Abstract
The essential but enigmatic functions of sleep1,2 must be reflected in molecular changes sensed by the brain's sleep-control systems. In Drosophila, two dozen sleep-inducing neurons3 with projections to the dorsal fan-shaped body (dFB) adjust their electrical output to sleep need4, via the antagonistic regulation of two potassium conductances: the leak channel Sandman imposes silence during waking, whereas augmented A-type currents through Shaker support tonic firing during sleep5. Here, we report that oxidative by-products of mitochondrial electron transport6,7 regulate the activity of dFB neurons through a nicotinamide adenine dinucleotide phosphate (NADPH) cofactor bound to the oxidoreductase domain8,9 of Shaker's KVβ subunit, Hyperkinetic10,11. Sleep loss elevates mitochondrial reactive oxygen species in dFB neurons, which register this rise by converting Hyperkinetic to the NADP+-bound form. The oxidation of the cofactor slows the inactivation of the A-type current and boosts the frequency of action potentials, thereby promoting sleep. Energy metabolism, oxidative stress, and sleep, three processes implicated independently in lifespan, aging, and degenerative disease6,12–14, are thus mechanistically connected. KVβ substrates8,15,16 or inhibitors capable of altering the NADP+:NADPH ratio (and hence the neurons' record of sleep debt or waking time) define novel prototypes of sleep-regulatory drugs.
Like sleep-active cells of the mammalian hypothalamus17, dFB neurons in sleeping flies tend to be electrically active. To cause awakening, dopamine, acting directly on these cells5, attenuates the voltage-gated A-type current18 (IA) carried by Shaker and upregulates a voltage-independent leak current through the two-pore domain channel Sandman5. Sandman translocates from an intracellular storage pool to the plasma membrane, where it opens a potassium shunt that short-circuits the spike generator and switches the sleep-inducing neurons OFF5. The reverse switch, of dFB neurons to their electrically active state, is at the core of the organism’s response to sleep loss4. We are therefore able to frame the question of sleep's biological role as a mechanistically well-defined problem: which signals or processes switch dFB neurons ON? Knowing the leading parts played in the excitability switch by Shaker and Sandman focuses the search for answers on two molecular events: the modulation of the Shaker current, and the internalization of Sandman. Our spotlight here is on Shaker.
Shaker coassembles with a β subunit called Hyperkinetic10 into a voltage-gated potassium channel with fourfold rotational symmetry9. The N-terminal domains of four Shaker subunits form a hanging platform, suspended below the voltage sensors of the channel, to which a Hyperkinetic tetramer docks. Hyperkinetic and other KVβ subunits are related in sequence10,19 and structure8,9 to aldo-keto-reductases, complete with a catalytic tyrosinate anion, charge-relay system, and NADPH cofactor in their active sites. Oxidation or reduction of the stably bound nicotinamide by small-molecule substrates8,15,16 can alter the voltage-dependent activation and/or inactivation kinetics of the channels10,15,20. The functional purpose of coupling a cell’s excitability to its metabolism, however, remains mysterious8,9.
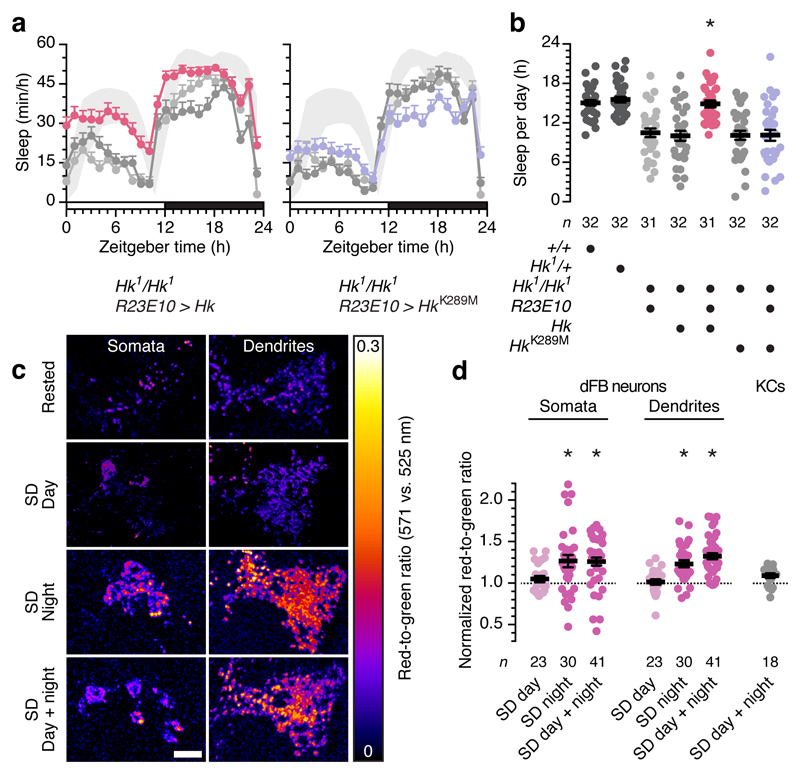
Mutations in Shaker or Hyperkinetic both cause insomnia11,13. Unsurprisingly, given the importance of IA for sustaining the sleep-promoting activity of these cells5, dFB neurons are a major sleep-relevant site of action for both channel subunits: the depletion of either gene product from these cells alone, using R23E10-GAL4-restricted4 RNA interference (RNAi), reproduces the sleep disruptions of the genomic mutations5. To complement these demonstrations of necessity with a test of sufficiency, we restored Hyperkinetic expression exclusively in the dFB of otherwise homozygous mutant flies. Sleep returned to wild-type levels, but only if Hyperkinetic’s active site was intact (Fig. 1a, b): a putative rescue transgene encoding a variant21 with a point mutation (K289M) that abolishes the protein's oxidoreductase activity15,20 but leaves its expression15,20,21 and the amplitude of IA unaltered (see later) proved ineffective. This finding has three implications. First, it suggests that Hyperkinetic's sleep-regulatory role is tied to its ability to sense changes in cellular redox state, which are therefore expected to accompany changes in sleep pressure. Second, it predicts that perturbing the redox chemistry of dFB neurons will have consequences for sleep. And third, it identifies a biophysical mechanism for coupling redox chemistry and sleep. Because redox reactions, oxygen use, and ATP synthesis are linked at the level of the flow of reducing equivalents through the mitochondrial electron transport chain, dFB neurons may monitor redox processes as a gauge of energy metabolism.
Figure 1. Hyperkinetic senses redox changes linked to sleep history.
a, R23E10-GAL4-driven expression of Hk (left), but not of HkK289M (right), in a homozygous Hk1 mutant background restores wild-type sleep (shaded bands: 95% confidence intervals) relative to parental controls (gray colours and sample sizes as in b). Two-way repeated-measures ANOVA with Holm-Šídák’s post-hoc test detected significant differences from both parental controls (P<0.0001) but not from wild-type (P=0.8599) in flies expressing Hk, and a significant difference from wild-type (P<0.0001) but not from either parental control (P>0.9741) in flies expressing HkK289M. b, Sleep in homozygous Hk1 mutants expressing R23E10-GAL4-driven Hk rescue transgenes and parental, wild-type, and heterozygous controls. One-way ANOVA with Holm-Šídák’s post-hoc test detected significant differences from both parental controls (P<0.0001) but not from wild-type (P=0.9763) in flies expressing Hk, and a significant difference from wild-type (P<0.0001) but not from either parental control (P>0.9704) in flies expressing HkK289M. c, Example maximum intensity projections of the somata and dendritic arbors of dFB neurons expressing MitoTimer under R23E10-GAL4 control, in rested and sleep-deprived (SD) flies (sample sizes in d). The red-to-green emission ratio is pseudocoloured according to the key on the right. Scale bar, 10 µm. d, Sleep deprivation during the night (SD effect: P<0.0001, Kruskal-Wallis ANOVA), but not during the day (P>0.6416, Mann-Whitney test), increases MitoTimer's red-to-green ratio in somata and dendrites of dFB neurons but not in Kenyon cells (KCs) (P=0.1328, t-test). Fluorescence ratios are normalized to those of unperturbed controls at the end of sleep deprivation (n=22 and 62 dFB controls for daytime and night-time deprivation; n=20 KC controls). Asterisks indicate significant differences (P<0.05) from both parental controls (b) or rested conditions (d) in pairwise post-hoc comparisons. Data are means ± s.e.m. n, number of flies. Statistical detail in Supplementary Table 1.
To examine the first of these implications, we compared the redox histories of flies that had been mechanically sleep-deprived with those of rested controls (Fig. 1c, d). The metabolic machinery of the inner mitochondrial membrane is the principal cellular source of oxidants, especially under conditions of ample NADH supply, large proton-motive force, and low ATP demand, when electrons stall in the transport chain and transfer directly to oxygen6,7, producing superoxide (O2-) that is subsequently dismuted to H2O2 (Fig. 2a). Chief conduits for electron leakage7 are a fully reduced ubiquinone pool and the resulting tailback of electrons onto complex I. Although some reactive oxygen species (ROS) generated in mitochondria could conceivably reach the active site of Hyperkinetic by diffusion, a more plausible scenario is that O2- and H2O2 react locally and release a longer-lived carbonyl substrate8 whose reduction by Hyperkinetic then causes the oxidation of NADPH. Lipid peroxidation products, such as the aldehyde 4-oxo-2-nonenal, serve as established hydride acceptors in KVβ subunits15,16 and may represent the ill-defined electron densities9 overlying their hydrophobic active sites.
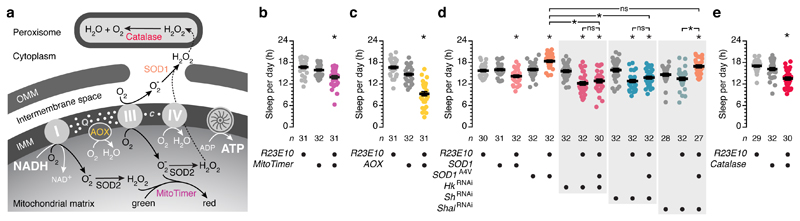
Figure 2. dFB-restricted perturbations of redox chemistry alter sleep.
a, Ubiquinone (Q) and cytochrome c (c) ferry electrons (white dots) between the proton-pumping complexes I, III, and IV. When more electrons enter the transport chain than can be used to fuel ATP synthesis, a backlog accumulates in the Q pool. These electrons react directly with O2, releasing O2- into the matrix and the space between the inner and outer mitochondrial membranes (IMM and OMM). Superoxide dismutases (SOD2 in the matrix, SOD1 in the intermembrane space and cytoplasm) convert O2- to membrane-permeant H2O2; catalase decomposes H2O2. AOX uses surplus Q electrons to reduce O2 to water. Coloured components were manipulated in b–e. b, Sleep in flies expressing R23E10-GAL4-driven MitoTimer and parental controls (genotype effect: P=0.0007, one-way ANOVA). c, Sleep in flies expressing R23E10-GAL4-driven AOX and parental controls (genotype effect: P<0.0001, one-way ANOVA). d, Sleep in flies expressing R23E10-GAL4-driven SOD1 or pro-oxidant SOD1A4V, with or without RNAi transgenes targeting KV channel subunits, and parental controls (genotype effect: P<0.0001, one-way ANOVA). e, Sleep in flies expressing R23E10-GAL4-driven catalase and parental controls (genotype effect: P<0.0001, one-way ANOVA). Asterisks indicate significant differences (P<0.05) from both parental controls or in relevant pairwise post-hoc comparisons (brackets). Data are means ± s.e.m. n, number of flies. Statistical detail in Supplementary Table 1.
For a cumulative estimate of ROS production, we labeled the mitochondria of dFB neurons with a matrix-targeted fluorescent protein (MitoTimer) whose green-emitting chromophore converts irreversibly to red when oxidized22. We then deprived age-matched flies of variable amounts of sleep and determined the fluorescence emission ratio by two-photon microscopy. Mitochondrial ROS production rose with the size of the imposed sleep deficit: a night of sleep deprivation red-shifted MitoTimer's fluorescence relative to rested controls, but applying the same sleep deprivation protocol during the day, when flies are mostly awake, or adding a day to a night of sleep disruption produced only insignificant effects (Fig. 1c, d). Because dFB neurons generate few energetically costly action potentials in the awake, fed state, when calories are plentiful but the Sandman detent blocks spiking5, the condition of a high ATP:ADP ratio known to favour mitochondrial O2- production6,7 from an abundance of reducing substrates is likely to be met. Consistent with this idea, mushroom body Kenyon cells, which are electrically active during waking, showed little evidence of oxidant exposure even after 24 h of sleep deprivation (Fig. 1d). In addition, or instead, dFB neurons may have an unusually low capacity for degrading ROS, making them canaries in the mine for their detection.
Curiously, flies expressing MitoTimer in dFB neurons lost ~2 h of baseline sleep per day (Fig. 2b). As the oxidation of MitoTimer will consume ROS, we interpret this finding as tentative evidence of a causal connection between mitochondrial oxidative burden and sleep. To strengthen this connection, we quantified sleep after three further dFB-neuron-specific interventions: manipulation of mitochondrial electron transport; chronic interference with antioxidant enzymes; and acute optogenetic induction of singlet oxygen (1O2) formation.
We first installed an electron overflow pathway in the inner mitochondrial membrane of dFB neurons by expressing the alternative oxidase AOX of Ciona intestinalis23,24. Like complex III, AOX taps into the ubiquinone pool, but instead of transferring an electron each to two cytochrome c carriers, it reduces molecular oxygen to water in a single four-electron transfer reaction (Fig. 2a). Alternative respiration thus siphons off electrons that would otherwise spill from the ubiquinone pool and produce ROS when the cytochrome branch of the transport chain is saturated or the availability of ADP is low6,7,23. Introducing AOX into the mitochondria of dFB neurons, which normally lack a capacity for alternative respiration, decreased daily sleep by nearly 7 h (Fig. 2c): clamping mitochondrial ROS production eased the pressure to sleep.
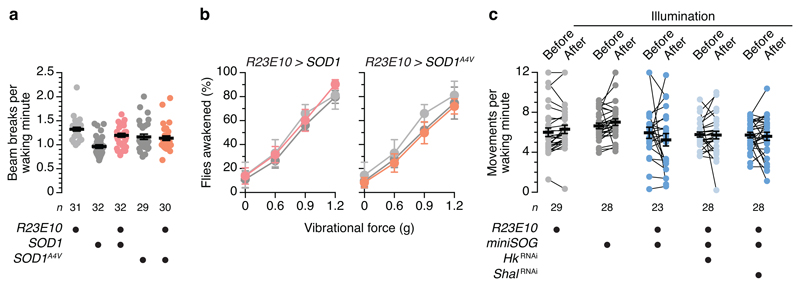
In animals without bifurcated electron transport chains, superoxide dismutases (SODs) and catalase, which acts as a sink for SOD-generated H2O2 and thereby pulls the dismutation reaction forward, form the first line of anti-oxidant defenses6,12 (Fig. 2a). Shoring up these defenses in dFB neurons reduced sleep (Fig. 2d, e), while breaching them with the help of a mutant enzyme (SOD1A4V) whose peroxidase activity is enhanced25,26 had the converse effect; it increased sleep (Fig. 2d) without inhibiting waking locomotion (Extended Data Fig. 1a) or arousability (Extended Data Fig. 1b). The crucial link between redox chemistry and sleep was the Shaker–Hyperkinetic complex: the RNAi-mediated depletion of either channel subunit from dFB neurons not only occluded the sleep-promoting effect of SOD1A4V but reduced sleep below wild-type levels (Fig. 2d). In contrast, interference with the expression of Shal, a KV channel without a sleep-regulatory function in dFB neurons5, proved innocuous (Fig. 2d).
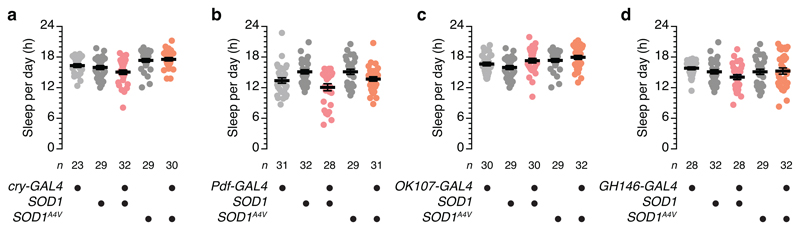
Analogous SOD1 manipulations in cryptochrome- or PDF-positive clock neurons or Kenyon cells (which all have demonstrated roles in sleep control27) or in olfactory projection neurons (for which no such role has been reported) failed to influence sleep (Extended Data Fig. 2a–d). dFB neurons thus appear unique, at least among this comparison group, in their ability to transduce oxidative stress into sleep.
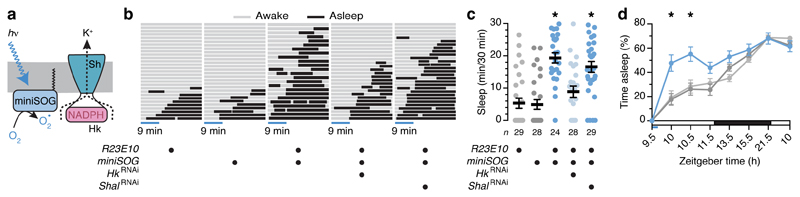
As a third test of the redox control of sleep, we anchored miniSOG, an engineered flavoprotein28 that photogenerates 1O2, via a myristoyl group at the cytoplasmic face of the plasma membrane29 (Fig. 3a). If the light-driven release of 1O2 near Hyperkinetic causes the oxidation of bound NADPH, either directly or via local lipid peroxidation, it should be possible to bypass the chain of events that couples this final transduction step to mitochondrial respiration and induce sleep acutely. Indeed, during a 30-min observation interval that began with a 9-min exposure to blue light, flies expressing miniSOG in dFB neurons fell quiescent in greater proportion, and for longer, than control flies did (Fig. 3b, c). Epochs of quiescence outlasted the illumination period by ~1 h (Fig. 3d), could be blocked by the removal of Hyperkinetic but not of Shal (Fig. 3b, c), and were not due to the suppression of waking movements (Extended Data Fig. 1c) or a confounding influence of cryptochrome photoreceptors21 (Extended Data Fig. 3).
Figure 3. Optogenetically controlled ROS production in dFB neurons induces sleep.
a, N-myristoylated miniSOG near the Hyperkinetic (Hk) gondola underneath Shaker (Sh). b, Periods of wake (gray) and sleep (black) during and after an initial 9-min exposure to blue light, in flies expressing R23E10-GAL4-driven miniSOG, with or without RNAi transgenes targeting KV channel subunits, and parental controls. Each row depicts one individual; all individuals were awake at the onset of illumination. The fraction of experimental flies falling asleep differed from both parental controls (P<0.0001) and from flies coexpressing HkRNAi (P=0.0030) but not ShalRNAi (P=0.3622, Fisher’s exact test throughout, sample sizes in c). c, Sleep in flies expressing R23E10-GAL4-driven miniSOG, with or without RNAi transgenes targeting KV channel subunits, and parental controls (genotype effect: P<0.0001, Kruskal-Wallis ANOVA). d, Cumulative sleep percentages after a 9-min exposure to blue light at zeitgeber time 9.5 h, in flies expressing R23E10-GAL4-driven miniSOG (n=19) and parental controls (n=25 each, gray colours as in c) (time × genotype interaction: P<0.0001, two-way repeated-measures ANOVA). Asterisks indicate significant differences (P<0.05) from both parental controls or in relevant pairwise post-hoc comparisons. Data are means ± s.e.m. n, number of flies. Statistical detail in Supplementary Table 1.
Whole-cell recordings from dFB neurons, before and after miniSOG-mediated photooxidation under sleep-inducing conditions, revealed some of the well-documented biophysical changes underpinning the wake–sleep switch4,5: the neurons’ action potential responses to depolarizing current became more vigorous (Fig. 4a–c); their membrane time constants lengthened (Fig. 4b); the interspike interval contracted (Fig. 4a, c); and fast IA inactivation slowed (Fig. 4d, e, Extended Data Table 1). Because A-type channels in the conducting state are the repolarizing force that returns the membrane potential to its resting level after a spike, changes in inactivation kinetics influence firing rates18. Both are regulated by KVβ subunits, with oxidation of NADPH to NADP+, slow inactivation10,15,20, and high-frequency spiking going hand-in-hand30.
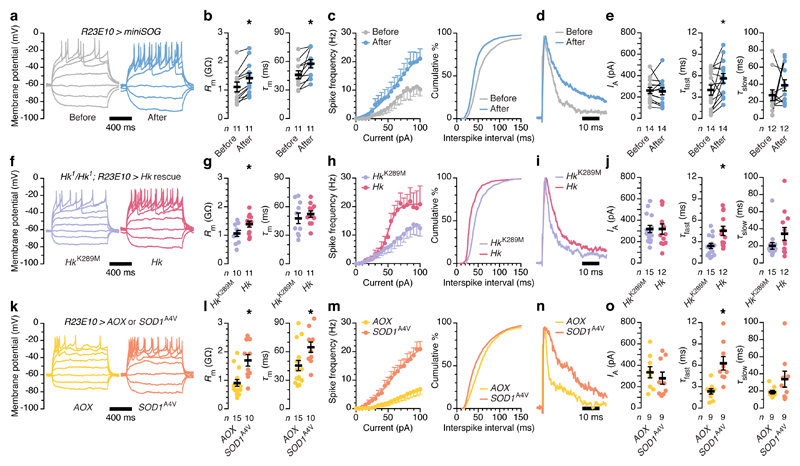
Figure 4. Changes in redox chemistry alter the spiking activity of dFB neurons via IA.
a–e, dFB neurons expressing R23E10-GAL4-driven miniSOG and CD8::GFP, before and after a 9-min exposure to blue light. Example voltage responses to current steps (a, sample sizes in b): illumination increases the input resistance (b, Rm; P<0.0001, paired t-test) and membrane time constant (b, τm; P=0.0041, paired t-test), steepens the current-spike frequency function (c, left; current × genotype interaction: P=0.0014, two-way repeated-measures ANOVA), and shifts the interspike interval distribution toward shorter values (c, right; P<0.0001, Kolmogorov-Smirnov test). Example IA (normalized to peak) evoked by voltage steps to +40 mV (d, sample sizes in e): illumination leaves the IA amplitude unchanged (e; P=0.7295, paired t-test) and increases the fast (e, τfast; P=0.0245, Wilcoxon test) but not the slow inactivation time constant (e, τslow; P=0.3804, Wilcoxon test). f–j, dFB neurons expressing R23E10-GAL4-driven HkK289M or Hk rescue transgenes in a homozygous Hk1 mutant background. Example voltage responses to current steps (f, sample sizes in g): catalytically competent Hk increases the input resistance (g, Rm; P=0.0467, t-test) but not the membrane time constant (g, τm; P=0.4962, t-test), steepens the current-spike frequency function (h, left; current × genotype interaction: P<0.0001, two-way repeated-measures ANOVA), and shifts the interspike interval distribution toward shorter values (h, right; P<0.0001, Kolmogorov-Smirnov test). Example IA (normalized to peak) evoked by voltage steps to +40 mV (i, sample sizes in j): catalytically competent Hk leaves the IA amplitude unchanged (j; P=0.9827, t-test) and increases the fast (j, τfast; P=0.0061, t-test) but not the slow inactivation time constant (j, τslow; P=0.1257, Mann-Whitney test). k–o, dFB neurons expressing R23E10-GAL4-driven AOX or SOD1A4V. Example voltage responses to current steps (k, sample sizes in l): pro-oxidant SOD1A4V increases the input resistance (l, Rm; P=0.0023, Mann-Whitney test) and membrane time constant (l, τm; P=0.0166, Mann-Whitney test), steepens the current-spike frequency function (m, left; current × genotype interaction: P<0.0001, two-way repeated-measures ANOVA), and shifts the interspike interval distribution toward shorter values (m, right; P<0.0001, Kolmogorov-Smirnov test). Example IA (normalized to peak) evoked by voltage steps to +40 mV (n, sample sizes in o): pro-oxidant SOD1A4V leaves the IA amplitude unchanged (o; P=0.4892, t-test) and increases the fast (o, τfast; P=0.0027, t-test) but not the slow inactivation time constant (o, τslow; P=0.3401, Mann-Whitney test). Asterisks indicate significant differences (P<0.05). Data are means ± s.e.m. n, number of cells. Statistical detail in Supplementary Table 1.
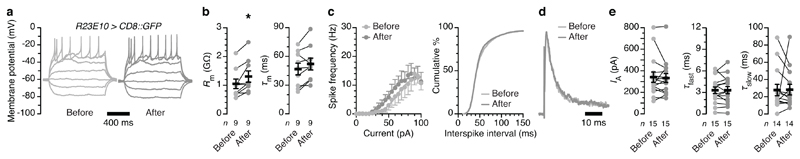
Like the induction of sleep (Fig. 3b, c), these biophysical changes required the abrupt burst of ROS production caused by the high 1O2 quantum yield28 of miniSOG. No cell physiological changes—apart from a modest increase in input resistance—were seen after equally intense and prolonged irradiation of dFB neurons expressing membrane-bound GFP, whose chromophore is encased in a protein shell that prevents the close apposition of O2 necessary for energy transfer (Extended Data Fig. 4a–e).
The coherent picture emerging from these within-cell analyses was mirrored in between-cell comparisons of neurons with chronically altered redox-sensing or redox-buffering capacity: the homozygous Hyperkinetic mutants carrying catalytically active or dead rescue transgenes that were our point of departure (Fig. 4f–j), or cells containing pro-oxidant25,26 SOD1A4V or anti-oxidant23 AOX (Fig. 4k–o). dFB neurons equipped with a functional Shaker β subunit expressed slowly inactivating A-type currents (Fig. 4i, j) that enabled high-frequency action potential trains (Fig. 4f, h). In flies forced to make do with the K289M mutant15,20,21, which cannot convert NADPH to NADP+, dFB neurons exhibited fast-inactivating IA (Fig. 4i, j), long interspike intervals (Fig. 4f, h), and shallow current-spike frequency functions (Fig. 4f, h) that can account for the insomnia of these animals (Fig. 1a, b). Profound shifts of Hyperkinetic's NADP+:NADPH ratio in opposite directions must also underlie the divergent interspike interval distributions (Fig. 4k, m), current-spike frequency functions (Fig. 4k, m), and A-type inactivation kinetics of dFB neurons expressing SOD1A4V or AOX (Fig. 4n, o), which parallel large and opposite changes in daily sleep (Fig. 2c, d).
Because KVβ subunits have extremely low cofactor exchange rates that limit catalysis 15,20, perhaps to a single hydride transfer8, even a fleeting oxidant exposure will form a lasting biochemical memory. The Shaker–Hyperkinetic complex therefore unites three discrete functions in a single device: Its redox sensitivity allows it to monitor a key process relevant to sleep—the generation of oxidative by-products of mitochondrial electron transport. Its catalytic inefficiency allows the protein to compute and store the time integral of the resulting oxidative burden, as would be required if sleep's purpose were to protect against oxidative stress14. And its ability to set the spike frequency via conformational coupling to the channel's inactivation gate allows it to titrate the commensurate corrective action.
Before dFB neurons can discharge the accumulated sleep pressure, however, the inhibitory clamp imposed by Sandman must be released. This almost certainly involves the regulated endocytosis of the channel5. We suspect that Sandman's movements between intracellular storage vesicles and the cell surface are essential for the conversion of continuously varying sleep pressure into discrete sleep–wake states. Sharp transitions, not slow drift through twilight states, are of course the desired output of any sleep-control system. The exo-endocytic cycling of Sandman acts as an ON–OFF switch in the homeostatic feedback loop, allowing dFB neurons to alternate between 'fill' and 'discharge' modes31. In fill mode (that is, during waking), Sandman is present at the cell surface5, action potentials are inhibited, and the Hyperkinetic pool fills with NADP+. Sleep pressure builds but cannot be discharged until an unidentified signal arrives that causes the extraction of Sandman from the plasma membrane. At this point, the neurons switch to discharge mode, electrical activity resumes at the level dictated by the NADP+:NADPH ratio of the Hyperkinetic pool, and the animal goes to sleep.
In order to dissipate the accumulated sleep pressure, the NADP+:NADPH ratio must return to baseline during sleep. An elegant way to accomplish this reset would be to gate Hyperkinetic's enzymatic activity by voltage8,20. Cofactor release from the active site would be impeded in fill mode because the membrane potential of dFB neurons remains below the activation threshold of Shaker, but in discharge mode, the action potential-driven rearrangements of the channel would open an escape path for NADP+. Bidirectional coupling of a redox-modulated ion channel and a voltage-modulated oxidoreductase may thus be the accounting principle at the heart of the somnostat.
Methods
Drosophila strains and culture
Fly stocks were grown on media of sucrose, yeast, molasses, and agar under a 12 h light : 12h dark cycle at 25 °C. All studies were performed on randomly selected females aged 2–6 days post eclosion. Experimental flies were heterozygous for all transgenes and homozygous for either a wild-type or mutant (Hk1) Hyperkinetic allele32,33, as indicated. Driver lines R23E10-GAL434, cry-GAL435, pdf-GAL436, OK107-GAL437, and GH146-GAL438 were used to target dFB neurons, cryptochrome- or PDF-expressing clock neurons, Kenyon cells, or olfactory projection neurons, respectively. Effector transgenes encoded a fluorescent marker for visually guided patch-clamp recordings (UAS-CD8::GFP39); wild-type or mutant (HkK289M) Hyperkinetic rescue transgenes21; an optical integrator of ROS exposure in the mitochondrial matrix (UAS-MitoTimer22); the mitochondrial alternative oxidase AOX24; wild-type40 and mutant41 (SOD1A4V) versions of human superoxide dismutase 1; catalase42; an N-myristoylated covalent hexamer29 (myr-MS6T2) of the singlet oxygen generator miniSOG28; and RNAi constructs43 for interference with the expression of Hyperkinetic, Shaker, Shal, or cryptochrome (101402KK, 104474KK, 103363KK, and 7238GD or 105172KK, respectively; Vienna Drosophila Resource Center).
Sleep measurements
In standard sleep assays, females aged 3–5 d were individually inserted into 65-mm glass tubes, loaded into the Trikinetics Drosophila Activity Monitor system, and housed under 12 h light : 12h dark conditions. Periods of inactivity lasting at least 5 minutes were classified as sleep44,45. Immobile flies (< 2 beam breaks per 24 h) were excluded from the analysis. In sleep deprivation experiments, a spring-loaded platform stacked with Trikinetics monitors was slowly tilted by an electric motor, released, and allowed to snap back to its original position46. The mechanical cycles lasted 12 s and were repeated continuously.
Arousal thresholds in standard sleep assays were determined with the help of mechanical stimuli generated by vibration motors (Precision Microdrives, model 310-113) 47. Stimuli were delivered once every hour, and the percentages of sleeping flies awakened within 5 minutes of each 5-s stimulation episode were quantified.
Sleep after light-induced ROS generation was measured at zeitgeber time 9.5 h. Female flies aged 3–5 d and expressing miniSOG in dFB neurons were individually inserted into 35-mm glass tubes and loaded into a custom-built array of light-tight chambers31. Each chamber was equipped with a high-power LED (Osram Opto Semiconductors LB W5SM-FZHX-35-0, 467 nm) running on an 80% duty cycle at 10 Hz and delivering 8 mW cm-2 at the distal and 80 mW cm-2 at the proximal end of the tube. In this intensity range, each miniSOG molecule in the central brain underwent an estimated 2–40 excitation cycles s-1, based on the measured optical transmission of 7 fly heads at 467 nm (4.8 ± 0.3% (mean ± s.e.m.), assumed to be isotropic) and a miniSOG absorption cross-section28 of 5.0 × 10-17 cm-2.
The apparatus was operated in a temperature-controlled incubator (Sanyo MIR-154) at 25 °C. Excess heat from the high-power LEDs was removed by a water-cooling device incorporating liquid heat exchangers (Thermo Electric Devices LI102), a centrifugal pump (RS 702-6891), Peltier module (Adaptive ETC-128-10-05-E), and CPU cooler (Corsair CW-9060007-WW). For movement tracking, the chambers were continuously illuminated by low-power infrared (850 nm) LEDs from below and imaged from above at 25 frames s-1 with a high-resolution CMOS camera (Thorlabs DCC1545M), using an 8 mm lens (Thorlabs MVL8M23) and a long-pass filter (Thorlabs, FEL800nm) to reject photostimulation light. A virtual instrument written in LabVIEW (National Instruments) extracted real-time position data from video images by subtracting the most recently acquired image from a temporally low-pass-filtered background31. Non-zero pixels in the difference image indicated that a movement had occurred, with the centroid of the largest cluster of non-zero pixels taken to represent the fly's new position. To eliminate noise, intensity and size thresholds were applied to pixel clusters in the difference image, and movements <2.5 mm (approximately one body length) were discarded. Periods of inactivity lasting at least 5 minutes were classified as sleep44,45. The flies were monitored for 10 min before the photooxidation of miniSOG, and subjects found asleep during that period were excluded from the analysis. Only individuals with a confirmed waking time >30 s were used to quantify waking movements, which were counted as distinct events if they were separated by >5 s of immobility.
Functional imaging
Single-housed females were analysed 2–6 days post eclosion after 12 or 24 h of mechanical sleep deprivation46, begun at zeitgeber times 0 h (daytime deprivation) or 12 h (night-time deprivation), and compared to age-matched controls at the end of sleep deprivation. After head-fixing the flies to a custom mount with eicosane (Sigma), cuticle, adipose tissue, and trachea were removed to create a small surgical window, and the brain was continuously superfused with extracellular solution equilibrated with 95% O2–5% CO2 and containing 103 mM NaCl, 3 mM KCl, 5 mM TES, 8 mM trehalose, 10 mM glucose, 7 mM sucrose, 26 mM NaHCO3, 1 mM NaH2PO4, 1.5 mM CaCl2, 4 mM MgCl2, pH 7.3.
MitoTimer fluorescence was imaged in vivo by two-photon laser-scanning microscopy. Excitation light pulses with 140 fs duration and a centre wavelength of 910 nm (Chameleon Ultra II, Coherent) were intensity-modulated with the help of a Pockels cell (302RM, Conoptics) and focused by a 20×, 1.0 NA water immersion objective (W-Plan-Apochromat, Zeiss) on a Movable Objective Microscope (Sutter Instruments). Emitted photons were separated from excitation light by a series of dichromatic mirrors and dielectric and coloured glass filters, split into red and green channels (Semrock BrightLine FF01-571/72 and FF01-525/45, respectively), and detected by GaAsP photomultiplier tubes (H10770PA-40 SEL, Hamamatsu Photonics). Photocurrents were passed through high-speed amplifiers (HCA-4M-500K-C, Laser Components) and custom-designed integrator circuits to maximize the signal-to-noise ratio. The microscope was controlled through ScanImage (Vidrio Technologies) via a PCI-6110 DAQ board (National Instruments). Images were acquired as z-stacks with an axial resolution of 1 µm.
Maximum-intensity projections of image stacks were analysed blind to sleep history, using a semi-automated script22 in MATLAB (The MathWorks). The algorithm rejected saturated or MitoTimer-negative pixels (fluorescence <1.5-fold above the mean of a manually defined background area) and calculated the average red-to-green ratio for the remaining image area.
Electrophysiology
For whole-cell patch-clamp recordings in vivo, female flies aged 3–5 days post eclosion were prepared as for functional imaging, but the perineural sheath was also removed for electrode access. The somata of GFP-labeled dFB neurons were visually targeted with borosilicate glass electrodes (12–14 MΩ). The internal solution contained 140 mM potassium aspartate, 10 mM HEPES, 1 mM KCl, 4 mM MgATP, 0.5 mM Na3GTP, 1 mM EGTA, pH 7.3. Signals were acquired with a Multiclamp 700B amplifier (Molecular Devices), filtered at 6–10 kHz, and digitised at 10–20 kHz using an ITC-18 data acquisition board (InstruTECH) controlled by the Nclamp/Neuromatic package. Data were analysed using Neuromatic software (www.neuromatic.thinkrandom.com) and custom procedures in Igor Pro (Wavemetrics).
For photostimulation of miniSOG during whole-cell recordings, a 455-nm LED (Thorlabs M455L3) was focused onto the head of the fly with a mounted f = 20.1 mm aspheric condenser lens (Thorlabs ACP2520-A) and controlled by a TTL-triggered dimmable constant-current LED driver (Thorlabs LEDD1B). The optical power at the sample was ~3.5 mW cm-2.
Membrane resistances were calculated from linear fits of the steady-state voltage changes elicited by 1-s steps of hyperpolarizing currents (5-pA increments) from a pre-pulse potential of –60 ± 5 mV. Membrane time constants were estimated by fitting a single exponential to the voltage deflection caused by a hyperpolarizing 10 pA current step lasting 200 ms. Interspike intervals were determined from voltage responses to a standard series of depolarizing current steps (5 pA increments from 0 to 100 pA, 1 s duration). Spikes were detected by finding minima in the time derivative of the membrane potential trace. Interspike intervals at all levels of injected current were pooled for the calculation of frequency distributions.
Voltage-clamp experiments were performed in the presence of 1 µM tetrodotoxin (Tocris) and 200 µM cadmium to block sodium and calcium channels, respectively. Neurons were stepped from holding potentials of –110 or –30 mV to a test potential of +40 mV. When the cells were held at –110 mV, depolarization steps (1 s duration) elicited the full complement of potassium currents; when the cells were held at –30 mV, voltage-gated channels inactivated and the evoked potassium currents lacked the IA (A-type or fast outward) component48. Digital subtraction of the non-A-type component from the full complement of potassium currents gave an estimate of IA. To determine the fast and slow inactivation time constants, double-exponential functions were fit to the decaying phase of currents elicited by 1-s depolarizing voltage pulses after digitally subtracting non-inactivating outward currents (Extended Data Table 1). In cases where the fits of slow inactivation time constants were poorly constrained, only the fast inactivation time constants were included in the analysis.
Statistics and reproducibility
Data were analysed in Prism 7 (GraphPad). Statistical detail, including test statistics, degrees of freedom, and exact P values (to four significant digits) is reported in Supplementary Tables 1 and 2.
All null hypothesis tests were two-sided. Group means were compared by two-sided t-test or one-way or two-way ANOVA, using repeated measures designs or the Welch correction where indicated, followed by planned pairwise post-hoc analyses using Holm-Šídák’s multiple comparisons test. Where the assumption of normality was violated (as indicated by Shapiro-Wilk test), group means were compared by two-sided Mann-Whitney test, two-sided Wilcoxon signed-rank test, or Kruskal-Wallis ANOVA; the latter was followed by Dunn’s multiple comparisons test. χ2 or Fisher's exact tests were performed on contingency tables of categorical data, as noted. Interspike interval distributions were evaluated by two-sided Kolmogorov-Smirnov test. Figure legends give P value ranges if multiple post-hoc tests were performed after ANOVA; the P values of individual comparisons are found in Supplementary Tables 1 and 2.
The investigators were blind to group allocation in MitoTimer imaging experiments but not otherwise. Sample sizes in behavioural experiments (typically n=32 flies per genotype) were chosen to detect 2-h differences in daily sleep with a power of 0.9. All behavioural experiments were run at least three times, on different days and with different batches of flies. The figures show representative examples.
Extended Data
Extended Data Figure 1. Chronic or acute dFB-restricted perturbations of redox chemistry have no impact on waking locomotor activity or arousability.
a, Locomotor counts per waking minute of flies expressing R23E10-GAL4-driven SOD1 or pro-oxidant variant SOD1A4V, in the Trikinetics Drosophila Activity Monitor system, do not differ from their respective parental controls (genotype effect: P>0.2612, Kruskal-Wallis ANOVA with Dunn's post-hoc test). b, Arousability of flies expressing R23E10-GAL4-driven SOD1 (left) or pro-oxidant SOD1A4V (right) and parental controls (gray colours as in a) (genotype effects: P>0.2487, vibrational force effects: P<0.0001, vibrational force × genotype interactions: P>0.9857, two-way ANOVA). Data are means ± s.e.m. of 6 trials per genotype (n=16–32 flies each). c, Locomotor counts per waking minute of flies expressing R23E10-GAL4-driven miniSOG, with or without RNAi transgenes targeting KV channel subunits, and parental controls, in a custom video-tracking system31. Activity was monitored for 10 min before the photooxidation of miniSOG and then for a 30-min interval that included a 9-min exposure to blue light (genotype effect: P=0.0827, illumination effect: P=0.8059, illumination × genotype interaction: P=0.3086, two-way repeated-measures ANOVA). Data are means ± s.e.m. n, number of flies (a, c) or trials (b). Statistical detail in Supplementary Table 2.
Extended Data Figure 2. Chronic perturbations of redox chemistry in cryptochrome- or Pdf-expressing clock neurons, Kenyon cells, or olfactory projection neurons have no impact on sleep.
a, Sleep in flies expressing cry-GAL4-driven SOD1 or SOD1A4V in clock neurons and parental controls. Kruskal-Wallis ANOVA with Dunn's post-hoc test failed to detect significant differences of experimental flies from both of their respective parental controls (P>0.1426). b, Sleep in flies expressing Pdf-GAL4-driven SOD1 or SOD1A4V in clock neurons and parental controls. Kruskal-Wallis ANOVA with Dunn's post-hoc test failed to detect significant differences of experimental flies from both of their respective parental controls (P>0.1732). c, Sleep in flies expressing OK107-GAL4-driven SOD1 or SOD1A4V in Kenyon cells and parental controls. One-way ANOVA with Holm-Šídák's post-hoc test failed to detect significant differences of experimental flies from both of their respective parental controls (P>0.0603). d, Sleep in flies expressing GH146-GAL4-driven SOD1 or SOD1A4V in olfactory projection neurons and parental controls. Kruskal-Wallis ANOVA with Dunn's post-hoc test failed to detect significant differences of experimental flies from both of their respective parental controls (P>0.6901). Data are means ± s.e.m. n, number of flies. Statistical detail in Supplementary Table 2.
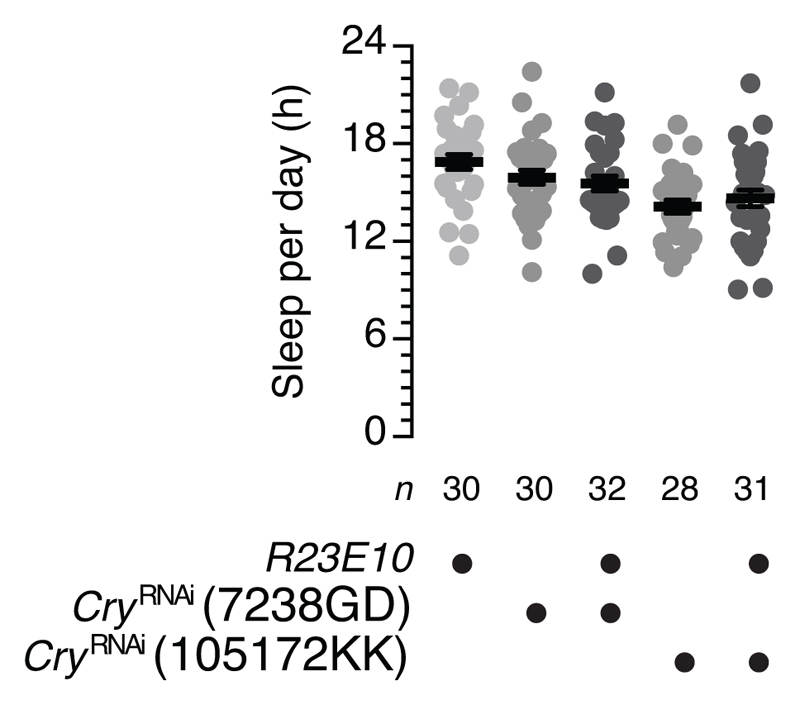
Extended Data Figure 3. Chronic dFB-restricted manipulations of cryptochrome have no impact on sleep.
Sleep in flies expressing two different R23E10-GAL4-driven cryRNAi transgenes and parental controls. One-way ANOVA with Holm-Šídák's post-hoc test failed to detect significant differences of experimental flies from both of their respective parental controls (P>0.1718). Date are means ± s.e.m. n, number of flies. Statistical detail in Supplementary Table 2.
Extended Data Figure 4. Blue illumination of miniSOG-negative dFB neurons has no impact on their electrical activity.
a–e, dFB neurons expressing R23E10-GAL4-driven CD8::GFP, before and after a 9-min exposure to blue light. Example voltage responses to current steps (a, sample sizes in b): illumination increases the input resistance (b, Rm; P=0.0098, paired t-test) but not the membrane time constant (b, τm; P=0.0723, paired t-test) and leaves unchanged the current-spike frequency function (c, left; current × genotype interaction: P=0.9982, two-way repeated-measures ANOVA) and interspike interval distribution (c, right; P=0.0947, Kolmogorov-Smirnov test). Example IA (normalized to peak) evoked by voltage steps to +40 mV (d, sample sizes in e): illumination leaves unchanged the IA amplitude (e; P=0.8040, Wilcoxon test) and both inactivation time constants (e, τfast: P=0.6387, τslow: P=0.2958, Wilcoxon tests). Asterisks indicate significant differences (P<0.05). Data are means ± s.e.m. n, number of cells. Statistical detail in Supplementary Table 2.
Extended Data Table 1. Parameters of IA inactivation.
Inactivation time constants of IA evoked by voltage steps to +40 mV were obtained by fitting double-exponential functions to the decaying phase. Afast/(Afast+Aslow) represents the fraction of the fast component of the total A-current. Data are means ± s.e.m. n, number of cells. All dFB neurons expressed CD8::GFP in addition to the indicated transgenes.
Supplementary Material
Acknowledgments
We are indebted to D. Pimentel for electrophysiology advice and thank C. Chintaluri and M. Murphy for discussion. N. Bonini, B. Dickson, B. Ganetzky, J. Hall, T. Holmes, K. Ito, H. Jacobs, L. Luo, J. Ng, J. Phillips, F. Rouyer, G. Rubin, R. Stocker, P. Taghert, Z. Yan, the Bloomington Stock Center, and the Vienna Drosophila Resource Center kindly provided flies. This work was supported by grants (to G.M.) from the Wellcome Trust and the Gatsby Charitable Foundation. A.K. held postdoctoral fellowships from the Swiss National Science Foundation and EMBO; S.M.S. was a Commonwealth Scholar.
Footnotes
Data availability. The datasets generated during this study are available from the corresponding author upon reasonable request.
Code availability. Custom instrument control and analysis code used in this study is available from the corresponding author upon reasonable request.
Author Contributions. G.M., S.M.S., and A.K. designed the study and analysed the data. A.K. performed electrophysiological recordings and carried out imaging experiments, molecular manipulations, and behavioural analyses with S.M.S. C.B.T. developed instrumentation and code. G.M. directed the research and wrote the paper.
Author Information. A patent application has been filed by G.M., A.K., S.M.S., and Oxford University Innovation Ltd. on the basis of work described in this study.
References
- 1.Rechtschaffen A. Current perspectives on the function of sleep. Perspect Biol Med. 1998;41:359–390. doi: 10.1353/pbm.1998.0051. [DOI] [PubMed] [Google Scholar]
- 2.Mignot E. Why we sleep: the temporal organization of recovery. PLoS Biol. 2008;6:e106. doi: 10.1371/journal.pbio.0060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science. 2011;332:1571–1576. doi: 10.1126/science.1202249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donlea JM, Pimentel D, Miesenböck G. Neuronal machinery of sleep homeostasis in Drosophila. Neuron. 2014;81:860–872. doi: 10.1016/j.neuron.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pimentel D, et al. Operation of a homeostatic sleep switch. Nature. 2016;536:333–337. doi: 10.1038/nature19055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gulbis JM, Mann S, MacKinnon R. Structure of a voltage-dependent K+ channel beta subunit. Cell. 1999;97:943–952. doi: 10.1016/s0092-8674(00)80805-3. [DOI] [PubMed] [Google Scholar]
- 9.Long SB, Campbell EB, MacKinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 10.Chouinard SW, Wilson GF, Schlimgen AK, Ganetzky B. A potassium channel beta subunit related to the aldo-keto reductase superfamily is encoded by the Drosophila Hyperkinetic locus. Proc Natl Acad Sci USA. 1995;92:6763–6767. doi: 10.1073/pnas.92.15.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bushey D, Huber R, Tononi G, Cirelli C. Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J Neurosci. 2007;27:5384–5393. doi: 10.1523/JNEUROSCI.0108-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orr WC, Sohal RS. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263:1128–1130. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- 13.Cirelli C, et al. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 14.Hill VM, et al. A bidirectional relationship between sleep and oxidative stress in Drosophila. PLoS Biol. 2018;16:e2005206. doi: 10.1371/journal.pbio.2005206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weng J, Cao Y, Moss N, Zhou M. Modulation of voltage-dependent Shaker family potassium channels by an aldo-keto reductase. J Biol Chem. 2006;281:15194–15200. doi: 10.1074/jbc.M513809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tipparaju SM, Barski OA, Srivastava S, Bhatnagar A. Catalytic mechanism and substrate specificity of the beta-subunit of the voltage-gated potassium channel. Biochemistry. 2008;47:8840–8854. doi: 10.1021/bi800301b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherin JE, Shiromani PJ, McCarley RW, Saper CB. Activation of ventrolateral preoptic neurons during sleep. Science. 1996;271:216–219. doi: 10.1126/science.271.5246.216. [DOI] [PubMed] [Google Scholar]
- 18.Connor JA, Stevens CF. Prediction of repetitive firing behaviour from voltage clamp data on an isolated neurone soma. J Physiol (Lond) 1971;213:31–53. doi: 10.1113/jphysiol.1971.sp009366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCormack T, McCormack K. Shaker K+ channel beta subunits belong to an NAD(P)H-dependent oxidoreductase superfamily. Cell. 1994;79:1133–1135. doi: 10.1016/0092-8674(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 20.Pan Y, Weng J, Cao Y, Bhosle RC, Zhou M. Functional coupling between the Kv1.1 channel and aldoketoreductase Kvbeta1. J Biol Chem. 2008;283:8634–8642. doi: 10.1074/jbc.M709304200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fogle KJ, et al. CRYPTOCHROME-mediated phototransduction by modulation of the potassium ion channel β-subunit redox sensor. Proc Natl Acad Sci USA. 2015;112:2245–2250. doi: 10.1073/pnas.1416586112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laker RC, et al. A novel MitoTimer reporter gene for mitochondrial content, structure, stress, and damage in vivo. J Biol Chem. 2014;289:12005–12015. doi: 10.1074/jbc.M113.530527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxwell DP, Wang Y, McIntosh L. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA. 1999;96:8271–8276. doi: 10.1073/pnas.96.14.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Ayala DJM, et al. Expression of the Ciona intestinalis alternative oxidase (AOX) in Drosophila complements defects in mitochondrial oxidative phosphorylation. Cell Metab. 2009;9:449–460. doi: 10.1016/j.cmet.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Wiedau-Pazos M, et al. Altered reactivity of superoxide dismutase in familial amyotrophic lateral sclerosis. Science. 1996;271:515–518. doi: 10.1126/science.271.5248.515. [DOI] [PubMed] [Google Scholar]
- 26.Yim MB, et al. A gain-of-function of an amyotrophic lateral sclerosis-associated Cu,Zn-superoxide dismutase mutant: An enhancement of free radical formation due to a decrease in Km for hydrogen peroxide. Proc Natl Acad Sci USA. 1996;93:5709–5714. doi: 10.1073/pnas.93.12.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Artiushin G, Sehgal A. The Drosophila circuitry of sleep-wake regulation. Curr Opin Neurobiol. 2017;44:243–250. doi: 10.1016/j.conb.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shu X, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng J, et al. Genetically targeted 3D visualisation of Drosophila neurons under electron microscopy and X-ray microscopy using miniSOG. Sci Rep. 2016;6 doi: 10.1038/srep38863. 38863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao WD, Wu CF. Auxiliary Hyperkinetic beta subunit of K+ channels: regulation of firing properties and K+ currents in Drosophila neurons. J Neurophysiol. 1999;81:2472–2484. doi: 10.1152/jn.1999.81.5.2472. [DOI] [PubMed] [Google Scholar]
- 31.Donlea JM, et al. Recurrent circuitry for balancing sleep need and sleep. Neuron. 2018;97:378–389.e4. doi: 10.1016/j.neuron.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaplan WD, Trout WE. The behavior of four neurological mutants of Drosophila. Genetics. 1969;61:399–409. doi: 10.1093/genetics/61.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stern M, Ganetzky B. Altered synaptic transmission in Drosophila hyperkinetic mutants. J Neurogenet. 1989;5:215–228. doi: 10.3109/01677068909066209. [DOI] [PubMed] [Google Scholar]
- 34.Jenett A, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneko M, Park JH, Cheng Y, Hardin PE, Hall JC. Disruption of synaptic transmission or clock-gene-product oscillations in circadian pacemaker cells of Drosophila cause abnormal behavioral rhythms. J Neurobiol. 2000;43:207–233. doi: 10.1002/(sici)1097-4695(20000605)43:3<207::aid-neu1>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 36.Renn SCP, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka NK, Tanimoto H, Ito K. Neuronal assemblies of the Drosophila mushroom body. J Comp Neurol. 2008;508:711–755. doi: 10.1002/cne.21692. [DOI] [PubMed] [Google Scholar]
- 38.Stocker RF, Heimbeck G, Gendre N, de Belle JS. Neuroblast ablation in Drosophila P[GAL4] lines reveals origins of olfactory interneurons. J Neurobiol. 1997;32:443–456. doi: 10.1002/(sici)1097-4695(199705)32:5<443::aid-neu1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 39.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 40.Parkes TL, et al. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- 41.Watson MR, Lagow RD, Xu K, Zhang B, Bonini NM. A Drosophila model for amyotrophic lateral sclerosis reveals motor neuron damage by human SOD1. J Biol Chem. 2008;283:24972–24981. doi: 10.1074/jbc.M804817200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson PR, Kirby K, Hilliker AJ, Phillips JP. RNAi-mediated suppression of the mitochondrial iron chaperone, frataxin, in Drosophila. Hum Mol Genet. 2005;14:3397–3405. doi: 10.1093/hmg/ddi367. [DOI] [PubMed] [Google Scholar]
- 43.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 44.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 45.Hendricks JC, et al. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 46.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 47.van Alphen B, Yap MHW, Kirszenblat L, Kottler B, van Swinderen B. A dynamic deep sleep stage in Drosophila. J Neurosci. 2013;33:6917–6927. doi: 10.1523/JNEUROSCI.0061-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Connor JA, Stevens CF. Voltage clamp studies of a transient outward membrane current in gastropod neural somata. J Physiol (Lond) 1971;213:21–30. doi: 10.1113/jphysiol.1971.sp009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.