direct or indirect use of the functional peptides and bioengineered phages could promote the regeneration of a variety of tissues, including bone, nerves, cartilage, skin, and heart.
2.2. Function as growth factor alternatives
Growth factors, the signaling small proteins/polypeptides capable of stimulating and regulating cellular proliferation, differentiation, migration, adhesion, and gene expression [136–138], play a critical role in promoting tissue repair processes. They have even been considered as one of the most important components (cells, scaffold, and growth factors) in tissue engineering [139]. In most current tissue regeneration strategies, growth factors are incorporated inside the scaffolds and gradually released during the regeneration process. The currently used growth factors are commercial recombinant exogenous growth factors produced through the cell-based or E. Coli-based recombinant technologies, but the production and purification costs are still high (about $4000-$6000/mg) and their half-lives are short (up to several hours) [15]. During the long period of tissue regeneration, a large amount of growth factors will be needed for clinical treatments and the total costs will be extremely high. Growth factors initiate their functions by binding to the specific cell surface receptors through the interaction between the receptor-binding domain of growth factors and the extracellular domain of the receptors. Besides the growth factors, cellular functional signaling can also be activated by growth factor alternatives, such as functional peptides [140]. For examples, RDG peptide interacts with integrins to promote cell adhesion and differentiation [141–144]. Osteogenic growth peptide (OGP) with a sequence of YGFGG could enhance cell adhesion and proliferation with low concentrations [145, 146]. Some other functional peptides that can enhance tissue regeneration have been summarized in a recent review article.[140] Recently, several peptides that can induce the differentiation of stem cells and enhance the tissue regeneration have even been identified directly by phage display technique [50, 54, 55, 68, 69, 147].
Although these functional peptides were mostly studied in the form of synthetic molecules, they could also be used as molecules displayed on the phage nanofibers [27, 113–115, 134]. Taking advantage of the phage display technologies, several groups have recently displayed functional peptides on the phage protein shell to form bioengineered phage nanofibers and demonstrated that these phage nanofibers could regulate ASC proliferation, differentiation, migration, adhesion, and gene expression [27, 112–117, 134]. They also found that when displayed on the phage surface, the functional peptides (e.g., RGD [120–125]) still retain the same biological functions for cell adhesion [27, 112–117, 134]. The advantages of using phage-based functional peptides as growth factor alternatives for tissue regeneration include: (1) Bioengineered phage nanofibers can be easily manufactured in large scale with high-cost effectiveness; (2) More than one peptide can be displayed on one single phage nanofiber using the phage double or triple display techniques so that displayed peptides could function collectively to achieve better regeneration efficiency; (3) By co-displaying both the functional peptide and scaffold material-binding peptide on phage surface, these bioengineered phage nanofibers could be conveniently introduced into the scaffolds without any chemical reactions. A potential problem of using phage nanofibers as growth factor alternatives include: (1) Filamentous phages may activate a range of immune responses [148]. However, the previous work has also demonstrated that the injection of phages didn’t cause serious side effects to the patients [128]. (2) Phages may be eliminated shortly (in 1 hour) after implantation in vivo by B cells, based on a study of the in vivo fate of T7 phages in mice [149]. However, one previous study has found that bioengineered M13 phages decorating the inside of a 3D printed bone scaffold successfully induced vascularized osteogenesis in diaphysis defect Sprague Dawley (SD) rat models [115]. Studies in the in vivo fate of bioengineered filamentous phages are still needed to further evaluate the direct use of phage in tissue regeneration.
2.3. Function as scaffolds
Scaffolds play a central role in tissue regeneration. An ideal scaffold should have good biocompatibility to support and control the proliferation and differentiation of ASCs on them, a unique architecture to load growth factors before implantation and release them during the regeneration, an appropriate porosity to allow ASCs and other molecules to reach the inside of the scaffold, a good wettability for the attachment, ingrowth, and proliferation of the cells, tissue-mimetic mechanical properties, and great biodegradability in accordance with the regeneration [150, 151]. The mostly used three types of biomaterials for constructing tissue regeneration scaffolds are ceramics (e.g. hydroxyapatite and tricalcium phosphate), synthetic polymers (e.g. poly-dl-lactic-co-glycolic acid and poly-l-lactic acid), and natural macromolecules (e.g. collagen fibers) [150]. Due to their inherent limitations [150], combinations of these materials have recently been used to build composite scaffolds, such as collagen-hydroxyapatite composite scaffolds for bone regeneration [152]. During or after the construction of these scaffolds, bioactive molecules, such as growth factors, are loaded or functionalized into the scaffolds through diffusion or chemical reactions [15]. As a non-toxic fiber-like natural macromolecule with a genetically modifiable surface [24, 103, 128–130], genetically engineered phage nanofibers that display functional peptides can not only self-assemble into scaffolds to support ASC proliferation and differentiation [27] but also decorate pre-formed scaffolds to introduce bioactive molecules for tissue regeneration [115]. Since the phage surface is actually a protein shell, it can be mixed or conjugated with a material or a chemical (including polymers) that does not denature proteins. Although the phages themselves can’t work as the main building block for generating 3D structures for hard tissue regeneration (e.g., bone) due to its softness, bioengineered phages can serve as an important component of regeneration scaffolds with two obvious advantages. First, the engineered phages display peptides may replace the traditional bioactive molecules, such as expensive growth factors, in decorating the scaffolds without any chemical conjugation [115]; Second, the scaffolds assembled from such engineered phage nanofibers is an ideal tool to systemically study how the physical and chemical cues collectively affect the ASC behaviors [27].
2.4. Improve gene/drug delivery
One strategy to introduce growth factors into regeneration scaffolds is to deliver the growth factor gene into the therapeutic ASCs, then implant ASCs into scaffolds, and finally allow the genetically modified ASCs to continually express the growth factors during regeneration. Although growth factor genes can be delivered into ASCs using viral vectors with a high transfection efficiency, there are still concerns about using the viral vectors for patient treatments. On the other hand, the transfection efficiencies using non-viral gene delivery vectors to stem cells are still low and need to be improved. To promote gene transfection efficiencie s into ASCs, phage biopanning technique has been used to identify ASC-binding peptides which in turn are conjugated onto non-viral gene delivery vectors to achieve targeted gene transfection [39, 58, 59].
2.5. Phage biopanning for tissue regeneration
In vitro phage biopanning has led to the discovery of peptide ligands that can target the tissue regeneration associated proteins and stem-cell targeting peptides that are capable of guiding stem cell adhesion, proliferation, and differentiation in scaffolds [153]. These peptides are usually functionalized onto the tissue regeneration scaffolds to enhance the in vitro therapeutic stem cell expansion and control the stem cell behaviors within the scaffolds [153]. For example, when the self-assembled monolayer (SAM) of alkanethiols on gold was functionalized to display the human embryonic stem cell (hESCs)-targeting peptides identified through phage display technique, it could support the adhesion and growth of the undifferentiated hESCs similar to the Matrigel [154]. In vivo phage biopanning has been used to identify organ/tissue-targeting peptides, including the injured organ targeting peptides (blood vessel) [155, 156]. These peptides could be integrated into injectable nanomedicines to achieve targeted drug delivery to target tissues [155, 156].
3. Phages for promoting tissue regeneration
3.1. Bone regeneration
For bone regeneration, the mostly used therapeutic stem cells are mesenchymal stem cells (MSCs) [157–159], preosteoblasts [111, 112], and recently developed induced pluripotent stem cells (iPSCs) [160–162]. The behaviors of the therapeutic stem cells can be controlled by growth factors, and/or scaffolds to achieve osteogenic differentiation during the regeneration process [163, 164]. Hydroxyapatite (HAP) is the main inorganic component of natural bones. Bone scaffold containing HAP shows good osteoconductivity and osteoinductivity [165–169], so HAP has been widely used in the development of bone regeneration scaffolds [170, 171]. Phage and phage display technique has been used for bone regeneration in the following directions: (1) Phage display derived functional peptides could be integrated into scaffolds to enhance the bone regeneration; (2) Bioengineered phage nanofibers could be used as a biomaterial component/substrate of the bone scaffolds; (3) Phage display derived peptides could promote the gene delivery efficiency into MSCs to form genetically modified MSCs for promoting tissue regeneration.
3.1.1. Phage display derived functional peptides for bone regeneration
HAP (Ca10(PO4)6(OH)2), the main inorganic component of bone, has been widely used for developing bone regeneration scaffolds [170, 171]. The availability of HAP-binding peptides could facilitate the introducing of HAP into bone scaffolds for better osteoconductivity and osteoinductivity [165–169]. Several groups have successfully identified HAP-binding peptides using the phage biopanning technique [42–49]. Lee et al. screened Ph.D.-12, Ph.D.-7, and Ph.D.-C7C phage libraries (from New England Biolabs) against the (001) faces of single-crystal HAP rods and identified three potential HAP-binding peptides, including NPYHPTIPQSVH (CLP12) from Ph.D.-12 library, HAPVQPQ (CLP7) from Ph.D.-7 library, and CNYPTLKSC (CLP7C) from Ph.D.-C7C library [42]. During the following tests, the CLP7C peptide showed the best HAP-binding affinity and CLP12 successfully induced the nucleation of HAP crystals from a precursor Ca2+/HPO42- solution. The periodic proline and hydroxyl residues (Y, T, and S) of the CLP12 peptide mimic the type I collagen repeated sequence and also match the periodic lattice of the HAP (001) face. These findings suggest that the binding of the CLP12 peptide to HAP (001) face depends on collective noncovalent interactions and native collagen may also have the HAP-nucleating ability. Backer et al. identified a HAP-binding peptide, SVSVGMKPSPRP (HA 6–1), through six rounds of phage display selection using the Ph.D.-12 phage library [43]. The HA 6–1 peptide showed both strong binding affinity and specificity to HAP crystals. Then the same group measured the binding affinity of the HA 6–1 (SVSVGMKPSPRPGGGK, termed HA-1) peptide and its derived peptide fragments, SVSVGGK (HA-2) and VSMKPSPGGGK (HA-3), to HAP crystals by using the surface plasmon resonance imaging technique [44]. The results showed that the binding affinity of the HA-1 peptide to HAP was relatively high (KD = 14.1 µM ± 3.8 µM) but those of the HA-2 (KD = 54.4 µM ± 7.6 µM) and HA-3 peptides (−) were low, suggesting the fragment SVSV contributed to the interaction of HA 6–1 and fragment MKPSP may function as a conformational dependent component for enhancing the binding affinity. Sarikaya et al. identified a HAP-binding peptide, CMLPHHGAC (HABP1), through phage biopanning using the Ph.D.-C7C phage library [45]. The HABP1 peptide could regulate the nucleation process of HAP crystals and the addition of HABP1 peptide in a precursor solution resulted in a slower nucleation process and larger HAP crystals. The same group [46] combined a self-assembly peptide, VKVKVKVKVDPPTKVEVKVKV (MAX8), with the HABP1 peptide form a dual functional peptide, VKVKVKVKVDPPTKVEVKVKV-MLPHHGA (MDG1). The MDG1 peptide could form hydrogels through the self-assembly of the MAX 8 fragment. The HAP-nucleating HABP1 fragment of the MDG1 peptide could direct the mineralization of HAP crystals on the formed hydrogel to form a HAP-hydrogel 3D network capable of supporting the cementoblasts (OCCM-30 cells). Kohn et al. identified three apatite-binding peptides, APWHLSSQYSRT (A), STLPIPHEFSRE (S) and VTKHLNQISQSY (VTK), through phage biopanning using the Ph.D.-12 phage library [47]. The phosphorylation of the two serine amino acids on the VTK peptide further enhanced the binding affinity of VTK to bone-like mineral [48]. The same group [49] identified a human bone marrow stromal cell (hBMSC)-specific peptide, DPIYALSWSGMA (DPI), through phage biopanning using the Ph.D.-12 phage library and combined the HAP-binding VTK and hBMSC-specific DPI peptides into a dual functional peptide, DPIYALSWSGMA-VTKHLNQISQSY (DPI-VTK). They found that the DPI-VTK peptide could be used to coat apatite substrates and thus improve hBMSC adhesion, spreading, proliferation and differentiation on the apatite substrates. All these results have demonstrated that phage display technique is a powerful tool to identify HAP-binding/nucleating peptide that can be decorated onto scaffolds/substrates for bone regeneration applications.
Bioactive small proteins such as BMP-2 play an important role in the bone regeneration process.[172] The peptides capable of specifically binding to these proteins can be identified by phage biopanning technique [50, 51, 53]. These peptides could regulate the functions of their targeting proteins [50] or help the immobilization of the proteins onto artificial bone scaffolds for tissue regeneration [51, 53]. Tartrate-resistant acid phosphatase (type V) (TRAP), a glycosylated monomeric metalloprotein enzyme, is critical for bone formation and development. Puzas et al. identified a TRAP-binding protein, TGFβ receptor interacting protein 1 (TRIP-1), from the osteoblasts by using an osteoblast cDNA T7 phage-display library [50]. They found that the TRAP could interact with the TRIP-1, activate the TGFβ differentiation pathway, and trigger the osteoblast differentiation process. Hodges et al. first selected 59 BMP-2-binding peptides from a 17-mer phage display library and identified two peptide patterns (W-X-X-F-X-X-L and L-X-F-P-L-K) that may contribute to the binding of peptides to BMP-2 [51]. Based on the two patterns, they built two 2nd generation focused phage display libraries and further selected two better BMP-2-binding peptides, GGGAWEAFSSLSGSRVG and GGALGFPLKGEVVEGWA, using the two libraries. Then, they combined one of the identified BMP-2-binding peptides with the collagen-binding peptide to form a dual functional peptide (SWWGFWNGSAAPVWSR-GSSG-AGAWEAFSSLSGSRV, BC-1) and found that BC-1 peptide could help the immobilization of free standing BMP-2 onto the collagen matrix. Finally, they found the injection of the collagen matrix with BMP-2 and BC-1 resulted in more osteogenic cellular activity, a larger area of bone formed, and better bone maturity than the collagen matrix with BMP-2 (but without BC-1). Stupp et al. identified a BMP-2-binding peptide, TSPHVPYGGGS, through phage biopanning using the Ph.D.-12 phage library [52]. The same group integrated the identified BMP-2-binding peptide with the peptide amphiphile (PA) or diluted PA nanofibers as building blocks (BMP2b-PA or D-BMP2b-PA, Figure 2-A and B) to form gel scaffolds with a high BMP-2-binding affinity for bone regeneration [53]. The in vitro osteogenesis-inducing study results showed that more ALP-active cells were found after treatment of the D-BMP2b-PA gel for four days on C2C12 pre-myoblasts in the presence of BMP-2 (1 and 10 µg/mL) (Figure 2-C), suggesting the D-BMP2b-PA gel captured more BMP-2 molecules and significantly enhanced the BMP-2-induced osteogenesis of C2C12 cells. The in vivo spinal fusion (bone forming) study results indicated that the D-BMP2b-PA gel greatly (10-fold) enhanced spinal fusion with less additional BMP-2 (0.1 or 1 µg) in a rat postero-lateral lumbar intertransverse spinal fusion model (Figure 2-D), indicating that less BMP-2 is needed for osteogenesis at the regeneration sites in the presence of the D-BMP2b-PA gel. All these results suggest that phage display technique is a powerful tool to identify a bioactive molecule-binding peptide that can be used to induce the osteogenic differentiation of therapeutic stem cells for bone regeneration. Moreover, the display and conjugation of such peptides onto the scaffolds will greatly improve their regeneration effects.
Figure 2.
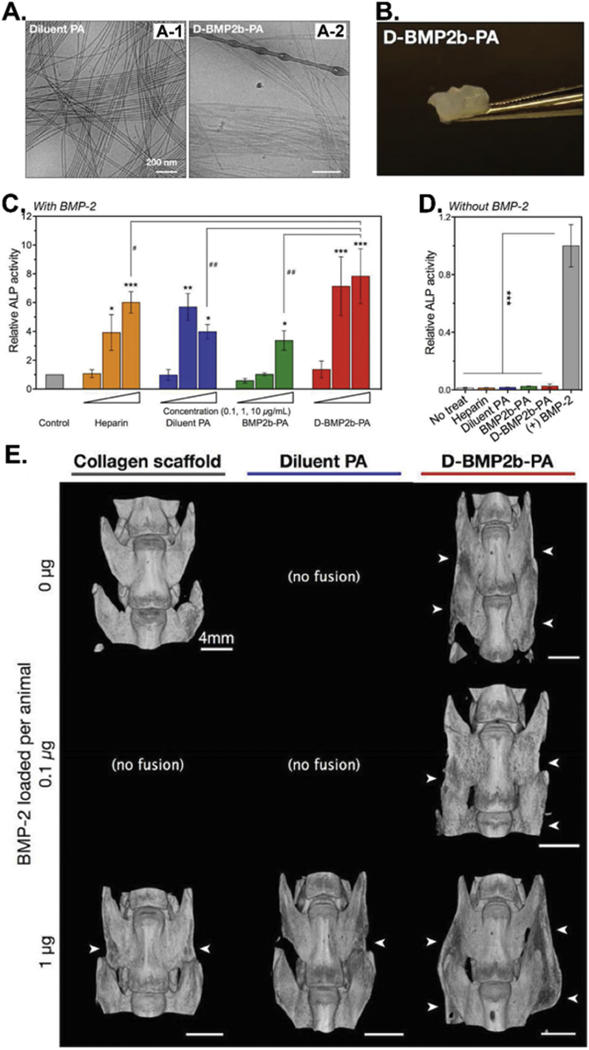
BMP-2-binding peptide functionalized amphiphile (PA) nanofibers (D-BMP2b-PA) as biomaterials for BMP-2 absorption and in vivo spinal fusion (bone forming). (A) Cryo-TEM images of the filamentous morphologies of the diluent PA (A-1) and the diluted BMP-2-binding PA (D-BMP2b-PA) (A-2). (B) Photograph of the self-supporting D-BMP2b-PA gel. (C&D) ALP enzyme activities of the BMP-2-induced osteoblast differentiation of C2C12 pre-myoblasts after 4 days of culturing in the presence (C) or absence (D) of BMP-2 or without BMP-2. Measurements were normalized to their respective DNA content, and the final average values from treatments are normalized to control treatment with BMP-2 alone. The results showed that D-BMP2b-PA gel could capture and retain BMP-2 molecules and significantly enhanced the BMP-2-induced osteogenesis of C2C12 cells in the presence or absence of BMP-2. (E) Micro-CT images of representative spine fusions showing that D-BMP2b-PA efficiently induced the fusion of spine. White arrows indicate the presence of fusion mass in the transverse processes. (Reprinted with permission from ref. [53], copyright 2015, Wiley)
Recently, the peptides that can specifically home to bone marrow have been successfully identified by in vivo phage biopanning and have shown great potential to induce the osteogenesis and heal bone defects.[54, 55, 147] Becker et al. identified a bone marrow-homing and primitive hematopoietic stem cell-binding peptide, GCPFSSTKTE (bone marrow homing peptide 1 or BMHP1), through an in vivo phage biopanning [55]. Zhang et al. found that the BMHP1 peptide and its circular form, cBMHP1, could not only enhance the adhesion and proliferation of bone marrow derived MSCs on peptide-coated quartz substrates but also induce the osteogenic differentiation of the stem cells either in an osteogenic differentiation medium or regular MSC culture medium without osteogenic supplements [54]. Balian et al. identified two bone marrow-homing peptides, L7 and R1, through an in vivo phage biopanning [147]. Mesenchymal cells treated with 5 nM of L7 or R1 showed higher ALP activity (2 fold for L7 and 3.5 folds for R1) and higher gene expression of bone cell transcription factors or bone matrix proteins (higher Osterix and Runx2 gene expression for R1, and higher BSP and osteocalcin gene expression for L7), suggesting that both peptides could enhance the osteogenesis in mesenchymal cell cultures. When the R1 peptide was introduced into the gel foam for bone repair, the best cortical repairs were achieved by using the gel foam-R1, suggesting the R1 peptide can promote cortical bone regeneration. These reported findings have demonstrated that in vivo phage biopanning technique can identify bone marrow-binding peptides that can induce the osteogenesis of therapeutic stem cells in vitro and promote bone regeneration in vivo.
3.1.2. Bioengineered phage nanofibers as a substrate for bone regeneration
Filamentous phages are genetically engineerable bio-nanofibers that mimic the collagen fibrils in natural bone.[25, 106] When displaying functional peptides (e.g. RGD) on the surface, the bioengineered phage bio-nanofibers could serve as a substrate/scaffold component for bone regeneration [27, 28, 106–115]. Our group has demonstrated that bioengineered phage bio-nanofibers could nucleate HAP on them to form HAP-phage bundle structures as a potential bone scaffold building block [27, 28, 106]. They found that positively charged Ca2+ triggered the self-assembly of the negatively charged wild type M13 phage into the Ca2+-loaded phage structures, which could further interact with phosphate and hydroxyl ions to induce the oriented nucleation of HAP along phage bundles to form the HAP-phage hybrid nanostructures [106]. They also used a bioengineered phage bio-nanofiber that surface displays a negatively charged peptide, EEEEEEEE, as a biotemplate to form phage bundles (Figure 3A-1) and found that oriented HAP crystals could be formed along phage bundles (Figure 3A-2). The c-axis direction of the HAP matched the direction of the long axis of phage bundles (Figure 3A-3), which mimics some features of the lowest level of bone structure. Moreover, they co-assembled collagen and phages into hybrid bundles which could also induce the oriented nucleation of HAP on them to form a HAP-collagen-HAP hybrid nanostructure (Figure 3B) [107]; Moreover, they genetically displayed two HAP-nucleating peptides, ESQES (pE) and QESQSEQDS (pQ), derived from the dentin matrix protein-1 on the M13 phage surface to form the E phage (displaying pE) and Q phage (displaying pQ), respectively. The E and Q phage could self-assemble into phage bundles through the formation of beta structure between the pE and pQ that are displayed on the phage surface. The formed beta structure induced the oriented nucleation and growth of HAP crystals along the phage bundles. The c-axis of HAP had the same direction as the long axis of phage bundles, mimicking the collagen-HAP hybrid structures in natural bone [28]. These results suggest that pE and pQ could be collectively decorated onto scaffolds to induce HAP nucleation for potantial bone regeneration applications.
Figure 3.
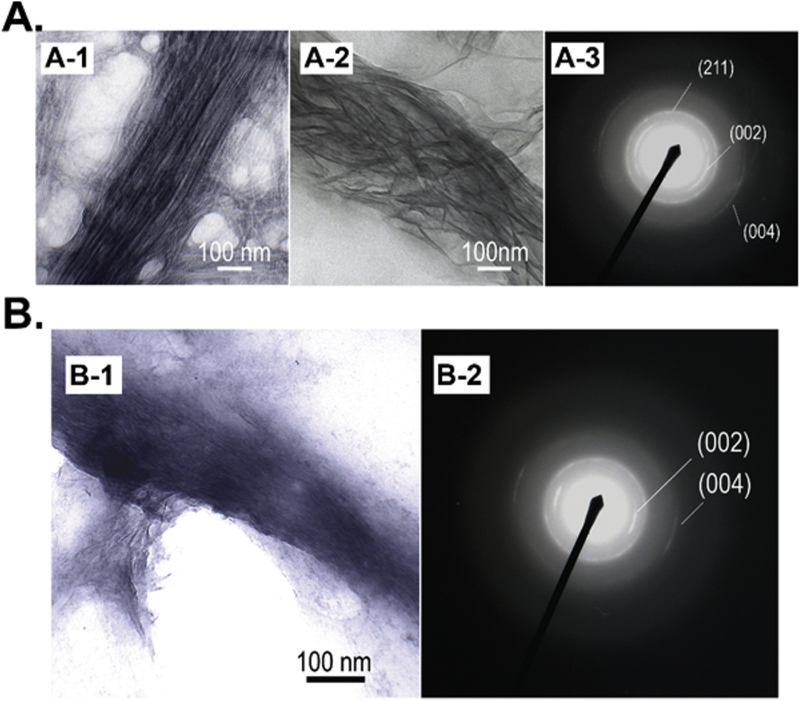
Bioengineered phage bio-nanofibers as biotemplates to enable the oriented nucleation of hydroxyapatite (HAP). (A-1) TEM image of the bioengineered phage (displaying EEEEEEEE peptide) formed bundle structures induced by calcium ions. (A-2) Oriented HAP crystals formed along Ca2+ induced phage bundles by adding phosphate ions. (A-3) The electron diffraction pattern of the oriented HAP crystals (A-2) showing the c-axis direction of the HAP matched the direction of the long axis of phage bundles. (B) Co-assembled collagen-phage hybrid bundles could induce the oriented nucleation of HAP on them. (Reprinted with permission from ref. [107], copyright 2010, Wiley)
Bioengineered phage bio-nanofibers have also been used as a substrate for enhancing bone cell adhesion and controlling their elongation [108–110]. Han et al. genetically displayed a cell-adhesive RGD peptide derived from the proteins in ECM for integrin receptors onto M13 phage surface to form the RGD-phage and then fabricated 3D porous ECM-mimicking nanofiber sheets composed of electrospinning poly(lactic-co-glycolic acid) (PLGA) and RGD-phage bionanofibers [108]. They found that the adhesion and proliferation behaviors of the preosteoblastic MC3T3-E1 cells on the RGD-phage-PLGA nanofiber sheets were significantly enhanced due to the decorated RGD peptides on the phage surface, suggesting the phage-PLGA nanofiber sheet is a great cell-adhesive substrate for bone scaffolds. Lee et al. fabricated smectic helicoidal or nematic hierarchical nanofilament (SHN and NHN) films using the RGD-phage nanofibers through a self-assembly-templating process with different pulling speeds [109]. They found that the SHN-NHN double patterned films made from RGD-phage could elongate the resident preosteoblastic MC3T3-E1 cells perpendicular to the long axis of the phage nanofibers on SHN regions but parallel to them on NHN regions (Figure 4), suggesting that the phage substrate could provide both the physical (pattern) and biochemical (RGD peptide) cues to cells for controlling cell morphology. This method could be used to regenerate the tissue consisting of multiple layers with each layer holding unique cell properties (type, growth direction and so on).
Figure 4.
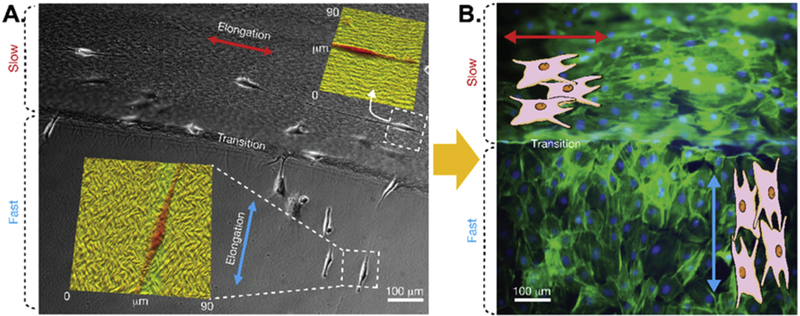
Control over the growth orientation of preosteoblastic MC3T3-E1 cells by bioengineered phage assembled films with different structures. (A) MC3T3-E1 cells cultured on the phage films with SHN (smectic helicoidal nanofilament, top) and NHN (nematic hierarchical nanofilament, bottom) structures. Insets: AFM images showing the directional guidance of cell elongation in response to the orientation of the underlying film microstructure. (B) Fluorescence image showing that the MC-3T3-E1 cells grew in the desired directions on the self-assembled RGD phage films, suggesting that phage assembled substrates could provide physical cues to cells for morphology controls. (Reprinted with permission from ref. [109] copyright 2011, Springer Nature)
Bioengineered phage bio-nanofiber based substrates can provide both the physical and biochemical cues to therapeutic stem cells for inducing their osteogenic differentiation [27, 110, 112–114]. Kim et al. fabricated a 3D multi-layered porous hybrid scaffold with a high modulus (9.4–12.9 MPa) using a mixture of RGD-M13 phage that was pre-conjugated with alginate and poly(ε-caprolactone) (PCL) through a low-temperature printing process [110]. The formed RGD-phage/PCL hybrid scaffold could absorb functional proteins via its pore structure, induce the formation of HAP on it, and support the adhesion and proliferation as well as osteoblastic differentiation of the osteoblast-like MG63 cells, suggesting its potential applications in various hard tissue regeneration. The same group constructed a tissue-mimetic 3D printing cell-laden scaffold using pre-osteoblast MC3T3-E1 cell-containing phage hydrogel as the bioink [111]. The phage hydrogel was made of a mixture of the phages displaying an integrin-binding peptide GRGDS, the phages displaying a calcium-binding peptide DDYD, and alginate cross-linked by calcium ions. The 3D cell-laden scaffold enhanced the cell viability, proliferation, and osteogenesis. Lee et al. genetically displayed a collagen-derived DGEA peptide onto M13 phage surface to form the DGEA-phage and prepared 2D phage films through a self-assembly process [112]. They found that the film derived from DGEA-phage could support the adhesion, proliferation and spreading of the bone progenitor cells (MC3T3) and induce their osteogenic differentiation.
Recently, our group [27, 113, 114] have developed a layer-by-layer (LBL) self-assembly strategy to fabricate phage films with variable patterns and systemically studied how the physical and biochemical cues, offered by the phage films, affected the osteogenic differentiation of MSCs and iPSCs. In the first study [27], they genetically fused an osteocalcin (OCN)-derived PDPLEPRREVCE (PD) peptide and an osteogenic growth peptide (OGP)-derived YGFGG (YG) peptide onto M13 phage surface to form the PD- and YG-phage, respectively. Then they prepared phage films through the LBL method. The resultant phage films showed a unique highly ordered ridge/groove topography where phage bundle ridges were separated by grooves and parallel to each other. The diameter of phage bundles (or ridges) could be controlled by the concentration of phage solutions used in the film fabrication. Higher concentration of phage (1014 pfu/mL) resulted in the larger diameter of the formed phage bundles on the film surface, while smaller diameter of phage bundles was formed when a lower concentration of phage solution (1012 pfu/mL) was used. When seeded on the phage films, the MSCs were elongated along the phage bundle direction, suggesting the topography of phage films could affect the morphologies of resident MSCs. Cell proliferation was affected by both the topography of phage films and the peptide sequences displayed on the phage surface. Phage films assembled from YG-phage at a lower concentration favored the proliferation of MSCs. In addition, both the phage films assembled from PD- and YG-phage could accelerate and enhance the osteogenic differentiation of MSCs in the osteogenic medium.
Based on these previous findings [27], we [113] further studied the fate of MSCs directed by the grooved phage films assembled from the RGD-phage that displays a fibronectin-derived adhesive peptide (RGD), the PHSRN-phage that displays a fibronectin-derived adhesive peptide (PHSRN), or their combination. We found that the phage films made from RGD-phage and a mixture of RGD- and PHSRN phages at a lower concentration (1012 pfu/mL) greatly favored the proliferation of MSCs. The films assembled from a mixture of RGD-phage and PHSRN-phage showed the greatest efficiency in promoting the osteogenic differentiation of MSCs in the primary media without osteogenic supplements. Besides the MSCs, our group [114] successfully applied the phage film system to control the behaviors of iPSCs. We constructed four bioengineered phages: (1) FN1-phage that displays RGD; (2) FN2-phage that is a mixture of RGD-phage and PHSRN-phage; (3) OGP-phage that displays the OGP (ALKRQGRTLYGFGG); and (4) BMP-2-phage that displays a BMP-2-derived peptide KIPKASSVPTELSAISTLYL. The resident iPSCs were elongated and aligned along the phage bundles. The morphology of the iPSCs-derived embryonic bodies (EBs) changed from nearly spherical to bipolar fibroblast-like and then to polygonal over time. These findings suggested that the topography of the phage films could control the cell morphologies and regulate their behaviors. Then we studied the osteoblastic differentiation of iPSCs on the phage films assembled from different bioengineered phages in the primary media without osteogenic supplements (Figure 5A). We found that iPSCs were differentiated into osteoblasts only on the phage films with ordered ridge/groove topography but not on those with random topography. We also found that iPSCs were more efficiently differentiated into osteoblasts on the ridge/groove phage films assembled from OGP-phage and BMP-2-phage than on the films assembled from other phages (wild-type phage, FN1-phage, FN2-phage), suggesting that the phage film system can efficiently regulate the osteoblastic differentiation of iPSCs by varying its topography (physical cue) and phage-displayed peptide (chemical cue) (Figure 5B-D). These results suggest that when the peptides derived from bone proteins are displayed on the phages, the resultant phages could induce the differentiation of stem cells [113, 114]. Such phages have a great potential to be used for controlling stem cell differentiation.
Figure 5.
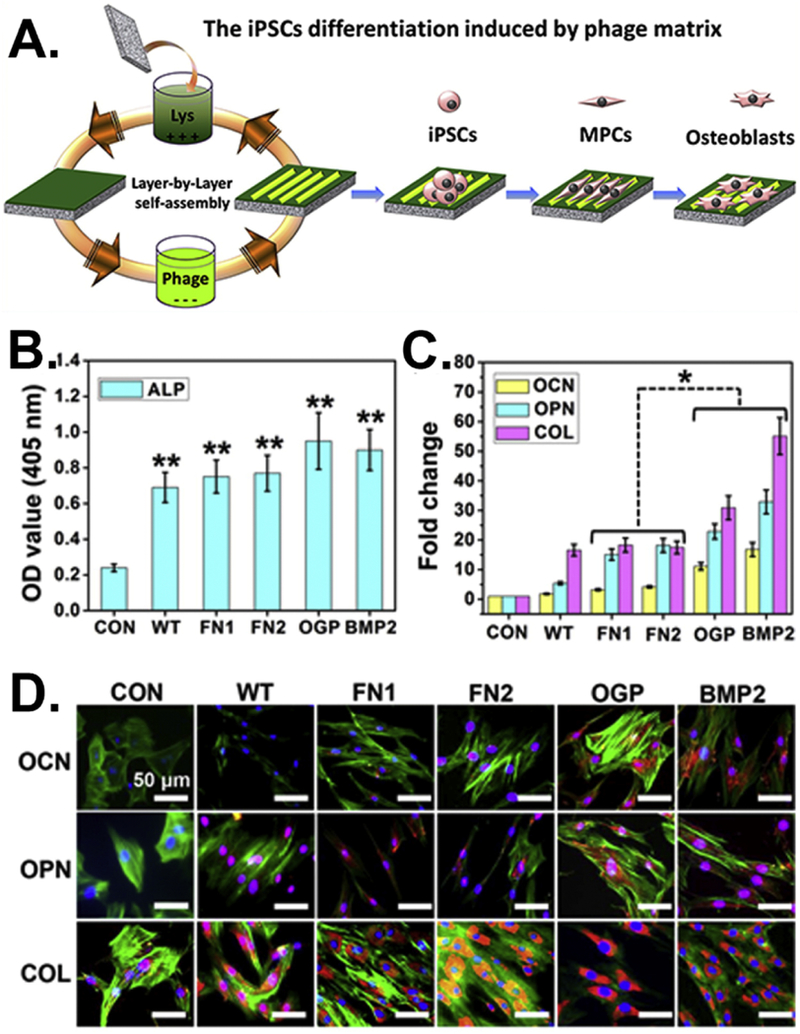
Efficient regulation of the osteoblastic differentiation of iPSCs by varying the peptides displayed on the side walls of filamentous phages. (A) Fabrication of bioengineered phage films through a layer-by-layer self-assembly method. The formed phage films could induce the osteoblastic differentiation of iPSCs. (B) The ALP activity of the cells cultured on different phage films showing a significant increase in all of the phage matrices compared with the control (**, p < 0.01). (C) Real-time PCR and (D) immunofluorescence staining assays for evaluating the expression levels of the osteogenic mark proteins, including osteoclacin (OCN) and osteopontin (OPN). Collagen (COL) was used as a reference protein and plates without phage films were the control (CON). (Reprinted with permission from ref. [114] copyright 2014, American Chemical Society)
Besides the in vitro studies, our group [115] has recently integrated the RGD-phage into a 3D printed bone scaffold for the regeneration of vascularized bone in vivo (Figure 6). We introduced the RGD-phage into the pores of a pre-printed 3D mineral scaffold along with chitosan and then seeded MSCs into the scaffold to form a virus-activated matrix (VAM). We found that the VAM containing the RGD-phage and regular MSCs induced bone tissues and blood vessels more efficiently than the control scaffolds (containing wildtype (WT) phage or without phage) but less efficiently than the VAM containing the RGD-phage and VEGF-expressing MSCs (Figure 6). These in vivo study results indicated that RGD-phage could induce both osteogenesis and angiogenesis for vascularized bone regeneration. The study suggests that the bioengineered phage is an ideal biological molecule to be introduced into 3D scaffolds for enhancing tissue regeneration.
Figure 6.
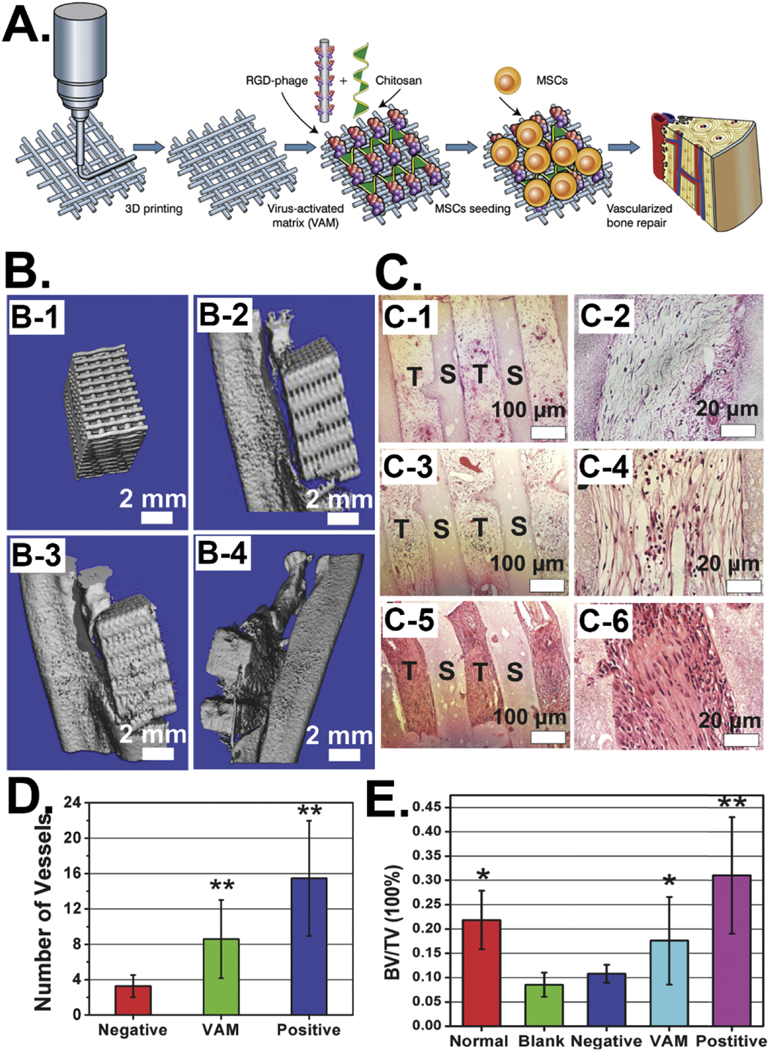
Integration of phages into 3D printed scaffolds for induce vascularized bone formation. (A) RGD-phage can induce both osteogenesis and angiogenesis for in vivo vascularized bone regeneration. (B) Micro-CT images of the bone regeneration by using the scaffold (B-1) filled with wild type phage (negative control, B-2), with RGD-phage (VAM, B-3), and with both RGD-phage and VEGF (positive control, B-4). (C) H&E staining of the sections of implants showing both VAM (C-3 & C-4) and its negative (C-1 & C-2) and positive (C-5 & C-6) controls could induce the formation of new bone tissue in the scaffolds. The VAM significantly promoted the formation of bone in comparison with negative control, but not as efficient as the positive control, suggesting the combination of VAM and VEGF was able to enhance the osteogenesis. (D) The number of newly formed blood vessels within the scaffolds showing significantly more blood vessels formed in VAM scaffold than the negative control. (E) The quantification of bone volume density (bone volume/tissue volume) within the scaffolds showing the bone volume density within the VAM scaffold is higher than the blank and negative control groups. (Reprinted with permission from ref. [115], copyright 2014, Wiley)
3.1.3. Phage display assisted gene delivery into MSCs
Genetically modified MSCs could continually express functional proteins, such as growth factors, inside the implanted 3D scaffolds to enhance in vivo bone regeneration [115]. However, genes are difficult to be efficiently transfected into MSCs using non-viral gene delivery vectors [39, 58, 59]. Phage biopanning technique has been used to identify a MSC-targeting peptide which could be integrated onto non-viral gene delivery vectors to enhance the gene delivery into MSCs [39, 56, 58, 59].
Balian et al. identified a high-affinity MSC-binding (HAB) peptide, SGHQLLLNKMPN, through both in vitro and in vivo phage biopanning against bone tissues [57]. They [56] found that the poly(amidoamine) (PAMAM) dendrimer-based gene delivery vectors modified with the HAB peptide could greatly enhance the Luc gene transfection efficiency, suggesting the high MSC-binding affinity of the peptide identified through phage biopanning could advance the non-viral gene delivery into MSCs for tissue regeneration purpose. Our group [39] identified a MSC-binding peptide, VTAMEPGQ (VT peptide), through an in vitro phage biopanning using an 8-mer landscape (pVIII) phage library. We [58] integrated the phage coat protein pVIII displaying the VT peptide into a gene delivery nanocluster to form a virus-mimetic magnetic/silica nanocluster (VMSNC), which could efficiently enter target cells in the presence of a magnetic field (1 T) and release the target gene by intracellular glutathione (GSH) inside cells. We demonstrated that the VMSNC reached a high gene transfection efficiency of 40.01% in MSCs (Figure 7A-C). In another study [59], we conjugated the VT peptide either alone or in the repeated form onto the surface of the lipid-based nanoparticles (LBNs) and found that the MSC-targeting VT peptide greatly improved the gene (VEGF and GFP) transfection efficiencies. Surprisingly, the LBNs modified with both triple reiterated VT peptide and nuclear localization signaling (NLS, DKKKRKV) peptide even reached a gene transfection efficiency of 52.52% (Figure 7D-F). The study demonstrated that the phage biopanning derived peptides are ideal targeting molecules for enhancing gene delivery into the stem cells.
Figure 7.
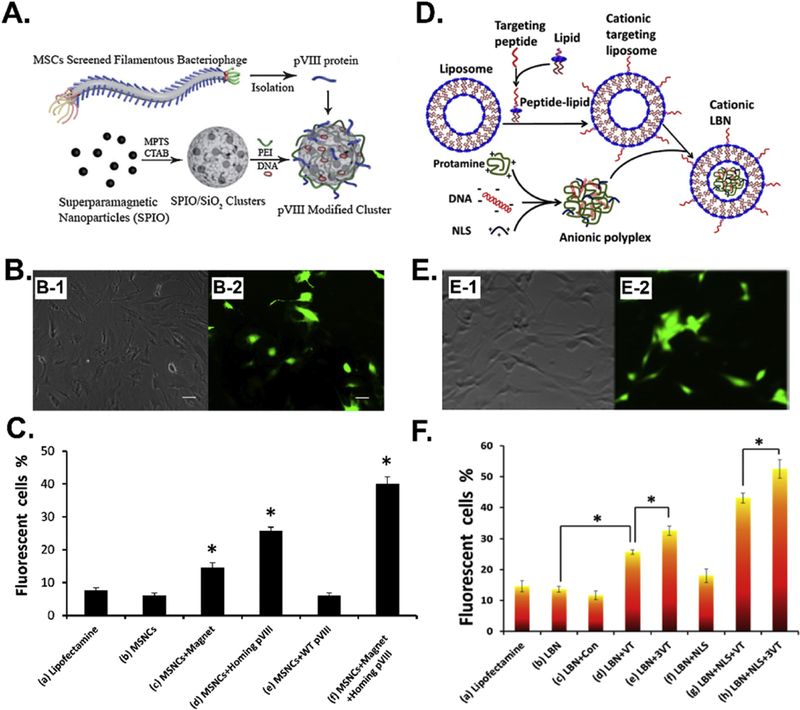
MSC-binding peptide selected from phage biopanning can promote the efficiency of gene delivery into MSCs by non-viral vectors, including (A) virus-mimetic magnetic/silica nanocluster (VMSNC) and (D) lipid-based nanoparticles (LBNs). (B) EGFP gene expression in MSCs transfected by VMSNCs under 1T magnetic field. (C) The EGFP gene transfection efficiency by VMSNCs and control vectors. (E) EGFP gene expression in MSCs transfected by LBNs modified with both triple reiterated VT peptide (VTAMEPGQ) and nuclear localization signaling (NLS, DKKKRKV) peptide. (F) The EGFP gene transfection efficiency by lipid-based nanoparticles (LBNs) modified with both triple reiterated VT peptide (VTAMEPGQ) and nuclear localization signaling (NLS, DKKKRKV) peptide and control vectors. (Reprinted with permission from ref. [58], copyright 2013, Wiley and ref. [59], copyright 2015, American Chemical Society)
3.2. Neural regeneration
For neural regeneration, the mostly used therapeutic stem cells are neural progenitor cells (NPCs), neural stem cells (NSCs), and embryonic stem cells (ESCs) [63–71, 73, 173]. The behaviors of the therapeutic stem cells can be controlled by functional peptides [174] or proteins to achieve their neuritogenesis and/or neural differentiation for the regeneration process [63–71, 73, 173]. Phage and phage display technique has been used for neural regeneration in a variety of ways. First, phage display derived functional peptides could be used to enhance the neural regeneration. Second, bioengineered phage nanofibers could be used as a biomaterial component/substrate to support neural regeneration. Third, phage display derived peptides could promote the gene delivery to neurons.
3.2.1. Phage display derived functional peptides/proteins for neural regeneration
Several groups have conducted phage biopanning against critical proteins in the central nervous system and identified several functional peptides that could promote neuritogenesis and/or neural differentiation.[60–62, 175] L1 cell adhesion molecule (L1CAM) is a cell surface glycoprotein involved in neural cell migration and neurite outgrowth. Schachner et al. identified two single-chain variable fragment antibodies (scFvs), I75 and I105, which can specifically bind to both the H3 peptide (GSQRKHSKRHIHKDHV) and the extracellular domain of L1CAM by using a phage display scFv Tomlinson I library [175]. They found that the I75 and I105 scFvs could increase neurite outgrowth of SK-N-SH cells by activating the signal transducers Erk and Src in these cells, suggesting their potential use in the therapy of neurodegenerative diseases. Nogo-A, an important myelin inhibitor, triggers the inhibition of CNS myelin and axonal regeneration by binding to its receptor NgR1. The neutralization of the 66 residues of the C terminal of the Nogo-A (termed Nogo-66) could reduce its inhibition of myelin and thus promote the regeneration of axons [60]. Su et al. identified a peptide, RRQTLSHQMRRP, that specifically binds to the 1–35 residues of the Nogo-66 (NEP1-35) through phage biopanning using the Ph.D.-12 phage library [60, 61]. Their results showed that the identified NEP1-35-binding peptide strongly bound to the Nogo-66, interrupted the interaction between Nogo-66 and its receptor NgR1, neutralized the inhibition of CNS myelin by Nogo-66, enhanced neuronal neurite outgrowth on CNS myelin substrates, and promoted CNS regeneration after spinal cord injury (SCI). Sun et al. [62] identified a NgR1-binding peptide, HIYTALV (NAP2), through phage biopanning using the Ph.D.-7 phage library and demonstrated the NAP2 peptide could also block the Nogo-66-induced inhibition of neurite growth to promote axonal regeneration.
Phage biopanning has successfully been applied to identify NSC-, NPC-binding peptides, which have shown great potential for neural regenerative applications [63–66, 73]. Arap et al. identified a murine NSC-specific peptide, CGLPYSSVC, through phage biopanning using the Ph.D.-C7C phage library [63]. The identified peptide shares a motif of netrin-4 (404–410) and could bind to laminin γ1 chain. Based on this finding, they found that the netrin-4, laminin γ1 chain, and α6β1 integrin could form a complex to promote the proliferation, adhesion, and migration of NSC, and the differentiation of MSCs is along with the decrease of netrin-4 receptors, indicating a molecular mechanism for affecting neural cell behaviors. Gelain et al. [64] identified a NSC-derived NPC-specific peptide, FAQRVPP, through phage biopanning using the Ph.D.-7 phage library. They also linked acetylated FAQRVPP to the LDLK12 self-assembly peptide (SAPeptide) to form Ac-FAQRVPP-GGG-(LDLK)3-CONH2 (Ac-FAQ peptide). They found that the Ac-FAQ peptide hydrogel could promote the proliferation and neural differentiation of human and mouse NSCs in vitro (Figure 8A-C) and improve neural tissue regeneration in acute contusive spinal cord injury in vivo (Figure 8D-G). The same group [65] identified a murine NSC-specific peptide, KLPGWSG, using the Ph.D.-7 phage library and linked acetylated KLPGWSG to the SAPeptide to form Ac-KLP peptide. They found that although the KLP peptide itself could not support the proliferation of NSCs, both the KLP peptide and the Ac-FAQ peptide hydrogel could enhance neuronal differentiation of NSCs. Zhao et al. identified a peptide, HGEVPRFHAVHL, which can specifically bind NSCs derived from rhesus monkey ESCs through phage biopanning using the Ph.D.-12 phage library [66]. The peptide was conjugated to quantum dots (QDs) for labeling NSCs. All these results have demonstrated the peptides identified from the phage biopanning against NSC/NPC could be displayed or conjugated onto scaffolds to promote neural regeneration.
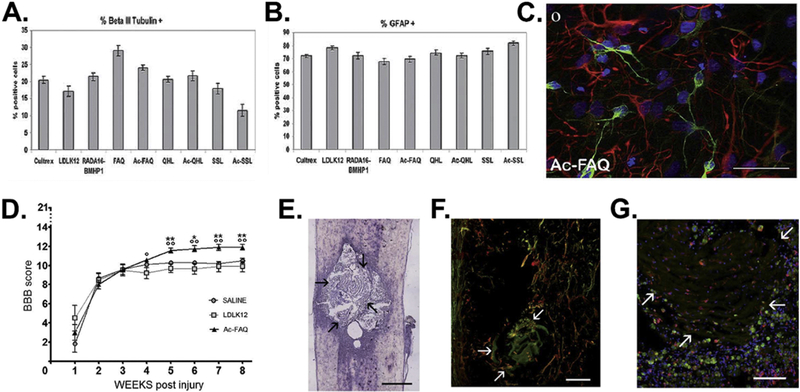
Figure 8.
Ac-FAQ peptide formed through linking of a NSC-derived NPC-specific peptide, FAQRVPP, and the LDLK12 self-assembly peptide (SAPeptide) as well as the hydrogels derived from the Ac-FAQ peptide for promoting the neural differentiation of human NSCs in vitro and neural tissue regeneration in acute contusive spinal cord injury in vivo. (A-C) Neural differentiation of human NSCs. Quantification of human neurons (A) and astrocytes (B) cultured on different scaffolds. (C) Fluorescence image of βIII-Tubulin positive (green) neurons and GFAP positive (red) astrocytes on the scaffold. (D) In vivo Ac-FAQ delivery into acute spinal cord injury. Hindlimb motor recovery of injured rats was assessed via BBB scale, showing Ac-FAQ treated rats achieved a better locomotor recovery than control groups. (E-G) Longitudinal spinal cord sections of Ac-FAQ-treated rats at 8 weeks post-injury/injection. Arrows point at the interface between nervous tissue and scaffold. (E) HE staining image. Scale bar: 800 µm (F) Immunofluorescence staining for GAP-43 (red) and βIII-Tubulin (green). Nerve fibers expressing both markers are colored in yellow. Scale bar: 50 µm. (G) Immunofluorescence staining for CD68 (green) and Iba1 (red), which reveal the markers of macrophages and microglia cells, respectively. Scale bar: 30 µm. (Reprinted with permission from ref. [64], copyright 2012, Royal Society of Chemistry)
Becker et al. identified two unique bone marrow-homing and primitive hematopoietic stem cell-binding peptides, PFSSTKT (BMHP1) and SKPPGTSS (BMHP2), through an in vivo phage biopanning [55]. Although the peptides were initially identified by phage biopanning against bone marrow tissue, they have shown great neural regenerative properties [67–71]. The possible reason is that bone marrow cells share some differentiating pathways and adhesion receptors with NSCs [176]. In the first study, Becker et al. linked a self-assembly peptide (SAP) RADA16 with several functional peptides, including the BMHP1, BMHP2, fibronectin-derived RGDS, collagen VI-derived RGDSGYRGDS, laminin derived motifs (YIGSR, IKVAV, PDSGR), and a myelo-peptide GFLGFPT, to form 3-D scaffolds and investigated the cell behaviors of mouse NSCs on them [67, 69]. The results showed that both the BMHP1 and BMHP2 peptides favored the adhesion, survival, and proliferation of NSCs and the BMHP1 peptide promoted the neural differentiation of NSCs. In the in vivo study [70], they injected the BMHP1-SAP scaffold into the rat spinal cord. They found that cellular infiltration, basement membrane deposition and axon regeneration/sprouting within the cyst were increased 8 weeks after spinal cord injury (SCI). They also found that the injected BMHP1-SAP scaffold partially filled the cavities and was compatible with the surrounding nervous tissue, and the scaffold also improved hindlimbs’ motor performance and forelimbs-hindlimbs coordination. These results demonstrated that the BMHP1-SAP scaffold could provide a physical and chemical support to the regeneration applications in the injured CNS. The same group [68] redesigned the SAP sequences and generated several novel BMHP1-SAP structures in the nano- or micro-scale, including tabular fibers, twisted ribbons, tubes and hierarchical self-assembled sheets. They demonstrated that these BMHP1-SAP 3D scaffolds could promote hNSC survival, proliferation, and differentiation in vitro and showed great in vivo biocompatibility. Besides the direct use of the BMHP1-SAP scaffold, Becker et al. also assembled the BMHP1-SAP into electrospun micro channel guidance constructs made of PLGA and PCL blended fibers and transplanted them for the regeneration in chronic injuries of the spinal cord [71]. Six months after the implantation, cavities were filled with the newly formed tissues, and a vascular network was well developed. In addition, immature and mature axons and myelination were regenerated throughout the lesion area. Moreover, astrocytes, neuronal fibers, myelin, microglial cells, fibroblasts, basement membrane components, and endothelial vessels were located around the degrading guidance channels (Figure 9). These results indicate a significant level of anatomical and functional regeneration by the BMHP1-SAP-electrospun PLGA/PCL hybrid structures in chronic SCI. All these results indicated that the BMHP1 peptide is a functional small molecule that could be introduced into the scaffolds for inducing NSC differentiation and promoting neural regeneration.
Figure 9.
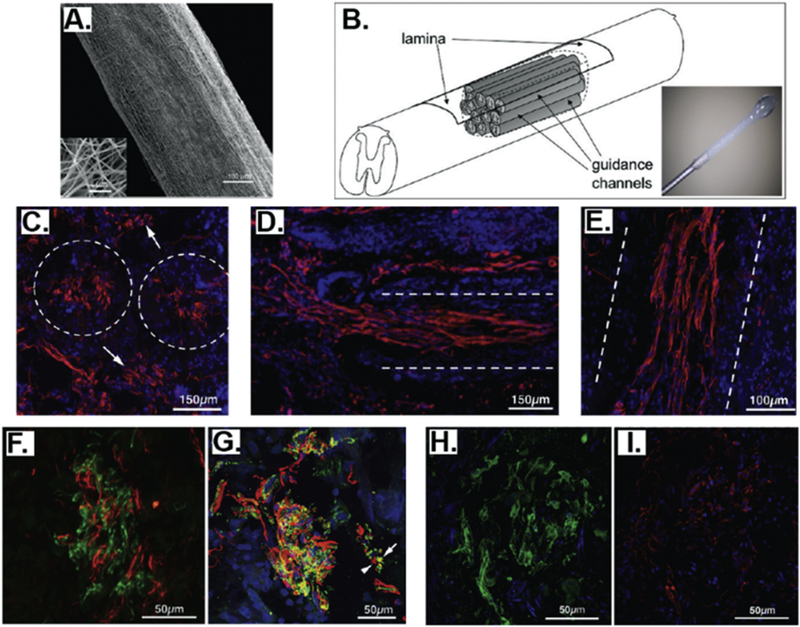
Assembly of identified primitive hematopoietic stem cell-binding peptide (BMHP1) and self-assembly peptide (SAP) into electrospun micro channel guidance constructs made of PLGA and PCL blended fibers and their transplantation into chronic injuries of the spinal cord. BMHP1-SAP-electrospun PLGA/PCL hybrid scaffolds can induce a significant level of anatomical and functional regeneration in chronic injuries of the spinal cord. (A) Electrospun PCL/PLGA microguidance channel construct. (B) Schematic of the implantation of PCL/PLGA microguidance channel with self-assembled BMHP1-SAP. (C-I) Nervous regeneration in the chronically injured rat spinal cord. (C-E) Transverse sections. Dashed lines outline channel walls. (C) GAP-43 positive fibers (red) inside and between transplanted tubes (arrows; coronal section). (D) NF200 positive fibers (red) crossing the top rostral interface of the lesion. (E) βIII tubulin positive fasciculi (red) stretched through the lumen of the conduits in a longitudinal spinal cord section. (F) NF200 (green) and GAP43 (red) are not overlapped inside the same fiber. (G) Myelinated (green, arrow) and unmyelinated (red, arrowhead) βIII tubulin positive fibers could be observed. Both SMI-32 (H, green) and SMI-31 (I, red) positive fibers could be detected within guidance conduits in adjacent coronal sections. (Reprinted with permission from ref. [71], copyright 2010, American Chemical Society)
3.2.2. Bioengineered phage nanofibers as a substrate to support NSCs/NPCs
Lee et al. have employed the bioengineered phage nanofiber assemblies as a biomaterial component/substrate to support NSCs/NPCs. They first constructed the bioengineered phages displaying RGD, IKVAV (a laminin motif known to promote neural cell adhesion and neurite extension) [177], and HPQ (a biotin-like peptide) to form GRD-, IKVAV-, and HPQ-phages and then assembled the phage nanofibers into cell-supporting long-range-ordered liquid crystalline phage films for controlling cell behaviors of NSCs and NPCs [116–118]. They found that RGD-phage formed films were biocompatible with neural cells and could direct the elongation of cell growth [118]. They also found that the films made of a mixture of HPQ-phage and RGD-phage could facilitate the immobilization of growth factors/ligands on them and direct orientation of the growth of NPCs [116]. In addition, they found that the films made of a mixture of IKVAV-phage and RGD-phage could support the proliferation and differentiation of NPCs and control their morphology and growth [117].
3.2.3. Phage display assisted drug/gene therapy
Neural cell- or tissue-targeting peptides identified by phage biopanning could be integrated onto delivery vectors to enable the targeted drug/gene therapy for treating neural disease and damages. Hou et al. identified a mouse cerebellar granule neuron specific peptide, D(E)–W(F)–I(N)–D–W, which may serve as a targeting molecule to promote selective drug/gene delivery to neurons [178]. Putzer et al. identified two neural precursor cell-specific peptides, QTRFLLH and VPTQSSG, and conjugated them onto the GFP/RFP gene carrying adenovirus [72]. The peptide-functionalized adenovirus could selectively bind, be internalized, and deliver genes into the neural precursor cells in vitro and in vivo, suggesting a novel strategy for manipulating neural precursor cells in CNS. Han et al. identified an ischemic stroke tissue-homing peptide, CLEVSRKNC, through in vivo phage biopanning using the Ph.D.-C7C phage library [73]. The identified CLEVSRKNC peptide could specifically detect the apoptotic neuronal cells, suggesting its potential use for targeted drug/gene delivery to stroke tissue. Solomen et al. used an AβP antibody-displaying scFv phage as both a probe and a delivery vector to scan brain Aβ deposits and successfully targeted the β amyloid deposition both in vitro and in vivo [179]. All these results indicate that phage biopanning derived peptides or bioengineered phage nanofibers could serve as a targeting probe and/or delivery vector for neural drug/gene therapies.
3.3. Cartilage regeneration
For cartilage regeneration, the mostly used therapeutic stem cells are MSCs [74, 76–87, 180, 181]. The behaviors of the therapeutic stem cells can be controlled by functional peptides or proteins to achieve the chondrogenic differentiation for the regeneration process [74, 76–87, 180, 181]. Phage-based techniques have been used for cartilage regeneration. Phage display derived functional peptides could be used to control the chondrogenic differentiation of MSCs and enhance the cartilage regeneration. Furthermore, phage display derived chondrocyte-homing peptides could promote the gene delivery to chondrocyte for regeneration.
3.3.1. Phage display derived TGF-β1- and collagen II-binding peptides for cartilage regeneration
Stupp et al. identified a TGF-β1-binding peptide, HSNGLPL, through phage display and integrated the peptide with the PA to form nanofiber gel materials with high TGF-β1-binding affinity [74, 75]. The formed gels showed the better capability of retaining TGF-β1 within gels. They found that the materials could induce the chondrogenic differentiation of human MSCs in vitro and promote the regeneration of articular cartilage in a chondral defect in a rabbit model with or without additional TGF-β1 growth factor (Figure 10). Hubbell et al. identified a collagen II α 1-binding peptide, WYRGRL, through phage biopanning using a fUSE5/6-mer phage library and functionalized the 38-nm-diameter-poly(propylene sulfide) (PPS) nanoparticles with the peptide [77]. The resultant WYRGRL functionalized PPS nanoparticles showed high collagen II α 1-binding affinity and could efficiently home to articular cartilage up to 72-fold more than control nanoparticles in the mouse, suggesting a great strategy to target a vascular tissue for drug therapy. Jonkeijm et al. immobilized both the TGF-β1- (HSNGLPL) and collagen II- (WYRGRL) binding peptides on a fluorogenic surface [76]. They found that the co-immobilized peptides were able to selectively capture TGF-β1 and collagen II, and the HSNGLPL coated surface supplemented with TGF-β1 could support the proliferation of human articular chondrocytes and also stimulate their expression of chondrogenic markers. The results suggest that a combination of biopanning derived TGF-β1- and collagen II-binding peptides can be used to functionalize the scaffolds for enhancing cartilage regeneration.
Figure 10.
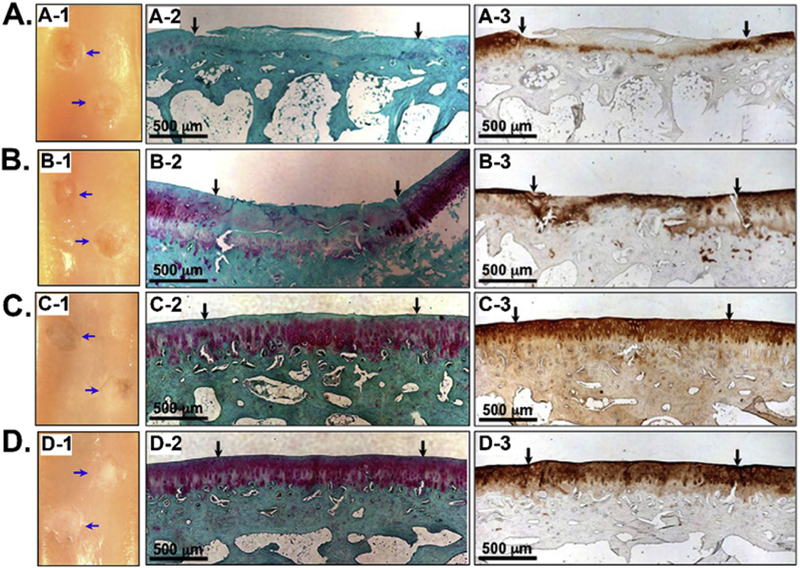
Integration of TGF-β1-binding peptide, HSNGLPL, with the peptide amphiphile (PA) to form nanofiber gel materials with high TGF-β1-binding affinity. The regeneration of articular cartilage was achieved in a chondral defect in a rabbit model with TGF-β1-binding PA (TGFBPA) nanofiber gel. The regeneration of articular cartilage 12 weeks after treatment with (A-1) 100 ng/mL TGF-β1, (B-1) filler PA+100 ng/mL TGF-β1, (C-1) 10%TGFBPA+100 ng/mL TGF-β1, and (D-1) 10%TGFBPA alone. Safranin-O staining for glycosaminoglycans (A-2, B-2, C-2 and D-2) and type II collagen staining (A-3, B-3, C-3, and D-3) in articular cartilage defects treated with (A-2&3) 100 ng/mL TGF-β1, (B-2&3) filler PA+100 ng/mL TGF-β1, (C-2&3) 10%TGFBPA+100 ng/mL TGF-β1, and (D-2&3) 10%TGFBPA alone. All the results showed that 10%TGFBPA could greatly promote the regeneration process. (Reprinted with permission from ref. [74], copyright 2010, National Academy of Sciences)
3.3.2. Phage display derived MSC-binding peptide, E7, for cartilage regeneration
Ao et al. identified a bone marrow derived MSC-specific peptide, EPLQLKM (E7), through phage biopanning using the Ph.D.-7 phage library and chemically conjugated the peptide onto polycaprolactone (PCL) electrospun meshes to construct E7-conjugated PCL electrospun meshes [79]. When implanted into a cartilage defect site of rat knee joints combined with a microfracture procedure, the E7-conjugated PCL electrospun meshes could trigger the homing of the endogenous MSCs and result in the significantly better MSC recruitment than RGD-modified meshes in vivo. In addition, they [80] conjugated the peptide and seeded MSCs onto the lattice-like PLLA mesh to promote healing of ligament bone after rabbit ACL reconstruction. They found that the scaffold could absorb endogenous MSCs and support MSCs proliferation and osteogenic differentiation. They also discovered that the regenerated insertion contained more abundant collagen I, II, and III, better fibrocartilage growth, and higher tensile strength than controls, indicating an enhanced ligament-bone healing using the E7-modified lattice-like PLLA meshes. In another study, they [81] fabricated coaxial electrospun fibers with poly(ε-caprolactone) (PCL) as the shell and PVP/BSA/TGF-β1 as the core and chemically functionalized them with the E7 peptide. They found that the E7-coated TGF-β1-releasing scaffolds could promote the MSCs adhesion, favor their proliferation, and induce their chondrogenic differentiation for cartilage repair. Moreover, the E7 peptide was integrated with demineralized bone matrix (DBM) and a chitosan (CS) hydrogel to form a composite scaffold for in vivo cartilage repair [82, 83]. The results showed that the composite scaffold stimulated the proliferation and chondrogenic differentiation of MSCs in vitro, and the conjugated E7 peptide facilitated the recruitment and homing of surrounding endogenous MSCs to further enhance cartilage regeneration and favor the integration within the host tissue in vivo (Figure 11). The results suggest that the biopanning derived MSC-binding peptide, E7, can be functionalized on the scaffolds for improved cartilage regeneration.
Figure 11.
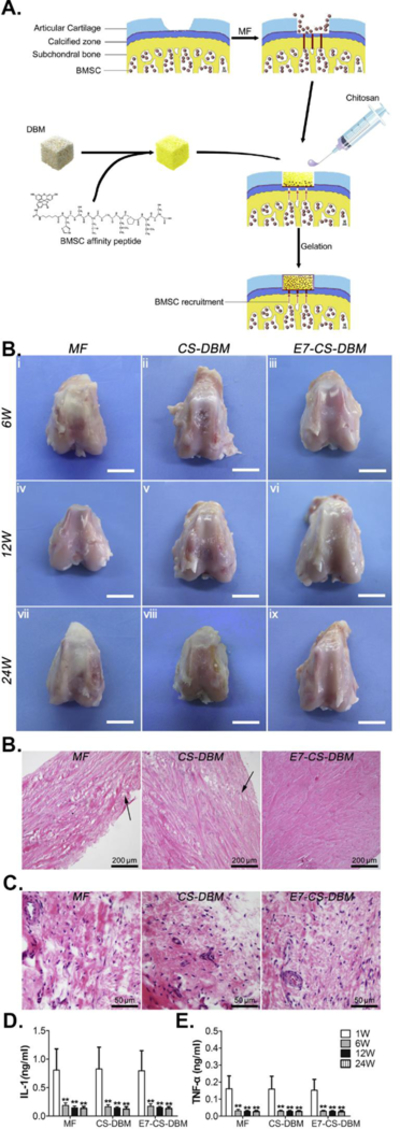
Efficient repair of cartilage defects by E7 peptide functionalized hydrogels. (A) Knee repair by E7-conjugated demineralized bone matrix (DBM) and chitosan (CS) hydrogels (E7-CS-DBM). (B) The repaired cartilage defects at various time points. Scale bar: 1 cm. (C) HE staining of the sections in affiliated load-bearing tissue in the joint with a histological observing of meniscus at week 24 (black arrow, wearing collagen fibers). (D) HE staining of the sections of the synovium at week 24; (E) Content of Interleukin-1 in joint fluid; (F) Content of tumor necrosis factor-a in joint fluid. (Reprinted with permission from ref. [82], copyright 2014, Elsevier)
3.3.3. Phage display derived hyaluronan-binding peptide, Pep-1 or HABPep, for cartilage regeneration
Hyaluronan (HA), a glycosaminoglycan produced in the ECM in large amount, could bind many adhesion molecules such as the receptor CD44 to regulate cell activities. Its main function in joints is to lubricate and protect cartilage [182]. Mummert et al. identified a HA-binding peptide, GAHWQFNALTVR (Pep-1 or HABPep), through phage biopanning using the Ph.D.-12 phage library and found that the Pep-1 could inhibit the HA-mediated “two-way” trafficking of leukocytes [84]. Elisseeff et al. conjugated the HABPep peptide onto the poly(ethylene glycol) (PEG) to form the HA-interacting hydrogel [85]. They found that the hydrogel containing exogenous HA could retain HA and promote the proliferation and chondrogenic differentiation of MSCs in vitro. They applied the hydrogels to an osteochondral defect model in the rat knee and found that the HA-binding hydrogels promoted the cartilage tissue production in defects and reduced the degeneration of cartilage surrounding the defects. These results indicated that the HA-binding peptide facilitated the hydrogels to concentrate local or exogenous HA for the regeneration of cartilage tissues. The same group [78] conjugated the HABPep peptide and phage display identified collagen II-binding peptide (WYRGRL, termed ColBpep) with PEG to form the HABPep-PEG-ColBpep and applied it to the joint environment. The in vivo study results showed that the HABPep-PEG-ColBpep coating could retain the HA for much longer time and increase the lubrication in the joint. They also constructed the HABPep-PEG-collagen I-binding peptide and applied it in eye drops to bind ocular tissue surface. The recruitment and retention of HA on damaged eye surface resulted in the decreased evaporation of water.
3.3.4. Phage display assisted gene delivery for cartilage regeneration
Ao et al. identified a chondrocyte-homing peptide, DWRVIIPPRPSA (CAP), through phage biopanning using the Ph.D.-12 phage library and then chemically conjugated the peptide with polyethyleneimine (PEI) to construct a non-viral gene vector [86]. They found that CAP-functionalized PEI vectors could retain in the cartilage and increase the gene transfection efficiency into knee joints, suggesting the CAP-functionalized vector is a cartilage-specific vector for cartilage therapy. They [87] also employed the chondrocyte-homing CAP-functionalized vector to deliver small interfering RNA (siRNA) into the cartilage of the knee joints of a mouse to silence the expression of Hif-2α, which is a key factor in triggering the cartilage degradation in osteoarthritis (OA). The results showed that the CAP was concentrated and retained siRNA in the cartilage and the cartilage degeneration and synovium inflammation in the knee joints were alleviated. Both studies demonstrated that the chondrocyte-homing peptide discovered by phage display, CAP, could serve as a chondrocyte targeting probe for cartilage regeneration gene therapies.
3.4. Skin regeneration and wound healing
One of the challenges in skin regeneration and wound healing is how to efficiently deliver drugs to skin cells and wounds. Phage display technique has been used for skin regeneration and wound healing in two major directions. One is to use phage display derived functional peptides to home to wound and recognize/inhibit HA in skin tissues [88–91, 181]. Another is to use phage display derived skin-penetrating peptides to promote the drug/siRNA delivery for skin regeneration [92–96].
3.4.1. Phage display derived functional peptides for wound healing
The expression level of HA in the skin is associated with several diseases. The phage display identified HA-binding peptide Pep-1 [84] could be used for the strong dermal staining but weak epidermis staining [181]. In another study, Turley et al. identified a HA-binding peptide, STMMSRSHKTRSHHV (P15-1), through phage biopanning using a 15-mer phage library [88]. The peptide could block the RHAMM/HA signaling to reduce the number of wound macrophages and fibroblasts as well as the blood vessel density, promoting the scarless healing of the skin. To facilitate the linking of drugs to a wound-healing matrix (Integra®), Baird et al. identified two matrix-binding peptides, CFKHSSHQC and SQIDYATGPRQA, through phage biopanning from the Ph.D.-C7C and Ph.D.-12 phage library, respectively [89]. They found both peptides could efficiently help the introducing of drugs into the matrix for improved performance. Ruoslahti et al. identified two peptides, CARSKNKDC (CAR) and CRKDKC (CRK), which can specifically target skin and tendon through phage biopanning using a CX7C T7 phage library [90]. In addition, they found that the two peptides could selectively home to wound sites in vivo. They also found that CAR favored early stages of wound healing and CRK favored later stages of wound healing. Decorin is a therapeutic molecule that can inhibit TGF-β to prevent tissue fibrosis and promote tissue regeneration. In the following study, Ruoslahti et al. genetically displayed the wound-homing CAR peptide on the decorin surface to form a recombinant CAR-decorin [91]. They found that the I.V. injected CAR-decorin could selectively accumulate in the wound sites, greatly promote wound healing, and efficiently inhibit scar formation in a mice-wounding model. All these studies indicate that phage display is a powerful tool to identify functional peptides for skin regeneration and wound healing.
3.4.2. Phage display assisted gene/drug delivery into skin
Mitragotri et al. [92] identified a skin penetrating and cell entering (SPACE) peptide, ACHSALTKHCG, through phage biopanning using a Ph.D.-C7C phage library. The SPACE peptide could enable vectors to penetrate across stratum corneum into epidermis and dermis layers and into cells in skin tissue such as keratinocytes, fibroblasts, and endothelial cells.[93] In addition, they found that when functionalized onto siRNAs, SPACE was able to selectively deliver the siRNA into the skin and reduce the expression level of the target genes in the skin (Figure 12) [92, 96]. Similarly, the SPACE peptide could also help the targeted delivery of drugs, including HA [94] and cyclosporine A [95], into the skin for potential regeneration applications. The results suggest that the SPACE peptide can be functionalized on vectors for improved drug delivery for skin regeneration.
Figure 12.
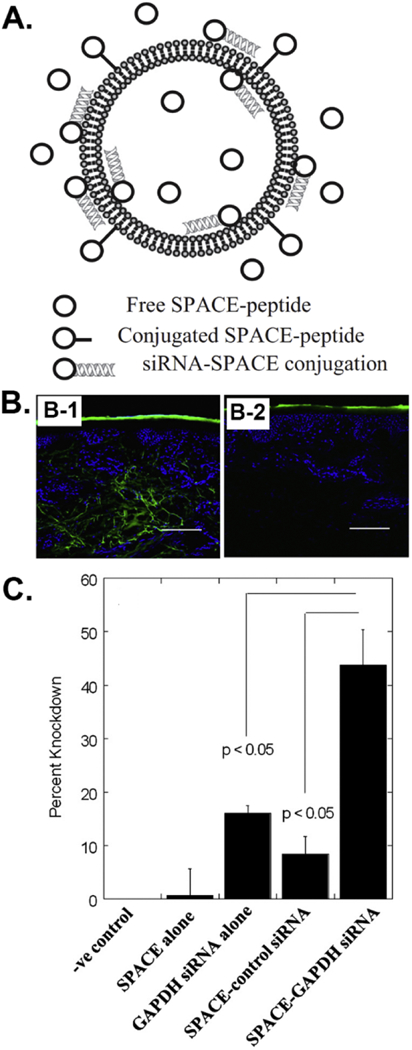
Enhanced delivery of siRNA into skin tissues by a skin penetrating and cell entering (SPACE) peptide identified from phage biopanning. (A) The structure of SPACE functionalized siRNA-carrying liposomes. (B) Confocal microscopy images of the skin penetration profiles of SPACE (B-1) and control peptide (B-2) into porcine skin, respectively, showing that the SPACE peptide could efficiently penetrate across stratum corneum into epidermis and dermis layers. (C) Percentage knockdown of GAPDH protein levels in mice 72 h after treatment, showing that SPACE could deliver siRNA into the skin and reduce the GAPDH expression level. (Reprinted with permission from ref. [96], copyright 2014, Elsevier and ref. [92], copyright 2011, National Academy of Sciences)
3.5. Cardiac regeneration
One of the challenges in cardiac regeneration and heart disease treatment is how to efficiently deliver drugs to injured vasculature, heart tissue, myocardial ischemia, cardiomyocyte and so on [97–102, 119, 183, 184]. Phage display technique has been used for cardiac regeneration. In vitro phage display derived functional peptides could be used to home to cardiomyocytes. Moreover, in vivo phage display derived injured vasculature and myocardial ischemia-homing peptides could promote the targeted drug delivery for cardiac regeneration and heart disease treatment.
3.5.1. In vitro phage display derived functional peptides and bioengineered phage nanofibers for cardiac regeneration
TGF-β1 plays a critical role in vascular smooth muscle cell (VSMC) proliferation and migration. Michon et al. [97] identified two peptides, CGLLPVGRPDRNVWRWLC (IM-1) and CKGQCDRFKGLPEWC (IM-2), which can specifically bind to the TGF-β type II receptor. Both IM-1 and IM-2 peptides could block the interaction between TGF-β1 and TGF-β type II receptor and serve as the antagonists of several TGF-β pathways to induce the VSMC migration and affect the expression of TGF-β1 related proteins. Han et al. [119] successfully used the RGD-phage functionalized PLGA to promote the proliferation and myogenic differentiation of CAC12 myoblasts in the presence of graphene oxide. Ma et al. identified a peptide, QPFTTSLTPPAR, which can specifically bind to the mouse ESC-derived cardiomyocytes through phage biopanning using a Ph.D.-12 phage library, providing a cardiomyocyte-targeting molecule for further regeneration applications [98].
3.5.2. In vivo phage display derived functional peptides for cardiac regeneration
Robbins et al. identified a cardiac targeting peptide, APWHLSSQYSRT, through in vivo phage biopanning using a Ph.D.-12 phage library, providing a cardiac-homing molecule to deliver drugs/genes to the heart [99]. Lee et al. identified a peptide, CSTSMLKAC, that can specifically home to ischemic heart tissue through in vivo phage biopanning using a Ph.D.-C7C phage library [100]. They genetically fused the peptide onto the surface of the protein Sumo–mCherry and I.V. injected the recombinant protein into a mouse model of myocardial ischemia–reperfusion injury. The biodistribution results showed that the fusion protein selectively homed to ischemic myocardium, suggesting that the peptide is a novel molecular tool for delivering drugs/genes into the injured myocardium. Kelly et al. identified the peptides that can specifically bind to the cardiomyocytes (clones I-1, B-42), endothelial cells (clones B-40, B-47), myofibroblasts (clones B-7, B-29), and c-Kit + cells (clones B-50, B-27) in the border zone of the remodeling infarct through in vivo phage biopanning using a Ph.D.-7 phage library [184]. They conjugated liposomes with the cardiomyocyte specific peptide (I-1) to deliver a PARP-1 (poly [ADP-ribose] polymerase 1) inhibitor, AZ7379, and found that more AZ7379 molecules aggregated in the infarct/border zone 24 h after I.V. injection. Langer et al. identified a vascular wall-binding peptide, KLWVLPK, through in vivo phage biopanning using a Ph.D.-7 phage library and conjugated the peptide with nanoparticles (NPs) [101]. They found that after I.V. injection, the KLWVLPK-NPs homed and retained to the injured carotid arteries in vivo and abdominal aortas ex vivo (Figure 13). Stupp et al. conjugated the vascular wall-binding peptide, KLWVLPK, to the PA that can self-assemble into spherical and fiber-like supramolecular structures [102]. They found that only the fiber-like PA conjugated with KLWVLPK could home to and retain at the site of arterial injury for up to two days after injection. All these results have demonstrated the cardiac or vascular wall targeting peptides identified from in vivo biopanning could work as a targeting ligand to be integrated into a drug delivery system for efficient bio-imaging and cardiac regeneration.
Figure 13.
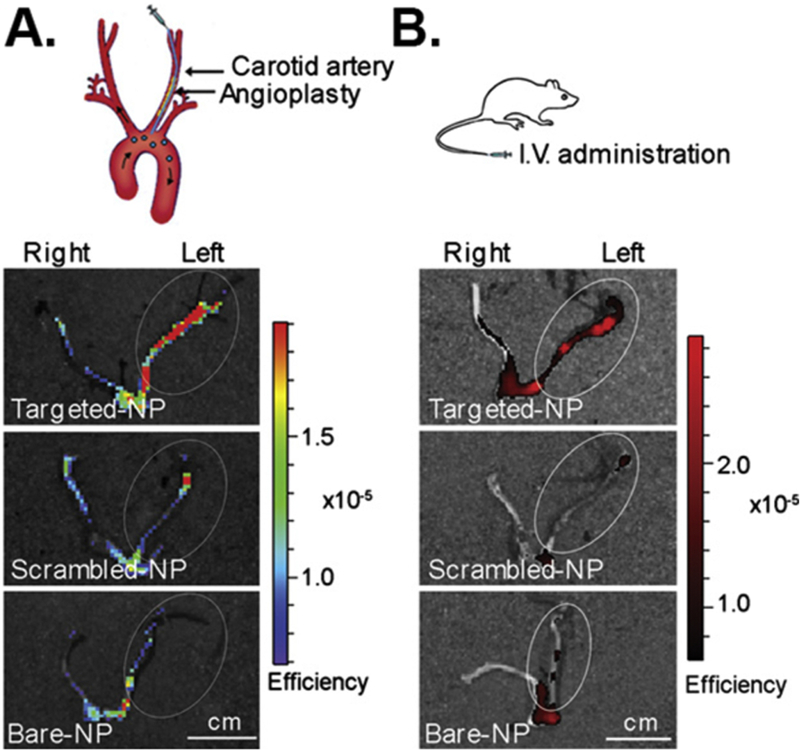
Selective binding to the sites of arterial injury by a vascular wall targeting peptide identified from in vivo phage biopanning. (A) In vivo IA infusion in a carotid injury model. A catheter was inserted via the external carotid into the common carotid and advanced into the aortic arch. Nanoparticles were allowed to circulate for 1 h. Fluorescence images overlaid on the photographs of carotid arteries incubated with nanoparticles. (B) In vivo systemic delivery in a carotid angioplasty model. Nanoparticles were I.V. injected through tail vein and allowed to circulate for 1 h. Fluorescence images overlaid on photographs of carotid arteries incubated with nanoparticles. For imaging, Alexa Fluor 647–PLGA dye conjugates were encapsulated in place of Ptxl–PLA drug conjugates. The results showed that targeted nanoparticles could selectively home to and retain at the injured carotid arteries (Scale bar, 1 cm.) (Reprinted with permission from ref. [101], copyright 2010, National Academy of Sciences)
3.6. Other applications
Fibroblast cells are a type of cells that generate the extracellular matrix and collagen to form the structural framework for various tissues. They are abundant in connective tissues and play an important role in the wound healing process [185]. Wang and Lee et al. assembled wild type and bioengineered phage nanofibers into liquid crystalline films to induce the directional growth of human NIH-3T3 fibroblasts, suggesting a novel strategy for controlling the orientation of fibroblasts and their generated ECM components [120–125]. ESC is a powerful stem cell source for tissue regeneration but the use of human ESC raised ethical concerns. Kiessling et al. identified two human ESC-binding peptides, TVKHRPDALHPQ and LTTAPKLPKVTR, through phage biopanning using a Ph.D.-12 phage library and found that both peptides could support the proliferation of ESCs without differentiation [103]. The same group [104] also identified two peptides, LTGKNFPMFHRN and MHRMPSFLPTTL, that could specifically bind to the extracellular domain of TGF-β receptor I and II through phage biopanning and cultured NMuMG, A549, and C2C12 cells on the peptide-coated substrate [105]. They found the peptides could concentrate endogenous TGF-β and regulate TGF-β responsive genes to induce the epithelial to mesenchymal transition, which has potential applications in tissue regeneration.
4. Summary and future perspectives
As summarized in this review article, bioengineered phages and phage display derived peptides have recently been employed to assist the regeneration of various tissues, including bone, neuron, cartridge, skin, heart, and so on (Table 1). The exciting findings have demonstrated the great potential of phages and phage display technique for efficient tissue regeneration. But there are some emerging problems in the field: (1) Due to the presence of various molecules on the cellular surface, different peptides targeting to the same cell type might be selected. It is necessary to identify the specific targeting molecules (e.g. cell surface receptor) corresponding to each selected peptide for the better use of the peptides; (2) Most groups preferred conjugating phage display identified functional peptides onto polymer/substrate/PA system for tissue regeneration applications. It might be better to directly use bioengineered human-safe phages which genetically display such peptides; (3) How to decide which phage library (e.g. Ph.D. M13 series, pVIII displayed M13, T7, and scFvs libraries) is better for a specific selection of a target molecule/cell/tissue; and (4) How to study the in vivo fate of bioengineered phages to reveal the direct use of phages in tissue regeneration. Researchers with multidisciplinary backgrounds will address these problems and continuously take advantage of phages and phage display technique to further advance this exciting field. We believe the future directions in this field include: (1) Different types of phages have their unique advantages, so more types of phages and phage libraries will be developed for tissue regeneration; (2) In vivo phage biopanning will be more often conducted to identify tissue-targeting peptides for in vivo tissue regeneration; (3) Due to the unique advantages of iPSCs in tissue regeneration, more studies will be focused on using bioengineered phage-based substrates to control the iPSC fate.
Acknowledgments
We would like to thank the financial support from National Institutes of Health (EB021339 and GM116116), Oklahoma Center for the Advancement of Science and Technology (HR17-043) and Oklahoma Center for Adult Stem Cell Research (434003). We also acknowledge the support of National Natural Science Foundation of China (21172194), Zhejiang Provincial Natural Science Foundation of China (LZ17C170002 and LZ16E030001), Projects of Zhejiang Provincial Science and Technology Plans (2012C12910), Silkworm Industry Science and Technology Innovation Team (2011R50028), China Agriculture Research System (CARS-22-ZJ0402), and National High Technology Research and Development Program 863 (2013AA102507).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Forbes SJ, Rosenthal N, Preparing the ground for tissue regeneration: from mechanism to therapy, Nature Medicine, 20 (2014) 857–869. [DOI] [PubMed] [Google Scholar]
- [2].Poss KD, Advances in understanding tissue regenerative capacity and mechanisms in animals, Nature Reviews Genetics, 11 (2010) 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Galliot B, Crescenzi M, Jacinto A, Tajbakhsh S, Trends in tissue repair and regeneration, Development, 144 (2017) 357–364. [DOI] [PubMed] [Google Scholar]
- [4].Lu ZF, Kleine-Nulend J, Li B, Bone Microenvironment, Stem Cells, and Bone Tissue Regeneration, Stem Cells International, (2017). [DOI] [PMC free article] [PubMed]
- [5].Scott EW, Stem Cell Reviews and Reports: Adult Stem Cells and Tissue Regeneration Section, Stem Cell Reviews and Reports, 13 (2017) 2–2. [DOI] [PubMed] [Google Scholar]
- [6].Montagnani S, Rueger MA, Hosoda T, Nurzynska D, Adult Stem Cells in Tissue Maintenance and Regeneration, Stem Cells International, (2016). [DOI] [PMC free article] [PubMed]
- [7].Caramella C, Conti B, Modena T, Ferrari F, Bonferoni MC, Genta I, Rossi S, Torre ML, Sandri G, Sorrenti M, Catenacci L, Dorati R, Tripodo G, Controlled delivery systems for tissue repair and regeneration, Journal of Drug Delivery Science and Technology, 32 (2016) 206–228. [Google Scholar]
- [8].Chua ILS, Kim HW, Lee JH, Signaling of extracellular matrices for tissue regeneration and therapeutics, Tissue Engineering and Regenerative Medicine, 13 (2016) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Silva EA, Mooney DJ, Synthetic extracellular matrices for tissue engineering and regeneration, Current Topics in Developmental Biology, Vol 64, 64 (2004) 181–205. [DOI] [PubMed] [Google Scholar]
- [10].Mao AS, Mooney DJ, Regenerative medicine: Current therapies and future directions, Proceedings of the National Academy of Sciences of the United States of America, 112 (2015) 14452–14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lee S, Yoo J, Atalt A, Fundamentals of In Situ Tissue Regeneration, in: A.A.a.J.J.Y. Lee Sang Jin (Ed.) In Situ Tissue Regeneration, Academic Press; 2016, pp. 3–17. [Google Scholar]
- [12].Lutolf MP, Blau HM, Artificial Stem Cell Niches, Advanced Materials, 21 (2009) 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Discher DE, Mooney DJ, Zandstra PW, Growth Factors, Matrices, and Forces Combine and Control Stem Cells, Science, 324 (2009) 1673–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Whitaker MJ, Quirk RA, Howdle SM, Shakesheff KM, Growth factor release from tissue engineering scaffolds, Journal of Pharmacy and Pharmacology, 53 (2001) 1427–1437. [DOI] [PubMed] [Google Scholar]
- [15].Babensee JE, McIntire LV, Mikos AG, Growth factor delivery for tissue engineering, Pharmaceutical Research, 17 (2000) 497–504. [DOI] [PubMed] [Google Scholar]
- [16].Caplan AI, Adult mesenchymal stem cells for tissue engineering versus regenerative medicine, Journal of Cellular Physiology, 213 (2007) 341–347. [DOI] [PubMed] [Google Scholar]
- [17].Tuan RS, Boland G, Tuli R, Adult mesenchymal stem cells and cell-based tissue engineering, Arthritis Research & Therapy, 5 (2003) 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lee K, Silva EA, Mooney DJ, Growth factor delivery-based tissue engineering: general approaches and a review of recent developments, Journal of the Royal Society Interface, 8 (2011) 153–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Goldring CEP, Duffy PA, Benvenisty N, Andrews PW, Ben-David U, Eakins R, French N, Hanley NA, Kelly L, Kitteringham NR, Kurth J, Ladenheim D, Laverty H, McBlane J, Narayanan G, Patel S, Reinhardt J, Rossi A, Sharpe M, Park BK, Assessing the Safety of Stem Cell Therapeutics, Cell Stem Cell, 8 (2011) 618–628. [DOI] [PubMed] [Google Scholar]
- [20].Amariglio N, Hirshberg A, Scheithauer BW, Cohen Y, Loewenthal R, Trakhtenbrot L, Paz N, Koren-Michowitz M, Waldman D, Leider-Trejo L, Toren A, Constantini S, Rechavi G, Donor-Derived Brain Tumor Following Neural Stem Cell Transplantation in an Ataxia Telangiectasia Patient, Plos Medicine, 6 (2009) 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sunderland KS, Yang MY, Mao CB, Phage-Enabled Nanomedicine: From Probes to Therapeutics in Precision Medicine, Angewandte Chemie-International Edition, 56 (2017) 1964–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Krom RJ, Bhargava P, Lobritz MA, Collins JJ, Engineered Phagemids for Nonlytic, Targeted Antibacterial Therapies, Nano Letters, 15 (2015) 4808–4813. [DOI] [PubMed] [Google Scholar]
- [23].Young R, Gill JJ, Phage therapy redux - What is to be done?, Science, 350 (2015) 1163–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cao BR, Yang MY, Mao CB, Phage as a Genetically Modifiable Supramacromolecule in Chemistry, Materials and Medicine, Accounts of Chemical Research, 49 (2016) 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Smith GP, Petrenko VA, Phage display, Chemical Reviews, 97 (1997) 391–410. [DOI] [PubMed] [Google Scholar]
- [26].Kehoe JW, Kay BK, Filamentous phage display in the new millennium, Chemical Reviews, 105 (2005) 4056–4072. [DOI] [PubMed] [Google Scholar]
- [27].Zhu HB, Cao BR, Zhen ZP, Laxmi AA, Li D, Liu SR, Mao CB, Controlled growth and differentiation of MSCs on grooved films assembled from monodisperse biological nanofibers with genetically tunable surface chemistries, Biomaterials, 32 (2011) 4744–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xu H, Cao BR, George A, Mao CB, Self-Assembly and Mineralization of Genetically Modifiable Biological Nanofibers Driven by beta-Structure Formation, Biomacromolecules, 12 (2011) 2193–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cao BR, Mao CB, Identification of Microtubule-Binding Domains on Microtubule-Associated Proteins by Major Coat Phage Display Technique, Biomacromolecules, 10 (2009) 555–564. [DOI] [PubMed] [Google Scholar]
- [30].Smith GP, Filamentous Fusion Phage - Novel Expression Vectors That Display Cloned Antigens on the Virion Surface, Science, 228 (1985) 1315–1317. [DOI] [PubMed] [Google Scholar]
- [31].Lee SW, Mao CB, Flynn CE, Belcher AM, Ordering of quantum dots using genetically engineered viruses, Science, 296 (2002) 892–895. [DOI] [PubMed] [Google Scholar]
- [32].Hufton SE, Moerkerk PT, Meulemans EV, de Bruine A, Arends JW, Hoogenboom HR, Phage display of cDNA repertoires: the pVI display system and its applications for the selection of immunogenic ligands, Journal of Immunological Methods, 231 (1999) 39–51. [DOI] [PubMed] [Google Scholar]
- [33].Mao CB, Solis DJ, Reiss BD, Kottmann ST, Sweeney RY, Hayhurst A, Georgiou G, Iverson B, Belcher AM, Virus-based toolkit for the directed synthesis of magnetic and semiconducting nanowires, Science, 303 (2004) 213–217. [DOI] [PubMed] [Google Scholar]
- [34].Wang YC, Ju ZG, Cao BR, Gao X, Zhu Y, Qiu PH, Xu H, Pan PT, Bao HZ, Wang L, Mao CB, Ultrasensitive Rapid Detection of Human Serum Antibody Biomarkers by Biomarker-Capturing Viral Nanofibers, Acs Nano, 9 (2015) 4475–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pande J, Szewczyk MM, Grover AK, Phage display: Concept, innovations, applications and future, Biotechnology Advances, 28 (2010) 849–858. [DOI] [PubMed] [Google Scholar]
- [36].Li Y, Cao BR, Yang MY, Zhu Y, Suh J, Mao CB, Identification of Novel Short BaTiO3-Binding/Nucleating Peptides for Phage-Templated in Situ Synthesis of BaTiO3 Polycrystalline Nanowires at Room Temperature, Acs Applied Materials & Interfaces, 8 (2016) 30714–30721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Petrenko VA, Smith GP, Gong X, Quinn T, A library of organic landscapes on filamentous phage, Protein Engineering, 9 (1996) 797–801. [DOI] [PubMed] [Google Scholar]
- [38].Sanghvi AB, Miller KPH, Belcher AM, Schmidt CE, Biomaterials functionalization using a novel peptide that selectively binds to a conducting polymer, Nature Materials, 4 (2005) 496–502. [DOI] [PubMed] [Google Scholar]
- [39].Ma K, Wang DD, Lin YY, Wang JL, Petrenko V, Mao CB, Synergetic Targeted Delivery of Sleeping-Beauty Transposon System to Mesenchymal Stem Cells Using LPD Nanoparticles Modified with a Phage-Displayed Targeting Peptide, Advanced Functional Materials, 23 (2013) 1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Babickova J, Tothova L, Boor P, Celec P, In vivo phage display - A discovery tool in molecular biomedicine, Biotechnology Advances, 31 (2013) 1247–1259. [DOI] [PubMed] [Google Scholar]
- [41].Teesalu T, Sugahara KN, Ruoslahti E, Mapping of Vascular Zip Codes by Phage Display, Methods in Enzymology: Protein Engineering for Therapeutics, Vol 203, Pt B, 503 (2012) 35–56. [DOI] [PubMed] [Google Scholar]
- [42].Chung WJ, Kwon KY, Song J, Lee SW, Evolutionary Screening of Collagen-like Peptides That Nucleate Hydroxyapatite Crystals, Langmuir, 27 (2011) 7620–7628. [DOI] [PubMed] [Google Scholar]
- [43].Roy MD, Stanley SK, Amis EJ, Becker ML, Identification of a highly specific hydroxyapatite-binding peptide using phage display, Advanced Materials, 20 (2008) 1830–+. [Google Scholar]
- [44].Weiger MC, Park JJ, Roy MD, Stafford CM, Karim A, Becker ML, Quantification of the binding affinity of a specific hydroxyapatite binding peptide, Biomaterials, 31 (2010) 2955–2963. [DOI] [PubMed] [Google Scholar]
- [45].Gungormus M, Fong H, Kim IW, Evans JS, Tamerler C, Sarikaya M, Regulation of in vitro calcium phosphate mineralization by combinatorially selected hydroxyapatite-binding peptides, Biomacromolecules, 9 (2008) 966–973. [DOI] [PubMed] [Google Scholar]
- [46].Gungormus M, Branco M, Fong H, Schneider JP, Tamerler C, Sarikaya M, Self assembled bi-functional peptide hydrogels with biomineralization-directing peptides, Biomaterials, 31 (2010) 7266–7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Segvich SJ, Smith HC, Kohn DH, The adsorption of preferential binding peptides to apatite-based materials, Biomaterials, 30 (2009) 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Addison WN, Miller SJ, Ramaswamy J, Mansouri A, Kohn DH, Mckee MD, Phosphorylation-dependent mineral-type specificity for apatite-binding peptide sequences, Biomaterials, 31 (2010) 9422–9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ramaraju H, Miller SJ, Kohn DH, Dual-functioning peptides discovered by phage display increase the magnitude and specificity of BMSC attachment to mineralized biomaterials, Biomaterials, 134 (2017) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sheu TJ, Schwarz EM, Martinez DA, O’Keefe RJ, Rosier RN, Zuscik MJ, Puzas JE, A phage display technique identifies a novel regulator of cell differentiation, Journal of Biological Chemistry, 278 (2003) 438–443. [DOI] [PubMed] [Google Scholar]
- [51].Hamilton PT, Jansen MS, Ganesan S, Benson RE, Hyde-DeRuyscher R, Beyer WF, Gile JC, Nair SA, Hodges JA, Gron H, Improved Bone Morphogenetic Protein-2 Retention in an Injectable Collagen Matrix Using Bifunctional Peptides, Plos One, 8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Behanna HA, Donners JJJM, Gordon AC, Stupp SI, Coassembly of amphiphiles with opposite peptide polarities into nanofibers, Journal of the American Chemical Society, 127 (2005) 1193–1200. [DOI] [PubMed] [Google Scholar]
- [53].Lee SSS, Hsu EL, Mendoza M, Ghodasra J, Nickoli MS, Ashtekar A, Polavarapu M, Babu J, Riaz RM, Nicolas JD, Nelson D, Hashmi SZ, Kaltz SR, Earhart JS, Merk BR, Mckee JS, Bairstow SF, Shah RN, Hsu WK, Stupp SI, Gel Scaffolds of BMP-2-Binding Peptide Amphiphile Nanofibers for Spinal Arthrodesis, Advanced Healthcare Materials, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cao FY, Yin WN, Fan JX, Zhuo RX, Zhang XZ, A novel function of BMHP1 and cBMHP1 peptides to induce the osteogenic differentiation of mesenchymal stem cells, Biomaterials Science, 3 (2015) 345–351. [DOI] [PubMed] [Google Scholar]
- [55].Nowakowski GS, Dooner MS, Valinski HM, Mihaliak AM, Quesenberry PJ, Becker PS, A specific heptapeptide from a phage display peptide library homes to bone marrow and binds to primitive hematopoietic stem cells, Stem Cells, 22 (2004) 1030–1038. [DOI] [PubMed] [Google Scholar]
- [56].Santos JL, Pandita D, Rodrigues J, Pego AP, Granja PL, Balian G, Tomas H, Receptor-Mediated Gene Delivery Using PAMAM Dendrimers Conjugated with Peptides Recognized by Mesenchymal Stem Cells, Molecular Pharmaceutics, 7 (2010) 763–774. [DOI] [PubMed] [Google Scholar]
- [57].Balian G, Bone targeting peptides US, 2008.
- [58].Gandra N, Wang DD, Zhu Y, Mao CB, Virus-Mimetic Cytoplasm-Cleavable Magnetic/Silica Nanoclusters for Enhanced Gene Delivery to Mesenchymal Stem Cells, Angewandte Chemie-International Edition, 52 (2013) 11278–11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wang DD, Yang MY, Zhu Y, Mao CB, Reiterated Targeting Peptides on the Nanoparticle Surface Significantly Promote Targeted Vascular Endothelial Growth Factor Gene Delivery to Stem Cells, Biomacromolecules, 16 (2015) 3897–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Deng QY, Cai WQ, Li SR, Su BY, Identification of a NEP1-35 recognizing peptide that neutralizes CNS myelin inhibition using phage display library, Neuroscience Letters, 536 (2013) 80–84. [DOI] [PubMed] [Google Scholar]
- [61].Deng QY, Cai WQ, Li SR, Zhang YQ, Su BY, Small Nogo-66-binding peptide promotes neurite outgrowth through RhoA inhibition after spinal cord injury, Brain Research Bulletin, 99 (2013) 140–144. [DOI] [PubMed] [Google Scholar]
- [62].Sun ZQ, Dai XY, Li Y, Jiang SW, Lou GF, Cao QY, Hu RD, Huang YD, Su ZJ, Chen MW, Luo HM, Lin X, Sun J, Xiao F, A novel Nogo-66 receptor antagonist peptide promotes neurite regeneration in vitro, Molecular and Cellular Neuroscience, 71 (2016) 80–91. [DOI] [PubMed] [Google Scholar]
- [63].Staquicinia FI, Dias-Neto E, Li JX, Snyder EY, Sidman RL, Pasqualini R, Arap W, Discovery of a functional protein complex of netrin-4, laminin gamma 1 chain, and integrin alpha 6 beta 1 in mouse neural stem cells, Proceedings of the National Academy of Sciences of the United States of America, 106 (2009) 2903–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gelain F, Cigognini D, Caprini A, Silva D, Colleoni B, Donega M, Antonini S, Cohen BE, Vescovi A, New bioactive motifs and their use in functionalized self-assembling peptides for NSC differentiation and neural tissue engineering, Nanoscale, 4 (2012) 2946–2957. [DOI] [PubMed] [Google Scholar]
- [65].Caprini A, Silva D, Zanoni I, Cunha C, Volonte C, Vescovi A, Gelain F, A novel bioactive peptide: assessing its activity over murine neural stem cells and its potential for neural tissue engineering, New Biotechnology, 30 (2013) 552–562. [DOI] [PubMed] [Google Scholar]
- [66].Zhao WX, Yuan H, Xu X, Ma L, Isolation and Initial Application of a Novel Peptide That Specifically Recognizes the Neural Stem Cells Derived from Rhesus Monkey Embryonic Stem Cells, Journal of Biomolecular Screening, 15 (2010) 687–694. [DOI] [PubMed] [Google Scholar]
- [67].Cunha C, Panseri S, Villa O, Silva D, Gelain F, 3D culture of adult mouse neural stem cells within functionalized self-assembling peptide scaffolds, International Journal of Nanomedicine, 6 (2011) 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gelain F, Silva D, Caprini A, Taraballi F, Natalello A, Villa O, Nam KT, Zuckermann RN, Doglia SM, Vescovi A, BMHP1-Derived Self-Assembling Peptides: Hierarchically Assembled Structures with Self-Healing Propensity and Potential for Tissue Engineering Applications, Acs Nano, 5 (2011) 1845–1859. [DOI] [PubMed] [Google Scholar]
- [69].Gelain F, Bottai D, Vescovi A, Zhang SG, Designer Self-Assembling Peptide Nanofiber Scaffolds for Adult Mouse Neural Stem Cell 3-Dimensional Cultures, Plos One, 1 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Cigognini D, Satta A, Colleoni B, Silva D, Donega M, Antonini S, Gelain F, Evaluation of Early and Late Effects into the Acute Spinal Cord Injury of an Injectable Functionalized Self-Assembling Scaffold, Plos One, 6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Gelain F, Panseri S, Antonini S, Cunha C, Donega M, Lowery J, Taraballi F, Cerri G, Montagna M, Baldissera F, Vescovi A, Transplantation of Nanostructured Composite Scaffolds Results in the Regeneration of Chronically Injured Spinal Cords, Acs Nano, 5 (2011) 227–236. [DOI] [PubMed] [Google Scholar]
- [72].Schmidt A, Haas SJP, Hildebrandt S, Scheibe J, Eckhoff B, Racek T, Kempermann G, Wree A, Putzer BM, Selective targeting of adenoviral vectors to neural precursor cells in the hippocampus of adult mice: New prospects for in situ gene therapy, Stem Cells, 25 (2007) 2910–2918. [DOI] [PubMed] [Google Scholar]
- [73].Hong HY, Choi JS, Kim YJ, Lee HY, Kwak W, Yoo J, Lee JT, Kwon TH, Kim IS, Han HS, Lee BH, Detection of apoptosis in a rat model of focal cerebral ischemia using a homing peptide selected from in vivo phage display, Journal of Controlled Release, 131 (2008) 167–172. [DOI] [PubMed] [Google Scholar]
- [74].Shah RN, Shah NA, Lim MMD, Hsieh C, Nuber G, Stupp SI, Supramolecular design of self-assembling nanofibers for cartilage regeneration, Proceedings of the National Academy of Sciences of the United States of America, 107 (2010) 3293–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].McLennan IS, Koishi K, The transforming growth factor-betas: multifaceted regulators of the development and maintenance of skeletal muscles, motoneurons and Schwann cells, International Journal of Developmental Biology, 46 (2002) 559–567. [PubMed] [Google Scholar]
- [76].Cabanas-Danes J, Nicosia C, Landman E, Karperien M, Huskens J, Jonkheijm P, A fluorogenic monolayer to detect the co-immobilization of peptides that combine cartilage targeting and regeneration, Journal of Materials Chemistry B, 1 (2013) 1903–1908. [DOI] [PubMed] [Google Scholar]
- [77].Rothenfluh DA, Bermudez H, O’Neil CP, Hubbell JA, Biofunctional polymer nanoparticles for intra-articular targeting and retention in cartilage, Nature Materials, 7 (2008) 248–254. [DOI] [PubMed] [Google Scholar]
- [78].Singh A, Corvelli M, Unterman SA, Wepasnick KA, McDonnell P, Elisseeff JH, Enhanced lubrication on tissue and biomaterial surfaces through peptide-mediated binding of hyaluronic acid, Nature Materials, 13 (2014) 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Shao ZX, Zhang X, Pi YB, Wang XK, Jia ZQ, Zhu JX, Dai LH, Chen WQ, Yin L, Chen HF, Zhou CY, Ao YF, Polycaprolactone electrospun mesh conjugated with an MSC affinity peptide for MSC homing in vivo, Biomaterials, 33 (2012) 3375–3387. [DOI] [PubMed] [Google Scholar]
- [80].Zhu JX, Zhang X, Shao ZX, Dai LH, Li L, Hu XQ, Wang XK, Zhou CY, Ao YF, In Vivo Study of Ligament-Bone Healing after Anterior Cruciate Ligament Reconstruction Using Autologous Tendons with Mesenchymal Stem Cells Affinity Peptide Conjugated Electrospun Nanofibrous Scaffold, Journal of Nanomaterials, (2013).
- [81].Man ZT, Yin L, Shao ZX, Zhang X, Hu XQ, Zhu JX, Dai LH, Huang HJ, Yuan L, Zhou CY, Chen HF, Ao YF, The effects of co-delivery of BMSC-affinity peptide and rhTGF-beta 1 from coaxial electrospun scaffolds on chondrogenic differentiation, Biomaterials, 35 (2014) 5250–5260. [DOI] [PubMed] [Google Scholar]
- [82].Huang HJ, Zhang X, Hu XQ, Shao ZX, Zhu JX, Dai LH, Man ZT, Yuan L, Chen HF, Zhou CY, Ao YF, A functional biphasic biomaterial homing mesenchymal stem cells for in vivo cartilage regeneration, Biomaterials, 35 (2014) 9608–9619. [DOI] [PubMed] [Google Scholar]
- [83].Meng QY, Man ZT, Dai LH, Huang HJ, Zhang X, Hu XQ, Shao ZX, Zhu JX, Zhang JY, Fu X, Duan XN, Ao YF, A composite scaffold of MSC affinity peptide-modified demineralized bone matrix particles and chitosan hydrogel for cartilage regeneration, Scientific Reports, 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Mummert ME, Mohamadzadeh M, Mummert DI, Mizumoto N, Takashima A, Development of a peptide inhibitor of hyaluronan-mediated leukocyte trafficking, Journal of Experimental Medicine, 192 (2000) 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Unterman SA, Gibson M, Lee JH, Crist J, Chansakul T, Yang EC, Elisseeff JH, Hyaluronic Acid-Binding Scaffold for Articular Cartilage Repair, Tissue Engineering Part A, 18 (2012) 2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Pi YB, Zhang X, Shi JJ, Zhu JX, Chen WQ, Zhang CG, Gao WW, Zhou CY, Ao YF, Targeted delivery of non-viral vectors to cartilage in vivo using a chondrocyte-homing peptide identified by phage display, Biomaterials, 32 (2011) 6324–6332. [DOI] [PubMed] [Google Scholar]
- [87].Pi Y, Zhang X, Shao Z, Zhao F, Hu X, Ao Y, Intra-articular delivery of anti-Hif-2 alpha siRNA by chondrocyte-homing nanoparticles to prevent cartilage degeneration in arthritic mice, Gene Therapy, 22 (2015) 439–448. [DOI] [PubMed] [Google Scholar]
- [88].Tolg C, Hamilton SR, Zalinska E, McCulloch L, Amin R, Akentieva N, Winnik F, Savani R, Bagli DJ, Luyt LG, Cowman MK, McCarthy JB, Turley EA, A RHAMM Mimetic Peptide Blocks Hyaluronan Signaling and Reduces Inflammation and Fibrogenesis in Excisional Skin Wounds, American Journal of Pathology, 181 (2012) 1250–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Sawada R, Peterson CY, Gonzalez AM, Potenza BM, Mueller B, Coimbra R, Eliceiri BP, Baird A, A phage-targeting strategy for the design of spatiotemporal drug delivery from grafted matrices, Fibrogenesis Tissue Repair, 4 (2011) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Jarvinen TAH, Ruoslahti E, Molecular changes in the vasculature of injured tissues, Am erican Journal of Pathology, 171 (2007) 702–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Jarvinen TAH, Ruoslahti E, Target-seeking antifibrotic compound enhances wound healing and suppresses scar formation in mice, Proceedings of the National Academy of Sciences of the United States of America, 107 (2010) 21671–21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Hsu T, Mitragotri S, Delivery of siRNA and other macromolecules into skin and cells using a peptide enhancer, Proceedings of the National Academy of Sciences of the United States of America, 108 (2011) 15816–15821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Kumar S, Zakrewsky M, Chen M, Menegatti S, Muraski JA, Mitragotri S, Peptides as skin penetration enhancers: Mechanisms of action, Journal of Controlled Release, 199 (2015) 168–178. [DOI] [PubMed] [Google Scholar]
- [94].Chen M, Gupta V, Anselmo AC, Muraski JA, Mitragotri S, Topical delivery of hyaluronic acid into skin using SPACE-peptide carriers, Journal of Controlled Release, 173 (2014) 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Chen M, Kumar S, Anselmo AC, Gupta V, Slee DH, Muraski JA, Mitragotri S, Topical delivery of Cyclosporine A into the skin using SPACE-peptide, Journal of Controlled Release, 199 (2015) 190–197. [DOI] [PubMed] [Google Scholar]
- [96].Chen M, Zakrewsky M, Gupta V, Anselmo AC, Slee DH, Muraski JA, Mitragotri S, Topical delivery of siRNA into skin using SPACE-peptide carriers, Journal of Controlled Release, 179 (2014) 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Michon IN, Penning LC, Molenaar TJ, van Berkel TJ, Biessen EA, Kuiper J, The effect of TGF-beta receptor binding peptides on smooth muscle cells, Biochem Biophys Res Commun, 293 (2002) 1279–1286. [DOI] [PubMed] [Google Scholar]
- [98].Li Z, Fan J, Zhao W, Jin L, Ma L, The specific binding of peptide ligands to cardiomyocytes derived from mouse embryonic stem cells, J Pept Sci, 17 (2011) 771–782. [DOI] [PubMed] [Google Scholar]
- [99].Zahid M, Phillips BE, Albers SM, Giannoukakis N, Watkins SC, Robbins PD, Identification of a cardiac specific protein transduction domain by in vivo biopanning using a M13 phage peptide display library in mice, PLoS One, 5 (2010) e12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kanki S, Jaalouk DE, Lee S, Yu AY, Gannon J, Lee RT, Identification of targeting peptides for ischemic myocardium by in vivo phage display, J Mol Cell Cardiol, 50 (2011) 841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Chan JM, Zhang L, Tong R, Ghosh D, Gao W, Liao G, Yuet KP, Gray D, Rhee JW, Cheng J, Golomb G, Libby P, Langer R, Farokhzad OC, Spatiotemporal controlled delivery of nanoparticles to injured vasculature, Proc Natl Acad Sci U S A, 107 (2010) 2213–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Moyer TJ, Kassam HA, Bahnson ES, Morgan CE, Tantakitti F, Chew TL, Kibbe MR, Stupp SI, Shape-Dependent Targeting of Injured Blood Vessels by Peptide Amphiphile Supramolecular Nanostructures, Small, 11 (2015) 2750–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Christiansen A, Kringelum JV, Hansen CS, Bogh KL, Sullivan E, Patel J, Rigby NM, Eiwegger T, Szepfalusi Z, de Masi F, Nielsen M, Lund O, Dufva M, High-throughput sequencing enhanced phage display enables the identification of patient-specific epitope motifs in serum, Scientific Reports, 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Li LY, Orner BP, Huang T, Hinck AP, Kiessling LL, Peptide ligands that use a novel binding site to target both TGF-beta receptors, Molecular Biosystems, 6 (2010) 2392–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Li LY, Klim JR, Derda R, Courtney AH, Kiessling LL, Spatial control of cell fate using synthetic surfaces to potentiate TGF-beta signaling, Proceedings of the National Academy of Sciences of the United States of America, 108 (2011) 11745–11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Wang FK, Cao BR, Mao CB, Bacteriophage Bundles with Prealigned Ca2+ Initiate the Oriented Nucleation and Growth of Hydroxylapatite, Chemistry of Materials, 22 (2010) 3630–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].He T, Abbineni G, Cao BR, Mao CB, Nanofibrous Bio-inorganic Hybrid Structures Formed Through Self-Assembly and Oriented Mineralization of Genetically Engineered Phage Nanofibers, Small, 6 (2010) 2230–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Shin YC, Lee JH, Kim MJ, Park JH, Kim SE, Kim JS, Oh JW, Han DW, Biomimetic Hybrid Nanofiber Sheets Composed of RGD Peptide-Decorated PLGA as Cell-Adhesive Substrates, J Funct Biomater, 6 (2015) 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chung WJ, Oh JW, Kwak K, Lee BY, Meyer J, Wang E, Hexemer A, Lee SW, Biomimetic self-templating supramolecular structures, Nature, 478 (2011) 364–368. [DOI] [PubMed] [Google Scholar]
- [110].Lee JY, Chung WJ, Kim G, A mechanically improved virus-based hybrid scaffold for bone tissue regeneration, Rsc Advances, 6 (2016) 55022–55032. [Google Scholar]
- [111].Lee DY, Lee H, Kim Y, Yoo SY, Chung WJ, Kim G, Phage as versatile nanoink for printing 3-D cell-laden scaffolds, Acta Biomaterialia, 29 (2016) 112–124. [DOI] [PubMed] [Google Scholar]
- [112].Yoo SY, Kobayashi M, Lee PP, Lee SW, Early Osteogenic Differentiation of Mouse Preosteoblasts Induced by Collagen-Derived DGEA-Peptide on Nanofibrous Phage Tissue Matrices, Biomacromolecules, 12 (2011) 987–996. [DOI] [PubMed] [Google Scholar]
- [113].Wang JL, Wang L, Li X, Mao CB, Virus activated artificial ECM induces the osteoblastic differentiation of mesenchymal stem cells without osteogenic supplements, Scientific Reports, 3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Wang JL, Wang L, Yang MY, Zhu Y, Tomsia A, Mao CB, Untangling the Effects of Peptide Sequences and Nanotopographies in a Biomimetic Niche for Directed Differentiation of iPSCs by Assemblies of Genetically Engineered Viral Nanofibers, Nano Letters, 14 (2014) 6850–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Wang JL, Yang MY, Zhu Y, Wang L, Tomsia AP, Mao CB, Phage Nanofibers Induce Vascularized Osteogenesis in 3D Printed Bone Scaffolds, Advanced Materials, 26 (2014) 4961–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Yoo SY, Merzlyak A, Lee SW, Facile growth factor immobilization platform based on engineered phage matrices, Soft Matter, 7 (2011) 1660–1666. [Google Scholar]
- [117].Merzlyak A, Indrakanti S, Lee SW, Genetically Engineered Nanofiber-Like Viruses For Tissue Regenerating Materials, Nano Letters, 9 (2009) 846–852. [DOI] [PubMed] [Google Scholar]
- [118].Chung WJ, Merzlyak A, Yoo SY, Lee SW, Genetically Engineered Liquid-Crystalline Viral Films for Directing Neural Cell Growth, Langmuir, 26 (2010) 9885–9890. [DOI] [PubMed] [Google Scholar]
- [119].Shin YC, Lee JH, Kim MJ, Hong SW, Kim B, Hyun JK, Choi YS, Park JC, Han DW, Stimulating effect of graphene oxide on myogenesis of C2C12 myoblasts on RGD peptide-decorated PLGA nanofiber matrices, J Biol Eng, 9 (2015) 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Shin YC, Lee JH, Jin L, Kim MJ, Oh JW, Kim TW, Han DW, Cell-adhesive RGD peptide-displaying M13 bacteriophage/PLGA nanofiber matrices for growth of fibroblasts, Biomater Res, 18 (2014) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Chung WJ, Merzlyak A, Lee SW, Fabrication of engineered M13 bacteriophages into liquid crystalline films and fibers for directional growth and encapsulation of fibroblasts, Soft Matter, 6 (2010) 4454–4459. [Google Scholar]
- [122].Yoo SY, Chung WJ, Kim TH, Le M, Lee SW, Facile patterning of genetically engineered M13 bacteriophage for directional growth of human fibroblast cells, Soft Matter, 7 (2011) 363–368. [Google Scholar]
- [123].Rong JH, Lee LA, Li K, Harp B, Mello CM, Niu ZW, Wang Q, Oriented cell growth on self-assembled bacteriophage M13 thin films, Chemical Communications, (2008) 5185–5187. [DOI] [PubMed]
- [124].Yoo SY, Oh JW, Lee SW, Phage-Chips for Novel Optically Readable Tissue Engineering Assays, Langmuir, 28 (2012) 2166–2172. [DOI] [PubMed] [Google Scholar]
- [125].Wu LY, Lee LA, Niu ZW, Ghoshroy S, Wang Q, Visualizing Cell Extracellular Matrix (ECM) Deposited by Cells Cultured on Aligned Bacteriophage M13 Thin Films, Langmuir, 27 (2011) 9490–9496. [DOI] [PubMed] [Google Scholar]
- [126].Li JH, Jia HJ, Cai XH, Zhong HZ, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, Juncker AS, Manichanh C, Chen B, Zhang WW, Levenez F, Wang J, Xu X, Xiao L, Liang SS, Zhang DY, Zhang ZX, Chen WN, Zhao HL, Al-Aama JY, Edris S, Yang HM, Wang J, Hansen T, Nielsen HB, Brunak S, Kristiansen K, Guarner F, Pedersen O, Dore J, Ehrlich SD, Bork P, Wang J, Consortium M, An integrated catalog of reference genes in the human gut microbiome, Nat Biotechnol, 32 (2014) 834–841. [DOI] [PubMed] [Google Scholar]
- [127].Miller-Ensminger T, Garretto A, Brenner J, Thomas-White K, Zambom A, Wolfe AJ, Putonti C, Bacteriophages of the urinary microbiome, J Bacteriol, (2018) DOI: 10.1128/JB.00738-00717. [DOI] [PMC free article] [PubMed]
- [128].Arap W, Kolonin MG, Trepel M, Lahdenranta J, Cardo-Vila M, Giordano RJ, Mintz PJ, Ardelt PU, Yao VJ, Vidal CI, Chen L, Flamm A, Valtanen H, Weavind LM, Hicks ME, Pollock RE, Botz GH, Bucana CD, Koivunen E, Cahill D, Troncoso P, Baggerly KA, Pentz RD, Do KA, Logothetis CJ, Pasqualini R, Steps toward mapping the human vasculature by phage display, Nature Medicine, 8 (2002) 121–127. [DOI] [PubMed] [Google Scholar]
- [129].Krag DN, Shukla GS, Shen GP, Pero S, Ashikaga T, Fuller S, Weaver DL, Burdette-Radoux S, Thomas C, Selection of tumor-binding ligands in cancer patients with phage display libraries, Cancer Research, 66 (2006) 7724–7733. [DOI] [PubMed] [Google Scholar]
- [130].Shukla GS, Krag DN, Peletskaya EN, Pero SC, Sun YJ, Carman CL, McCahill LE, Roland TA, Intravenous infusion of phage-displayed antibody library in human cancer patients: enrichment and cancer-specificity of tumor-homing phage-antibodies, Cancer Immunology Immunotherapy, 62 (2013) 1397–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Grieco SHH, Lee S, Dunbar WS, MacGillivray RTA, Curtis SB, Maximizing filamentous phage yield during computer-controlled fermentation, Bioprocess and Biosystems Engineering, 32 (2009) 773–779. [DOI] [PubMed] [Google Scholar]
- [132].Bakhshinejad B, Sadeghizadeh M, Bacteriophages and development of nanomaterials for neural regeneration, Neural Regeneration Research, 9 (2014) 1955–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Moon JS, Kim WG, Kim C, Park GT, Heo J, Yoo SY, Oh JW, M13 Bacteriophage-Based Self-Assembly Structures and Their Functional Capabilities, Mini-Reviews in Organic Chemistry, 12 (2015) 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Yoo SY, Merzlyak A, Lee SW, Synthetic Phage for Tissue Regeneration, Mediators of Inflammation, (2014). [DOI] [PMC free article] [PubMed]
- [135].Zhao X, Lin Y, Wang Q, Virus-based scaffolds for tissue engineering applications, Wiley; [DOI] [PubMed] [Google Scholar]
- [136].Koria P, Delivery of Growth Factors for Tissue Regeneration and Wound Healing, Biodrugs, 26 (2012) 163–175. [DOI] [PubMed] [Google Scholar]
- [137].Pierce GF, Growth-Factors in Soft-Tissue Repair and Regeneration, Faseb Journal, 7 (1993) A193–A193. [Google Scholar]
- [138].Tabata Y, Tissue regeneration based on growth factor release, Tissue Engineering, 9 (2003) S5–S15. [DOI] [PubMed] [Google Scholar]
- [139].Howard D, Buttery LD, Shakesheff KM, Roberts SJ, Tissue engineering: strategies, stem cells and scaffolds, Journal of Anatomy, 213 (2008) 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Visser R, Rico-Llanos GA, Pulkkinen H, Becerra J, Peptides for bone tissue engineering, Journal of Controlled Release, 244 (2016) 122–135. [DOI] [PubMed] [Google Scholar]
- [141].Le Guillou-Buffello D, Bareille R, Gindre M, Sewing A, Laugier P, Amedee J, Additive effect of RGD coating to functionalized titanium surfaces on human osteoprogenitor cell adhesion and spreading, Tissue Engineering Part A, 14 (2008) 1445–1455. [DOI] [PubMed] [Google Scholar]
- [142].Visser R, Arrabal PM, Santos-Ruiz L, Fernandez-Barranco R, Becerra J, Cifuentes M, A Collagen-Targeted Biomimetic RGD Peptide to Promote Osteogenesis, Tissue Engineering Part A, 20 (2014) 34–44. [DOI] [PubMed] [Google Scholar]
- [143].Jager M, Boge C, Janissen R, Rohrbeck D, Hulsen T, Lensing-Hohn S, Krauspe R, Herten M, Osteoblastic potency of bone marrow cells cultivated on functionalized biometals with cyclic RGD -peptide, Journal of Biomedical Materials Research Part A, 101 (2013) 2905–2914. [DOI] [PubMed] [Google Scholar]
- [144].Cao X, Yu WQ, Qiu J, Zhao YF, Zhang YL, Zhang FQ, RGD peptide immobilized on TiO2 nanotubes for increased bone marrow stromal cells adhesion and osteogenic gene expression, Journal of Materials Science-Materials in Medicine, 23 (2012) 527–536. [DOI] [PubMed] [Google Scholar]
- [145].Policastro GM, Becker ML, Osteogenic growth peptide and its use as a bio-conjugate in regenerative medicine applications, Wiley Interdisciplinary Reviews-Nanomedicine and Nanobiotechnology, 8 (2016) 449–464. [DOI] [PubMed] [Google Scholar]
- [146].Moore NM, Lin NJ, Gallant ND, Becker ML, The use of immobilized osteogenic growth peptide on gradient substrates synthesized via click chemistry to enhance MC3T3-E1 osteoblast proliferation, Biomaterials, 31 (2010) 1604–1611. [DOI] [PubMed] [Google Scholar]
- [147].Balian G, Beck G, Madhu V, Sikes R, Cui Q, Liang H, Bush J, Peptides from phage display library modulate gene expression in mesenchymal cells and potentiate osteogenesis in unicortical bone defects, J Vis Exp, (2010). [DOI] [PMC free article] [PubMed]
- [148].Henry KA, Arbabi-Ghahroudi M, Scott JK, Beyond phage display: non-traditional applications of the filamentous bacteriophage as a vaccine carrier, therapeutic biologic, and bioconjugation scaffold, Front Microbiol, 6 (2015) 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Srivastava A, Kaido T, Carrier E, Immunological factors that affect the in vivo fate of T7 phage in the mouse, Journal of Virological Methods, 115 (2004) 99–104. [DOI] [PubMed] [Google Scholar]
- [150].O’Brien FJ, Biomaterials & scaffolds for tissue engineering, Materials Today, 14 (2011) 88–95. [Google Scholar]
- [151].Guo B, Lei B, Li P, Ma PX, Functionalized scaffolds to enhance tissue regeneration, Regen Biomater, 2 (2015) 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Villa MM, Wang LP, Huang JP, Rowe DW, Wei M, Bone tissue engineering with a collagen-hydroxyapatite scaffold and culture expanded bone marrow stromal cells, Journal of Biomedical Materials Research Part B-Applied Biomaterials, 103 (2015) 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Martins IM, Reis RL, Azevedo HS, Phage Display Technology in Biomaterials Engineering: Progress and Opportunities for Applications in Regenerative Medicine, ACS Chem Biol, 11 (2016) 2962–2980. [DOI] [PubMed] [Google Scholar]
- [154].Derda R, Musah S, Orner BP, Klim JR, Li LY, Kiessling LL, High-Throughput Discovery of Synthetic Surfaces That Support Proliferation of Pluripotent Cells, Journal of the American Chemical Society, 132 (2010) 1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Moyer TJ, Kassam HA, Bahnson ESM, Morgan CE, Tantakitti F, Chew TL, Kibbe MR, Stupp SI, Shape-Dependent Targeting of Injured Blood Vessels by Peptide Amphiphile Supramolecular Nanostructures, Small, 11 (2015) 2750–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Chan JM, Zhang LF, Tong R, Ghosh D, Gao WW, Liao G, Yuet KP, Gray D, Rhee JW, Cheng JJ, Golomb G, Libby P, Langer R, Farokhzad OC, Spatiotemporal controlled delivery of nanoparticles to injured vasculature, Proceedings of the National Academy of Sciences of the United States of America, 107 (2010) 2213–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Meinel L, Karageorgiou V, Fajardo R, Snyder B, Shinde-Patil V, Zichner L, Kaplan D, Langer R, Vunjak-Novakovic G, Bone tissue engineering using human mesenchymal stem cells: Effects of scaffold material and medium flow, Annals of Biomedical Engineering, 32 (2004) 112–122. [DOI] [PubMed] [Google Scholar]
- [158].Pelled G, Turgeman G, Aslan H, Gazit Z, Gazit D, Mesenchymal Stem Cells for bone gene therapy and tissue engineering, Current Pharmaceutical Design, 8 (2002) 1917–1928. [DOI] [PubMed] [Google Scholar]
- [159].Schneider RK, Neuss S, Knuchel R, Perez-Bouza A, Mesenchymal stem cells for bone tissue engineering, Pathologe, 31 (2010) 138–146. [DOI] [PubMed] [Google Scholar]
- [160].Jin GZ, Kim TH, Kim JH, Won JE, Yoo SY, Choi SJ, Hyun JK, Kim HW, Bone tissue engineering of induced pluripotent stem cells cultured with macrochanneled polymer scaffold, Journal of Biomedical Materials Research Part A, 101 (2013) 1283–1291. [DOI] [PubMed] [Google Scholar]
- [161].Lou XX, Induced Pluripotent Stem Cells as a new Strategy for Osteogenesis and Bone Regeneration, Stem Cell Reviews and Reports, 11 (2015) 645–651. [DOI] [PubMed] [Google Scholar]
- [162].Takahashi K, Yamanaka S, Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors, Cell, 126 (2006) 663–676. [DOI] [PubMed] [Google Scholar]
- [163].Dimitriou R, Jones E, McGonagle D, Giannoudis PV, Bone regeneration: current concepts and future directions, Bmc Medicine, 9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Henkel J, Woodruff MA, Epari DR, Steck R, Glatt V, Dickinson IC, Choong PFM, Schuetz MA, Hutmacher DW, Bone Regeneration Based on Tissue Engineering Conceptions - A 21st Century Perspective, Bone Research, 1 (2013) 216–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Hu J, Zhou Y, Huang L, Liu J, Lu H, Effect of nano-hydroxyapatite coating on the osteoinductivity of porous biphasic calcium phosphate ceramics, BMC Musculoskelet Disord, 15 (2014) 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].He P, Sahoo S, Ng KS, Chen K, Toh SL, Goh JC, Enhanced osteoinductivity and osteoconductivity through hydroxyapatite coating of silk-based tissue-engineered ligament scaffold, J Biomed Mater Res A, 101 (2013) 555–566. [DOI] [PubMed] [Google Scholar]
- [167].Jiang J, Hao W, Li Y, Yao J, Shao Z, Li H, Yang J, Chen S, Hydroxyapatite/regenerated silk fibroin scaffold-enhanced osteoinductivity and osteoconductivity of bone marrow-derived mesenchymal stromal cells, Biotechnol Lett, 35 (2013) 657–661. [DOI] [PubMed] [Google Scholar]
- [168].Huang Y, He J, Gan L, Liu X, Wu Y, Wu F, Gu ZW, Osteoconductivity and osteoinductivity of porous hydroxyapatite coatings deposited by liquid precursor plasma spraying: in vivo biological response study, Biomed Mater, 9 (2014) 065007. [DOI] [PubMed] [Google Scholar]
- [169].Lin L, Chow KL, Leng Y, Study of hydroxyapatite osteoinductivity with an osteogenic differentiation of mesenchymal stem cells, J Biomed Mater Res A, 89 (2009) 326–335. [DOI] [PubMed] [Google Scholar]
- [170].Polo-Corrales L, Latorre-Esteves M, Ramirez-Vick JE, Scaffold design for bone regeneration, J Nanosci Nanotechnol, 14 (2014) 15–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Cholas R, Padmanabhan SK, Gervaso F, Udayan G, Monaco G, Sannino A, Licciulli A, Scaffolds for bone regeneration made of hydroxyapatite microspheres in a collagen matrix, Materials Science & Engineering C-Materials for Biological Applications, 63 (2016) 499–505. [DOI] [PubMed] [Google Scholar]
- [172].de Guzman RC, Saul JM, Ellenburg MD, Merrill MR, Coan HB, Smith TL, Van Dyke ME, Bone regeneration with BMP-2 delivered from keratose scaffolds, Biomaterials, 34 (2013) 1644–1656. [DOI] [PubMed] [Google Scholar]
- [173].Chen N, Tian LL, He LM, Ramakrishna S, Nanobiomaterials for neural regeneration, Neural Regeneration Research, 11 (2016) 1372–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI, Selective differentiation of neural progenitor cells by high-epitope density nanofibers, Science, 303 (2004) 1352–1355. [DOI] [PubMed] [Google Scholar]
- [175].Tang DY, Yu Y, Zhao XJ, Schachner M, Zhao WJ, Single chain fragment variable antibodies developed by using as target the 3rd fibronectin type III homologous repeat fragment of human neural cell adhesion molecule L1 promote cell migration and nemitogenesis, Experimental Cell Research, 330 (2015) 336–345. [DOI] [PubMed] [Google Scholar]
- [176].Bjornson CRR, Rietze RL, Reynolds BA, Magli MC, Vescovi AL, Turning brain into blood: A hematopoietic fate adopted by adult neural stem cells in vivo, Science, 283 (1999) 534–537. [DOI] [PubMed] [Google Scholar]
- [177].Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI, Selective differentiation of neural progenitor cells by high-epitope density nanofibers, Science, 303 (2004) 1352–1355. [DOI] [PubMed] [Google Scholar]
- [178].Hou ST, Dove M, Anderson E, Zhang HB, MacKenzie CR, Identification of polypeptides with selective affinity to intact mouse cerebellar granule neurons from a random peptide-presenting phage library, Journal of Neuroscience Methods, 138 (2004) 39–44. [DOI] [PubMed] [Google Scholar]
- [179].Frenkel D, Solomon B, Filamentous phage as vector-mediated antibody delivery to the brain, Proceedings of the National Academy of Sciences of the United States of America, 99 (2002) 5675–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Campo GM, Micali A, Avenoso A, D’Ascola A, Scuruchi M, Pisani A, Bruschetta A, Calatroni A, Puzzolo D, Campo S, Inhibition of small HA fragment activity and stimulation of A(2A) adenosine receptor pathway limit apoptosis and reduce cartilage damage in experimental arthritis, Histochemistry and Cell Biology, 143 (2015) 531–543. [DOI] [PubMed] [Google Scholar]
- [181].Zmolik JM, Mummert ME, Pep-1 as a novel probe for the in situ detection of hyaluronan, Journal of Histochemistry & Cytochemistry, 53 (2005) 745–751. [DOI] [PubMed] [Google Scholar]
- [182].Laurent TC, Laurent UB, Fraser JR, The structure and function of hyaluronan: An overview, Immunol Cell Biol, 74 (1996) A1–7. [DOI] [PubMed] [Google Scholar]
- [183].Kean TJ, Duesler L, Young RG, Dadabayev A, Olenyik A, Penn M, Wagner J, Fink DJ, Caplan AI, Dennis JE, Development of a peptide-targeted, myocardial ischemia-homing, mesenchymal stem cell, J Drug Target, 20 (2012) 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Dasa SSK, Suzuki R, Gutknecht M, Brinton LT, Tian Y, Michaelsson E, Lindfors L, Klibanov AL, French BA, Kelly KA, Development of target-specific liposomes for delivering small molecule drugs after reperfused myocardial infarction, J Control Release, 220 (2015) 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Wong T, McGrath JA, Navsaria H, The role of fibroblasts in tissue engineering and regeneration, British Journal of Dermatology, 156 (2007) 1149–1155. [DOI] [PubMed] [Google Scholar]