Abstract
Inhibiting MYC has long been considered unfeasible, although its key role in human cancers makes it a desirable target for therapeutic intervention. One reason for its perceived undruggability was the fear of catastrophic side effects in normal tissues. However, we previously designed a dominant-negative form of MYC called Omomyc and used its conditional transgenic expression to inhibit MYC function both in vitro and in vivo. MYC inhibition by Omomyc exerted a potent therapeutic impact in various mouse models of cancer, causing only mild, well-tolerated, and reversible side effects. Nevertheless, Omomyc has been so far considered only a proof of principle. In contrast with that preconceived notion, here, we show that the purified Omomyc mini-protein itself spontaneously penetrates into cancer cells and effectively interferes with MYC transcriptional activity therein. Efficacy of the Omomyc mini-protein in various experimental models of non–small cell lung cancer harboring different oncogenic mutation profiles establishes its therapeutic potential after both direct tissue delivery and systemic administration, providing evidence that the Omomyc mini-protein is an effective MYC inhibitor worthy of clinical development.
Introduction
MYC is a transcription factor that instructs multiple intracellular and extracellular programs including proliferation, metabolism, and apoptosis, as well as immune checkpoint regulation (1–4). To coordinate the transcription of its target genes, MYC heterodimerizes with MAX (MYC Associated Factor X) and binds the E-box (enhancer box) element CACGTG (or variants thereof) through its basic-helix–loop–helix leucine-zipper (b-HLH-LZ) domain (5, 6).
In adult individuals, MYC expression is normally low and tightly regulated and is restricted to proliferating tissues. In contrast, aberrantly high and/or deregulated MYC activity is causally implicated in most cancers and often correlates with the aggressiveness of the disease (7, 8). Hyperactive MYC function in tumor cells is generally a consequence of the genetic amplification or translocation of the MYC gene downstream of strong promoters, its relentless induction by upstream signals, or impaired turnover (9). All these mechanisms result in the uncoupling of cellular proliferation from normal growth factor regulation and contribute to many of the phenotypic hallmarks of cancer (10).
Numerous studies have substantiated the crucial role of MYC in governing tumorigenesis and tumor maintenance. In mouse models of MYC-driven malignancies, tumors develop addiction to MYC, and the shutdown of its function results in growth arrest, apoptosis, and differentiation (11–15). Even in models of malignancies driven by other oncogenes such as KRasG12D, Ha-RasV12, or SV40 viral antigen, tumors become addicted to tonic MYC function (16–18), supporting the therapeutic value of targeting MYC in most, if not all, oncological diseases.
However, despite this undisputable therapeutic opportunity, a MYC inhibitor has yet to become clinically available (19–21), and there are various caveats to its successful design and use: First, a complete inhibition of MYC function could result in important side effects (22, 23). Second, MYC is a nuclear protein, which is consequently more difficult to reach than membrane or cytoplasmic molecular targets. Third, the MYC family includes three different proteins, c-Myc, N-Myc, and L-Myc, which in certain conditions are functionally redundant, so ideally, all of them require simultaneous inhibition (24). Fourth, MYC is an intrinsically disordered molecule, which lacks an enzymatic “active site” that could be efficiently targeted by common small-molecule design. This latter point has posed a particularly hefty challenge in the successful design of a specific yet efficiently delivered MYC inhibitor. Most small chemicals designed to date have aimed at impeding the dimerization between MYC and its obligate partner MAX (direct inhibitors) or the binding of the MYC/MAX heterodimer to other interacting partners (indirect inhibitors) or have instead aimed at nonrelated synthetically lethal targets (19–21). However, so far, none of these approaches has resulted in any clinically viable therapy safely targeting MYC.
We previously modeled systemic inhibition of MYC using Omomyc, a dominant negative mutant consisting of the MYC dimerization domain with four mutations in the leucine zipper (25). These mutations alter Omomyc’s dimerization specificity and allow it to prevent binding of all MYC family members to their target promoters (25, 26). When expressed in a switchable transgenic mouse model (TRE-Omomyc;CMVrtTA), Omomyc demonstrated a therapeutic window independent of the driving oncogenic lesion or tissue of origin and caused only mild and reversible side effects in normal tissues (16–18, 27, 28).
In general, Omomyc has been used in its transgenic form as a proof of concept, and the Omomyc mini-protein has been considered too bulky and unfit for intracellular delivery (29, 30). However, other b-HLH-LZ–containing proteins can act as protein transduction domains (PTD) (30, 31), and Omomyc harbors an amphipathic helical basic region, a recurrent feature of cell-penetrating peptides (CPPs) (25, 32). Hence, we decided to test whether the purified Omomyc mini-protein itself could have cell-penetrating activity and evaluated its amenability to pharmaceutical development.
To do so, we used well-characterized and validated non–small cell lung cancer (NSCLC) in vitro and in vivo models. Despite recent clinical progresses in the treatment of this disease, NSCLC remains one of the deadliest cancers for both men and women, and a large population of patients still lacks an effective therapeutic option due to either poor effectiveness of available therapies for the initial mutational profile of their disease (for instance, for KRas-mutated tumors) or de novo mutations acquired alongside the development of resistance to therapy. The MYC gene itself is amplified in up to 30% of NSCLCs, which are frequently EGFR (epidermal growth factor receptor)–mutated or KRas-mutated (9, 33–36).
Results
The purified Omomyc mini-protein displays high affinity for MAX and for E-box DNA sequences
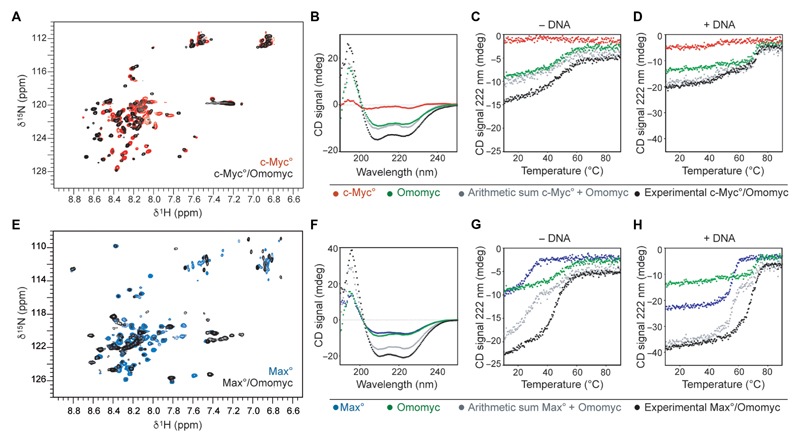
As a first step toward evaluating the capacity of the recombinantly produced and purified Omomyc mini-protein to bind MYC and MAX and interfere with their binding to DNA, we characterized its dimerization with the b-HLH-LZ domains of MYC (c-Myc°) and MAX (Max°) and the DNA binding of the complexes in the solution using circular dichroism (CD) and nuclear magnetic resonance (NMR) (Fig. 1, A to H). The chemical shift displacements observed in the 15N-HSQC (heteronuclear single-quantum coherence) spectra of each 15N-labeled c-Myc° (Fig. 1A) and Max° (Fig. 1E) upon addition of Omomyc in the absence of DNA demonstrate the change in conformation caused by the assembling of heterodimers in each mixture and indicate that the molecular interactions stabilizing the heterodimers involve a large portion of the b-HLH-LZ domain in both cases.
Fig. 1. The Omomyc mini-protein forms highly stable DNA binding homodimers and heterodimers with MAX in solution.
(A) Overlay of the 1H-15N-HSQC of 15N-Myc° in the absence (red) and in the presence (black) of Omomyc. (B) Far–ultraviolet (UV) CD spectra of c-Myc° (red, 8 μM monomer units), Omomyc (green, 8 μM monomer units), the arithmetic sum of both red and green spectra (gray), and the spectrum of an equimolar mix of c-Myc° and Omomyc at a total concentration of 16 μM in monomer units (black) recorded at 25°C. (C) Thermal denaturation of the solutions described in (B). (D) Thermal denaturation of the solutions described in (B) to which equimolar amounts (in dimer units) of an E-box DNA duplex were added. Contribution of the DNA to the denaturation curves was removed. (E) Overlay of the 1H-15N-HSQC of 15N-Max° in the absence (blue) and in the presence (black) of Omomyc. (F) Far-UV CD spectra of Max° (blue, 8 μM monomer units), Omomyc (green, 8 μM monomer units), arithmetic sum of both blue and green spectra (gray), and experimental equimolar mix of Max° and Omomyc at a total concentration of 16 μM in monomer units (black) recorded at 25°C. (G) Thermal denaturation of the solutions described in (F). (H) Thermal denaturation of the solutions described in (F) to which equimolar amounts (in dimer units) of E-box DNA duplex were added. All thermal denaturations were monitored at 222 nm.
The CD spectrum of Omomyc in the absence of DNA (Fig. 1, B and F) depicts deep minima of about 208 and 222 nm, characteristic of a homodimer with a helical content comparable to that of the folded, homodimeric MAX b-HLH-LZ domain (Fig. 1F) (25, 37) and in contrast to the shallow minima observed in the spectrum of c-Myc° (Fig. 1B), which instead is consistent with the structure of a protein displaying mainly intrinsically disordered features. Perhaps unexpectedly, Omomyc forms a more thermodynamically stable homodimer than Max° (Fig. 1G, fig. S1A, and table S1 in data file S1). The gain in helical-specific signal intensity of the experimental mixtures compared to the arithmetic sum of each component’s curves (Fig. 1, B to D, and F to H) confirms in the low micromolar range the heterodimerization observed by NMR in the high micromolar range (Fig. 1, A and E). Furthermore, in the absence of DNA, the c-Myc°/Omomyc heterodimer is thermodynamically stable and comparable to the Omomyc homodimer (Fig. 1C). In addition, the thermal denaturation of Omomyc in the presence of Max° reveals their rapid heterodimerization and provides evidence that the thermodynamic stability of the Max°/Omomyc heterodimer is comparable to that of the Omomyc homodimer (Fig. 1G and table S1 in data file S1).
To evaluate the affinities of the different dimers for DNA, we then performed thermal denaturation of equimolar mixtures of c-Myc°/Omomyc (Fig. 1D) or Max°/Omomyc (Fig. 1H) and a canonical E-box duplex. Both c-Myc° and Omomyc homodimers formed a stable complex with the DNA probe (Fig. 1D), but the homodimeric Omomyc/DNA complex was more thermodynamically stable than the c-Myc°/DNA complex, consistent with recently reported observations from electrophoretic mobility shift assays (38). Consistent with early observations (25), no additional complex was detected in the c-Myc°/Omomyc/DNA mixture by CD or by fluorescence anisotropy of labeled E-box probes (fig. S1B and table S2 in data file S1), indicating that the c-Myc°/Omomyc heterodimer does not bind DNA at physiological temperatures or that its affinity for DNA is much weaker than that of either homodimer. In contrast, in the Max°/Omomyc/DNA mixture (Fig. 1H), in addition to the two transitions corresponding to the homodimeric Max° or Omomyc complexes with DNA, there is a third transition present, corresponding to the heterodimeric Max°/Omomyc/DNA complex. The relative amplitudes of these three transitions approach a 1:2:1 ratio distribution, as would be expected for dimers with similar affinities for DNA. Hence, in physiological conditions, both homodimeric Omomyc and heterodimeric MAX/Omomyc are likely to be the predominant dimers on DNA; the dominant dimeric species will depend on the relative nuclear concentration of each protein. Globally, these data confirm that, as shown previously by distinct methods with transgenic Omomyc (25, 39, 40), at physiologically relevant temperatures and concentrations, the Omomyc mini-protein has the potential to act as a MYC/Omomyc heterodimer, sequestering MYC away from DNA, as well as an Omomyc homodimer and MAX/Omomyc heterodimer that competitively displace MYC from its target genes.
Omomyc is a bioactive PTD
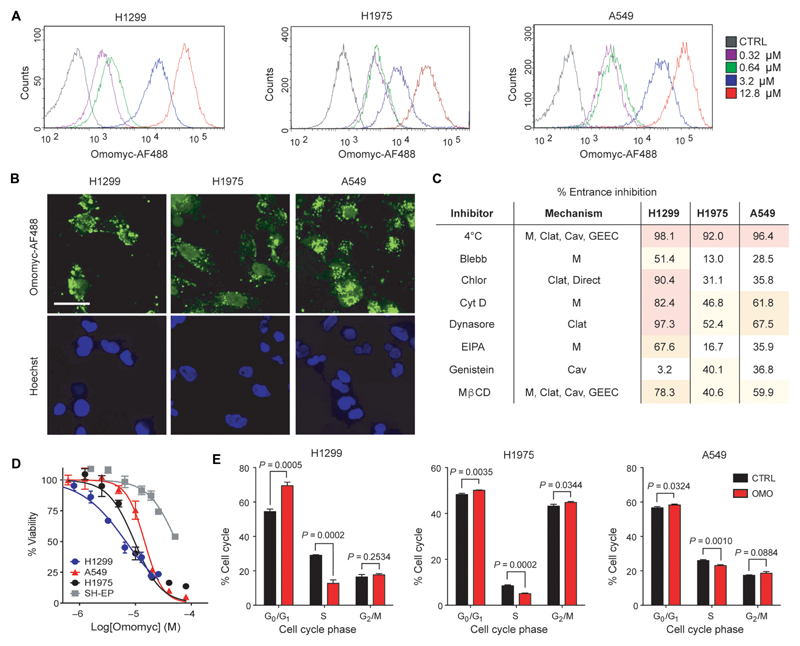
To investigate the potential of the Omomyc mini-protein to act as a PTD, we purified the 91–amino acid Omomyc mini-protein and labeled it with a fluorescent Alexa Fluor 488 (AF488)–maleimide group. We then treated various cancer cell lines with increasing concentrations of Omomyc-AF488 for 15 min and, after trypsinization to remove extracellularly bound mini-protein, analyzed intracellular fluorescence by flow cytometry (Fig. 2A and fig. S2). Internalization of Omomyc-AF488 into live cells was observed at concentrations as low as 0.32 μM in each cell line. Fluorescence microscopy revealed an uneven cellular distribution, with Omomyc-AF488 partially localized in the nuclei, in addition to showing a punctate membrane signal typical of endosomal localization (Fig. 2B).
Fig. 2. The Omomyc mini-protein spontaneously penetrates into human NSCLC cells.
(A) Lung adenocarcinoma H1299, H1975, and A549 cell lines were treated with 0.32, 0.64, 3.2, or 12.8 μM Omomyc-AF488 for 15 min in serum-free medium, trypsinized, and analyzed by flow cytometry. (B) H1299, H1975, and A549 cells were treated with 3.2 μM Omomyc-AF488 for 4 hours, stained with Hoechst 3342, washed, mounted, and analyzed by confocal microscopy. Scale bar, 10 μm. (C) NSCLC cell lines were preincubated at 4° or 37°C and treated with 0.64 μM Omomyc-AF488 for 15 min at the same temperature, trypsinized, and immediately analyzed by flow cytometry or pretreated with inhibitors (Blebb, blebbistatin; Chlor, chlorpromazine; Cyt D, cytochalasin D; EIPA, 5-ethylisopropylamiloride; Mβ methyl-β-cyclodextrin) of endocytosis or lipid raft-mediated macropinocytosis (M, macropinocytosis; Cav, caveolin-mediated; Clat, clathrin-mediated) and then treated with 0.64 μM Omomyc-AF488 for 15 min in the presence of the inhibitor followed by trypsinization and analysis by flow cytometry. Inhibition of entrance (%) compared to vehicle-treated cells at 37°C is shown for each cell line. (D) Dose response of the NSCLC panel of cells and of MYC-independent SH-EP cells to increasing concentrations of Omomyc as measured by resazurin dye colorimetric assay. The median inhibitory concentration (IC50) was 5.9 μM for H1299, 8.2 μM for H1975, 11.4 μM for A549, and 25.6 μM for SH-EP cells. (E) Quantification of cell cycle phase populations from flow cytometric analysis of PI incorporation after 3 days of treatment with 12.8 μM Omomyc (OMO) compared to vehicle control (CTRL). All experiments were performed at least twice for each condition. Mean and SD are shown in (D) and (E), and statistical significance was calculated by a two-tailed unpaired Student’s t test.
CPPs and PTDs enter cells via a variety of uptake pathways including clathrin- or caveolin-mediated endocytosis, glycosyl-phosphatidylinositol (GPI)–enriched early endosomal compartment (GEEC) entry, and lipid raft macropinocytosis (M), often using more than one pathway at a time (32). Incubation of NSCLC cells at 4°C efficiently blocked the entry of 0.64 μM Omomyc-AF488, suggesting involvement of an adenosine triphosphate–dependent uptake mechanism (Fig. 2C). Treatment with a panel of endocytosis inhibitors before incubation with Omomyc-AF488 and analysis by flow cytometry indicated that the contributions of each of these mechanisms vary across cell lines and that clathrin-mediated endocytosis and macropinocytosis appear to account for most of its cell-penetrating properties in the cell lines tested, although a contribution of caveolin-dependent entrance cannot be excluded (Fig. 2C). Enhanced macropinocytosis by KRas-mutated cells has been previously reported (41), suggesting that these cells would be particularly permeable to Omomyc.
The cell-penetrating properties of Omomyc appear to be dependent on its DNA binding basic region and/or the arginine content of this region because an Omomyc-mutant construct bearing three arginine-to-alanine substitutions within this region (termed OmoRA) could not efficiently enter cells in the same experimental conditions (fig. S3, A and B).
Treatment of NSCLC cell lines with increasing concentrations of Omomyc from 390 nM to 50 μM resulted in progressively fewer live cells as measured by resazurin dye quantification, with the IC50 ranging from 6.2 to 13.6 μM (Fig. 2D). In contrast, the IC50 measured in a MYC-independent cancer cell line (42–44) was 27.7 μM. Cell cycle analysis by flow cytometry showed a reduction of proliferating cells, along with an increase of the G0/G1 population for all cell lines tested (Fig. 2E). In the case of H1975 cells, an increase in G2/M was also measured.
Omomyc reverts MYC-driven transcriptional programs
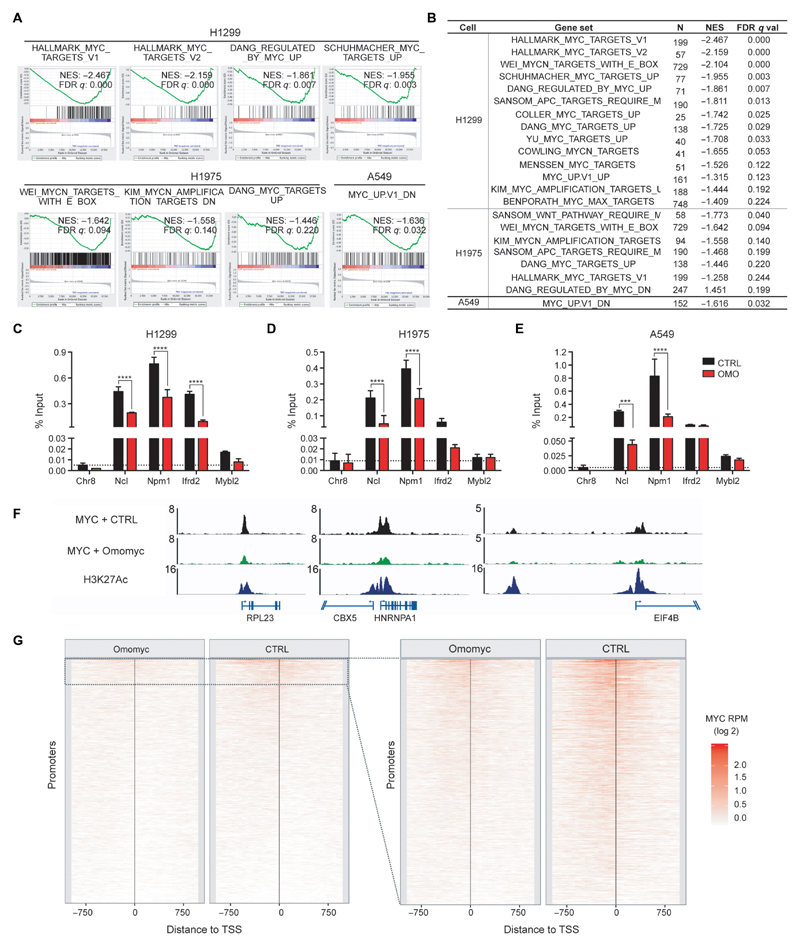
To evaluate the biological outcome of the mini-protein treatment, we performed microarray and gene set enrichment analysis (GSEA). These studies revealed that Omomyc efficiently reverts the expression of MYC-related gene signatures in the three cell lines tested (GSE126455; Fig. 3, A and B, and tables S3 to S6 in data file S1). The amplitude of this MYC-inhibiting activity, as reflected by the number of MYC-driven gene sets altered, was greater in H1299 and H1975 cells compared to A549 cells. This is in accordance with the relative MYC protein expression detected for each cell line (fig. S4). Despite the heterogeneous oncogenic-driving mutation profiles across the three cell lines tested, several programs associated with typical MYC functions such as RNA biogenesis, translational control (including ribosome biogenesis), cellular metabolism, and cell cycle (2, 45, 46) were consistently shut down after treatment with Omomyc (figs. S5 to S7). In addition, we also found that E2F transcription signature modules were reversed, in accordance with the established role of MYC in regulating E2F activity and its binding to E2F-specific promoters in the context of cell cycle regulation (figs. S5 to S7) (47). Last, gene signatures associated with poor prognosis in lung cancer (for example, SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6) were also down-regulated, consistent with the central role of MYC in transformation and tumor maintenance (figs. S5 and S6). In contrast, Omomyc treatment did not suppress gene sets for other transcription factors involved in the pathophysiology of lung cancer, such as ARNT, NFY, SMAD4, STAT1, USF, or YY1 (table S3 in data file S1), supporting the specificity of the Omomyc mini-protein for MYC function.
Fig. 3. Omomyc disrupts MYC transcriptional regulation and binding to promoters of its target genes.
(A and B) GSEA comparing gene expression of vehicle-treated versus Omomyc-treated NSCLC cells. Representative plots of MYC signatures enriched in the vehicle samples compared to the treated samples are shown in (A). Normalized enrichment scores (NES) and q values of well-characterized MYC signature gene sets are listed in (B) and were calculated from technical triplicates. (C to E) MYC chromatin immunoprecipitation (ChIP)–quantitative polymerase chain reaction (qPCR) from H1299 (C), H1975 (D), and A549 (E) cells treated for 72 hours with 12.8 μM Omomyc (red) or vehicle (black) is shown for typical MYC target proximal promoter regions [Chromosome 8 “gene desert” region (Chr8), nucleolin (Ncl), nucleophosmin (Npm1), interferon-related developmental regulator 2 (Ifrd2), and MYB proto-oncogene–like 2 (Mybl2); vehicle-treated (black) and Omomyc-treated (red)]. (C to E) Mean and SD are shown, and statistical significance was calculated by a two-tailed unpaired Student’s t test. ***P < 0.005 and ****P < 0.001. (F and G) MYC ChIP sequencing (ChIP-seq) from A549 cells treated for 48 hours with 12.8 μM Omomyc or vehicle (CTRL). (F) MYC occupancy at selected MYC target proximal promoter regions: ribosomal protein L23 (RPL23), chromobox protein homolog 5 (CBX5), heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1), and eukaryotic translation initiation factor 4B (EIF4B). The ChIP-seq enrichment is displayed as reads per million (RPM). (G) Global analysis: MYC ChIP-seq RPM were calculated in the region (±1 kb) around the transcriptional start site (TSS) of all active promoters (n = 32292), in 50–base pair bins. The promoters are sorted by the RPM in the full 2-kb promoter region in the vehicle sample. The panel on the right shows promoters with a MYC ChIP-seq peak in the vehicle condition (n = 3014).
To gain more insight into the mechanism of action of the Omomyc mini-protein and confirm its MYC inhibitory activity, we used ChIP assays. We first verified that the treatment with Omomyc could displace MYC from specific bona fide target promoter locations previously identified in other cellular systems (38, 48, 49) and/or down-regulated in the three NSCLC cell lines (tables S4 to S7 in data file S1). Consistently, upon Omomyc treatment, all cell lines displayed a marked reduction in MYC binding to those promoters (Fig. 3, C to E), regardless of the differences in their mutational profile. This displacement of MYC was accompanied by changes in the epigenetic mark H3K27Ac characteristic of active transcription (fig. S8, A to C), indicating a potential long-lasting effect of the Omomyc treatment on the expression of these target genes.
We then extended our analysis of MYC occupancy across the whole genome of A549 cells (Fig. 3, F and G). Despite the relatively low MYC protein expression in these cells (fig. S4) (50), in vehicle-treated cells, MYC was detected at numerous target locations that coincided with open and active chromatin regions near transcriptional start sites and enhancers (Fig. 3, F and G). In contrast, less MYC occupancy was detected in Omomyc-treated cells throughout the genome, encompassing both MYC-specific strong and weak motifs and super-enhancers (fig. S8, D and E). At the gene level, complete displacement of MYC peaks surveyed in gene promoters was observed in 77.4% (854 of 1104) of genes upon Omomyc treatment, and at least partial displacement was observed in as many as 97.3% (1055 of 1084) of MYC peaks found in active promoter regions (Fig. 3G).
The Omomyc mini-protein is safe and effective after direct pulmonary administration in a mouse model of lung adenocarcinoma
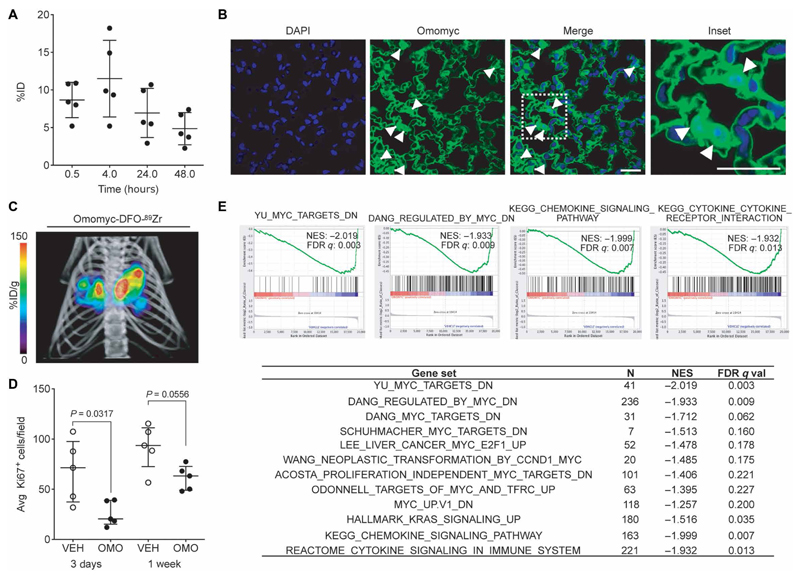
To evaluate the potential therapeutic utility of the Omomyc mini-protein in vivo, we first analyzed its tissue distribution after intranasal administration, a technique that enables direct pulmonary delivery of macromolecule formulations in mice (51). We first covalently attached a deferoxamin-maleimide (DFO) group to Omomyc and radiolabeled this with 89Zr and then measured biodistribution and pharmacokinetic properties in healthy mice by ex vivo radiocounting (Fig. 4A). On average, 8% of the Omomyc-DFO-89Zr dose (2.37 mg/kg) readily (within 30 min) reached the lungs after intranasal administration and persisted there for at least 48 hours (Fig. 4A). Immuno-fluorescence with a specific anti-Omomyc antibody confirmed the detection (and partial nuclear localization) of unlabeled Omomyc mini-protein within the pulmonary epithelium of the treated mice at 4 hours after intranasal instillation (Fig. 4B and fig. S9). To confirm that this method was applicable to mice bearing lung adenocarcinoma, we repeated the procedure using the well-characterized KRasLSL-G12D/+-induced lung adenocarcinoma mouse model (52). The micro–positron emission tomography /micro-computed tomography (mPET)/mCT imaging of the mice revealed that, 24 hours after intranasal instillation, Omomyc-DFO-89Zr localized mainly in the lung tumors (Fig. 4C and movie S1). Although the exact cause for this preferential tumor retention is unknown, it could be related to the altered tumor vasculature or metabolism typically observed in cancer or possibly to the enhanced use of macropinocytosis by KRas-driven tumors (53).
Fig. 4. Omomyc displays efficacy in a lung adenocarcinoma mouse model upon intranasal administration.
(A) Quantification of Omomyc-DFO-89Zr detected in the lungs of healthy mice as a function of time is represented as %ID. Mean, SD, and number of animals are shown. (B) Immunofluorescence of lung tissue from mice treated with the Omomyc mini-protein. A specific anti-Omomyc antibody confirms the presence of Omomyc in the lung cells 4 hours after administration. Arrowheads indicate positively stained nuclei. Scale bars, 10 μm. Higher magnification of the area surrounded by a white dashed line is shown in the right panel. (C) 3D rendering of mPET/mCT imaging of lungs of a tumor-bearing mouse 24 hours after intranasal administration of Omomyc-DFO-89Zr (2.37 mg/kg). CT data are displayed in gray scale and Omomyc-DFO-89Zr mPET data in color scale (n = 2 mice were analyzed). The color scale is expressed as %ID/g for Omomyc-DFO-89Zr uptake. See the accompanying supplementary movie for a rotating representation (Movie S1). (D) Graphical representation of total cells scored as proliferating (Ki67-positive) cells in the lung of mice treated for 3 days or 1 week with Omomyc (2.37 mg/kg) or vehicle (VEH). Median, interquartile range (IQR), and number of animals are shown. Two-tailed unpaired Mann-Whitney test was used to analyze statistical differences between the groups. (E) GSEA comparing gene expression in lung tumors from vehicle-treated versus Omomyc-treated mice. NES and false discovery rate (FDR) q values of MYC signatures and other relevant gene sets are shown. Four representative plots are shown.
Knowing that Omomyc was able to reach its target tissue, we next tested its short-term therapeutic impact on established KRasLSL-G12D/+-driven lung adenocarcinomas. Both 3-day and 1-week treatments with mini-protein (2.37 mg/kg) markedly reduced Ki67 positivity of the tumor tissue (Fig. 4D). The 3-day treatment sufficed to alter the transcriptional profile of the lung tumors, again shutting down several MYC-driven modules along with other gene sets related to the pathophysiology of lung adenocarcinoma, including several gene signatures associated with poor lung cancer prognosis or KRAS activity (Fig. 4E, fig. S10, and table S7 in data file S1). This in vivo treatment also caused changes in chemokine and cytokine profiles, consistent with MYC’s role in modulating the cross-talk between tumor and microenvironment (3, 17).
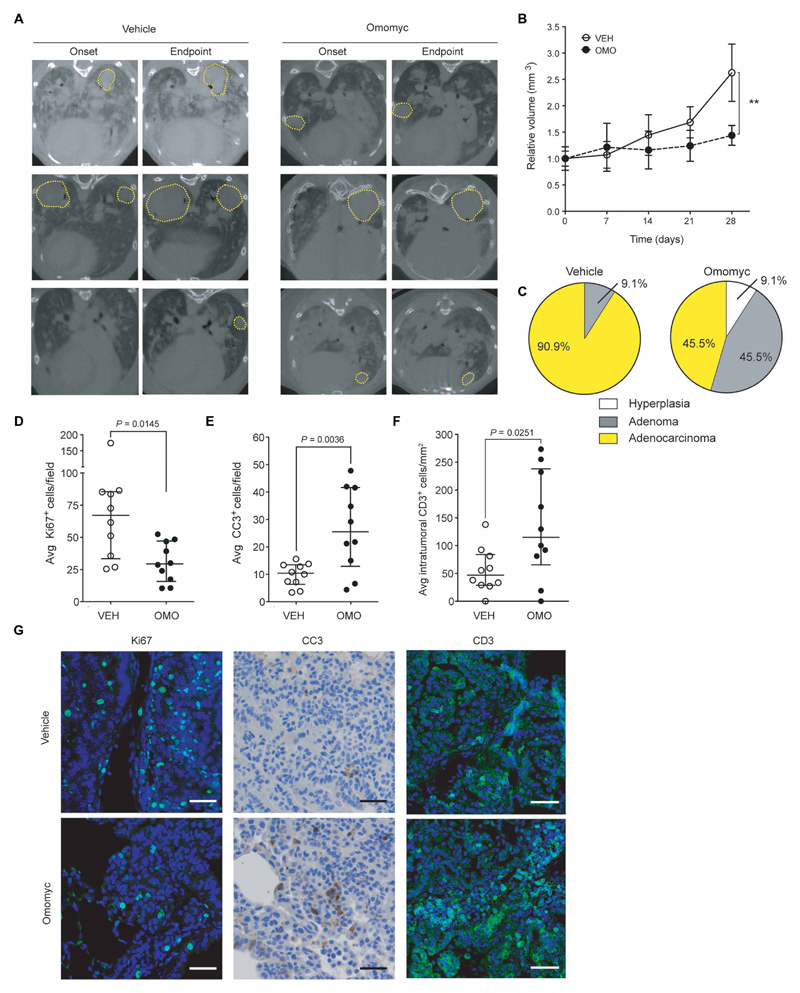
To evaluate longer-term treatment efficacy in the same mouse model of lung adenocarcinoma, we followed the tumor burden progression longitudinally by mCT. After a 4-week treatment with Omomyc (2.37 mg/kg) administered three times per week, tumors of the treated mice did not show any progression, whereas tumors from vehicle-treated mice had more than doubled in size (Fig. 5, A and B, and fig. S11). Furthermore, tumor grade was markedly different between the groups (Fig. 5C): At the experimental endpoint, the Omomyc-treated group included 7.7% of hyperplastic foci, 46.1% of adenomas, and only 46.4% of adenocarcinomas, whereas most vehicle-treated foci were adenocarcinomas (90.9%), suggesting that Omomyc is causing much more than a mere cytostatic effect. Ki67 and cleaved caspase 3 (CC3) immunostaining confirmed that the therapeutic impact of Omomyc is the result of both its antiproliferative and proapoptotic effects (Fig. 5, D to G). Last, immunostaining with anti-CD3 revealed that the Omomyc treatment increased recruitment of T lymphocytes specifically to the tumor site (Fig. 5, F and G), suggesting a possible immune contribution as part of the mechanism of action of the Omomyc mini-protein. No deleterious side effects or weight loss was observed over the course of the 28-day treatment (fig. S11).
Fig. 5. Omomyc reduces tumor burden in a KRasG12D-driven lung adenocarcinoma mouse model.
Mice bearing KRasLSL-G12D/+-induced lung tumors were treated for 4 weeks with Omomyc (2.37 mg/kg) or vehicle administered intranasally. (A) Representative transverse planar CT images from each experimental group taken at treatment onset and endpoint with tumors circled by yellow dotted lines. (B) Relative tumor volumes (RTVs) of vehicle-treated (open circles) and Omomyc-treated (black dots) mice were measured weekly. Mean normalized to size at treatment onset and SEM are shown. Longitudinal growth within a group was analyzed by Kruskal-Wallis test. In contrast to the vehicle-treated group (P = 0.0002), the Omomyc-treated tumors did not show significant growth throughout treatment (P = 0.1579). For analysis at endpoint, two-tailed unpaired Mann-Whitney test was used to assess statistical significance of the difference between the groups (**P < 0.01). (C) Tumor grade was blindly assessed by histological and pathological analysis of hematoxylin and eosin–stained slides of lungs of treated mice and is represented as a pie chart for each group. (D to G) Ki67 (D), CC3 (E), and intratumoral CD3-positive cells (F) of representative animals were quantified by immunostaining at endpoint. Median, IQR, and number of mice are shown. Statistical significance was calculated using a two-tailed Mann-Whitney test. (G) Representative images are shown. Scale bars, 20 μm for Ki67 and CC3 panels. Scale bars, 50 μm for CD3 panel.
Systemic administration of Omomyc and combination with a microtubule-targeting agent increases survival of a xenograft model of NSCLC
To further evaluate the safety and therapeutic potential of the Omomyc mini-protein in vivo administered systemically as a single agent, we made use of a subcutaneous xenograft mouse model of human H1975 cells, which are erlotinib-resistant and EGFR-mutated, PI3K (phosphatidylinositol 3-kinase)–mutated, and P53-mutated. After intravenous administration of Omomyc-DFO-89Zr (2.6 mg/kg), the plasma clearance was 2.85 ml/kg per min (fig. S12), a value compatible with the development of a drug with a reasonable dosage regimen.
Once the tumors were established (average tumor size of 130 ± 59 mm3), mice were randomized for systemic treatment with Omomyc (60 mg/kg) or vehicle, administered four times per week by intravenous injection. During the whole experiment, tumor-volume measurements confirmed a reduction in tumor growth with Omomyc treatment (Fig. 6A), already evident at 8 days after treatment onset. In addition, Omomyc was well tolerated at this dose and schedule, as monitored through changes in body weight, necropsy, complete cell count, biochemistry data, and light microscopy pathology reports from the heart, lung, liver, spleen, kidney, stomach, small and large intestine, muscle, peripheral nerve, bone marrow, brain, and skin of healthy mice (fig. S13).
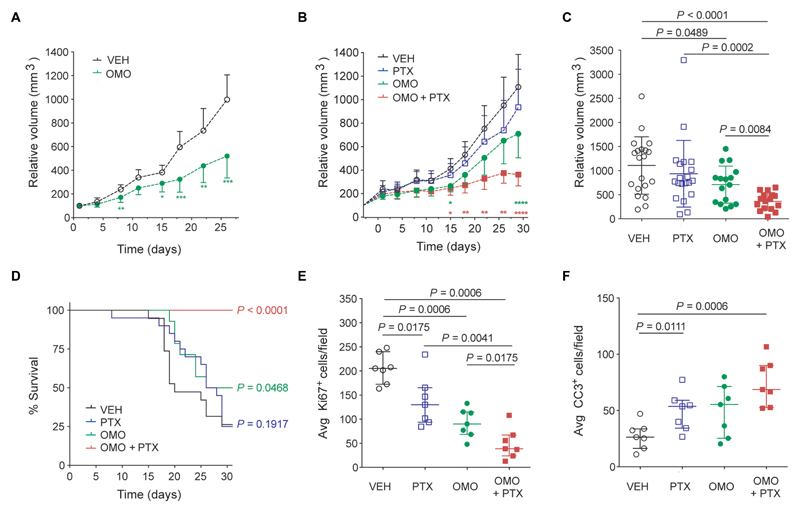
Fig. 6. Intravenous treatment with Omomyc and combination with paclitaxel display superiority to chemotherapy in a lung adenocarcinoma xenograft model.
(A) RTV is reduced in mice with established disease (H1975 xenografts) treated with Omomyc (60 mg/kg) four times per week (OMO, green dots) (n = 14 per group). (B) RTV of vehicle-treated (open circles), paclitaxel (PTX)–treated (open squares), Omomyc-treated (120 mg/kg; green dots), and combination-treated (OMO + PTX; red squares) mice measured twice per week (n = 20 per group). Mean and 95% confidence interval are shown, and statistical significance compared to vehicle-treated group at each time point was calculated by a two-tailed unpaired Student’s t test (A and B). *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001. (C) Tumor volume at experimental endpoint (30 days). Mean and SD are shown. (D) A Kaplan-Meier survival curve is shown, and the statistical significance was determined by a log-rank test. (E and F) Ki67 (E) and CC3 (F) positivity was quantified by immunostaining at endpoint for seven representative animals per group. Median and IQR are shown.
Chemotherapeutics constitute one of the main drug options for treatment of patients with EGFR-mutated NSCLC resistant to tyrosine kinase inhibitors (54). We thus compared the Omomyc mini-protein’s efficacy to the microtubule-stabilizing agent paclitaxel and tested their combination in the same mouse model. In this case, mice bearing H1975 tumors were randomized (at an average tumor size of 257 ± 142 mm3) and subjected to intravenous injection with a high dose of Omomyc (120 mg/kg, to check for its tolerability) four times per week, intraperitoneal injection with paclitaxel (5 mg/kg) twice per week, a combination of both, or vehicle. Once again, Omomyc slowed down tumor progression in established tumors, even more than paclitaxel itself (Fig. 6, B and C). Combination therapy of Omomyc with paclitaxel proved superior to both stand-alone therapies and almost completely abrogated tumor growth, promoting prolonged mouse survival (Fig. 6D). No adverse effects were detected in any of the groups during the treatment period (fig. S13). Immunostaining of Ki67 and CC3 confirmed the antiproliferative effect of each therapeutic regimen and the increased proapoptotic effects of the combination therapy (Fig. 6, E and F).
Discussion
MYC constitutes a key cellular node that is frequently deregulated in cancer. In NSCLC in particular, MYC amplification is found in up to 30% of malignancies (9), and in the rest of the cases, MYC function is likely deregulated owing to the high prevalence of mutations in upstream oncogenes such as KRas and EGFR.
The relevance of attacking MYC in cancer is already well established, but no drugs are clinically available for its direct targeting yet (19). Indirect inhibition by bromodomain and extraterminal protein inhibitors constitutes the most clinically advanced strategy so far, enabling the down-regulation of MYC expression in some specific contexts but not in others, for instance, in lung adenocarcinomas that display LKB1 mutations (55).
On the other hand, attempts to directly target MYC with small molecules have yielded disappointing results overall, partly due to poor bioavailability and to the difficulty in disrupting the MYC/MAX heterodimer, which has a wide interface devoid of binding pockets that hinder development of highly selective small molecules (19–21).
We previously showed that the use of a dominant negative such as Omomyc addresses these shortcomings. Omomyc enables the efficient displacement of MYC from its target genes through a combined molecular mechanism that involves interference both with MYC/MAX heterodimerization (by sequestering either protein partner) and with MYC’s binding to DNA (25–27, 38). In accordance, our results show how the purified Omomyc mini-protein achieves potent interference with MYC function by displacing it from its target genes and binding to DNA in the form of inactive competitors, either as a homodimer or as a heterodimer with MAX. The relative contribution of each dimer to the mechanism of action will depend on the relative concentrations of Omomyc, MYC, and MAX [and other members of the extended Myc network; (56)] in any given cellular context. Our spectroscopic analyses show that the dissociation constant (Kd) of the Omomyc homodimer for the E-box DNA sequences is in the nanomolar range, indicating that even a low nuclear concentration of Omomyc could efficiently displace MYC from its DNA binding sites, making it effective in tumor types displaying a wide range of MYC concentrations. Our results in vitro and in vivo support this hypothesis, although future studies including larger panels of cell lines would enable a more thorough validation.
However, the key finding here is the intrinsic cell-penetrating property of the Omomyc mini-protein, which prompts its direct delivery in vivo. Direct use of such a large polypeptide has long been deemed therapeutically unviable, and recent data suggested that the naked Omomyc peptide was not cell penetrating (57). Here, we show, however, that the purified Omomyc mini-protein displays unexpected cell-penetrating properties and constitutes a PTD capable of cellular entry and target engagement, blocking MYC transcriptional function in vitro and in vivo in both human and murine model systems.
Peptides and proteins have not been typically considered as drug candidates because their large molecular size and sometimes hydrophilic nature often impaired their permeation across biological membranes. However, they have finally become interesting biopharmaceutical candidates since the discovery in 1988 that some cationic peptides derived from the human immunodeficiency virus Tat transactivator protein can penetrate living cells (58, 59). Since then, multiple CPPs and PTDs have been identified or engineered and have been used to deliver various types of cargos ranging from nucleic acids (up to 1 kb) to proteins and even microparticles (60–62). This class of therapeutics opened the valuable opportunity to target the “undruggable” landscape of intracellular protein domains lacking structured binding pockets for small-molecule ligands. Given the increasing need for highly efficient and nontoxic carriers to facilitate permeation of therapeutic peptides and proteins across biomembranes and to enhance the pharmacological effects of biopharmaceuticals, CPPs have the potential to become an important tool in pharmaceutical research.
Although a portion of the fluorescently labeled Omomyc mini-protein appears to remain trapped in the endosomes (or in the perinuclear region), the amount of Omomyc reaching the nuclei is sufficient to cause MYC displacement from its promoter binding sites and to shut down multiple MYC-specific transcriptional programs, as well as other programs typically driven by MYC, such as cell cycle, ribosomal biogenesis, and metabolism. Consistent with the established role of MYC in histone modifications on genomic target locations (45), our data also show that the displacement of MYC from DNA target promoters is accompanied by a modulation of the epigenetic mark H3K27Ac. Future studies should enable a more comprehensive characterization of the effect of Omomyc treatment on epigenetic marks. This transcriptional reprogramming also occurs in vivo, in KRasG12D tumors isolated from mice treated by intranasal administration with the Omomyc mini-protein, independently of changes in the amount of MYC protein. Furthermore, consistent with the more benign phenotype observed in the residual tumors in Omomyc-treated groups at the experimental endpoint in vivo, Omomyc also caused the shutdown of gene signatures indicative of KRAS activity and of poor prognosis in lung cancer. Several active transcriptional programs driven by other transcription factors involved in the pathology of the disease, including other E-box–binding transcription factors, were not modulated by Omomyc treatment, supporting the specificity of the observed effect.
We also observed modulation of cytokines and chemokine gene signatures of the tumors and found an active recruitment of CD3-positive cells specifically at the tumor site. These observations are in line with recent studies validating the role of MYC in the regulation of immune checkpoints, such as cluster of differentiation 47 and programmed death ligand 1 (4), and suggest that the successful inhibition of MYC in tumors could promote autologous or heterologous regulation of such immune checkpoints and reactivate an anergic antitumor immune response. A better characterization of this response elicited by the Omomyc treatment is required to clarify the mini-protein’s mechanism of action and might pave the way to future studies evaluating the therapeutic potential of combination with immune checkpoint inhibitors to overcome primary, adaptive, or acquired resistance to immune therapy (53, 54). Further studies will also be needed to address the specific relationship between the mutational profile of tumors and their response to treatment.
Most relevant to clinical application, the Omomyc mini-protein displayed sufficient delivery in vivo to elicit specific therapeutic impact in tumors while avoiding toxicity. Treatment with the Omomyc mini-protein was safe even at relatively high systemic doses while abrogating the progression of murine and human lung adenocarcinomas, with no detectable side effects in normal tissues. Intravenous administration of Omomyc displayed superiority to the tubulin-targeting agent paclitaxel in an EGFR-mutated and erlotinib-resistant tumor model, and the combination of both agents more than doubled the survival of the mice.
In conclusion, the data presented here reveal several features and unforeseen properties of the Omomyc mini-protein, which overcome some important obstacles related to the design of a clinically viable MYC inhibitor, opening an opportunity to pharmacologically inhibit MYC in NSCLC and potentially other malignant diseases and supporting its further preclinical development. Although some skepticism still persists regarding the therapeutic utility of CPPs in the clinic, an increasing number of clinical trials demonstrate their therapeutic applicability, in particular, in oncology (63), recognizing their potential to hit previously undruggable targets.
Materials and Methods
Study design
This study was designed to validate the Omomyc mini-protein as a therapeutic strategy for NSCLC. We assessed its efficacy using in vitro and in vivo assays, combining cell lines and mouse models of NSCLC. For in vitro studies, a minimum of two biological replicates were performed. For in vivo models, we used the following: (i) a transgenic KRasLSL-G12D/+-driven lung adenocarcinoma model and (ii) a subcutaneous xenograft mouse model of human H1975 cells, which are EGFR-, PI3K-, and P53-mutated. All the animal studies were performed in accordance with the ARRIVE (Animal research: Reporting of in vivo experiments) guidelines and the 3R rule of replacement, reduction and refinement principles. Mice were housed and treated following the protocols approved by the CEEA (Ethical Committee for the Use of Experimental Animals) at the Vall d’Hebron Institute of Oncology (VHIO), Barcelona.
For the in vivo experiments, 5 (for the biodistribution and safety experiments) to 11 (for the efficacy experiments) mice were used per treatment group, and they were randomized by tumor size and weight. When female and male animals were used (such as with the transgenic model of NSCLC), mice were also randomized to achieve similar numbers of each in all groups. Mice that died before the end of the experiment for reasons unrelated to the treatment were excluded. For ethical reasons, we ended experiments before mice developed respiratory distress or when tumor volume surpassed 1750 mm3. Experiments were not performed in a blinded fashion.
Mice
KRasLSL-G12D/+ mice were genotyped by Transnetyx, and generation of lung tumors in both males and females was performed as previously described (52). Animals were maintained in a mixed C57BL/6J × FVB/N background. A minimum of five mice per time point and condition were randomized, and treatment started 16 weeks after adenoviral Cre recombinase infection (two biological replicates of the experiment were performed). Animals were anesthetized by inhaled isoflurane (AbbVie Farmaceutica, S.L.U.) and were intranasally administered with either Omomyc or vehicle [10 mM sodium acetate (pH 6.5)] in 30 μl of total volume.
For the experiments using the xenograft mouse model of NSCLC, 2 × 106 H1975 cells were inoculated subcutaneously to the dorsal flank of 6-week-old athymic nude-Foxn1 mice (JANVIER LABS). Once the tumors were established, mice were randomized for treatment. Intravenous and intraperitoneal administrations were performed in a volume of 200 μl.
Pharmacokinetic and biodistribution study
For the pharmacokinetic and biodistribution studies by mPET/mCT, female 8-week-old FVB/NRj mice were purchased from JANVIER LABS. Experiments were carried out in compliance with National Guidelines for Animal Protection and the approval of the regional animal care committee and Animal Ethical Committee of Centro de Investigaciones Energéticas, Medioambientales y Tecnológicas (CIEMAT). Mice were housed in an animal facility at the CIEMAT.
For the intranasal administration studies, one group of five mice receiving isoflurane anesthesia was removed from the induction chamber, and intranasal administration was performed immediately by pipetting 30 μl of Omomyc-DFO-89Zr [2.9 ± 0.4 megabecquerel (MBq), 2.37 ± 0.5 mg Omomyc-DFO/kg body weight] onto the outer edge of the nares. Mice were euthanized 48 hours after injection by cervical dislocation under anesthesia with isoflurane in O2, and blood was immediately collected by cardiac puncture. For biodistribution studies, organ tissues were excised, wet-weighed, and counted for radioactivity with a gamma counter (2470 Wizard2, PerkinElmer), along with a standard sample of the injected dose. Grubbs’ test was applied to detect the presence of one outlier animal in the global dataset, and the data from this outlier were discarded.
For the intravenous administration studies, one group of five BALB/c nude mice bearing established subcutaneous xenografts of H1975 cells (average size of 430 mm3) was treated with Omomyc-DFO-89Zr (3.2 ± 0.1 MBq, 2.62 ± 0.3 mg Omomyc-DFO/kg body weight) in the tail vein. Mice were euthanized at 72 hours after injection by cervical dislocation under anesthesia with isoflurane in O2, and blood was immediately collected by cardiac puncture. For biodistribution studies, mice were imaged at the indicated time points by mPET/mCT imaging as described below. At the endpoint (72 hours), organ tissues were excised, wet-weighed, and counted for radioactivity with a gamma counter (2470 Wizard2, PerkinElmer), along with a standard sample of the injected dose at 48 hours after administration.
Tissue activity was expressed as percentage injected dose per gram of tissue (%ID/g) and percentage of injected dose per organ (%ID/organ). For the pharmacokinetic study, blood samples from the tail vein were collected in heparinized tubes and centrifuged to obtain plasma. Plasma concentrations of radioactivity were calculated as ng of Omomyc-DFO/ml and plotted versus the time after injection. Pharmacokinetic parameters were estimated by noncompartmental analysis.
mPET/mCT imaging
After intranasal instillation, mice were scanned immediately with a small-animal Argus PET-CT scanner (SEDECAL). The PET studies (energy window, 400 to 700 KeV; static acquisition, 20 min) and CT (voltage, 45 kV; current, 150 μA; shots, 8; projections, 360; standard resolution) were performed at various time points after injection (30 min and 4, 24, and 48 hours) in mice anesthetized by inhalation of 2 to 2.5% isoflurane. The PET images were reconstructed using a two-dimensional (2D)–ordered subset expectation maximization algorithm (16 subsets and two iterations), with random and scatter correction. A calibration factor predetermined by scanning a cylindrical phantom containing a known activity of 89Zr was used to convert counts per pixel/s to kBq/cm3. Manually drawn regions of interest (ROIs) in PET images (intranasal delivery, oral cavity and oropharyngeal region, esophagus, and gut) or ROIs selected from PET images using CT anatomical guidelines (for the lung, liver, and kidneys) were used to determine the mean radiotracer accumulation in units of %ID/g tissue (decay-corrected to the time of injection) by dividing the obtained average tracer concentration (kBq/cm3) in the region by the total ID (kBq). Percentage-injected dose in a ROI was calculated by multiplying the obtained average tracer concentration (kBq/cm3) by the ROI volume (cm3). Separate image calibration factors were also determined for the lung, kidneys, and liver by comparing the final scans (48 hours) with direct assays of organs performed after the animals were euthanized. These calibration factors were used to normalize the %ID/g obtained from PET imaging to activity concentrations at different time points after injection. Images were analyzed using the image analysis software ITK-SNAP (www.itksnap.org). Movie S1 (the rotating representation) was assembled using the AMIDE software, version 1.0.4 (http://amide.sourceforge.net).
Therapeutic impact studies
For the therapeutic validation, mice were randomized according to weight, sex, and tumor burden at treatment onset. For the assessment of the therapeutic potential of Omomyc after topical administration, mice were intranasally treated with Omomyc (2.37 mg/kg) three times per week for four consecutive weeks or every second day for the short-term 3-day (two doses) and 1-week (three doses) treatments. To evaluate the therapeutic potential of Omomyc after systemic administration, mice were treated intravenously with the indicated dose of the Omomyc mini-protein four times per week for four consecutive weeks. Paclitaxel was administered intraperitoneally twice per week at 5 mg/kg. Investigators responsible for monitoring the mice were blinded to their treatment groups. Ki67, CC3, and CD3 positivity was measured from representative fluorescent microscopy images for each animal and captured at 20× magnification.
Statistical analysis
All analyses and graphs were performed using the GraphPad Prism 5 software. Normal distribution of the data was assessed for each group using D’Agostino-Pearson test. Differences in samples’ average values were analyzed using Student’s t test or analysis of variance (parametric) for normally distributed data or Mann-Whitney or Kruskal-Wallis test otherwise. F test was used to calculate the difference in the variances of the groups. We did not use statistical methods to predetermine sample size in animal studies, but we did make efforts to achieve scientific goals using the minimum number of animals. Original data are provided in data file S2.
Supplementary Material
Acknowledgments
We thank I. Ringshausen for critical reading of the manuscript, the “Whitehead Institute Genome Technology Core” and D. Orlando (Syros) for computational support, and S. P. Tenbaum for expert advice on the microarray analysis. The SH-EP cell line was a gift from A. Sala. We also acknowledge the Cellex Foundation for providing research facilities and equipment.
Funding: This work was funded by the Worldwide Cancer Research (WCR/AICR grant no. 13-1182), the European Research Council (CoG grant no. 617473), the Instituto de Salud Carlos III (FIS grant no. PI13/01705), the BBVA Foundation (no. 67_2015), and the FERO Foundation. M.-E.B. was supported by the Fonds de Recherche du Québec en Santé. M.F.Z.-F. is supported by the Juan de la Cierva Programme of the Spanish Ministry of Economy and Competitiveness (IJCI-2014-22403).
Footnotes
Author contributions: M.-E.B. and L.S. designed the study. M.-E.B., T.J., D.M.-V., M.F.Z.-F., S.M.-M., P.R., C.F., S.C.-S., E.S.d.P., L.M., M.A.M.A., E.R.S., M.S.-H., C.T., M.O., D.L., L.F., and M.G. performed and contributed to the analysis of the experiments. M.M., S.M.-M., J.R.W., D.L., G.M., V.C.C., and P.L. helped in the cloning and mini-proteins’ production and anti-Omomyc antibody characterization and provided feedback and interpretation of structural and biological data. M.-E.B., T.J., and L.S. wrote the manuscript with comments from all authors.
Competing interests: L.S. and M.-E.B. are founders and shareholders of Peptomyc S.L. and are inventors on patent application WO2014180889 A8 that covers the use of the Omomyc mini-protein in medicine, held by VHIO and licensed to Peptomyc. All other authors declare that they have no competing interests.
Data and materials availability: Microarray data were deposited under accession number GSE126455. Researchers may obtain the Omomyc mini-protein formulation used for this study with a material transfer agreement from VHIO and Peptomyc. Primary rabbit polyclonal anti-Omomyc antibody is available from L.S.’s laboratory. All reasonable requests for collaboration involving materials used in the research will be fulfilled provided that a written agreement is executed in advance between Peptomyc/VHIO and the requester (and his or her affiliated institution). Such inquiries or requests should be directed to the corresponding author. All data associated with this study are present in the paper or Supplementary Materials.
References
- 1.Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harb Perspect Med. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 3.Whitfield JR, Soucek L. Tumor microenvironment: Becoming sick of Myc. Cell Mol Life Sci. 2011;69:931–934. doi: 10.1007/s00018-011-0860-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gütgemann I, Eilers M, Felsher DW. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwood EM, Eisenman RN. Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 6.Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 7.Wolfer A, Wittner BS, Irimia D, Flavin RJ, Lupien M, Gunawardane RN, Meyer CA, Lightcap ES, Tamayo P, Mesirov JP, Liu XS, et al. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc Natl Acad Sci U S A. 2010;107:3698–3703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee KS, Kwak Y, Nam KH, Kim D-W, Kang S-B, Choe G, Kim WH, Lee HS. C-MYC copy-number gain is an independent prognostic factor in patients with colorectal cancer. PLOS ONE. 2015;10:e0139727. doi: 10.1371/journal.pone.0139727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalkat M, De Melo J, Hickman K, Lourenco C, Redel C, Resetca D, Tamachi A, Tu W, Penn L. MYC deregulation in primary human cancers. Genes. 2017;8:E151. doi: 10.3390/genes8060151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol. 2006;16:313–317. doi: 10.1016/j.semcancer.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 13.Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 14.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 15.Flores I, Murphy DJ, Swigart LB, Knies U, Evan GI. Defining the temporal requirements for Myc in the progression and maintenance of skin neoplasia. Oncogene. 2004;23:5923–5930. doi: 10.1038/sj.onc.1207796. [DOI] [PubMed] [Google Scholar]
- 16.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sodir NM, Swigart LB, Karnezis AN, Hanahan D, Evan GI, Soucek L. Endogenous Myc maintains the tumor microenvironment. Genes Dev. 2011;25:907–916. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Annibali D, Whitfield JR, Favuzzi E, Jauset T, Serrano E, Cuartas I, Redondo-Campos S, Folch G, Gonzàlez-Juncà A, Sodir NM, Massò-Vallés D, et al. Myc inhibition is effective against glioma and reveals a role for Myc in proficient mitosis. Nat Commun. 2014;5 doi: 10.1038/ncomms5632. 4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitfield JR, Beaulieu M-E, Soucek L. Strategies to inhibit myc and their clinical applicability. Front Cell Dev Biol. 2017;5:10. doi: 10.3389/fcell.2017.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fletcher S, Prochownik EV. Small-molecule inhibitors of the Myc Oncoprotein. Biochim Biophys Acta. 2014;1849:525–543. doi: 10.1016/j.bbagrm.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKeown MR, Bradner JE. Therapeutic strategies to inhibit MYC. Cold Spring Harb Perspect Med. 2014;4:a014266. doi: 10.1101/cshperspect.a014266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prochownik EV, Eagle Grove L, Deubler D, Zhu XL, Stephenson RA, Rohr LR, Yin X, Brothman AR. Commonly occurring loss and mutation of the MXI1 gene in prostate cancer. Genes Chromosomes Cancer. 1998;22:295–304. [PubMed] [Google Scholar]
- 23.Morton JP, Sansom OJ. MYC-y mice: From tumour initiation to therapeutic targeting of endogenous MYC. Mol Oncol. 2013;7:248–258. doi: 10.1016/j.molonc.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malynn BA, de Alboran IM, O’Hagan RC, Bronson R, Davidson L, DePinho RA, Alt FW. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–1399. [PMC free article] [PubMed] [Google Scholar]
- 25.Soucek L, Helmer-Citterich M, Sacco A, Jucker R, Cesareni G, Nasi S. Design and properties of a Myc derivative that efficiently homodimerizes. Oncogene. 1998;17:2463–2472. doi: 10.1038/sj.onc.1202199. [DOI] [PubMed] [Google Scholar]
- 26.Soucek L, Jucker R, Panacchia L, Ricordy R, Tatò F, Nasi S. Omomyc, a potential Myc dominant negative, enhances Myc-induced apoptosis. Cancer Res. 2002;62:3507–3510. [PubMed] [Google Scholar]
- 27.Soucek L, Nasi S, Evan GI. Omomyc expression in skin prevents Myc-induced papillomatosis. Cell Death Differ. 2004;11:1038–1045. doi: 10.1038/sj.cdd.4401443. [DOI] [PubMed] [Google Scholar]
- 28.Soucek L, Whitfield JR, Sodir NM, Massò-Vallés D, Serrano E, Karnezis AN, Swigart LB, Evan GI. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Eyss B, Eilers M. Addicted to Myc—But why? Genes Dev. 2011;25:895–897. doi: 10.1101/gad.2053311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montagne M, Beaudoin N, Lavoie CL, Klinck R, Lavigne P. The max b-HLH-LZ can transduce into cells and inhibit c-Myc transcriptional activities. PLOS ONE. 2012;7:e32172. doi: 10.1371/journal.pone.0032172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noguchi H, Bonner-Weir S, Wei F-Y, Matsushita M, Matsumoto S. BETA2/NeuroD protein can be transduced into cells due to an arginine- and lysine-rich sequence. Diabetes. 2005;54:2859–2866. doi: 10.2337/diabetes.54.10.2859. [DOI] [PubMed] [Google Scholar]
- 32.Koren E, Torchilin VP. Cell-penetrating peptides: Breaking through to the other side. Trends Mol Med. 2012;18:385–393. doi: 10.1016/j.molmed.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 33.Cappuzzo F, Varella-Garcia M, Rossi E, Gajapathy S, Valente M, Drabkin H, Gemmil R. MYC and EIF3H coamplification significantly improve response and survival of non-small cell lung cancer patients (NSCLC) treated with gefitinib. J Thorac Oncol. 2015;4:472–478. doi: 10.1097/JTO.0b013e31819a5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwakawa R, Kohno T, Kato M, Shiraishi K, Tsuta K, Noguchi M, Ogawa S, Yokota J. MYC amplification as a prognostic marker of early-stage lung adenocarcinoma identified by whole genome copy number analysis. Clin Cancer Res. 2011;17:1481–1489. doi: 10.1158/1078-0432.CCR-10-2484. [DOI] [PubMed] [Google Scholar]
- 35.Richardson GE, Johnson BE. The biology of lung cancer. Semin Oncol. 1993;20:105–127. [PubMed] [Google Scholar]
- 36.Kubokura H, Tenjin T, Akiyama H, Koizumi K, Nishimura H, Yamamoto M, Tanaka S. Relations of the c-myc gene and chromosome 8 in non-small cell lung cancer: Analysis by fluorescence in situ hybridization. Ann Thorac Cardiovasc Surg. 2001;7:197–203. [PubMed] [Google Scholar]
- 37.J-Naud F, Frédéric G, Raymund W, Chabot B, Lavigne P. Improving the thermodynamic stability of the leucine zipper of max increases the stability of its b-HLH-LZ:E-box complex. J Mol Biol. 2003;326:1577–1595. doi: 10.1016/s0022-2836(03)00029-9. [DOI] [PubMed] [Google Scholar]
- 38.Jung LA, Gebhardt A, Koelmel W, Ade CP, Walz S, Kuper J, von Eyss B, Letschert S, Redel CL, d’Artista, Biankin A, et al. OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors. Oncogene. 2017;36:1911–1924. doi: 10.1038/onc.2016.354. [DOI] [PubMed] [Google Scholar]
- 39.Savino M, Annibali D, Carucci N, Favuzzi E, Cole MD, Evan GI, Soucek L, Nasi S. The action mechanism of the myc inhibitor termed omomyc may give clues on how to target Myc for cancer therapy. PLOS ONE. 2011;6:e22284. doi: 10.1371/journal.pone.0022284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiorentino FP, Tokgün E, Solé-Sánchez S, Giampaolo SO, Tokgün, Jauset T, Kohno T, Perucho M, Soucek L, Yokota J. Growth suppression by MYC inhibition in small cell lung cancer cells with TP53 and RB1 inactivation. Oncotarget. 2016;7:31014–31028. doi: 10.18632/oncotarget.8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, Rabinowitz JD, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ushmorov A, Hogarty MD, Liu X, Knauss H, Debatin KM, Beltinger C. N-myc augments death and attenuates protective effects of Bcl-2 in trophically stressed neuroblastoma cells. Oncogene. 2008;27:3424–3434. doi: 10.1038/sj.onc.1211017. [DOI] [PubMed] [Google Scholar]
- 43.Boon K, Caron HN, van Asperen R, Valentijn L, Hermus M-C, van Sluis P, Roobeek I, Weis I, Voûte PA, Schwab M, Versteeg R. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qing G, Li B, Vu A, Skuli N, Walton ZE, Liu X, Mayes PA, Wise DR, Thompson CB, Maris JM, Hogarty MD, et al. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell. 2012;22:631–644. doi: 10.1016/j.ccr.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kress TR, Sabò A, Amati B. MYC: Connecting selective transcriptional control to global RNA production. Nat Rev Cancer. 2015;15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- 46.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H-Z, Tsai S-Y, Leone G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marinkovic D, Marinkovic T, Kokai E, Barth T, Möller P, Wirth T. Identification of novel Myc target genes with a potential role in lymphomagenesis. Nucleic Acids Res. 2004;32:5368–5378. doi: 10.1093/nar/gkh877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rajbhandari P, Lopez G, Capdevila C, Salvatori B, Yu J, Rodriguez-Barrueco R, Martinez D, Yarmarkovich M, Weichert-Leahey N, Abraham BJ, Alvarez MJ, et al. Cross-cohort analysis identifies a TEAD4–MYCN positive feedback loop as the core regulatory element of high-risk neuroblastoma. Cancer Discov. 2018;8:582–599. doi: 10.1158/2159-8290.CD-16-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ciribilli Y, Singh P, Inga A, Borlak J. C-Myc targeted regulators of cell metabolism in a transgenic mouse model of papillary lung adenocarcinoma. Oncotarget. 2016;7:65514–65539. doi: 10.18632/oncotarget.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Southam DS, Dolovich M, O’Byrne PM, Inman MD. Distribution of intranasal instillations in mice: Effects of volume, time, body position, and anesthesia. Am J Physiol Lung Cell Mol Physiol. 2002;282:L833–L839. doi: 10.1152/ajplung.00173.2001. [DOI] [PubMed] [Google Scholar]
- 52.Jackson EL. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berger KN, Pu JJ. PD-1 pathway and its clinical application: A 20 year journey after discovery of the complete human PD-1 gene. Gene. 2018;638:20–25. doi: 10.1016/j.gene.2017.09.050. [DOI] [PubMed] [Google Scholar]
- 54.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimamura T, Chen Z, Soucheray M, Carretero J, Kikuchi E, Tchaicha JH, Gao Y, Cheng KA, Cohoon TJ, Qi J, Akbay E, et al. Efficacy of BET bromodomain inhibition in Kras-mutant non–small cell lung cancer. Clin Cancer Res. 2013;19:6183–6192. doi: 10.1158/1078-0432.CCR-12-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaub FX, Dhankani V, Berger AC, Trivedi M, Richardson AB, Shaw R, Zhao W, Zhang X, Ventura A, Liu Y, Ayer DE, et al. Pan-cancer alterations of the MYC oncogene and its proximal network across the cancer genome atlas. Cell Syst. 2018;6:282–300.e2. doi: 10.1016/j.cels.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang E, Sorolla A, Cunningham PT, Bogdawa HM, Beck S, Golden E, Dewhurst RE, Florez L, Cruickshank MN, Hoffmann K, Hopkins RM, et al. Tumor penetrating peptides inhibiting MYC as a potent targeted therapeutic strategy for triple-negative breast cancers. Oncogene. 2019;38:140–150. doi: 10.1038/s41388-018-0421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 59.Green M, Loewenstein PM. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 60.Jones AT, Sayers EJ. Cell entry of cell penetrating peptides: Tales of tails wagging dogs. J Control Release. 2012;161:582–591. doi: 10.1016/j.jconrel.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 61.vanTorchilin VP. Cell penetrating peptide-modified pharmaceutical nanocarriers for intracellular drug and gene delivery. Biopolymers. 2008;90:604–610. doi: 10.1002/bip.20989. [DOI] [PubMed] [Google Scholar]
- 62.Foged C, Nielsen HM. Cell-penetrating peptides for drug delivery across membrane barriers. Expert Opin Drug Deliv. 2007;5:105–117. doi: 10.1517/17425247.5.1.105. [DOI] [PubMed] [Google Scholar]
- 63.Guidotti G, Brambilla L, Rossi D. Cell-penetrating peptides: From basic research to clinics. Trends Pharmacol Sci. 2017;38:406–424. doi: 10.1016/j.tips.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 64.Mcduff F-O, Naud J-F, Montagne M, Sauvé S, Lavigne P. The max homodimeric b-HLH-LZ significantly interferes with the specific heterodimerization between the c-Myc and max b-HLH-LZ in absence of DNA: A quantitative analysis. J Mol Recognit. 2009;22:261–269. doi: 10.1002/jmr.938. [DOI] [PubMed] [Google Scholar]
- 65.Sauvé S, Tremblay L, Lavigne P. The NMR solution structure of a mutant of the max b/HLH/LZ free of DNA: Insights into the specific and reversible DNA binding mechanism of dimeric transcription factors. J Mol Biol. 2004;342:813–832. doi: 10.1016/j.jmb.2004.07.058. [DOI] [PubMed] [Google Scholar]
- 66.Al Soraj M, He L, Peynshaert K, Cousaert J, Vercauteren D, Braeckmans K, De Smedt SC, Jones AT. siRNA and pharmacological inhibition of endocytic pathways to characterize the differential role of macropinocytosis and the actin cytoskeleton on cellular uptake of dextran and cationic cell penetrating peptides octaarginine (R8) and HIV-tat. J Control Release. 2012;161:132–141. doi: 10.1016/j.jconrel.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 67.Rahl PB, Lin CY, Seila AC, Flynn RA, Mccuine S, Burge CB, Sharp PA, Young RA. C-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maltais L, Montagne M, Bédard M, Tremblay C, Soucek L, Lavigne P. Biophysical characterization of the b-HLH-LZ of ∆Max, an alternatively spliced isoform of max found in tumor cells: Towards the validation of a tumor suppressor role for the max homodimers. PLOS ONE. 2017;12:e0174413. doi: 10.1371/journal.pone.0174413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.