Abstract
Membrane proteins reside in lipid bilayers and are typically extracted from this environment for study, which often compromises their integrity. Here we eject intact assemblies from membranes, without chemical disruption, and use mass spectrometry to define their composition. From E. coli outer membranes, we identify a chaperone-porin association and lipid interactions in the beta-barrel assembly machinery. Bridging inner and outer membranes we observe efflux pumps, and from inner membranes a pentameric pore of TonB, and the protein-conducting channel Sec YEG, in association with F1FO ATP-synthase. Intact mitochondrial membranes from Bos taurus yield respiratory complexes and fatty acid-bound dimers of the ADP/ATP transporter (ANT-1). These results highlight the importance of native membrane environments for retaining small-molecule binding, subunit interactions and associated chaperones of the membrane proteome.
Genes encoding membrane proteins comprise between 20-30% of the genome of all living cells and perform critical processes ranging from mediating drug resistance in bacteria through to the complex mitochondrial respiratory chain in humans. Recent developments in structural biology, including high-resolution cryo-electron microscopy (EM), are uncovering new structures and roles of membrane proteins (1). Often, subunit stoichiometry and lipid binding properties of complexes extracted in detergent micelles has been controversial prompting development of native mass spectrometry (nMS) methods. Now broadly accepted for retaining the stoichiometry of soluble complexes (2) recent developments in nMS of membrane proteins assemblies have not only uncovered subunit stoichiometries but have also found roles for lipids in modulating structures (3, 4). To reveal stoichiometry and lipid binding in the absence of detergents alternative nMS approaches have been developed, including bicelles (5), amphipols (6), nanodiscs (7), and styrene maleic acid copolymer lipid particles (SMALPs) (8). All these approaches require some chemical intervention and high levels of protein expression, thereby restricting their use primarily to proteins of bacterial origin. Here we overcome these limitations and show that we can obtain mass spectra for protein assemblies ejected directly from native membranes of prokaryotic and eukaryotic organisms and uncover many previously uncharacterized interactions in the process.
To develop this approach we first used membrane protein enriched extracellular vesicles (MPEEVs) overexpressing the Epithelial Fusion Failure protein 1 (EFF-1), reported to be monomeric, or the Anchor Cell Fusion Failure protein 1 (AFF-1) with unknown stoichiometry (9, 10). MPEEVs of both proteins from Syrian hamster BHK21 cultured cells were prepared and characterised as described previously (9). The presence of either EFF-1 or AFF-1 increased the diversity of cardiolipins (CDL) as was confirmed by standard approaches (fig. S1A and C). Subjecting these vesicles to sonication, to destabilize their integrity (see Online Methods and fig. S2), and to high energy across a modified Orbitrap MS (11), we released monomeric EFF-1 and intact dimeric AFF-1 directly from vesicles (fig. S1B).
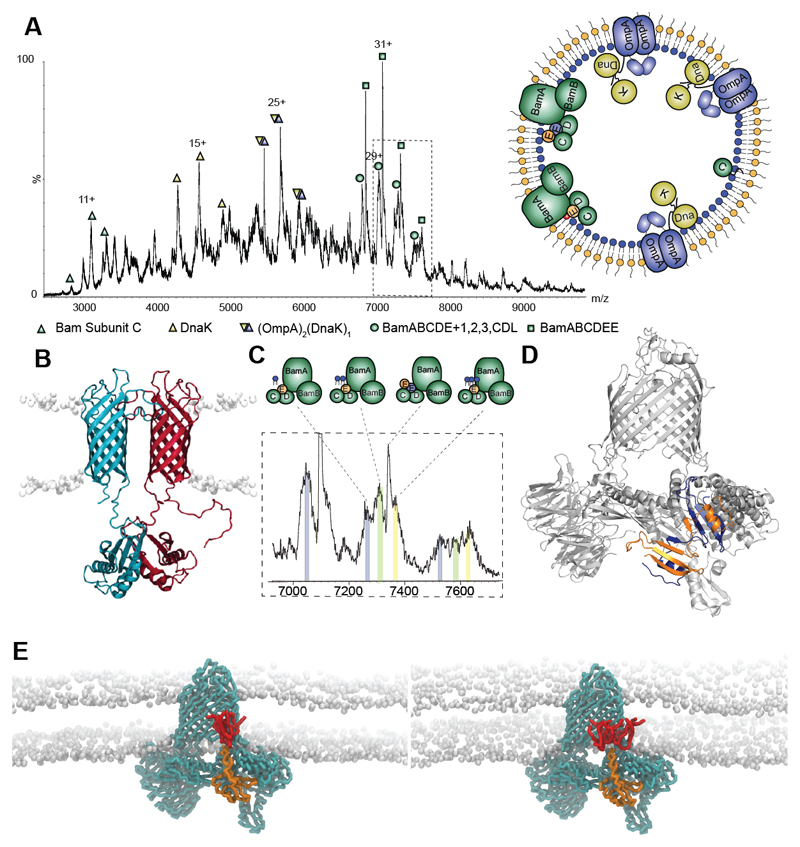
Having established the feasibility of our approach, we investigated its application to additional native membranes. We separated outer and inner membranes of E. coli via a sucrose gradient, prepared vesicles and used proteomics to identify membrane proteins (12) (fig. S3). To interpret the mass spectra we developed and applied a protocol that takes into account peak width, collision induced dissociation (CID) and accurate mass only accepting solutions within ± ~ 0.3 % of calculated masses (fig. S2 and Tables S1-3). Starting from the low m/z range of the spectrum recorded for E. coli outer membranes we assigned BamC with a lipid anchor, a component of the beta barrel assembly machinery (BAM) (fig. 1A) (13). Moving to higher m/z we assign DnaK, implicated previously in the assembly of outer membrane porins (14) and confirmed via CID of a 143 kDa complex together with OmpA (fig. S4). Previous reports that DnaK co-immunoprecipitates with full-length pro-OmpA, but not with pro-OmpA(Δ3) (14), implied that sequences outside the β-barrel are required to maintain accessibility of DnaK binding sites. Our measured mass (within 0.10 %) is consistent with ADP-bound DnaK binding to proOmpA and associating with a second OmpA, likely through the C-terminal dimerisation domain (15), to form OmpA:proOmpA:DnaK:ADP (fig. 1B).
Fig. 1. Protein complexes ejected directly from E. coli outer membranes.
A Mass spectrum recorded at 400 V assigned to BamC, DnaK, DnaK:OmpA:pro-OmpA, and two states of the Bam complex with inset an outer membrane vesicle with complexes observed. B Model of the OmpA dimer (15) with the hydrophobic pro-sequence (red) a potential binding site for DnaK. C Expansion of the mass spectrum assigned to the Bam complex with monomeric BamE (Bam ABCDE) binding to one, two and three cardiolipins (grey, green, yellow respectively). D Atomic structure of BamE dimer (PDB: 2YH9) (orange and blue) docked into the Bam complex (PDB: 5D0O) with BamE monomer removed. E MD simulations of the BamABCDE complex (cyan) with monomeric BamE (orange) and two (lhs) and three (rhs) CDL molecules (red).
Turning to the high m/z region of the mass spectrum recorded for outer membranes, a predominant series of peaks was assigned to BAM (13). Structural studies, following detergent extraction and overexpression of all five subunits on a single plasmid yielded primarily a 1:1:1:1:1 stoichiometry for Bam subunits (A-E) (13, 16, 17). From native membranes however, a hexamer was ejected with a subunit composition of ABCD(E)2 (fig. 1A and D), a small population of this complex had been observed previously from recombinant preparations (17). Since a domain swapped dimer was observed by x-ray analysis of Bam E alone (18), and NMR solution studies were consistent with a population of BamE dimers (19), we docked into Bam ABCD a BamE dimer and used MD simulation to test its stability (20). The complex remained stable over 5 μs, following heating to 323 K, consistent with its viability in the E. coli outer membrane lipid environment. A second series was assigned to pentameric Bam ABCDE complex, its diffuse peaks consistent with binding of up to three CDL molecules (fig. 1C and fig. S5). Preferential binding of phosphatidylglycerol (PG) over CDL had been reported previously (19) prompting us to explore lipid binding preferences of BamE using MD simulations (3 x 5 μs) (fig. 1E and fig. S6). Up to three CDL lipids made contact implying CDL attachment, through BamE, which likely anchors the complex to a region of the membrane high in CDL and may contribute to a membrane targeting mechanism.
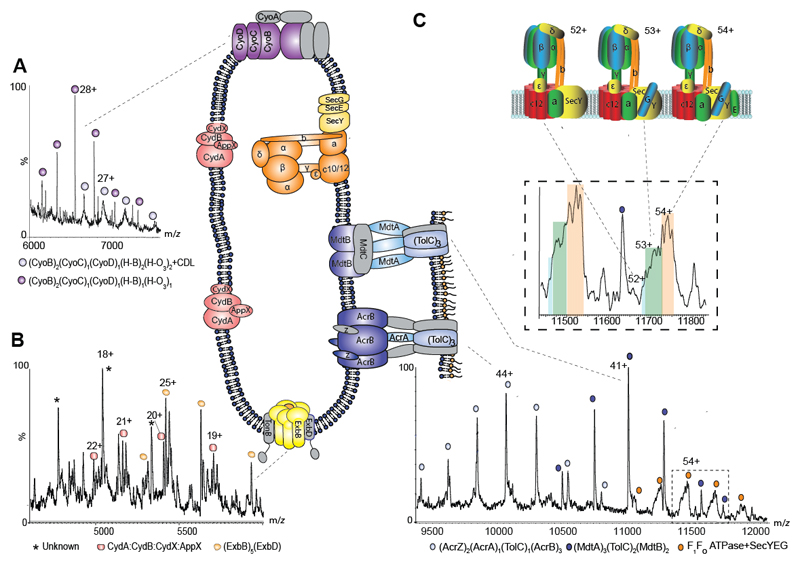
Inner membrane vesicles prepared from E. coli represent a significant challenge as together they contain a minimum of 42 different proteins (12) and yield complex spectra for assignment (fig. S7). We used the heterogeneity of cofactor binding to first identify cytochromes bo3 and CydAB bd oxidase. Peaks corresponding to (CyoB)2(CyoC)1(CyoD)1, with one or two HemeO3/HemeB factors and additional CDL binding (diffuse peaks) imply that lipid binding stabilizes structures with the full-heme complement, supported by reduced charge state (fig. 2A) and in-line with the proposed dimer association for cytochrome bo3 from native membranes (21). Extensive peak splitting attributed to different Heme groups (B558, B595) and ubiquinol helped identify the CydAB cytochrome bd oxidase complex (fig. 2B and fig. S8). Both CydX and the paralogous small transmembrane protein AppX have the potential to interact with the CydAB complex and have overlapping cellular functions (22). From the native membrane we found that CydX and AppX were able to interact simultaneously with CydAB to form a heterotetramer.
Fig. 2. Regions of the mass spectrum recorded for inner membranes from E. coli yield cytochromes, the Ton complex multi-drug transporters and the intact ATP synthase in complex with the SecYEG translocon.
A and B Expanded regions of the spectrum assigned to the cytochromes bo3 and cytochrome bd oxidase showing peak splitting due to binding of quinol and heme groups (fig. S8). The pentameric ExbB complex, with one copy of ExbD located in the center of the pore, forming part of the TonB complex is also observed (yellow). C High m/z region of the mass spectrum assigned to multidrug efflux pumps AcrAB and MdtAB and the intact ATP synthase. Expansion of the peaks assigned to the ATPase reveals binding of SecY (blue) SecYG (green) and SecYEG (orange) charge states 52+, 53+ and 54+ shown schematically.
We next assigned, on the basis of mass, parts of the energy-transducing Ton complex located within the inner membrane. In the inner membrane three integral membrane proteins reside: ExbB, ExbD and TonB. From x-ray crystallography a second copy of ExbD was located within the pentameric ExbB pore (23) while from EM both hexameric and pentameric assemblies were defined (24). Our results confirm the existence of only the pentameric pore within the native membrane with measured charge states implying trapping of one ExbD protomer within the compact globular complex (fig. 2B).
At the higher m/z region sub-assemblies of multidrug efflux pumps, including AcrAB-TolC and the less well-characterised but related pump MdtABTolC (25), were uncovered spanning both membranes (fig. 2C). For AcrAB-TolC all three inner membrane subunits (AcrB) are preserved in mass spectra bound to the small subunit (AcrZ) discovered only recently, thought to modulate substrate preference (26) and modelled into cryo-EM structures (27). One copy of the outer membrane protein TolC is bridged by a single copy of the periplasmic subunit AcrA to the inner membrane complex yielding AcrB3:AcrZ2:AcrA:TolC. In the case of MdtABTolC dimeric MdtB remains assembled with (MdtA)3 and (TolC)2 in the outer membrane (MdtB2MdtA3:TolC2). Since all three MdtA subunits remain attached they are likely supported by dimeric MdtB in the inner membrane, consistent with the role of MdtC in substrate binding (28) and not in supporting periplasmic subunits. AcrABZ-TolC has undergone more extensive disassembly during the solution/sonication process than MdtAB-TolC which remains largely intact with charge states (fig. S9) indicative of highly-charged subunits from AcrABZ-TolC undergoing CID (29). The fact that both complexes survive at least in part, however, points to new ways of studying the effects of antibiotics on the assembly and conformational change of these multidrug resistance pumps.
We also observed peak splitting due to ATP/ADP binding, at the highest m/z values which, together with dissociation of subunits with the mass of the c-subunit, is indicative of ATP synthase (fig. 2C, figs. S10 and S11). Mass differences between populations were assigned to binding of SecE, Y and G, consistent with SecYEG remaining in contact with the F1FOATP synthase as reported previously for insertion of subunit a (30). We next considered the stoichiometry of the FO ring. Early reports had suggested a variable stoichiometry depending upon metabolic conditions (31), our data are consistent with 12 c subunits in the FO ring. We conclude therefore that in the native membrane interactions between FOF1 ATPase and SecYEG are maintained following insertion in the membrane, together with c-subunits in FO, which are either lost during detergent extraction (32) or filtered out in other methods.
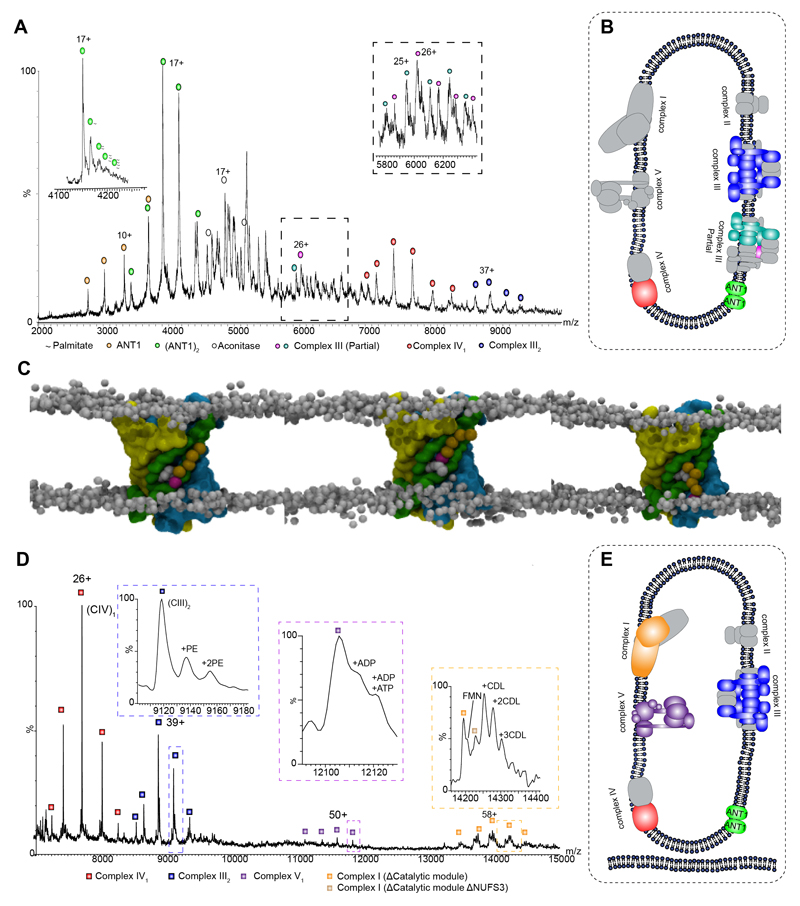
Observation of E. coli ATPase prompted us to consider inner mitochondrial membranes, densely populated with protein complexes responsible for control of the proton gradient and oxidative phosphorylation between the intermembrane space and the inner mitochondrial matrix (33). Surprisingly inner mitochondrial membranes from Bos taurus yielded only a single subunit of the ATPase (β) and no significant subassemblies of complex I (fig. 3A). Other complexes in the respiratory chain that were observed include complex II, monomeric complex IV with lipid and cofactor occupancy (34) and dimeric complex III, with seven core subunits confirmed by dissociation of cytochrome b and UQCRB (fig. 3A and fig. S12).
Fig. 3. Intact Mitochondria and inner membranes yield complexes I, III, IV and V as well as the adenine nucleotide translocator 1 (Ant1) with palmitate transport through the dimer interface.
A Mass spectrum of bovine mitochondrial inner membranes recorded at 600 V reveals the ANT-1 dimer, complexes III and IV, as well as aconitase. The left inset, recorded at 400 V, shows an expanded view of the spectrum at 4100 to 4300 m/z revealing multiple palmitate anions bounds to the dimer of ANT-1. The right inset shows an expansion of the boxed area in the main panel. B Depiction of the protein assemblies ejected from sonicated mitochondrial inner membranes. Subunits shown in gray have dissociated. C MD simulation of ANT-1 after 2.1, 2.5, and 2.6 μs (left to right) in an asymmetric membrane containing phosphatidylcholine (PC), phosphatidylethanolamine (PE), and CDL (only in the matrix leaflet). The protein surface is colored according to the three pseudo-repeats (R1, yellow; R2, green; R3, cyan), and the charged palmitate headgroup (magenta) is buried between helices three and four. D Complexes I, III, IV, and V are expelled from intact mitochondria. Intact complex V is observed with associated nucleotides, together with a dimer of complex IV (fig. S15) and partial assemblies of complex I (the charge state of the subassembly lacking NUFS3 is z = 56+), in the absence of the catalytic core, bound to FMN. Inset schematics represent assigned membrane complexes, color coded according to labels on the peaks. E Depiction of the protein assemblies ejected from sonicated intact mitochondrial membranes color coded according to the labels on the peaks. Gray subunits were not observed.
Interestingly the most abundant protein in the mass spectrum of the inner mitochondrial membrane was the adenine nucleotide translocase 1 (ANT-1) (fig. 3A). The stoichiometry of ANT-1 has remained controversial with monomeric structures of ANT-1 and UCP2 solved by X-ray crystallography and NMR, and proposed to be functional (35–37). However, in-vivo and biochemical experiments were consistent with the functional unit of ANTs and UCPs being dimeric (38), a proposal supported by short timescale by MD simulations (39). MS of ANT-1 revealed that it is predominantly dimeric with low occupancy binding of a number of saturated fatty acids (palmitate anions) indicative of a transport mechanism rather than specific binding interaction (fig. 3A and S13). MD simulations in lipid bilayers showed that within 10 μs tightly bound dimers formed only if CDL was present in the inner leaflet of the membrane (fig. S14). Fatty acid binding was also observed with the palmitate head group buried between two helices in each subunit (Y132 and F177) (fig. 3B, S14D and movie S1). In situ binding of multiple palmitate anions within the dimer ejected from the native mitochondrial membrane provides direct evidence in support of the role of this fatty acid in the control of uncoupling through ANT-1 transport (40).
Since complexes I and V were largely absent from spectra of inner mitochondrial membranes we applied our protocol to intact mitochondria, without prior separation of inner and outer membranes, reasoning that the outer membrane and non-inverted protein orientation may protect complexes exposed during sonication. Resulting mass spectra again revealed ANT-1 dimers bound to palmitate (fig. S13), but also in this case lipid bound sub-assemblies of Complex I bound to its FMN cofactor and lipids, a Complex IV dimer (fig. 3C and S15), together with intact Complex V bound to nucleotides.
The release of complexes I and V, only when protected by the outer membrane, prompts consideration of the mechanism of direct ejection from native membranes. The three steps to consider are (i) formation of vesicles (ii) sonication in ammonium acetate to disrupt vesicles and (iii) mass spectrometry under high electric fields from -400 to -700 V. During the first step vesicles are formed from membrane preparations with protein complexes in both orientations. For purified inner mitochondrial membranes, we observe primarily inside-out vesicles with ATPase F1 heads visible in cryo-EM images (fig. S16). We anticipate that sonication may form defects in vesicles allowing ingress of the ammonium acetate buffer used for electrospray. The third stage, in which high voltage is applied, favors expulsion of complexes charged by the electrospray process and attracted by the high electric field. Striking differences are observed between the two preparations from mitochondria. For inner membranes we attribute the virtual absence of intact mitochondrial ATP synthase to exposure to shearing forces during sonication. Similar arguments could be made for the non-appearance of complex I from inner membranes; its exposed hydrophilic peripheral arm may have sheared during sonication leaving the hydrophobic membrane embedded complex less susceptible to charge. By contrast for intact mitochondria complexes I and V are ejected, largely intact, supporting the hypothesis that exposure to shearing forces during sonication is a key determinant in survival of complexes ejected from native membranes.
While full details of the mechanism of ejection from native membranes into vacuum are the subject of ongoing research the data presented here establish a detergent and chemical free MS approach that overcomes potential artefacts introduced by the use of these reagents. The number of new interactions of membrane proteins uncovered, with lipids, chaperones and cofactors in association, is testament to the stability endowed by the native membrane. Significantly, access to the protein ensemble of different membrane compartments, at unparalleled mass resolution, will enable a new perspective on the effects of drugs and disease associated mutations on target complexes within the context of their native membrane environments.
Supplementary Material
One Sentence Summary.
Mass spectra reveal the composition of complexes ejected directly from native membrane environments.
Acknowledgments
The authors thank C. J. Schofield for access to a probe sonicator and the Robinson group for helpful discussions.
Funding
DSC and CVR. are supported by the European Research Council grant no. 69551-ENABLE and a Wellcome Trust Investigator Award (104633/Z/14/Z). LAB was supported by a Human Frontier Science Program Long Term Fellowship and a Canadian Institutes for Health Research Postdoctoral Fellowship. KG was supported by a Wellcome Trust Senior Research Fellowship (090895/Z/09/Z) and a core award (090532/Z/09/Z). C.G. is supported by the JST (JPMJCR13M4), BINDS and MEXT (Kiban B: 17H03647). F.S. and S.K. are supported by the BBSRC (BB/M029573/1). A.G.S was supported by the NHMRC fellowship (1090408).
Footnotes
Author contributions:
D.S.C. produced the MPEEVs and preformed MS experiments. L.A.B and K.G. purified E. coli membranes and visualized vesicles by EM. D.W. established and executed the lipidomics. V.B.E. provided critical advice for proteomics. S.L.R., S.J.M, S.K and F.S performed MD simulations. TZBM provided critical support in establishing the MPEEV system. C.J. and C.G purified intact mitochondria and inner membrane samples. A.G.S provided critical review. D.S.C and C.V.R designed experiments, preformed data analysis and wrote the manuscript.
Competing interests:
Authors declare no competing interests.
Data and materials availability
All data is available in the main text or the supplementary materials.
References
- 1.Vinothkumar KR, Henderson R. Single particle electron cryomicroscopy: trends, issues and future perspective. Q Rev Biophys. 2016 Jan;49:e13. doi: 10.1017/S0033583516000068. [DOI] [PubMed] [Google Scholar]
- 2.Heck AJ. Native mass spectrometry: a bridge between interactomics and structural biology. Nat Methods. 2008 Nov;5:927. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 3.Gupta K, et al. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017 Jan 19;541:421. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laganowsky A, et al. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014 Jun 5;510:172. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hopper JT, et al. Detergent-free mass spectrometry of membrane protein complexes. Nat Methods. 2013 Dec;10:1206. doi: 10.1038/nmeth.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bechara C, et al. MALDI-TOF mass spectrometry analysis of amphipol-trapped membrane proteins. Analytical chemistry. 2012 Jul 17;84:6128. doi: 10.1021/ac301035r. [DOI] [PubMed] [Google Scholar]
- 7.Marty MT, Hoi KK, Gault J, Robinson CV. Probing the Lipid Annular Belt by Gas-Phase Dissociation of Membrane Proteins in Nanodiscs. Angew Chem Int Ed Engl. 2016 Jan 11;55:550. doi: 10.1002/anie.201508289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Postis V, et al. The use of SMALPs as a novel membrane protein scaffold for structure study by negative stain electron microscopy. Biochim Biophys Acta. 2015 Feb;1848:496. doi: 10.1016/j.bbamem.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeev-Ben-Mordehai T, Vasishtan D, Siebert CA, Whittle C, Grunewald K. Extracellular vesicles: a platform for the structure determination of membrane proteins by Cryo-EM. Structure. 2014 Nov 04;22:1687. doi: 10.1016/j.str.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeev-Ben-Mordehai T, Vasishtan D, Siebert CA, Grunewald K. The full-length cell-cell fusogen EFF-1 is monomeric and upright on the membrane. Nature communications. 2014 May 28;5:3912. doi: 10.1038/ncomms4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van de Waterbeemd M, et al. High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat Methods. 2017 Mar;14:283. doi: 10.1038/nmeth.4147. [DOI] [PubMed] [Google Scholar]
- 12.Stenberg F, et al. Protein complexes of the Escherichia coli cell envelope. J Biol Chem. 2005 Oct 14;280 doi: 10.1074/jbc.M506479200. 34409. [DOI] [PubMed] [Google Scholar]
- 13.Bakelar J, Buchanan SK, Noinaj N. The structure of the beta-barrel assembly machinery complex. Science. 2016 Jan 8;351:180. doi: 10.1126/science.aad3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi HY, Hyndman JB, Bernstein HD. DnaK promotes the selective export of outer membrane protein precursors in SecA-deficient Escherichia coli. J Biol Chem. 2002 Dec 27;277:51077. doi: 10.1074/jbc.M209238200. [DOI] [PubMed] [Google Scholar]
- 15.Marcoux J, et al. Mass spectrometry defines the C-terminal dimerization domain and enables modeling of the structure of full-length OmpA. Structure. 2014 May 6;22:781. doi: 10.1016/j.str.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu Y, et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature. 2016 Mar 03;531:64. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 17.Iadanza MG, et al. Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nature communications. 2016 Sep;7:12865. doi: 10.1038/ncomms12865. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albrecht R, Zeth K. Structural basis of outer membrane protein biogenesis in bacteria. J Biol Chem. 2011 Aug 5;286:27792. doi: 10.1074/jbc.M111.238931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knowles TJ, et al. Structure and function of BamE within the outer membrane and the beta-barrel assembly machine. EMBO Rep. 2011 Feb;12:123. doi: 10.1038/embor.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu PC, Samsudin F, Shearer J, Khalid S. It Is Complicated: Curvature, Diffusion, and Lipid Sorting within the Two Membranes of Escherichia coli. J Phys Chem Lett. 2017 Nov 16;8:5513. doi: 10.1021/acs.jpclett.7b02432. [DOI] [PubMed] [Google Scholar]
- 21.Stenberg F, von Heijne G, Daley DO. Assembly of the cytochrome bo3 complex. J Mol Biol. 2007 Aug 17;371:765. doi: 10.1016/j.jmb.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 22.VanOrsdel CE, et al. The Escherichia coli CydX protein is a member of the CydAB cytochrome bd oxidase complex and is required for cytochrome bd oxidase activity. J Bacteriol. 2013 Aug;195:3640. doi: 10.1128/JB.00324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Celia H, et al. Structural insight into the role of the Ton complex in energy transduction. Nature. 2016 Oct 6;538:60. doi: 10.1038/nature19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maki-Yonekura S, et al. Hexameric and pentameric complexes of the ExbBD energizer in the Ton system. Elife. 2018 Apr 17;7 doi: 10.7554/eLife.35419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anes J, McCusker MP, Fanning S, Martins M. The ins and outs of RND efflux pumps in Escherichia coli. Front Microbiol. 2015;6:587. doi: 10.3389/fmicb.2015.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc Natl Acad Sci U S A. 2012 Oct 9;109:16696. doi: 10.1073/pnas.1210093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, et al. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. Elife. 2017 Mar 29;6 doi: 10.7554/eLife.24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HS, Nikaido H. Different functions of MdtB and MdtC subunits in the heterotrimeric efflux transporter MdtB(2)C complex of Escherichia coli. Biochemistry. 2012 May 22;51:4188. doi: 10.1021/bi300379y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall Z, Hernandez H, Marsh JA, Teichmann SA, Robinson CV. The role of salt bridges, charge density, and subunit flexibility in determining disassembly routes of protein complexes. Structure. 2013 Aug 6;21:1325. doi: 10.1016/j.str.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kol S, et al. Subunit a of the F(1)F(0) ATP synthase requires YidC and SecYEG for membrane insertion. J Mol Biol. 2009 Jul 31;390:893. doi: 10.1016/j.jmb.2009.05.074. [DOI] [PubMed] [Google Scholar]
- 31.Schemidt RA, Qu J, Williams JR, Brusilow WS. Effects of carbon source on expression of F0 genes and on the stoichiometry of the c subunit in the F1F0 ATPase of Escherichia coli. J Bacteriol. 1998 Jun;180:3205. doi: 10.1128/jb.180.12.3205-3208.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobti M, et al. Cryo-EM structures of the autoinhibited E. coli ATP synthase in three rotational states. Elife. 2016 Dec 21;5 doi: 10.7554/eLife.21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhlbrandt W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015 Oct 29;13:89. doi: 10.1186/s12915-015-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liko I, et al. Dimer interface of bovine cytochrome c oxidase is influenced by local posttranslational modifications and lipid binding. Proc Natl Acad Sci U S A. 2016 Jul 19;113:8230. doi: 10.1073/pnas.1600354113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pebay-Peyroula E, et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003 Nov 6;426:39. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 36.Berardi MJ, Shih WM, Harrison SC, Chou JJ. Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature. 2011 Jul 24;476:109. doi: 10.1038/nature10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunji ER, Crichton PG. Mitochondrial carriers function as monomers. Biochim Biophys Acta. 2010 Jun-Jul;1797:817. doi: 10.1016/j.bbabio.2010.03.023. [DOI] [PubMed] [Google Scholar]
- 38.Klingenberg M. The ADP and ATP transport in mitochondria and its carrier. Biochim Biophys Acta. 2008 Oct;1778:1978. doi: 10.1016/j.bbamem.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 39.Hedger G, et al. Lipid-Loving ANTs: Molecular Simulations of Cardiolipin Interactions and the Organization of the Adenine Nucleotide Translocase in Model Mitochondrial Membranes. Biochemistry. 2016 Nov 15;55:6238. doi: 10.1021/acs.biochem.6b00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sparks LM, et al. ANT1-mediated fatty acid-induced uncoupling as a target for improving myocellular insulin sensitivity. Diabetologia. 2016 May;59:1030. doi: 10.1007/s00125-016-3885-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.