Abstract
Observational epidemiological studies are prone to confounding, measurement error, and reverse causation, undermining robust causal inference. Mendelian randomization (MR) uses genetic variants to proxy modifiable exposures to generate more reliable estimates of the causal effects of these exposures on diseases and their outcomes. MR has seen widespread adoption within cardio-metabolic epidemiology, but also holds much promise for identifying possible interventions for cancer prevention and treatment. However, some methodological challenges in the implementation of MR are particularly pertinent when applying this method to cancer aetiology and prognosis, including reverse causation arising from disease latency and selection bias in studies of cancer progression. These issues must be carefully considered to ensure appropriate design, analysis, and interpretation of such studies.
In this review, we provide an overview of the key principles and assumptions of MR focusing on applications of this method to the study of cancer aetiology and prognosis. We summarize recent studies in the cancer literature that have adopted a MR framework to highlight strengths of this approach compared to conventional epidemiological studies. Lastly, limitations of MR and recent methodological developments to address them are discussed, along with the translational opportunities they present to inform public health and clinical interventions in cancer.
Keywords: Mendelian randomization, causal inference, review, genetic epidemiology
Introduction
Obtaining reliable evidence of causal relationships from observational epidemiological studies remains a pervasive challenge1–3. While observational studies have made fundamental contributions to understanding the primary environmental causes of various cancers (e.g., smoking and lung cancer, hepatitis B and liver cancer, asbestos and mesothelioma)4–6, recent decades have seen numerous instances of apparently robust observational associations being subsequently contradicted by large chemoprevention trials7–15. Notable translational failures include the ineffectiveness of beta-carotene supplementation to prevent lung cancer among smokers in the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study and vitamin E supplementation to prevent prostate cancer in the Selenium and Vitamin E Cancer Prevention Trial. Contrary to expectations from observational data, findings from both trials suggested that supplementation may increase rather than reduce the incidence of cancer8,16.
Part of the difficulty in translating observational findings into effective cancer prevention and treatment strategies lies in the susceptibility of conventional observational designs to various biases, such as residual confounding (due to unmeasured or imprecisely measured confounders) and reverse causation17,18. These biases frequently persist despite energetic statistical and methodological efforts to address them19–21, making it difficult for observational studies to reliably conclude that a risk factor is causal, and thus a potentially effective intervention target. This issue is likely further compounded by the modern epidemiological pursuit of risk factors that confer increasingly modest effects on disease risk, which can contribute to a ubiquity of spurious findings in the literature22–24.
Despite these challenges, observational studies remain crucial for informing cancer prevention and treatment policy given issues in translating basic science to human populations and because intervention trials are expensive, time-consuming, and often unfeasible in a primary prevention setting. The development of novel analytical tools that can help address some of the limitations of conventional observational studies therefore remains an important field of research. One such approach known as Mendelian randomization (MR) which uses genetic variants to proxy potentially modifiable exposures has seen increased adoption within population health research and offers much promise to generate a more reliable evidence-base for cancer prevention and treatment.
What is Mendelian randomization?
MR uses germline genetic variants as instruments (i.e., proxies) for exposures (e.g., environmental factors, biological traits, or druggable pathways) to examine the causal effects of these exposures on health outcomes (e.g., disease incidence or progression)25–31. The use of genetic variants as proxies exploits their random allocation at conception (Mendel’s first law of inheritance) and the independent assortment of parental variants at meiosis (Mendel’s second law of inheritance). These natural randomization processes mean that, at a population level, genetic variants that are associated with levels of a specific modifiable exposure will generally be independent of other traits and behavioural or lifestyle factors, although several caveats exist (see Table 1). Analyses using genetic variants as instruments to examine associations with outcomes have a number of advantages: i) effect estimates should be less prone to the confounding that typically distorts conventional observational associations32, ii) because germline genetic variants are fixed at conception, they cannot be modified by subsequent factors, thus overcoming possible issues of reverse causation, and iii) measurement error in genetic studies is often low as modern genotyping technologies provide relatively precise measurement of genetic variants, unlike the substantial (and at times differential) exposure measurement error which can accompany observational studies (e.g., due to self-report).
Table 1. Limitations of Mendelian randomization and techniques available to address them.
| Limitation | Description | Techniques to Address Limitation |
|---|---|---|
| Limitations to robust causal inference | ||
| Horizontal pleiotropy | A genetic variant affecting an outcome via a biological pathway independent of the exposure under investigation, violating the “exclusion restriction criterion” | Assessment of heterogeneity across individual SNP estimates MR-Egger regression and intercept test median-approaches Mode-based approaches Sensitivity analysis removing potentially pleiotropic SNPs Restrict risk score to SNPs in well-characterized genes Stratification by exposure status (e.g., ALDH2 and self-reported alcohol intake) |
| Linkage disequilibrium | Linkage disequilibrium (LD) is the non-random association of alleles at different loci that are close in proximity on a chromosome. If a certain SNP is being used as an instrument for an exposure in a MR analysis, and this SNP is in LD with another SNP that affects the outcome via an independent pathway, then the assumptions for MR will be violated | LD pruning of SNPs prior to MR analysis Weighted generalized linear regression Perform studies in populations with different LD structures |
| Population stratification | Allele frequencies vary among populations of different genetic ancestry, and similarly, disease risk often varies among populations of different genetic ancestry, which could introduce genetic confounding into a MR analysis, potentially resulting in spurious causal estimates | Restricting analyses to individuals of a homogenous genetic ancestry Genomic inflation factor calculation Adjusting MR analysis by genetic ancestry or ancestry-informative principal components |
| Trait heterogeneity | For a given trait (e.g., adiposity), SNPs may influence various dimensions of this trait (e.g., both overall and visceral adiposity) but GWAS have only examined associations with a subset of these dimensions (e.g., solely BMI). This may produce misleading inferences if the aim of an analysis is to ascertain the causal effect of a particular dimension of a trait. | Better understanding of complex phenotypes Multivariable MR |
| Limitations that complicate interpretation | ||
| Canalization | Developmental compensation against the effect of a genetic variant being used as an instrument that could attenuate the magnitude of an observed MR association towards the null | Knowledge of the period of life when the influence of a genetic variant(s) on an exposure may emerge can help guide whether developmental compensatory processes are plausible. For example, behavioural exposures that typically occur after fetal development (e.g., alcohol, smoking) will be unlikely to be influenced by canalization whereas in utero exposure may. There are currently no approaches for evaluating suspected canalization in MR analyses. |
| Complexity of association | Misinterpretation of MR results can arise from limited biological understanding of genetic variants utilised as IVs. Examples include interpretation of the effect of the heterozygous ALDH2 genotype on oesophageal cancer risk (discussed in “Illustrative examples”) and previous MR analyses that have examined the effects of interleukin-6 42 and extracellular superoxide dismutase 176 on CHD risk (discussed in more detail elsewhere 49). | Improved biological understanding of genetic variants with functional annotation, pathway analysis, and gene set enrichment |
| Dynastic effects | In certain circumstances, it is possible that parental genotype can confound an association of offspring genotype with offspring disease risk. For example, genetic variants influencing parental height will not only influence offspring height genotype but could also influence offspring disease risk via an independent effect of maternal height-raising alleles on the in utero environment of the offspring 177,178). | Between-sibling MR design Within-family MR design |
| Critical period effects | If a biomarker primarily influences disease risk over a critical or sensitive period of the life course, a MR estimate should capture the causal effect of this biomarker but may not be able to distinguish period effects | Negative exposure control design |
| Weak instrument bias | If IV is not robustly associated with the exposure, estimates will be biased towards the observational estimate in a one-sample setting and towards the null in a two-sample setting | Increase sample size Genetic risk scores or combining summarized data from multiple genetic variants Two-sample MR analysis |
| “Winner’s Curse” | Chance correlation between genetic variants and confounders can introduces an overestimation of the effect of a “lead” genetic variant on an exposure of interest in the discovery stage of a GWAS. The effect of this phenomenon will depend on the degree of overlap of participants in the GWAS discovery dataset and subsequent MR analyses. In a one-sample MR setting with a binary outcome, winner’s curse should not lead to bias if control participants were used in the discovery GWAS. If both cases and controls were used in the discovery dataset, this will lead to weak instrument bias. If the instrument is identified in a sample independent to the one in which MR analysis is performed, this will lead to an underestimate of the causal effect. | Two-sample MR analysis Split-sample MR analysis |
| Low statistical power | Genetic variants typically explain a small amount of variance for a given exposure, thus MR requires large sample sizes to test hypotheses with adequate power. Furthermore, in finite samples, confounders may not be perfectly balanced between genotypic groups | Large GWAS and GWAS consortia Genetic risk scores or combining summarized data from multiple genetic variants Two-sample MR analysis |
Comparison of Mendelian randomization to Randomized Controlled Trials
Due to the random allocation of alleles at conception it can be useful to compare the structure of a MR analysis to the design of a randomized trial, where individuals are randomly allocated at baseline to an intervention or control group (Figure 1). Groups defined by genotype should be comparable in all respects (e.g., approximately equal distribution of potential confounding factors) except for the exposure of interest. It follows that any observed differences in outcomes between these genotypic groups can be attributed to differences in long-term exposure to the trait of interest. This latter point is an important distinction when interpreting results from a MR analysis as compared to a randomized controlled trial: MR will generally estimate the effect of life-long “allocation” to an exposure on an outcome, unless an exposure typically occurs only from a certain age - e.g., alcohol consumption and smoking - and the genetic proxy affects metabolism of that exposure33. If the effect of this exposure on an outcome is cumulative over time, a MR analysis may generate a larger effect estimate than that which would be obtained from a randomized trial examining an intervention over a limited duration of time. Additionally, if the effect of an exposure on an outcome operates primarily or exclusively over a critical or sensitive period of the life course (e.g., early childhood), a MR analysis should be able to “capture” a causal effect of this exposure but will not be able to distinguish such period effects. In contrast, a randomized trial will have the flexibility to test certain interventions over restricted periods of follow-up and in individuals who may be within narrow age ranges. These distinctions are discussed in more detail in “Cancer Latency and Reverse Causation – benefits of MR”.
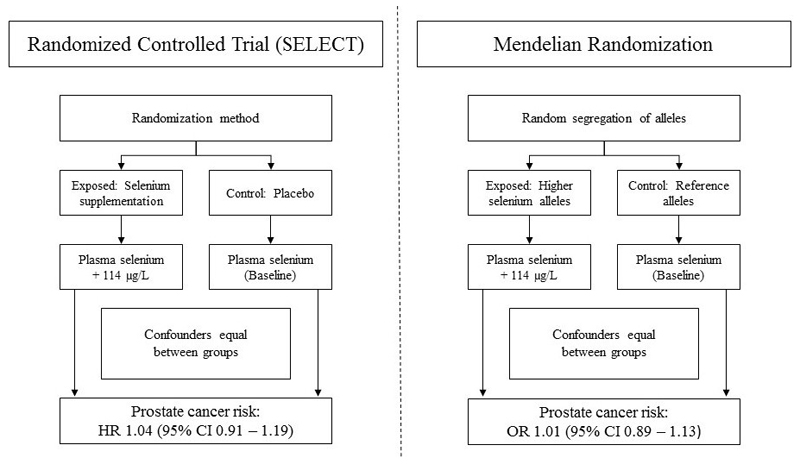
Figure 1. Schematic comparison of the structure of a randomized controlled trial (SELECT) and a Mendelian randomization analysis (PRACTICAL).
In SELECT (left), individuals were randomly allocated to the intervention (200 μg daily selenium supplementation, which lead to a 114μg/L increase in blood selenium) or control group (placebo). In PRACTICAL (right), the additive effects of selenium-raising alleles at eleven SNPs, randomly allocated at conception, were scaled to mirror a 114μg/L increase in blood selenium. If an RCT trial is adequately sized, randomization should ensure that intervention and control groups are comparable in all respects (e.g., distribution of potential confounding factors) except for the intervention being tested. In an intention-to-treat analysis, any observed differences in outcomes between intervention and control groups can then be attributed to the trial arm to which they were allocated. Likewise, in a MR analysis, groups defined by genotype should be comparable in all respects (e.g., distribution of both genetic and environmental confounding factors) except for their exposure to a trait of interest. Any observed differences in outcomes between groups defined by genotype can then be attributed to differences in life-long exposure to the trait of interest under study.
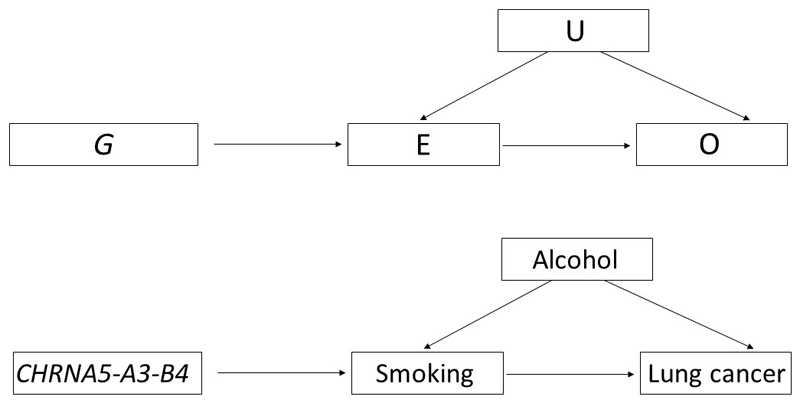
More formally, MR is a form of instrumental variable (IV) analysis that relies on three key assumptions: the IV (here, one or more genetic variants) should (i) be reliably associated with the exposure of interest; (ii) not be associated with any confounding factor(s) that would otherwise distort the association between the exposure and outcome; and (iii) should not be independently associated with the outcome, except through the exposure of interest (known as the “exclusion restriction criterion”) (Figure 2a). If all assumptions are met, MR can provide an unbiased causal estimate of the effect of an exposure on disease or a health-related outcome. Violation of one or more of these assumptions means that instruments are invalid and, consequently, that findings from such an analysis may yield a biased effect estimate.
Figure 2. Illustration of MR methodology.
(A) A genetic variant (G) is used as a proxy for a modifiable exposure (E) to assess the association between E and an outcome of interest (O) without the issues of reverse causation, and confounding (U). MR methodology relies on three main assumptions, in that G must (i) be reliably associated with E; (ii) not be associated with U; and (iii) not be independently associated with O, except through E. This method is exemplified in the context of assessing the association of smoking and lung cancer (B), using the CHRNA5-A3-B4 SNP as a genetic instrument for heaviness of smoking.
Previous success of Mendelian randomization approaches and potential for cancer research
Over the past decade, MR has been increasingly adopted as an analytical approach within population health research, particularly the fields of metabolic and cardiovascular disease (CVD), where there are several notable examples of important causal inferences. For example, MR has suggested a likely causal role of statins on type 2 diabetes (T2D) risk34,35; likely non-causal roles of circulating levels of high-density lipoprotein cholesterol (HDL-C) in myocardial infarction36 and C-reactive protein (CRP) in T2D37; pointed to the efficacy of proprotein convertase subtilisin/kexin type 2 (PCSK9) inhibitors for CHD prevention prior to the publication of confirmatory long-term trial results34,38; and prioritized further examination of apolipoprotein B39,40, lipoprotein(a)41 and interleukin-642 and de-prioritized fibrinogen43 and secretory phospholipase A(2)-IIA44 as intervention targets for CVD. Although this approach has scope to test the effects of an increasing number of exposures relevant to cancer through the continued growth in large-scale genome-wide association study (GWAS) output, to date there remains a noticeable gap in the MR literature with regard to cancer compared to other outcomes (Supplementary Figure 1).
Here, we provide an overview of some recent studies that have applied MR to cancer outcomes, highlighting both the potential strengths compared to conventional epidemiological studies and the unique challenges of performing MR studies in cancer. Recent methodological extensions to the original MR paradigm are presented, with emphasis on the translational opportunities that they may offer to inform drug target validation and public health strategies to reduce the burden of cancer.
Considerations for MR in cancer
Both the principal strengths of MR and important limitations of this method have been discussed in detail previously25–31,45–49. The latter are presented in Table 1 with some methodological and statistical approaches that have been developed to address them outlined in Table 2 and Table 3. Considerations which are specific to investigating causality in the setting of cancer are outlined below.
Table 2. Summarized data and two-sample MR.
| Methodological approaches and related considerations | Description |
|---|---|
| Two-sample MR | Historically, both gene-exposure and gene-outcome estimates in MR analyses had to be obtained from a single sample which relied upon the availability of information on genotype, exposure, and outcome among all participants in that dataset. In practice, this not only posed a challenge in that large-scale measurement of a given exposure of interest (e.g., many molecular traits) may not only be prohibitively expensive but also that measurement of certain exposures may not be possible (e.g., if adequate blood sample collection or preservation has not taken place)50. An extension to the original MR paradigm that has allowed MR analyses to overcome some of these challenges is the integration of gene-exposure and gene-outcome estimates from two independent (non-overlapping) datasets into a single analysis, an approach called “two-sample MR” analysis50,51. |
| Two-sample MR with summarized genetic association data | It is possible and increasingly common practice to perform MR analyses exclusively using summarized data on gene-exposure and gene-outcome estimates51,52. A strength of two-sample MR with summary data is that the scope of possible MR analysis can be expanded significantly by exploiting the growing amount of publicly-available summary data from large genome-wide association study (GWAS) consortia53 and is aided by the development of a harmonised MR platform that has collated these datasets (MR-Base)54. Utilizing data from separate exposure and outcome samples can help to bolster statistical power in MR analyses by increasing the overall sample size of an analysis, particularly when testing effects on binary disease outcomes like cancer, and also reduces the likelihood of “winner’s curse” bias (see Table 1)51. This increased power also means that sensitivity analyses to test pleiotropy assumptions (see Table 3: Genetic risk scores and pleiotropy) which are often statistically inefficient are better-powered to detect violations of these assumptions. Furthermore, whereas in a one-sample MR setting weak instruments can bias effect estimates towards the observational effect, resulting in potential false positive associations, in a two-sample setting weak instrument bias distorts findings towards the null. Thus, conducting both analyses is a form of sensitivity analysis that provides bounds to a possible causal effect. To test whether height has a causal effect on risk of colorectal, lung, and prostate cancer, Khankari et al. used a two-sample MR approach. This employed: i) summarized gene-exposure estimates from a panel of 423 single-nucleotide polymorphisms (SNPs) previously found to be associated with height in a large GWAS meta-analysis (GIANT consortium; N=253,288) and collectively explaining approximately 16% of variance in height; and ii) summarized gene-outcome estimates from a total of 47,800 cancer cases (across the three outcomes ascertained) and 81,533 controls from the Genetic Associations and Mechanisms in Oncology (GAME-ON) consortium55. This approach allowed robust causal inference with adequate statistical power. While Khankari et al. did not examine the effects of height across stage/grade or histological sub-type of the three cancers examined, two-sample approaches enable statistically efficient examination of risk factors across such stratified groups which may have limited sample sizes. |
| Limitations of two-sample MR | While two-sample MR offers some clear advantages over a conventional one-sample approach, it also introduces additional assumptions. One important assumption is that the separate datasets from which gene-exposure and gene-outcome associations are obtained are representative of the same underlying population, for example with regard to sex, age, ethnicity, or genetic profile. While most GWAS that have examined sex-specific associations of traits have often reported at most modest evidence of sexual dimorphism56,57, given the sex-specific nature of certain cancers, care should be taken to ensure that instruments are obtained from sex-stratified GWAS for analyses of these cancers when available. For example, in examining the effect of waist-hip-ratio (WHR) on endometrial or ovarian cancer this could involve using the 34 SNPs associated with WHR in women exclusively as a primary instrument, then comparing results with those obtained using the 47 SNPs associated with WHR across both sexes as a sensitivity analysis58,59. Concordance of findings between both approaches may suggest that directionally-consistent SNPs associated with WHR at genome-significance in women, but not men, simply reflected reduced statistical power in sex-stratified GWAS analyses and not genuine heterogeneity in SNP-effects between sexes. A second challenge when performing two-sample MR using summary data is the difficulty in examining the IV assumption that an instrument used is independent of exposure-outcome confounders. While restriction of analyses to ethnically homogenous gene-exposure and gene-outcome datasets will reduce the possibility of confounding through population stratification, in lieu of data on measured potential confounders, this assumption cannot be directly tested. While one way of approximately testing this assumption is performing look-up of associations of SNPs with suspected potential confounders in curated GWAS databases, this would not preclude chance confounding relationships arising in the dataset(s) from which summary data were obtained. Third, with the use of summary data from large GWAS consortia, it is possible that there may be some participant overlap in the datasets from which gene-exposure and gene-outcome associations are obtained. If overlap is small, this should not substantially bias effect estimates, however substantial overlap will bias MR toward the observational effect60. |
Table 3. Genetic risk scores and pleiotropy.
| Methodological approaches and related considerations | Description |
|---|---|
| Using multiple genetic variants as an instrument | While GWAS over the past decade have been successful at identifying robust associations between common genetic variants (usually SNPs) and thousands of phenotypes, the effects of individual variants on traits are often modest61. Consequently, statistical power for MR analyses using single variants as instruments can be limited. A common approach of overcoming limited statistical power is to combine multiple variants into a genetic risk score (GRS) or combine summary data across multiple SNPs, which increases the variance explained for a trait of interest, improving instrument strength62,63. A GRS or instrument with summarized data from multiple SNPs can consist of an unweighted summation of risk-factor increasing alleles across variants but, more commonly, a weighted approach is used (e.g., weighted by the estimated SNP-exposure effect size or, in settings with summary data, by the inverse of the standard error of the gene-outcome association – called the “inverse-variance weighted (IVW) method”). In a two-sample setting (see Table 2: Summarized data and two-sample MR), an instrument consisting of summarized data from multiple variants will typically be constructed by combining SNPs that are independent (i.e., not in LD with each other). However, it is also possible to combine correlated SNPs in low to moderate LD into an instrument, using weighted generalized linear regression for example62. This requires the creation of a weighting matrix which takes into account correlations between SNPs, often with use of a reference panel like the Hapmap or the 1,000 Genomes Project64,65, which is then used to correctly inflate standard error estimates. The latter method may be preferable to overcome weak instrument issues when few independent SNPs are available. |
| Vertical vs horizontal pleiotropy | While construction of a GRS can help to enhance statistical power in MR analyses, increasing the number of variants included in a score is accompanied by an increased probability that any of these variants could be pleiotropic (i.e., one variant having effects on two or more traits). In a genetic epidemiological context, an important distinction is made between vertical and horizontal pleiotropy, each having different effects on the interpretation of MR findings. Vertical pleiotropy occurs when one variant has an effect on two or more traits that both influence an outcome through the same biological pathway. For example, variants in FTO that not only associate with BMI, but also with fasting insulin and glucose concentrations would be consistent with a causal effect of BMI on these downstream traits66. In this case, a MR analysis examining the effect of BMI on T2D risk using these FTO variants would be consistent with an instrument (genetic variants associated with BMI) influencing an outcome (T2D) exclusively through the exposure of interest (BMI). This form of pleiotropy would be expected in complex biological systems and does not pose a threat to the validity of a MR analysis67. In contrast, horizontal pleiotropy occurs when one variant has an effect on two or more traits that influence an outcome through independent biological pathways. For example, genetic variants associated with triglyceride levels also show substantial overlap with variants associated with LDL-C and HDL-C68. As a putative effect of triglyceride-increasing variants on CHD risk may not only operate through elevation of triglycerides but through alternate cholesterol pathways, a naïve MR analysis using all triglyceride-increasing variants without addressing pleiotropy in this instance could invalidate the “exclusion restriction criterion” IV assumption. The presence of horizontal pleiotropy thus poses a direct threat to the validity of MR findings. |
| Assessment of horizontal pleiotropy | When using either a single or a small number of genetic variants as IVs, the presence of horizontal pleiotropy for any individual variant can be assessed through SNP look-ups in curated GWAS databases with complete summary data (e.g., MR-Base54, PhenoScanner69, dbGap70) to examine whether associations for a given SNP have been reported for traits other than the exposure of interest. Sensitivity analyses can then be performed by dropping variants that are suspected to be horizontally pleiotropic and then carefully interpreting pooled causal estimates with and without suspected horizontally pleiotropic SNPs. When an instrument consists of multiple genetic variants, an important first step in examining the presence of horizontal pleiotropy in analyses is to assess heterogeneity in causal estimates across individual IVs (including visually examining heterogeneity using a funnel plot). While substantial heterogeneity in causal estimates may be indicative of the presence of horizontal pleiotropy, if there is overall symmetry in the funnel plot, pleiotropic effects will be balanced (termed “balanced pleiotropy”) and the overall causal estimate generate will be unbiased. In contrast, if there is considerable asymmetry in a funnel plot, this will suggest that horizontal pleiotropic effects of individual IVs are not balanced and that overall causal estimates will be biased (termed “directional pleiotropy”). MR-Egger regression and the weighted median estimator (WME) are two widely implemented approaches for detecting and accounting for directional pleiotropy, and are applicable to analyses utilizing individual-level and summary-level data71,72. An additional approach called the mode-based estimate (MBE) has also recently been proposed as a method to examine horizontal pleiotropy in MR analyses73. All of these methods can help to detect IV violations while making different assumptions about the nature of horizontal pleiotropy and thus, when feasible, using all approaches as sensitivity analyses in a given MR analysis can serve as an important mechanism to assess the robustness of findings to pleiotropic bias. |
| Sensitivity analyses to examine horizontal pleiotropy when using multiple genetic variants | MR-Egger regression provides a consistent causal effect estimate even when all genetic variants are invalid IVs because they violate the exclusion restriction criterion. This approach performs a weighted linear regression of the gene-outcome coefficients on the gene-exposure coefficients with an unconstrained intercept term. If the IV assumption that the association of each variant with the outcome is mediated exclusively through the exposure of interest is met, this intercept term should be zero. An intercept term that differs from zero would suggest the presence of unbalanced pleiotropy, thus providing a test for directional pleiotropy. In turn, the slope coefficient in MR-Egger regression will provide an estimate of a causal effect adjusted for directional pleiotropy. An important consideration when using MR-Egger is that it works under the InSIDE (instrument strength independent of direct effect) assumption. In essence, InSIDE assumes that no association exists between the strength of gene-exposure associations and the strength of bias due to horizontal pleiotropy. Intuitively, if multiple genetic variants in an MR analysis have horizontally pleiotropic effects through unrelated intermediate variables, it would be expected that this assumption should hold. However, this assumption is unlikely to be satisfied in situations where all pleiotropic effects are due to the presence of a single confounder. As such, in lieu of an established method of formally testing the InSIDE assumption, interpretation of intercept terms and slope coefficients generated through MR-Egger should be made with this assumption in mind. A complementary sensitivity analysis to MR-Egger is the weighted median estimator. This approach provides an estimate of the weighted median of a distribution in which individual IV causal estimates in a risk score are ordered and weighted by the inverse of their variance. Unlike MR-Egger which can provide an unbiased causal effect even when all IVs are invalid, WME requires that at least 50% of the information in a risk score is coming from IVs that are valid in order to provide a consistent estimate of a causal effect in a MR analysis. However, an advantage of WME is that it provides improved precision as compared to MR-Egger and does not rely on the InSIDE assumption. The mode based estimator generates a causal effect using the mode of a smoothed empirical density function of individual IV causal estimates in a risk score. This approach operates under the assumption that the most common effect estimate of individual IVs in a risk score arises from valid instruments (called the Zero Modal Pleiotropy Assumption, or ZEMPA). If this assumption holds, the mode can provide a consistent causal estimate even if most of the (non-modal) IVs are invalid. Both simple and weighted mode approaches (weighted by the inverse variance of the SNP-outcome association) can be utilized. Mode-based approaches have less power to detect a causal effect than the weighted median estimator but greater power than MR-Egger regression under the condition of no invalid instruments. Similar to the weighted median estimator, mode-based approaches are also (by default) less susceptible to bias from outlying variants in a risk score. |
Cancer Latency and Reverse Causation – benefits of MR
Given long latency periods for many cancers, spurious findings resulting from reverse causation are an important concern in cancer epidemiology. Reverse causation has been suspected in several instances of ambiguous74–76 or paradoxical findings77 in the cancer literature. For example, early studies documenting an association between higher circulating cholesterol and lower cancer incidence were variably interpreted as plausible evidence of a protective effect of raised cholesterol on cancer risk or as latent cancer leading to a reduction in cholesterol levels78–80. With the introduction and widespread usage of low-density lipoprotein cholesterol (LDL-C) lowering medications for the prevention and treatment of CVD, concern arose that such measures could thus be increasing cancer rates81,82.
In an early proposal of the use of genetics as a tool to circumvent issues of reverse causation in observational data, Katan et al.83 suggested examining the association of genetic variants in APOE, determinants of circulating cholesterol levels, with cancer risk. As germline APOE genotype was fixed at conception, it was argued that it would not be influenced by subsequent cancer development and could therefore be used to establish whether cholesterol had a causal effect on cancer incidence. Subsequent MR analyses testing the effect of lifelong elevated cholesterol through genetic variation in APOE, NPC1L1, PCSK9, and ABCG8 have reported null associations with overall cancer risk84–86. These findings alongside secondary analyses of statin trials showing no effect on cancer rates87 suggest that – a potential explanatory role of confounding aside - early observational findings supporting a protective effect of cholesterol on cancer risk likely reflected undiagnosed cancer or early carcinogenic processes causing a reduction in cholesterol levels in pre-diagnostic samples.
Long-term exposure – benefits of MR
The advantages of exploiting the fixed nature of germline genotype extends beyond addressing reverse causation in observational studies. Large cancer prevention trials are often constrained to examining interventions over a limited duration in time and over a particular period in the life-course (e.g., middle and/or late adulthood)88. Given the length of time required for solid tumor development89, randomized trials will often not allow sufficient follow-up for the effect of an intervention to be detected. In turn, long-term chemoprevention trials that are conducted may suffer from issues of non-compliance in the intervention arm, contamination in the control arm, and attrition during follow-up.
Further, the optimal timing of an exposure to prevent cancer may be early in the life-course and therefore may not be adequately addressed in randomized trials90. For example, it has been proposed that certain carcinogenic agents or processes may confer an effect, or a particularly pronounced effect, only over ‘critical periods’ of early life or adolescence (e.g., the influence of inadequate childhood nutrient intake on adult cancer risk or the pubertal period as a window of breast cancer susceptibility)91–95. Interrogating the long-term effect on cancer of a given intervention in a prevention trial among children or adolescents would be unfeasible.
Examining the effect of genetic variants allocated at conception can therefore offer an important first step in identifying risk factors that may be sensitive to duration or timing of an exposure over the life course. Inferences made from promising MR findings to plausible intervention effects in a subsequent randomized trial would then need to carefully consider the possibility that effect estimates obtained in a MR analysis could be sensitive to critical period effects (in which case intervening on an exposure outside of this period may not alter disease risk) or represent the cumulative effect of lifelong exposure to a biomarker (in which case a relatively short-term trial may generate a smaller effect estimate than that obtained from MR). Adopting a “triangulation” framework where evidence from different epidemiological approaches with non-overlapping sources of bias are integrated can then be used to further examine durations of intervention necessary to confer an effect or ‘pinpoint’ possible critical windows of susceptibility to carcinogenic agents96. For example, multivariable regression analyses examining the association of an exposure, with some evidence of causality from MR studies, over different lengths of follow-up may help to identify the duration of exposure required to confer an effect. A negative control study with repeat measures of an exposure both within and outside of hypothesized critical periods (e.g., dietary fat intake before, during, and after pubertal development), in relation to subsequent disease risk (e.g., breast cancer)97 could be used to help refine periods of increased vulnerability to cancer-causing exposures.
Cancer Latency and Reverse Causation – limitations of MR
Genetic variants known to directly affect an exposure will in some cases be well-characterized (e.g., variants in APOE), and it will be established whether or not the variant-exposure associations are influenced by the outcome of interest. The biological understanding of other variants associated with risk factors that are identified in GWAS, however, is often more limited. In some situations in which genetic variants are associated with both an exposure and outcome of interest, the association between a variant and outcome might be via the exposure (i.e., a valid IV analysis) but it is also possible that, under certain circumstances, there may be a primary effect of the variant on the outcome which in turn causes a change in the exposure.
This situation has been illustrated previously in the context of body mass index (BMI) and CRP where an erroneous causal effect can be generated if a genetic variant that primarily influences BMI, which in turn influences CRP levels because BMI has a causal effect on CRP, is mistaken as being a variant with a primary influence on CRP25. Use of such a variant as an instrument for CRP in a MR analysis of the effect of CRP on BMI would then lead to biased results.
This introduction of reverse causation into a MR analysis may be problematic for common cancers with long latency periods between tumour initiation and diagnosis (e.g., breast and prostate)98. Reverse causation in this context could be mitigated by obtaining gene-exposure estimates in a healthy population where the prevalence of undiagnosed, latent cancer is likely to be low. These estimates could then be used to generate IV estimates in a two-sample MR framework. Additionally, steps could be taken to construct an instrument solely consisting of genetic variants that plausibly act directly on a trait. For example, in constructing an instrument for CRP levels, this could include solely using variants within CRP itself as these variants are more likely to be exclusively associated with CRP levels than variants in other genes99. However, it should be noted that a trade-off of using few, biologically-informed SNPs as an instrument is that sensitivity analyses examining horizontal pleiotropy – when feasible to perform – will have limited statistical power.
Selection bias in cancer progression analyses
A particular concern in cancer epidemiology is that exposures that influence cancer incidence may not influence cancer progression or survival. For example, although smoking is a robust risk factor for breast cancer incidence, smoking cessation upon development of breast cancer seems to have little effect on subsequent survival100. There has been some suggestion that folate may play a dual role in prostate and colorectal carcinogenesis: protective against DNA damage prior to the development of neoplasia, but promoting tumour progression via enhanced tumour proliferation and tissue invasion once cancer has developed101,102.
Some MR studies have begun to examine the effect of risk factors on both cancer incidence and progression103. In a recent analysis examining the effect of alcohol on prostate cancer risk in 46,919 men in the PRACTICAL consortium, alcohol consumption was not associated with overall prostate cancer risk but increased risk of prostate cancer mortality among men with low-grade disease104. Such MR studies exploit the fact that GWAS are being increasingly used to identify genetic variants associated with cancer progression or survival 105,106.
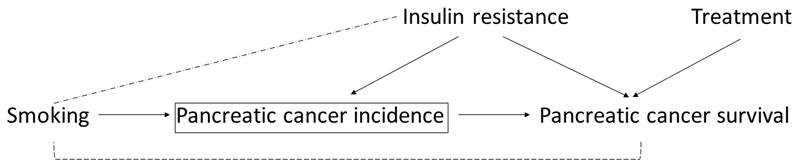
However, there are important methodological considerations in investigating factors causing cancer progression. This is because prognostic studies can suffer from selection bias due to the fact that any factors that cause disease incidence (or diagnosis) will tend to be correlated with each other in a sample of only cases, even when they are not correlated in the source population. Thus if at least one factor causes both incidence and disease survival (hypothetically, insulin resistance in Figure 3), all the other factors which cause disease incidence (hypothetically, smoking in Figure 3) will appear to be associated with survival, unless the true prognostic factor is conditioned upon. Thus, the estimated effect on progression for any factor that is associated with incidence is likely to be biased. However, any factor that is not associated with incidence will not suffer from selection bias by studying only cases in a MR analysis.
Figure 3. Directed acyclic graph for selection bias in prognostic studies.
In this example, the square bracket indicates that we are conditioning on pancreatic cancer incidence in a survival study by only studying pancreatic cancer cases, thus inducing an association between smoking (a factor that is otherwise independent of pancreatic cancer survival) and pancreatic cancer survival. This link is broken when conditioning on the factor that influences both cancer incidence and survival (e.g., insulin resistance), which can otherwise be seen as a confounder of the association between smoking and cancer survival. If a factor appears to influence pancreatic cancer survival that is not associated with pancreatic cancer incidence (e.g., treatment for pancreatic cancer), selection bias in such an MR analysis would not be expected.
When conducting prognostic studies, care should be taken to examine and (where possible) overcome the selection bias due to studying only cases103. First, the observed data could also be used to help identify plausible directed acyclic graphs (DAGs) including both disease incidence and progression. For example, if a risk score for a phenotype, and an environmental variable, are correlated in cases, but not in the source population this would suggest that both factors influence disease incidence, diagnosis, or self-selection into the study. However, lack of evidence for such correlations does not imply that there is no selection bias, and expert or external knowledge should be used in constructing the DAG, as is usual practice. The DAG can then be used to help inform sensitivity analyses. Additional data on factors that predict incidence could be combined with observed data in cases, to minimise selection bias, either by conditioning or by inverse probability weighting. If more than one DAG are considered plausible a priori, then they can be used to conduct sensitivity analyses by examining how robust the conclusions are to the causal assumptions made. The DAG can also be used to identify which assumptions are being made that are untestable given the observed data, and then sensitivity analyses can be conducted by examining plausible values for those relationships.
Illustrative examples
To illustrate the use of MR in analyses examining cancer outcomes, we have outlined three studies that have employed this approach to understand the causal role of various exposures on cancer incidence.
Selenium and prostate cancer risk
Prospective studies reporting inverse associations of dietary, blood, and toenail selenium with risk of prostate cancer107–113, along with findings from in vitro studies114,115, led to development of the Selenium and Vitamin E Cancer Prevention Trial (SELECT)116. SELECT was a 2x2 factorial trial of 35,533 healthy middle-aged men that examined the effect of daily supplementation with selenium, vitamin E, or both agents combined, as an intervention for prostate cancer prevention. The trial was stopped after 5.5 of a planned 12 years follow-up due to a lack of efficacy compounded by possible carcinogenic (increased rates of high-grade prostate cancer) and adverse metabolic (some evidence of increased rates of T2D) effects in the selenium supplementation group8,9. It is plausible that residual confounding may have accounted for conflicting results between prospective studies and SELECT117,118, though others have suggested that these differences may have reflected differences in baseline levels of selenium of participants in some observational studies as compared to SELECT119.
To test whether a MR approach could have predicted the results of SELECT, a two-sample MR analysis (Table 2) was performed using summary data on 72,729 individuals from the PRACTICAL consortium120,121. Eleven single-nucleotide polymorphisms (SNPs) robustly associated with blood selenium in previous GWAS122,123 (P<5x10-8) were combined into a genetic instrument (Table 3) to proxy circulating levels of selenium (Figure 1). To allow for direct comparison of effect estimates with SELECT, the authors investigated the odds ratio (OR) per 114 μg/L increase in circulating selenium, scaled to match the measured differences in blood selenium between supplementation and control arms in SELECT.
Consistent with results from SELECT, a 114 μg/L life-long increase in blood selenium in MR analyses was not associated with overall prostate cancer risk (OR:1.01, 95% CI:0.89-1.13; P=0.93; SELECT: Hazard Ratio (HR):1.04, 95% CI:0.91-1.19). MR analysis of selenium on advanced prostate cancer (OR:1.21, 95% CI:0.98-1.49; P=0.07) was concordant with weak evidence for an increased risk of high-grade prostate cancer in the selenium supplementation arm of SELECT (HR:1.21, 95% CI:0.97-1.52; P=0.20). Likewise, the effect of selenium on T2D (OR:1.18, 95% CI:0.97-1.43; P=0.11) was consistent with weak evidence for an increased risk of T2D in the selenium arm of SELECT (HR:1.07, 95% CI:0.97-1.18; P=0.16).
A limitation of this analysis is that the authors did not test the hypothesis that the effect of selenium on prostate cancer risk varied by baseline selenium status. One way to investigate this in an MR framework would be to test for interaction in effect estimates by study location – whether the study was conducted in selenium replete (e.g. USA) versus selenium deficient (e.g. Europe) countries. If differences in baseline levels of selenium do impact on the effect of selenium on prostate cancer, we would expect different effect estimates in these different settings.The overall similarities in findings between this MR analysis and that of SELECT, as compared to results from conventional observational studies, thus provides some support for the utility of an MR approach in approximating experimental results using observational data. Further, these results suggest that performing a MR analysis may be an important time-efficient and inexpensive step in predicting both efficacy and possible adverse effects of an intervention before an RCT is performed.
Alcohol and oesophageal cancer risk
Regular alcohol consumption is associated with a substantial increased risk of oesophageal squamous cell carcinoma in observational studies, with an approximate two-fold increased risk for moderate drinkers and five-fold increased risk for heavy drinkers when compared to occasional/non-drinkers124. However, alcohol consumption is often associated with other lifestyle and behavioural factors (e.g., smoking and dietary intake), which may themselves predispose toward oesophageal cancer125,126. Further, most studies that examined this hypothesis have used case-control designs, which may introduce reporting bias if cases recall alcohol consumption differently from controls124.
The ability to metabolize acetaldehyde, the principal metabolite of alcohol and a carcinogen127, is encoded by ALDH2, which is polymorphic in some East Asian populations. Specifically, the ALDH2 *2 allele produces an inactive protein subunit that is unable to metabolize acetaldehyde, resulting in markedly higher peak blood alcohol levels in *2*2 homozygotes compared to *1*1 homozygotes128. Individuals with the *2*2 genotype experience a flushing reaction to alcohol, along with dysphoria, nausea, and tachycardia, and therefore have very low levels of alcohol consumption129. Consequently, genetic variation in ALDH2 is robustly associated with both acetaldehyde levels and alcohol consumption (via differences in physiological response to levels of acetaldehyde). This satisfies the instrumental variable assumption that an instrument is robustly associated with an exposure of interest and ALDH2 can be utilized as an instrument for examining both regular alcohol consumption and blood acetaldehyde levels among alcohol consumers130.
In a meta-analysis of seven studies with a total of 905 oesophageal cancer cases of East Asian descent, individuals with the ALDH2 *2*2 genotype were found to have an approximately 3-fold reduced risk of oesophageal cancer, as compared to the ALDH2 *1*1 genotype (OR:0.36, 95% CI:0.16-0.80), suggesting a protective effect of reduced alcohol on oesophageal cancer131. However, when comparing individuals with a heterozygous *1*2 genotype to *1*1 individuals, the former were shown to have a (seemingly paradoxical) overall increased oesophageal cancer risk (OR:3.19, 95% CI:1.86-5.47). A naïve interpretation of this finding, without consideration of the effect of the ALDH2 *2 allele on blood acetaldehyde, would suggest that individuals with moderate alcohol intake had the highest risk of oesophageal cancer.
When this association was stratified by self-reported alcohol intake, the effect of *1*2 genotype on oesophageal cancer was shown to differ markedly by alcohol intake. Among non-drinkers, there was no strong evidence for an increase in risk among heterozygotes (OR:1.31, 95% CI:0.70-2.47) relative to *1*1 individuals. However, among heavy drinkers there was a 7-fold increase in risk (OR:7.07, 95% CI:3.67-13.6). Similarly, meta-regression analysis showed evidence that level of alcohol intake influenced the effect of the *1*2 genotype on oesophageal cancer risk (P=0.008) (i.e., the larger the amount of alcohol intake, the greater the OR of *1*2 versus *1*1 genotypes). As the possession of an ALDH2 *2 allele only appeared to increase risk of oesophageal cancer among heterozygotes who reported alcohol intake, this suggested that the substantially elevated acetaldehyde levels in these heterozygotes may mediate the effect of alcohol intake on oesophageal cancer.
More generally, this example illustrates how interpretation of MR findings can be challenging when there is limited biological understanding of the genetic variant used as a proxy for a given exposure. MR results that appear to be strongly discordant with underlying biology should be followed-up alongside available functional understanding of genetic variants employed as instruments to help resolve ambiguous or paradoxical results and avoid naïve interpretation of findings.
Body mass index and lung cancer risk
In contrast to the relationship of adiposity with risk of most cancers, BMI has shown consistent inverse associations with incidence of lung cancer, particularly among current and former smokers132,133. As smoking is a robust risk factor for lung cancer and has an inverse effect on BMI134, some have argued that residual confounding by smoking could account for this apparent protective association135. Reverse causation (i.e., undiagnosed lung cancer or disease processes leading up to lung cancer prior to study entry influencing subsequent weight loss), especially in cohorts with insufficient follow-up time, has also been proposed as an explanation for this observational finding136.
Attempts to address these possible sources of bias have failed to provide clarity. For example, studies that reported finely stratifying associations across various dimensions and classifications of smoking behaviour (e.g., number of cigarettes smoked per day, “cigarette-years” smoked, and time since quitting smoking) have found little evidence to support residual confounding by smoking influencing this association132,133. Further, studies removing individuals with inadequate follow-up have reported little effect on overall findings132,133,137,138, interpreted as suggesting that reverse causation is unlikely to be a major contributor to this association.
Given that germline genetic variants associated with BMI cannot be influenced by prevalent disease and should not be associated with potential confounding factors, a MR approach could be used to assess whether increased BMI is protective against lung cancer139,140. For example, Carreras-Torres et al. performed a MR analysis using GWAS results on 16,572 lung cancer cases and 21,480 controls of European descent141. 97 SNPs previously associated with BMI in a GWAS of 339,224 individuals were compiled into an instrument to proxy for anthropometrically measured BMI. This instrument was associated with measured BMI but not with available measures of tobacco exposure, including pack-years, cigarettes smoked per day, or cotinine levels, providing some evidence against confounding through measured smoking variables134. In two-sample MR analyses, a 1-SD increase in genetically-predicted BMI was weakly associated with an increased risk of lung cancer (OR:1.13, 95% CI:0.98-1.30; P=0.10), with strong heterogeneity across histological sub-types (Pheterogeneity<3x10-5). Notably, genetically-predicted BMI was positively associated with risk of both squamous cell (OR:1.45, 95% CI:1.16-1.62; P=1.2x10-3) and small cell carcinoma (OR:1.81, 95% CI:1.14-2.88;P=0.01) but showed weak evidence for a protective effect for adenocarcinoma (OR:0.82, 95% CI:0.66-1.01;P=0.06). These findings thus help to clarify a likely positive risk relationship of BMI with two major histosubtypes of lung cancer. Alongside some genetic evidence to suggest that elevated BMI may influence subsequent smoking uptake142, which itself reduces BMI while increasing lung cancer risk134, these findings collectively suggest a possible mechanism that could help to reconcile seemingly conflicting MR and observational findings. Further interrogation of a possible mediating role of smoking on the causal pathway between BMI and lung cancer risk using “two-step MR” (discussed in "MR for mediation") may be able to help shed further light on the possible intricate relationship between smoking and BMI in the aetiology of lung cancer.
Recent methodological extensions and future applications
In recent years, the development of various methodological extensions to the original MR paradigm have helped to enhance the scope of MR analyses, several of which are discussed below with reference to possible applications in cancer epidemiology.
MR for mediation
Over the past decade, high through-put “omics” technologies have begun to permit exhaustive profiling of the epigenome, metabolome, and proteome (as examples), allowing the collection of high-dimensional molecular data on increasingly large number of individuals143. Such omics measures may serve as important mediators on causal pathways linking macro-level risk factors with cancer incidence or progression. While conventional mediation analyses exist to examine possible exposure-mediator-outcome relationships, the validity of these approaches relies upon strong assumptions which are unlikely to be met in practice, such as no measurement error and no unmeasured confounding144.
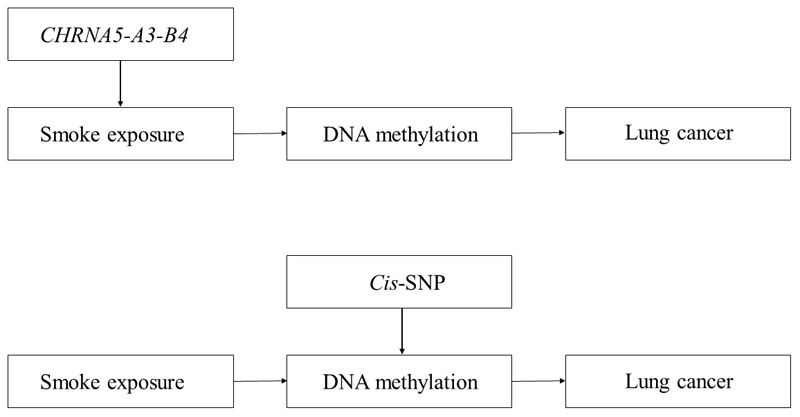
With the performance of GWAS on large collections of metabolites and other omic measures145,146, this will create opportunity to develop instruments for these traits. To establish whether a particular molecular intermediate is on the causal pathway between an exposure and cancer, genetic variants can be used as instruments for both exposures and putative mediators that influence a disease outcome in a two-step MR framework (Figure 4)147.
Figure 4. Two-step Mendelian randomization analysis examining the mediating effect of methylation on the association between smoke exposure and lung cancer.
In the first step, a SNP within CHRNA5-A3-B4 is used as an instrument for smoke exposure to assess the causal association between smoking and DNA methylation. In the second step, an independent cis-SNP is used as an instrument for DNA methylation to assess the causal association of DNA methylation with lung cancer risk. The two-step method allows interrogation of the mediation effect of DNA methylation in the association between smoking and lung cancer risk.
For example, a method of testing the mediating role of methylation changes on cancer outcomes would be to exploit the fact that genetic variants (e.g., methylation quantitative trait loci, mQTLs) are robustly associated with methylation at CpG sites across the epigenome, providing possible instruments for MR analyses148. Two-step MR could then used to examine the potential mediating role of DNA methylation sites associated with exposures such as tobacco smoke149 which have also been found to be strongly associated with lung cancer risk150. To test whether methylation is causally mediating (some, or all of) the effect of tobacco exposure on lung cancer risk, in the first step, a SNP could be used to proxy smoking behaviour in order to investigate its effect on the intermediate phenotype (DNA methylation). In the second step, an independent SNP could then be used to proxy the intermediate phenotype (DNA methylation) which could then be examined in relation to the disease outcome (lung cancer)144. This approach has the potential to be scaled up within the context of high dimensional ‘omic datasets to integrate multiple tiers of molecular data in a causal framework 151,152. While statistical and computational challenges arise with increasingly complex networks of molecular mediators, numerous data reduction and variable selection techniques may be used to identify informative causal molecular pathways to disease, including pathway analysis, penalised regression, machine learning, and data mining techniques which are increasingly being applied in an automated fashion153,154 (see Hypothesis-free MR).
Factorial MR
Akin to a factorial RCT, factorial MR is a method of testing the independent and additive effects of two or more exposures on disease outcomes. This approach was adopted by Ference et al. who performed a 2x2 factorial MR analysis to examine the effect of the LDL cholesterol-lowering drug ezetimibe on risk of coronary heart disease (CHD), as compared to the effect of statins alone or when combined with statins155. Ference et al. examined the effect of genetically-lower LDL-C on the risk of CHD through SNPs in NPC1L1 (a target of ezetimibe) alone, HMGCR (a target of statins) alone, or variants in both gene regions combined. The authors reported that natural randomization to lower LDL-C through SNPs in NPC1L1 and HMGCR alone showed similar decreases in LDL-C and CHD and that randomization to lower LDL-C in both groups combined had a linearly additive effect on LDL-C lowering and a log-linearly additive effect on CHD risk. These results were corroborated by the ‘Improved Reduction of Outcomes: Vytorin Efficacy International Trial,’ which allocated 18,144 participants to ezetimibe, statins, both, or placebo156.
An important caveat of this approach is that it relies on access to individual-level data and requires very large sample sizes to have adequate statistical power to reliably detect differences in effect across groups.
Hypothesis-free MR
A novel extension to a conventional “hypothesis-driven” MR analysis is a phenome-wide, “hypothesis-free” MR analysis (termed “MR-PhEWAS”)153. This approach makes use of genotyped datasets with high-dimensional phenotypic data or summary GWAS association statistics to perform hundreds or thousands of statistical tests simultaneously in an agnostic manner. For example, the approach can be used to examine the effect of a single exposure across multiple outcomes or multiple exposures across a single outcome. In contrast to hypothesis-driven analyses, hypothesis-free approaches allow for testing hypotheses that may not have been considered or tested previously, thus identifying novel risk relationships, and can help to address issues of publication bias as all analyses are openly specified and all results are presented157.
For example, using a two-sample MR framework with summary data, Haycock et al. performed a MR-PheWAS examining the effect of telomere length on risk of 35 cancers and 48 non-cancer diseases in 420,081 cases and 1,093,105 controls158. After correction for multiple-testing, they found that telomere length increased cancer risk across most sites and histological sub-types but reduced CVD risk. An important consideration when performing hypothesis-free MR analyses using summary data is the need to follow-up any putative findings in subsequent independent datasets. This can be a challenge when using summary GWAS data to perform such analyses if a large proportion of the available GWAS literature was used to provide causal estimates in the original “discovery phase” of an analysis.
MR for identifying causality of mutational signatures
Large-scale analysis of the genomes of thousands of cancer patients has helped to reveal somatic “mutational signatures” (distinctive somatic mutational patterns left by unique carcinogenic agents) involved in the development of their tumours159,160. To date, mutational signatures have been identified across more than 30 different cancer types, with anywhere from two to six distinction mutational processes for each cancer type. Knowledge of the causes of somatic mutations within tumour tissue can improve understanding of the mechanisms by which endogenous and exogenous exposures promote the development of a cancer. Of the mutational signatures identified across cancer types, a putative cause has been proposed for approximately half159; MR may offer particular promise in helping to identify the aetiology of other mutational signatures identified161.
Robles-Espinoza et al. examined the effect of germline MC1R status, associated with red hair, freckling, and sun sensitivity, on somatic mutation burden in melanoma. Such an analysis can be viewed as a MR appraisal of the effect of this sensitivity phenotype on somatic mutation burden in melanoma162. For all six mutational types assessed, there was evidence of an increased burden of somatic single nucleotide variants in individuals carrying one or two MC1R R alleles (disruptive variants). For one of the six mutational signatures characterized by an abundance of somatic C>T single nucleotide variants, each additional R allele at MC1R was associated with a 42% (95% CI:15-76%) increase in the C>T single nucleotide variant count. This approach therefore highlights the possibility of testing the causal effect of suspected carcinogenic agents on mutational burden for various mutational signatures across cancer tissues and sub-types.
Drug repurposing and adverse drug effects
Drug repurposing, applying known drugs to novel indications, can provide a rapid, cost-effective mechanism for drug discovery and may hold promise for the development of pharmacological interventions for cancer prevention163,164. In turn, for well-tolerated drugs that are considered candidates for repurposing, MR may offer an attractive approach for testing their potential chemopreventive efficacy. For example, it is currently possible to reliably instrument drugs for which there is a broad understanding of the biological mechanism of action (e.g., HMG Co-A reductase inhibitors, PCSK9 inhibitors, CETP inhibitors, and sPLA2 inhibitors in cardiovascular disease165). For the primary or tertiary prevention of certain cancers, aspirin, metformin, and bisphosphonates have all been proposed as possible candidate pharmaceutical agents for repurposing166–168. Using MR as a first step to test drug efficacy for novel cancer indications could help to prioritize or deprioritize which drugs should be taken forward to testing in RCTs for re-purposing.
MR may also provide a useful approach for predicting adverse effects of pharmaceuticals169. Pre-approval trials are often not able to adequately capture development of adverse effects due to the comparatively small number of individuals typically exposed to a drug in such trials (unless drug effects are very common or very large), the limited duration of most trials, and unknown generalizability of trial participants to the broader population. While many of these issues can be addressed post-approval of a drug through spontaneous reporting systems, these introduce their own limitations including confounding, for example by indication, environmental factors, or lifestyle traits. MR studies should be able to overcome these limitations and have been employed in some instances to test or anticipate adverse effects of interventions in ongoing trials (e.g., adverse effects of statins on T2D as proxied by variants in HMGCR)34,35,170–172.
While knowledge of biological pathways can help to anticipate some adverse drug effects pre-approval of a drug, it may not be possible to correctly predict all such effects173. One possible approach to resolve this would be to use MR-PhEWAS to perform a phenotypic scan of a genetically-instrumented drug exposure across hundreds or thousands of potential outcomes, as outlined previously. The identification of possible adverse effects of a drug through this approach could then be used to pre-specify and adequately power secondary outcome measures or, alternately, to de-prioritize further investigation of a therapeutic target.
Conclusion
Observational epidemiological studies are prone to various intractable biases which can undermine robust causal inference. Mendelian randomization offers a promising approach to generate a more reliable evidence-base for cancer prevention and treatment. The advent of MR methods using summarized data means that such analyses can now be performed more efficiently, rapidly, and with greater statistical power than previously possible. Further, the range of methodological extensions to the original MR paradigm now available have greatly expanded the scope of this approach, enabling increasingly sophisticated causal questions to be interrogated174. Despite this, there are inherent constraints on the types of epidemiological questions that can be answered with this approach as compared to conventional observational analyses. For example, MR is restricted to examining exposures that have a heritable component and suitable genetic proxies for these exposures; MR cannot isolate critical period effects for exposures; and MR will usually only represent the effect of lifelong exposure to a biomarker. These limitations mean that inferences made from MR will be most informative when integrated alongside insights gained from other epidemiological approaches and study designs. Given optimism surrounding use of the method in helping to strengthen evidence for public health and pharmacological interventions175, it is likely that there will be a continued proliferation of MR analyses in the literature in the near future. Careful design, analysis, and interpretation of such studies with consideration of the limitations of the method will provide the greatest opportunity for such studies to inform cancer prevention and treatment strategies.
Supplementary Material
Grant support
This work was supported by a Cancer Research UK programme grant (C18281/A19169) to KH Wade, RC Richmond, CL Relton, SJ Lewis, and RM Martin, including Cancer Research UK Research PhD studentships (C18281/A20988) to J Yarmolinsky and RJ Langdon. This work was also supported by a Wellcome Trust 4-year studentship (WT083431MA) to CJ Bull. All authors are members of the MRC IEU which is supported by the Medical Research Council and the University of Bristol (MC_UU_12013/1-9).
Abbreviations
- MR
Mendelian randomization
- IV
instrumental variable
- CVD
cardiovascular disease
- T2D
type 2 diabetes
- HDL-C
high-density lipoprotein cholesterol
- CRP
C-reactive protein
- PCSK9
proprotein convertase subtilisin/kexin type 2
- GWAS
genome-wide association study
- LDL-C
low-density lipoprotein cholesterol
- BMI
body mass index
- DAGs
directed acyclic graphs
- SELECT
Selenium and Vitamin E Cancer Prevention Trial
- GWAS
genome-wide association study
- SNPs
single-nucleotide polymorphisms
- mQTLs
methylation quantitative trait loci
- CHD
coronary heart disease
- MR-PheWAS
Mendelian randomization phenome-wide association study
- LD
linkage disequilibrium
- GAME-ON
Genetic Associations and Mechanisms in Oncology
- WHR
waist-hip ratio
- GRS
genetic risk score
- IVW
inverse-variance weighted
- WME
weighted median estimator
- MBE
mode-based estimate
- InSIDE
instrument strength independent of direct effects
- ZEMPA
Zero Modal Pleiotropy Assumption
Footnotes
Conflicts of interest
All authors declare no potential conflicts of interest.
References
- 1.Taubes G. Epidemiology faces its limits. Science. 1995;269(5221):164–169. doi: 10.1126/science.7618077. [DOI] [PubMed] [Google Scholar]
- 2.Davey Smith G, Ebrahim S. Epidemiology--is it time to call it a day? Int J Epidemiol. 2001;30(1):1–11. doi: 10.1093/ije/30.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Schoenfeld JD, Ioannidis JP. Is everything we eat associated with cancer? A systematic cookbook review. Am J Clin Nutr. 2013;97(1):127–134. doi: 10.3945/ajcn.112.047142. [DOI] [PubMed] [Google Scholar]
- 4.Vineis P, Alavanja M, Buffler P, et al. Tobacco and cancer: recent epidemiological evidence. J Natl Cancer Inst. 2004;96(2):99–106. doi: 10.1093/jnci/djh014. [DOI] [PubMed] [Google Scholar]
- 5.Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45(4):529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 6.McDonald JC, McDonald AD. The epidemiology of mesothelioma in historical context. Eur Respir J. 1996;9(9):1932–1942. doi: 10.1183/09031936.96.09091932. [DOI] [PubMed] [Google Scholar]
- 7.Gaziano JM, Glynn RJ, Christen WG, et al. Vitamins E and C in the prevention of prostate and total cancer in men: the Physicians' Health Study II randomized controlled trial. JAMA. 2009;301(1):52–62. doi: 10.1001/jama.2008.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein EA, Thompson IM, Tangen CM, Jr, et al. Vitamin E and the risk of prostate cancer: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA. 2011;306(14):1549–1556. doi: 10.1001/jama.2011.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lippman SM, Klein EA, Goodman PJ, et al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA. 2009;301(1):39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee IM, Cook NR, Gaziano JM, et al. Vitamin E in the primary prevention of cardiovascular disease and cancer: the Women's Health Study: a randomized controlled trial. JAMA. 2005;294(1):56–65. doi: 10.1001/jama.294.1.56. [DOI] [PubMed] [Google Scholar]
- 11.Omenn GS, Goodman GE, Thornquist MD, et al. Effects of a combination of beta carotene and vitamin A on lung cancer and cardiovascular disease. N Engl J Med. 1996;334(18):1150–1155. doi: 10.1056/NEJM199605023341802. [DOI] [PubMed] [Google Scholar]
- 12.Zhang SM, Cook NR, Albert CM, Gaziano JM, Buring JE, Manson JE. Effect of combined folic acid, vitamin B6, and vitamin B12 on cancer risk in women: a randomized trial. JAMA. 2008;300(17):2012–2021. doi: 10.1001/jama.2008.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole BF, Baron JA, Sandler RS, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA. 2007;297(21):2351–2359. doi: 10.1001/jama.297.21.2351. [DOI] [PubMed] [Google Scholar]
- 14.Schatzkin A, Lanza E, Corle D, et al. Lack of effect of a low-fat, high-fiber diet on the recurrence of colorectal adenomas. Polyp Prevention Trial Study Group. N Engl J Med. 2000;342(16):1149–1155. doi: 10.1056/NEJM200004203421601. [DOI] [PubMed] [Google Scholar]
- 15.Prentice RL, Caan B, Chlebowski RT, et al. Low-fat dietary pattern and risk of invasive breast cancer: the Women's Health Initiative Randomized Controlled Dietary Modification Trial. JAMA. 2006;295(6):629–642. doi: 10.1001/jama.295.6.629. [DOI] [PubMed] [Google Scholar]
- 16.The Alpha-Tocopherol, Beta Carotene Cancer Prevention Study Group. The effect of vitamin E and beta carotene on the incidence of lung cancer and other cancers in male smokers. N Engl J Med. 1994;330(15):1029–1035. doi: 10.1056/NEJM199404143301501. [DOI] [PubMed] [Google Scholar]
- 17.Lawlor DA, Davey Smith G, Kundu D, Bruckdorfer KR, Ebrahim S. Those confounded vitamins: what can we learn from the differences between observational versus randomised trial evidence? Lancet. 2004;363(9422):1724–1727. doi: 10.1016/S0140-6736(04)16260-0. [DOI] [PubMed] [Google Scholar]
- 18.Sattar N, Preiss D. Reverse Causality in Cardiovascular Epidemiological Research: More Common Than Imagined? Circulation. 2017;135(24):2369–2372. doi: 10.1161/CIRCULATIONAHA.117.028307. [DOI] [PubMed] [Google Scholar]
- 19.Phillips AN, Davey Smith G. How independent are "independent" effects? Relative risk estimation when correlated exposures are measured imprecisely. J Clin Epidemiol. 1991;44(11):1223–1231. doi: 10.1016/0895-4356(91)90155-3. [DOI] [PubMed] [Google Scholar]
- 20.Davey Smith G, Phillips AN. Confounding in epidemiological studies: why "independent" effects may not be all they seem. BMJ. 1992;305(6856):757–759. doi: 10.1136/bmj.305.6856.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fewell Z, Davey Smith G, Sterne JA. The impact of residual and unmeasured confounding in epidemiologic studies: a simulation study. Am J Epidemiol. 2007;166(6):646–655. doi: 10.1093/aje/kwm165. [DOI] [PubMed] [Google Scholar]
- 22.Bracken M. Risk, chance, and causation: investigating the origins and treatment of disease. New Haven: Yale University Press; 2013. [Google Scholar]
- 23.Kabat GC. Hyping Health Risks: Environmental Hazards in Daily Life and the Science of Epidemiology. New York: Columbia University Press; 2008. [Google Scholar]
- 24.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2(8):e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davey Smith G, Ebrahim S. “Mendelian randomisation”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiology. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 27.Davey Smith G, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epi. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 28.Evans DM, Davey Smith G. Mendelian Randomization: New Applications in the Coming Age of Hypothesis-Free Causality. Annu Rev Genomics Hum Genet. 2015;16:327–350. doi: 10.1146/annurev-genom-090314-050016. [DOI] [PubMed] [Google Scholar]
- 29.Haycock PC, Burgess S, Wade KH, Bowden J, Relton C, Davey Smith G. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. Am J Clin Nutr. 2016;103(4):965–978. doi: 10.3945/ajcn.115.118216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 31.Timpson NJ, Wade KH, Davey Smith G. Mendelian randomization: application to cardiovascular disease. Curr Hypertens Rep. 2012;14(1):29–37. doi: 10.1007/s11906-011-0242-7. [DOI] [PubMed] [Google Scholar]
- 32.Davey Smith G, Lawlor DA, Harbord R, Timpson N, Day I, Ebrahim S. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med. 2007;4(12):e352. doi: 10.1371/journal.pmed.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanson SA, Tiemeier H, Ikram MA, Hernan MA. Nature as a Trialist?: Deconstructing the Analogy Between Mendelian Randomization and Randomized Trials. Epidemiology. 2017;28(5):653–659. doi: 10.1097/EDE.0000000000000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ference BA, Robinson JG, Brook RD, et al. Variation in PCSK9 and HMGCR and Risk of Cardiovascular Disease and Diabetes. N Engl J Med. 2016;375(22):2144–2153. doi: 10.1056/NEJMoa1604304. [DOI] [PubMed] [Google Scholar]
- 35.Swerdlow DI, Preiss D, Kuchenbaecker KB, et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet. 2015;385(9965):351–361. doi: 10.1016/S0140-6736(14)61183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voight BF, Peloso GM, Orho-Melander M, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380(9841):572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunner EJ, Kivimaki M, Witte DR, et al. Inflammation, insulin resistance, and diabetes--Mendelian randomization using CRP haplotypes points upstream. PLoS Med. 2008;5(8):e155. doi: 10.1371/journal.pmed.0050155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabatine MS, Giugliano RP, Keech AC, et al. Evolocumab and Clinical Outcomes in Patients with Cardiovascular Disease. N Engl J Med. 2017;376(18):1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 39.Ference BA, Kastelein JJP, Ginsberg HN, et al. Association of Genetic Variants Related to CETP Inhibitors and Statins With Lipoprotein Levels and Cardiovascular Risk. JAMA. 2017;318(10):947–956. doi: 10.1001/jama.2017.11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The HPS3/TIMI55-REVEAL Collaborative Group. Effects of Anacetrapib in Patients with Atherosclerotic Vascular Disease. N Engl J Med. 2017;377(13):1217–1227. doi: 10.1056/NEJMoa1706444. [DOI] [PubMed] [Google Scholar]
- 41.Kamstrup PR, Tybjaerg-Hansen A, Steffensen R, Nordestgaard BG. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA. 2009;301(22):2331–2339. doi: 10.1001/jama.2009.801. [DOI] [PubMed] [Google Scholar]
- 42.Interleukin-6 Receptor Mendelian Randomisation Analysis (IL6R MR) Consortium. Swerdlow DI, Holmes MV, et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet. 2012;379(9822):1214–1224. doi: 10.1016/S0140-6736(12)60110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keavney B, Danesh J, Parish S, et al. Fibrinogen and coronary heart disease: test of causality by 'Mendelian randomization'. Int J Epidemiol. 2006;35(4):935–943. doi: 10.1093/ije/dyl114. [DOI] [PubMed] [Google Scholar]
- 44.Holmes MV, Simon T, Exeter HJ, et al. Secretory phospholipase A(2)-IIA and cardiovascular disease: a mendelian randomization study. J Am Coll Cardiol. 2013;62(21):1966–1976. doi: 10.1016/j.jacc.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheehan NA, Didelez V, Burton PR, Tobin MD. Mendelian randomisation and causal inference in observational epidemiology. PLoS Med. 2008;5(8):e177. doi: 10.1371/journal.pmed.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glynn RJ. Promises and limitations of mendelian randomization for evaluation of biomarkers. Clin Chem. 2010;56(3):388–390. doi: 10.1373/clinchem.2009.142513. [DOI] [PubMed] [Google Scholar]
- 47.Nitsch D, Molokhia M, Smeeth L, DeStavola BL, Whittaker JC, Leon DA. Limits to causal inference based on Mendelian randomization: a comparison with randomized controlled trials. Am J Epidemiol. 2006;163(5):397–403. doi: 10.1093/aje/kwj062. [DOI] [PubMed] [Google Scholar]
- 48.VanderWeele TJ, Tchetgen Tchetgen EJ, Cornelis M, Kraft P. Methodological challenges in mendelian randomization. Epidemiology. 2014;25(3):427–435. doi: 10.1097/EDE.0000000000000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmes MV, Ala-Korpela M, Smith GD. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol. 2017 doi: 10.1038/nrcardio.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol. 2013;178(7):1177–1184. doi: 10.1093/aje/kwt084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG. EPIC-InterAct Consortium. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30(7):543–552. doi: 10.1007/s10654-015-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hartwig FP, Davies NM, Hemani G, Davey Smith G. Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. 2016;45(6):1717–1726. doi: 10.1093/ije/dyx028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pasaniuc B, Pric AL. Dissecting the genetics of complex traits using summary association statistics. Nat Rev Genet. 2017;18(2):117–127. doi: 10.1038/nrg.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hemani G, Zheng J, Wade KH, Laurin C, Elsworth B, Burgess S, Bowden J, Langdon R, Tan V, Yarmolinsky J, Shihab HA, et al. MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations. bioRxiv.078972 [Google Scholar]
- 55.Khankari NK, Shu XO, Wen W, et al. Association between Adult Height and Risk of Colorectal, Lung, and Prostate Cancer: Results from Meta-analyses of Prospective Studies and Mendelian Randomization Analyses. PLoS Med. 2016;13(9):e1002118. doi: 10.1371/journal.pmed.1002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Randall JC, Winkler TW, Kutalik Z, et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9(6):e1003500. doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilks WP, Abbott JK, Morrow EH. Sex differences in disease genetics: evidence, evolution, and detection. Trends Genet. 2014;30(10):453–463. doi: 10.1016/j.tig.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 58.Heid IM, Jackson AU, Randall JC, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(11):949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shungin D, Winkler TW, Croteau-Chonka DC, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol. 2016;40(7):597–608. doi: 10.1002/gepi.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35(11):1880–1906. doi: 10.1002/sim.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol. 2013;42(4):1134–1144. doi: 10.1093/ije/dyt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.International HapMap Consortium. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 65.1000 Genomes Project Consortium. Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Freathy RM, Timpson NJ, Lawlor DA, et al. Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes. 2008;57(5):1419–1426. doi: 10.2337/db07-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tyler AL, Asselbergs FW, Williams SM, Moore JH. Shadows of complexity: what biological networks reveal about epistasis and pleiotropy. Bioessays. 2009;31(2):220–227. doi: 10.1002/bies.200800022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kathiresan S, Melander O, Guiducci C, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40(2):189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Staley JR, Blackshaw J, Kamat MA, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32(20):3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tryka KA, Hao L, Sturcke A, et al. NCBI's Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014;42(Database issue):D975–979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40(4):304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. International Journal of Epidemiology. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Giovannucci E, Harlan DM, Archer MC, et al. Diabetes and cancer: a consensus report. Diabetes Care. 2010;33(7):1674–1685. doi: 10.2337/dc10-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Collin SM, Metcalfe C, Refsum H, et al. Circulating folate, vitamin B12, homocysteine, vitamin B12 transport proteins, and risk of prostate cancer: a case-control study, systematic review, and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2010;19(6):1632–1642. doi: 10.1158/1055-9965.EPI-10-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poulsen AH, Christensen S, McLaughlin JK, et al. Proton pump inhibitors and risk of gastric cancer: a population-based cohort study. Br J Cancer. 2009;100(9):1503–1507. doi: 10.1038/sj.bjc.6605024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lennon H, Sperrin M, Badrick E, Renehan AG. The Obesity Paradox in Cancer: a Review. Curr Oncol Rep. 2016;18(9):56. doi: 10.1007/s11912-016-0539-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Williams RR, Sorlie PD, Feinleib M, McNamara PM, Kannel WB, Dawber TR. Cancer incidence by levels of cholesterol. JAMA. 1981;245(3):247–252. [PubMed] [Google Scholar]
- 79.Kark JD, Smith AH, Hames CG. The relationship of serum cholesterol to the incidence of cancer in Evans County, Georgia. J Chronic Dis. 1980;33(5):311–332. doi: 10.1016/0021-9681(80)90026-0. [DOI] [PubMed] [Google Scholar]
- 80.Wallace RB, Rost C, Burmeister LF, Pomrehn PR. Cancer incidence in humans: relationship to plasma lipids and relative weight. J Natl Cancer Inst. 1982;68(6):915–918. [PubMed] [Google Scholar]
- 81.Newman TB, Hulley SB. Carcinogenicity of lipid-lowering drugs. JAMA. 1996;275(1):55–60. [PubMed] [Google Scholar]
- 82.Wysowski DK, Kennedy DL, Gross TP. Prescribed use of cholesterol-lowering drugs in the United States, 1978 through 1988. JAMA. 1990;263(16):2185–2188. [PubMed] [Google Scholar]
- 83.Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1986;1(8479):507–508. doi: 10.1016/s0140-6736(86)92972-7. [DOI] [PubMed] [Google Scholar]
- 84.Trompet S, Jukema JW, Katan MB, et al. Apolipoprotein e genotype, plasma cholesterol, and cancer: a Mendelian randomization study. Am J Epidemiol. 2009;170(11):1415–1421. doi: 10.1093/aje/kwp294. [DOI] [PubMed] [Google Scholar]
- 85.Benn M, Tybjaerg-Hansen A, Stender S, Frikke-Schmidt R, Nordestgaard BG. Low-density lipoprotein cholesterol and the risk of cancer: a mendelian randomization study. J Natl Cancer Inst. 2011;103(6):508–519. doi: 10.1093/jnci/djr008. [DOI] [PubMed] [Google Scholar]
- 86.Benn M, Tybjærg-Hansen A, Stender S, Frikke-Schmidt R, Nordestgaard BG. Using genetics to explore whether the cholesterol-lowering drug ezetimibe may cause an increased risk of cancer. International Journal of Epidemiology. 2017;46(6):1777–1785. doi: 10.1093/ije/dyx096. [DOI] [PubMed] [Google Scholar]
- 87.Peto R, Emberson J, Landray M, et al. Analyses of cancer data from three ezetimibe trials. N Engl J Med. 2008;359(13):1357–1366. doi: 10.1056/NEJMsa0806603. [DOI] [PubMed] [Google Scholar]
- 88.Colditz GA, Taylor PR. Prevention trials: their place in how we understand the value of prevention strategies. Annu Rev Public Health. 2010;31:105–120. doi: 10.1146/annurev.publhealth.121208.131051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nadler DL, Zurbenko IG. Developing a Weibull Model Extension to Estimate Cancer Latency. ISRN Epidemiology. 2013;2013(750857) [Google Scholar]
- 90.Colditz GA. Overview of the epidemiology methods and applications: strengths and limitations of observational study designs. Crit Rev Food Sci Nutr. 2010;50(Suppl 1):10–12. doi: 10.1080/10408398.2010.526838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Uauy R, Solomons N. Diet, nutrition, and the life-course approach to cancer prevention. J Nutr. 2005;135(12 Suppl):2934S–2945S. doi: 10.1093/jn/135.12.2934S. [DOI] [PubMed] [Google Scholar]
- 92.Band PR, Le ND, Fang R, Deschamps M. Carcinogenic and endocrine disrupting effects of cigarette smoke and risk of breast cancer. Lancet. 2002;360(9339):1044–1049. doi: 10.1016/S0140-6736(02)11140-8. [DOI] [PubMed] [Google Scholar]
- 93.Macon MB, Fenton SE. Endocrine disruptors and the breast: early life effects and later life disease. J Mammary Gland Biol Neoplasia. 2013;18(1):43–61. doi: 10.1007/s10911-013-9275-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maynard M, Gunnell D, Emmett P, Frankel S, Davey Smith G. Fruit, vegetables, and antioxidants in childhood and risk of adult cancer: the Boyd Orr cohort. J Epidemiol Community Health. 2003;57(3):218–225. doi: 10.1136/jech.57.3.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van der Pols JC, Bain C, Gunnell D, Smith GD, Frobisher C, Martin RM. Childhood dairy intake and adult cancer risk: 65-y follow-up of the Boyd Orr cohort. Am J Clin Nutr. 2007;86(6):1722–1729. doi: 10.1093/ajcn/86.5.1722. [DOI] [PubMed] [Google Scholar]
- 96.Lawlor DA, Tilling K, Davey Smith G. Triangulation in aetiological epidemiology. Int J Epidemiol. 2016;45(6):1866–1886. doi: 10.1093/ije/dyw314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.MacLennan M, Ma DW. Role of dietary fatty acids in mammary gland development and breast cancer. Breast Cancer Res. 2010;12(5):211. doi: 10.1186/bcr2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hall F. Screening mammography-potential problems on the horizon. N Engl J Med. 1986;314:53–55. doi: 10.1056/NEJM198601023140111. [DOI] [PubMed] [Google Scholar]
- 99.Elliott P, Chambers JC, Zhang W, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA. 2009;302(1):37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.The Health Consequences of Smoking-50 Years of Progress: A Report of the Surgeon General. Atlanta (GA): 2014. [Google Scholar]
- 101.Rycyna KJ, Bacich DJ, O'Keefe DS. Opposing roles of folate in prostate cancer. Urology. 2013;82(6):1197–1203. doi: 10.1016/j.urology.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim YI. Role of folate in colon cancer development and progression. J Nutr. 2003;133(11 Suppl 1):3731S–3739S. doi: 10.1093/jn/133.11.3731S. [DOI] [PubMed] [Google Scholar]
- 103.Paternoster L, Tilling KM, Davey Smith G. Genetic Epidemiology And Mendelian Randomization For Informing Disease Therapeutics: Conceptual And Methodological Challenges. Plos Genet. 2017 Oct 5;13(10):e1006944. doi: 10.1371/journal.pgen.1006944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brunner C, Davies NM, Martin RM, et al. Alcohol consumption and prostate cancer incidence and progression: A Mendelian randomisation study. Int J Cancer. 2017;140(1):75–85. doi: 10.1002/ijc.30436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Berndt SI, Wang Z, Yeager M, et al. Two Susceptibility Loci Identified for Prostate Cancer Aggressiveness. Nature communications. 2015;6:6889–6889. doi: 10.1038/ncomms7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Szulkin R, Karlsson R, Whitington T, et al. Genome-Wide Association Study of Prostate Cancer–Specific Survival. Cancer Epidemiology Biomarkers & Prevention. 2015;24(11):1796. doi: 10.1158/1055-9965.EPI-15-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jain MG, Hislop GT, Howe GR, Ghadirian P. Plant foods, antioxidants, and prostate cancer risk: findings from case-control studies in Canada. Nutr Cancer. 1999;34:173–184. doi: 10.1207/S15327914NC3402_8. [DOI] [PubMed] [Google Scholar]
- 108.West DW, Slattery ML, Robison LM, French TK, Mahoney AW. Adult dietary intake and prostate cancer risk in Utah: a case-control study with special emphasis on aggressive tumors. Cancer Causes Control. 1991;2:85–94. doi: 10.1007/BF00053126. [DOI] [PubMed] [Google Scholar]
- 109.Helzlsouer KJ, Huang HY, Alberg AJ, et al. Association Between α-Tocopherol, γ-Tocopherol, Selenium, and Subsequent Prostate Cancer. Journal of the National Cancer Institute. 2000;92(24):2018–2023. doi: 10.1093/jnci/92.24.2018. [DOI] [PubMed] [Google Scholar]
- 110.Li H, Stampfer MJ, Giovannucci EL, et al. A Prospective Study of Plasma Selenium Levels and Prostate Cancer Risk. Journal of the National Cancer Institute. 2004;96(9):696–703. doi: 10.1093/jnci/djh125. [DOI] [PubMed] [Google Scholar]
- 111.Nomura AM, Lee J, Stemmermann GN, Combs GF. Serum Selenium and Subsequent Risk of Prostate Cancer. Cancer Epidemiology Biomarkers & Prevention. 2000;9(9):883–887. [PubMed] [Google Scholar]
- 112.Yoshizawa K, Willett WC, Morris SJ, et al. Study of prediagnostic selenium level in toenails and the risk of advanced prostate cancer. J Natl Cancer Inst. 1998;90(16):1219–1224. doi: 10.1093/jnci/90.16.1219. [DOI] [PubMed] [Google Scholar]
- 113.van den Brandt PA, Zeegers MPA, Bode P, Goldbohm RA. Toenail Selenium Levels and the Subsequent Risk of Prostate Cancer: A Prospective Cohort Study. Cancer Epidemiology Biomarkers & Prevention. 2003;12(9):866–871. [PubMed] [Google Scholar]
- 114.Redman C, Scott JA, Baines AT, et al. Inhibitory effect of selenomethionine on the growth of three selected human tumor cell lines. Cancer Letters. 1998;125(1–2):103–110. doi: 10.1016/s0304-3835(97)00497-7. [DOI] [PubMed] [Google Scholar]
- 115.Menter DG, Sabichi AL, Lippman SM. Selenium Effects on Prostate Cell Growth. Cancer Epidemiology Biomarkers & Prevention. 2000;9(11):1171. [PubMed] [Google Scholar]
- 116.Lippman SM, Klein EA, Goodman PJ, et al. Effect of Selenium and Vitamin E on Risk of Prostate Cancer and Other Cancers: The Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA : the journal of the American Medical Association. 2009;301(1):39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vinceti M, Crespi CM, Malagoli C, Del Giovane C, Krogh V. Friend or foe? The current epidemiologic evidence on selenium and human cancer risk. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2013;31(4):305–341. doi: 10.1080/10590501.2013.844757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dennert G, Zwahlen M, Brinkman M, Vinceti M, Zeegers MP, Horneber M. Selenium for preventing cancer. Cochrane Database Syst Rev. 2011;(5) doi: 10.1002/14651858.CD005195.pub2. CD005195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nicastro HL, Dunn BK. Selenium and prostate cancer prevention: insights from the selenium and vitamin E cancer prevention trial (SELECT) Nutrients. 2013;5(4):1122–1148. doi: 10.3390/nu5041122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yarmolinsky J, Bonilla C, Haycock PC, et al. Circulating Selenium and Prostate Cancer Risk: A Mendelian Randomization Analysis. J Natl Cancer Inst. 2018 May 17; doi: 10.1093/jnci/djy081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schumacher FR, Al Olama AA, Berndt SI, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018 doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Evans DM, Zhu G, Dy V, et al. Genome-wide association study identifies loci affecting blood copper, selenium and zinc. Human Molecular Genetics. 2013;22(19):3998–4006. doi: 10.1093/hmg/ddt239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cornelis MC, Fornage M, Foy M, et al. Genome-wide association study of selenium concentrations. Hum Mol Genet. 2015;24(5):1469–1477. doi: 10.1093/hmg/ddu546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bagnardi V, Rota M, Botteri E, et al. Alcohol consumption and site-specific cancer risk: a comprehensive dose–response meta-analysis. British Journal of Cancer. 2015;112(3):580–593. doi: 10.1038/bjc.2014.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Munoz N, Day NE. Cancer epidemiology and prevention. New York: Oxford University; 1996. Esophagus. [Google Scholar]
- 126.Ference BA, Julius S, Mahajan N, Levy PD, Williams KA, Sr, Flack JM. Clinical effect of naturally random allocation to lower systolic blood pressure beginning before the development of hypertension. Hypertension. 2014;63(6):1182–1188. doi: 10.1161/HYPERTENSIONAHA.113.02734. [DOI] [PubMed] [Google Scholar]
- 127.Secretan B, Straif K, Baan R, et al. A review of human carcinogens--Part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncol. 2009;10(11):1033–1034. doi: 10.1016/s1470-2045(09)70326-2. [DOI] [PubMed] [Google Scholar]
- 128.Enomoto N, Takase S, Yasuhara M, Takada A. Acetaldehyde Metabolism in Different Aldehyde Dehydrogenase-2 Genotypes. Alcoholism: Clinical and Experimental Research. 1991;15(1):141–144. doi: 10.1111/j.1530-0277.1991.tb00532.x. [DOI] [PubMed] [Google Scholar]
- 129.Peng GS, Yin SJ. Effect of the allelic variants of aldehyde dehydrogenase ALDH2*2 and alcohol dehydrogenase ADH1B*2 on blood acetaldehyde concentrations. Human Genomics. 2009;3(2):121–127. doi: 10.1186/1479-7364-3-2-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Au Yeung SL, Jiang C, Cheng KK, et al. Is aldehyde dehydrogenase 2 a credible genetic instrument for alcohol use in Mendelian randomization analysis in Southern Chinese men? International Journal of Epidemiology. 2013;42(1):318–328. doi: 10.1093/ije/dys221. [DOI] [PubMed] [Google Scholar]
- 131.Lewis SJ, Davey Smith G. Alcohol, ALDH2, and esophageal cancer: a meta-analysis which illustrates the potentials and limitations of a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev. 2005;14(8):1967–1971. doi: 10.1158/1055-9965.EPI-05-0196. [DOI] [PubMed] [Google Scholar]
- 132.Bhaskaran K, Douglas I, Forbes H, dos-Santos-Silva I, Leon DA, Smeeth L. Body-mass index and risk of 22 specific cancers: a population-based cohort study of 5·24 million UK adults. The Lancet. 384(9945):755–765. doi: 10.1016/S0140-6736(14)60892-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Smith L, Brinton LA, Spitz MR, et al. Body Mass Index and Risk of Lung Cancer Among Never, Former, and Current Smokers. JNCI Journal of the National Cancer Institute. 2012;104(10):778–789. doi: 10.1093/jnci/djs179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Åsvold BO, Bjørngaard JH, Carslake D, et al. Causal associations of tobacco smoking with cardiovascular risk factors: a Mendelian randomization analysis of the HUNT Study in Norway. International Journal of Epidemiology. 2014;43(5):1458–1470. doi: 10.1093/ije/dyu113. [DOI] [PubMed] [Google Scholar]
- 135.Rigotti NA. Cigarette Smoking and Body Weight. New England Journal of Medicine. 1989;320(14):931–933. doi: 10.1056/NEJM198904063201409. [DOI] [PubMed] [Google Scholar]
- 136.El-Zein M, Parent ME, Nicolau B, Koushik A, Siemiatycki J, Rousseau M-C. Body mass index, lifetime smoking intensity and lung cancer risk. International Journal of Cancer. 2013;133(7):1721–1731. doi: 10.1002/ijc.28185. [DOI] [PubMed] [Google Scholar]
- 137.Koh WP, Yuan JM, Wang R, Lee HP, Yu MC. Body mass index and smoking-related lung cancer risk in the Singapore Chinese Health Study. British Journal of Cancer. 2010;102(3):610–614. doi: 10.1038/sj.bjc.6605496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kabat GC, Miller AB, Rohan TE. Body mass index and lung cancer risk in women. Epidemiology. 2007;18:607–612. doi: 10.1097/ede.0b013e31812713d1. [DOI] [PubMed] [Google Scholar]
- 139.Gao C, Patel CJ, Michailidou K, et al. Mendelian randomization study of adiposity-related traits and risk of breast, ovarian, prostate, lung and colorectal cancer. Int J Epidemiol. 2016;45(3):896–908. doi: 10.1093/ije/dyw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Carreras-Torres R, Johansson M, Haycock PC, et al. Obesity, metabolic factors and risk of different histological types of lung cancer: A Mendelian randomization study. PLoS One. 2017;12(6):e0177875. doi: 10.1371/journal.pone.0177875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carreras-Torres R, Haycock PC, Relton CL, et al. The causal relevance of body mass index in different histological types of lung cancer: A Mendelian randomization study. Scientific Reports. 2016;6 doi: 10.1038/srep31121. 31121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Thorgeirsson TE, Gudbjartsson DF, Sulem P, et al. A common biological basis of obesity and nicotine addiction. Transl Psychiatry. 2013;3:e308. doi: 10.1038/tp.2013.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lopez de Maturana E, Pineda S, Brand A, Van Steen K, Malats N. Toward the integration of Omics data in epidemiological studies: still a “long and winding road”. Genet Epidemiol. 2016;40(7):558–569. doi: 10.1002/gepi.21992. [DOI] [PubMed] [Google Scholar]
- 144.Richmond RC, Hemani G, Tilling K, Davey Smith G, Relton CL. Challenges and novel approaches for investigating molecular mediation. Hum Mol Genet. 2016 Oct 1;25(R2):R149–R156. doi: 10.1093/hmg/ddw197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kettunen J, Tukiainen T, Sarin AP, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44(3):269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shin SY, Fauman EB, Petersen AK, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Relton CL, Davey Smith G. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol. 2012;41(1):161–176. doi: 10.1093/ije/dyr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gaunt TR, Shihab HA, Hemani G, et al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016;17:61. doi: 10.1186/s13059-016-0926-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zeilinger S, Kuhnel B, Klopp N, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One. 2013;8(5):e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fasanelli F, Baglietto L, Ponzi E, et al. Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nat Commun. 2015;6 doi: 10.1038/ncomms10192. 10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shin SY, Petersen AK, Wahl S, et al. Interrogating causal pathways linking genetic variants, small molecule metabolites, and circulating lipids. Genome Med. 2014;6(3):25. doi: 10.1186/gm542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hemani G, Tilling K, Smith GD. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. Plos Genetics. 2017;13(11) doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Millard LA, Davies NM, Timpson NJ, Tilling K, Flach PA, Davey Smith G. MR-PheWAS: hypothesis prioritization among potential causal effects of body mass index on many outcomes, using Mendelian randomization. Sci Rep. 2015;5 doi: 10.1038/srep16645. 16645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hemani G, Bowden J, Haycock PC, et al. Automating Mendelian randomization through machine learning to construct a putative causal map of the human phenome. bioRxiv. 2017 [Google Scholar]
- 155.Ference BA, Majeed F, Penumetcha R, Flack JM, Brook RD. Effect of naturally random allocation to lower low-density lipoprotein cholesterol on the risk of coronary heart disease mediated by polymorphisms in NPC1L1, HMGCR, or both: a 2 x 2 factorial Mendelian randomization study. J Am Coll Cardiol. 2015;65(15):1552–1561. doi: 10.1016/j.jacc.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cannon CP, Blazing MA, Giugliano RP, et al. Ezetimibe Added to Statin Therapy after Acute Coronary Syndromes. N Engl J Med. 2015;372(25):2387–2397. doi: 10.1056/NEJMoa1410489. [DOI] [PubMed] [Google Scholar]
- 157.Millard LAC, Davies NM, Gaunt TR, Davey Smith G, Tilling K. Software Application Profile: PHESANT: a tool for performing automated phenome scans in UK Biobank. Int J Epidemiol. 2017 Oct 5; doi: 10.1093/ije/dyx204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Telomeres Mendelian Randomization Consortium. Haycock PC, Burgess S, et al. Association Between Telomere Length and Risk of Cancer and Non-Neoplastic Diseases: A Mendelian Randomization Study. JAMA Oncol. 2017;3(5):636–651. doi: 10.1001/jamaoncol.2016.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Alexandrov LB, Ju YS, Haase K, et al. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;354(6312):618–622. doi: 10.1126/science.aag0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev. 2014;24:52–60. doi: 10.1016/j.gde.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Robles-Espinoza CD, Roberts ND, Chen S, et al. Germline MC1R status influences somatic mutation burden in melanoma. Nat Commun. 2016;7 doi: 10.1038/ncomms12064. 12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gronich N, Rennert G. Beyond aspirin-cancer prevention with statins, metformin and bisphosphonates. Nat Rev Clin Oncol. 2013;10(11):625–642. doi: 10.1038/nrclinonc.2013.169. [DOI] [PubMed] [Google Scholar]
- 164.Gupta SC, Sung B, Prasad S, Webb LJ, Aggarwal BB. Cancer drug discovery by repurposing: teaching new tricks to old dogs. Trends Pharmacol Sci. 2013;34(9):508–517. doi: 10.1016/j.tips.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 165.Mokry LE, Ahmad O, Forgetta V, Thanassoulis G, Richards JB. Mendelian randomisation applied to drug development in cardiovascular disease: a review. J Med Genet. 2015;52(2):71–79. doi: 10.1136/jmedgenet-2014-102438. [DOI] [PubMed] [Google Scholar]
- 166.Van Acker HH, Anguille S, Willemen Y, Smits EL, Van Tendeloo VF. Bisphosphonates for cancer treatment: Mechanisms of action and lessons from clinical trials. Pharmacol Ther. 2016;158:24–40. doi: 10.1016/j.pharmthera.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 167.Thun MJ, Jacobs EJ, Patrono C. The role of aspirin in cancer prevention. Nat Rev Clin Oncol. 2012;9(5):259–267. doi: 10.1038/nrclinonc.2011.199. [DOI] [PubMed] [Google Scholar]
- 168.Quinn BJ, Kitagawa H, Memmott RM, Gills JJ, Dennis PA. Repositioning metformin for cancer prevention and treatment. Trends Endocrinol Metab. 2013;24(9):469–480. doi: 10.1016/j.tem.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 169.Walker VM, Davey Smith G, Davies NM, Martin RM. Mendelian randomization: a novel approach for the prediction of adverse drug events and drug repurposing opportunities. Int J Epidemiol. 2017 Oct 11; doi: 10.1093/ije/dyx207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schmidt AF, Swerdlow DI, Holmes MV, et al. PCSK9 genetic variants and risk of type 2 diabetes: a mendelian randomisation study. Lancet Diabetes Endocrinol. 2017;5(2):97–105. doi: 10.1016/S2213-8587(16)30396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Preiss D, Seshasai SR, Welsh P, et al. Risk of incident diabetes with intensive-dose compared with moderate-dose statin therapy: a meta-analysis. JAMA. 2011;305(24):2556–2564. doi: 10.1001/jama.2011.860. [DOI] [PubMed] [Google Scholar]
- 172.Sattar N, Preiss D, Murray HM, et al. Statins and risk of incident diabetes: a collaborative meta-analysis of randomised statin trials. Lancet. 2010;375(9716):735–742. doi: 10.1016/S0140-6736(09)61965-6. [DOI] [PubMed] [Google Scholar]
- 173.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 174.Burgess S, Timpson NJ, Ebrahim S, Davey Smith G. Mendelian randomization: where are we now and where are we going? Int J Epidemiol. 2015;44(2):379–388. doi: 10.1093/ije/dyv108. [DOI] [PubMed] [Google Scholar]
- 175.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013;12(8):581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 176.Juul K, Tybjaerg-Hansen A, Marklund S, et al. Genetically reduced antioxidative protection and increased ischemic heart disease risk: The Copenhagen City Heart Study. Circulation. 2004;109(1):59–65. doi: 10.1161/01.CIR.0000105720.28086.6C. [DOI] [PubMed] [Google Scholar]
- 177.Gray L, Davey Smith G, McConnachie A, et al. Parental height in relation to offspring coronary heart disease: examining transgenerational influences on health using the west of Scotland Midspan Family Study. Int J Epidemiol. 2012;41(6):1776–1785. doi: 10.1093/ije/dys149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nuesch E, Dale C, Palmer TM, et al. Adult height, coronary heart disease and stroke: a multi-locus Mendelian randomization meta-analysis. Int J Epidemiol. 2016;45(6):1927–1937. doi: 10.1093/ije/dyv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.