Abstract
The big-headed turtle, Platysternon megacephalum, as the sole member of the monotypic family Platysternidae, has a number of distinct characteristics including an extra-large head, long tail, flat carapace, and a preference for low water temperature environments. We performed whole genome sequencing, assembly, and gene annotation of an adult male big-headed turtle based on the Illumina HiSeq X genomic sequencing platform. We generated ~497.1 Gb of raw sequencing data (×208.9 depth) and produced a draft genome with a total length of 2.32 Gb and contig and scaffold N50 sizes of 41.8 kb and 7.22 Mb, respectively. We also identified 924 Mb (39.84%) of repetitive sequences, 25,995 protein-coding genes, and 19,177 non-coding RNAs. We generated the first de novo genome of the big-headed turtle; these data will be essential to the further understanding and exploration of the genomic innovations and molecular mechanisms contributing to its unique morphology and physiological features.
Subject terms: Next-generation sequencing, Sequence annotation, Genome, Herpetology, DNA sequencing
| Design Type(s) | sequence annotation objective • sequence assembly objective |
| Measurement Type(s) | whole genome sequencing assay |
| Technology Type(s) | next generation DNA sequencing |
| Factor Type(s) | |
| Sample Characteristic(s) | Platysternon megacephalum • stream |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Background & Summary
The big-headed turtle, Platysternon megacephalum (NCBI Taxonomy ID: 55544), as the sole member of the family Platysternidae, is native to Southeast Asian countries, including China, Cambodia, Laos, Myanmar Thailand and Vietnam1. The species inhabits flowing cool rocky mountain streams with water temperatures usually between 12–28 °C2. It is an aquatic predator with strong climbing abilities, preying on lizard, frogs, fish, shrimps, crabs, snails, earthworms, insects, and even small birds and mammals, along with consuming some fruit and plant matter3. The big-headed turtle has some defining features, such as an extra-large head, long tail, and flat carapace. Its head width is approximately equal to half of its carapace width and therefore cannot be retracted into the shell. This species has the longest tail relative to body size of any turtle, with tail length being more than half of the carapace length4. In addition, it has a very flat carapace and an eagle-like hooked upper jaw (Fig. 1). The species prefers its body temperature to be around 25.3 °C5. While big-headed turtles are unique in many morphological and physiological characteristics compared to other turtle species, they are not unique in the anthropogenic threats they face.
Fig. 1.

A representative big-headed turtle, Platysternon megacephalum in China.
The Asian turtle crisis has resulted in population declines in many species, including the big-headed turtle. Over harvesting for the pet trade, traditional medicine, and food represent the main drivers of the population decline and have resulted in an over 89% reduction in population density in South China6. The species was listed in the CITES (Convention on International Trade in Endangered Species of Wild Fauna and Flora) Appendix I in 2013 and as an endangered species in the IUCN (International Union for Conservation of Nature) Red List of Threatened Species in 20001. Conservation and restoration measures are needed to protect this endangered species, and a genomic resource will be an essential tool for conservation efforts. However, there is currently little knowledge about the big-headed turtle’s genetic information, with previous research mainly focusing on mitochondrial DNA and the development of microsatellite markers7–11.
In this study, we report the first sequencing, assembly, and annotation of the big-headed turtle genome. The final draft genome assembly was approximately 2.32 Gb with a contig N50 of 41.8 kb and scaffold N50 of 7.22 Mb. A total of 25,995 protein-coding genes and 19,177 non-coding RNAs (409 rRNAs, 2,089 tRNAs, 16,050 miRNAs, and 629 snRNAs) were predicted from the genome assembly. The genomic resource of the big-headed turtle will be a key tool in the study of conservation genetics for this species.
Methods
Sample collection and sequencing
The genomic DNA of the big-headed turtle was extracted from the leg muscles of a single adult male obtained from Heyuan, Guangdong Province, China. The sampled turtle was one of a number that the law enforcement agencies confiscated from the black market and then transferred to scientific research institutions for study and captive breeding. Our institute has government permission to use confiscated big-headed turtles for scientific research (e.g. conservation genetics, ecology). The sampled turtle was euthanized and the animal collection and utility protocols were reviewed and approved by the Animal Ethics Committee at the Guangdong Institute of Applied Biological Resources (No: GIABR20170103). Ten paired-end libraries including four short-insert libraries (250 bp × 2, and 500 bp × 2) and six long-insert libraries (2 kb × 2, 5 kb × 2, and 10 kb × 2) were constructed and sequenced on an Illumina HiSeq X platform according to the manufacturer’s instructions (Illumina, San Diego, California, USA). The sequenced read length was 150 bp for each library. A total of 497.1 Gb (208.9×) of raw sequences were eventually obtained (Table 1). Prior to assembly, quality control was performed for raw reads using a SOAPfilter to filter out the adaptor sequences, the reads containing more than 10% unidentified nucleotides, and low-quality reads containing more than 50% bases with Illumina phred quality score ≤8. We obtained approximately 469.2 Gb of clean reads for further assembly.
Table 1.
Statistics of big-headed turtle genome sequencing data.
| Insert size | Libraries | Read length (bp) | Raw data | Clean data | ||
|---|---|---|---|---|---|---|
| Total data (Gb) | Sequence coverage (×)* | Total data (Gb) | Sequence coverage (×)* | |||
| 250 bp | 2 | 150 | 120.9 | 50.8 | 112.6 | 47.3 |
| 500 bp | 2 | 150 | 103.6 | 43.5 | 96.5 | 40.5 |
| 2 Kbp | 2 | 150 | 98.9 | 41.6 | 92.2 | 38.7 |
| 5 Kbp | 2 | 150 | 78.6 | 33.0 | 75.5 | 31.7 |
| 10 Kbp | 2 | 150 | 95.1 | 40.0 | 92.4 | 38.8 |
| Total | 10 | — | 497.1 | 208.9 | 469.2 | 197.1 |
*Sequence coverage was calculated based on the genome size of 2.38 Gb according to k-mer analysis.
Assembly and evaluation
A total of 170 Gb high-quality reads from the short-insert reads (350 bp) were used to estimate the genomic information of the big-headed turtle, and 17-mer frequency information was generated based on the K-mer analysis as implemented in the software GEC12. The heterozygous ratio was also evaluated based on the frequency of the heterozygous k-mers and homozygous k-mers using GCE software12,13. According to 17-mer analysis the estimated genome size of big-headed turtle was 2,383.87 Mb (~2.38 Gb), and the estimated heterozygous and repeat sequencing ratios were calculated to be 0.33 and 53%, respectively (Fig. 2 and Table 2).
Fig. 2.
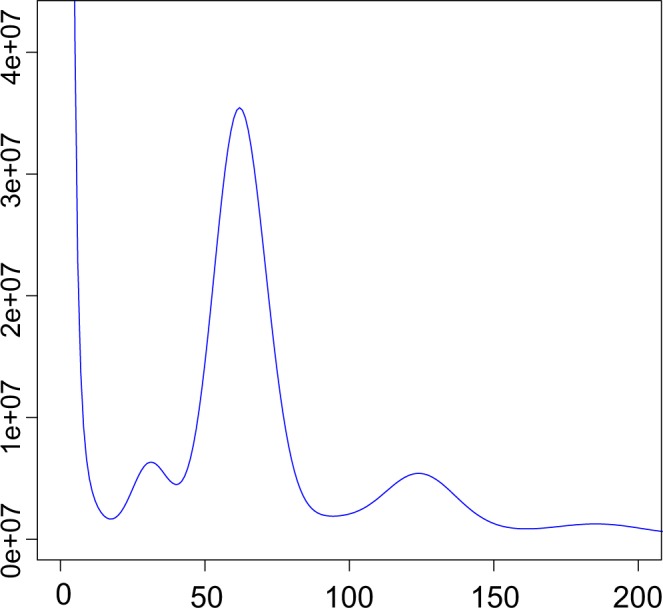
Distribution of 17-mer frequency. In total 170.2 Gb of high-quality short-insert reads (350 bp) were used to generate the 17-mer depth distribution curve frequency information.
Table 2.
Estimation of the genome size using K-mer analysis.
| Kmer | K-mer number | K-mer Depth | Estimated genome size (Mb) | Heterozygous Rate (%) | Repeat Rate (%) |
|---|---|---|---|---|---|
| 17 | 134,817,220,976 | 56 | 2,383.87 | 0.33 | 53 |
De novo assembly was performed from the generated clean reads using SOAPdenovo14, a de Bruijn graph algorithm-based de novo genome assembler. We assembled the big-headed turtle genome in three steps: contig construction, scaffolding, and gap filling. First, three K (45, 59, 71) values were used to assemble the genome, according to the N50 length of contig and the BUSCO assessments of three genomes. The genome of 59-mer was chosen as the final genome for subsequent analysis. The clean reads of short-insert libraries (250 bp and 500 bp) were used to construct the contigs with 59-mer. Then reads of long-insert libraries (2 kb, 5 kb and 10 kb) were implemented to link the contig sequences into scaffold sequences. To further improve assembly quality, GAPcloser14 and SSPACE15 were applied to reduce gap regions and raise scaffold length using a genome sketch assembled by SOAPdenovo. This last step improved the contig N50 and N90 sizes to 41,757 and 5,528 bp, and the scaffold N50 and N90 sizes to 7,221,511 and 257,323 bp, respectively, with the fragments being longer than 100 bp (Table 3). The final assembly of the big-headed turtle genome had a total length of 2.32 Gb, which was similar to the three previously published turtle genomes: Chrysemys picta bellii16, Chelonia mydas and Pelodiscus sinensis17. The key assembly statistics of the big-headed turtle genome are comparable to or better than those of previously published turtle genomes (Table 4).
Table 3.
Summary of the genome assembly.
| Sample ID | Length | Number | ||
|---|---|---|---|---|
| Contig (bp) | Scaffold (bp) | Contig | Scaffold | |
| Total | 2,282,988,448 | 2,319,520,870 | 470,184 | 360,291 |
| Max | 453,655 | 40,162,337 | — | — |
| Number >= 2000 | — | — | 100,848 | 13,027 |
| N50* | 41,757 | 7,221,511 | 15,524 | 84 |
| N60 | 32,248 | 5,116,999 | 21,745 | 121 |
| N70 | 23,516 | 3,324,563 | 30,018 | 177 |
| N80 | 15,027 | 1,848,865 | 42,079 | 269 |
| N90 | 5,528 | 257,323 | 65,609 | 576 |
*N50 referred to the scaffold larger than half the genome size which was added up from large to small.
Table 4.
Summary statistics of four turtle genomes.
| Chrysemys picta bellii | Chelonia mydas | Pelodiscus sinensis | Platysternon megacephalum | |
|---|---|---|---|---|
| Sequencing technology | Sanger + NGS | NGS | NGS | NGS |
| Assembly size (Gb) | 2.59 | 2.24 | 2.21 | 2.32 |
| Sequence coverage (×) | 18.0 | 82.3 | 105.6 | 204.2 |
| Contig N50 (kb) | 11.9 | 20.4 | 21.9 | 41.8 |
| Scaffold N50 (kb) | 5,212 | 3,778 | 3,331 | 7,222 |
| GC content (%) | 43 | 43.5 | 44.4 | 44.63 |
| Gene number | 21,796 | 19,633 | 19,327 | 22,400 |
Annotation
Repeat annotation
There are two major types of repetitive sequences: tandem repeats and interspersed repeats18. For the repeat annotation of the big-headed turtle genome, both homology-based predictions and de novo methods were performed. In the homolog-based methods, interspersed repeats were identified using RepeatMasker19 and RepeatProteinMask to search against the published RepBase sequences. In the de novo method, RepeatMasker and RepeatModeler (http://www.repeatmasker.org/RepeatModeler.html) were used to detect interspersed repeats in the genome. Tandem Repeats Finder (TRF)20 was subsequently used to search for tandem repeats. Overall, the results identified a total of 924 Mb of non-redundant repetitive sequences in the big-headed turtle genome, which account for 39.84% of the whole genome (Table 5). The most predominant elements were long interspersed nuclear elements (LINEs), which accounted for 27.47% (637 Mb) of the genome (Table 6).
Table 5.
Prediction of repeat elements in the big-headed turtle genome.
| Type | Repeat Size (bp) | % of genome |
|---|---|---|
| Trf | 47,338,094 | 2.04 |
| Repeatmasker | 874,588,835 | 37.71 |
| Proteinmask | 267,323,903 | 11.52 |
| Total | 924,094,854 | 39.84 |
Table 6.
Statistics of repeat elements in the big-headed turtle genome.
| Type | Length(bp) | % in Genome |
|---|---|---|
| DNA | 125,220,062 | 5.40 |
| LINE | 637,155,697 | 27.47 |
| SINE | 9,369,097 | 0.40 |
| LTR | 217,629,821 | 9.38 |
| Other | 52 | 0.00 |
| Satellite | 1,062,050 | 0.05 |
| Simple_repeat | 990,682 | 0.04 |
| Unknown | 15,561,759 | 0.67 |
| Total | 903,544,627 | 38.95 |
Structural annotation of genes
Three methods (homology-based, ab initio and transcriptome-based predictions) were used to predict gene structure in the big-headed turtle genome. In the homology-based method, the protein repertoires of Alligator sinensis, Chelonia mydas, Chrysemys picta bellii, Deinagkistrodon acutus, Gallus gallus, Gekko japonicus, Nanorana parkeri, Parus major, Pelodiscus sinensis, Philomachus pugnax and Xenopus laevis were downloaded from the NCBI database and mapped onto the big-headed turtle genome using TBLASTn21 with an E-value cutoff of 1 E−5. Then, homologous genome sequences were aligned against the matching proteins using GeneWise22 to define gene models. In the ab initio prediction, Augustus23, GlimmerHMM24 and SNAP25 were used to predict the coding regions of genes. To optimize the genome annotation, seven RNA tissue libraries (liver, skin, lung, intestine, heart, muscle and stomach) were constructed according to the manufacturer’s instructions (Illumina, San Diego, California, USA) and a total of 61.27 Gb of sequence data was generated. RNA-seq reads were used in de novo assembly with Trinity26. The unique transcriptional sequences were employed to predict gene models using PASA27. In total, 25,995 non-redundant protein-coding genes were annotated in the big-headed turtle genome (Table 7).
Table 7.
Statistics of predicted genes.
| Gene set | Number | Average transcript length (bp) | Average CDS length (bp) | Average exons per gene | Average exon length (bp) | Average intron length (bp) | |
|---|---|---|---|---|---|---|---|
| Ab initio prediction |
Augustus | 29,895 | 20,126.36 | 1,083.78 | 5.51 | 196.7 | 4,222.41 |
| GlimmerHMM | 163,252 | 12,105.64 | 451.4 | 3.91 | 115.37 | 4,001.41 | |
| SNAP | 83,162 | 36,326.76 | 549.05 | 3.61 | 152.28 | 1,3731.58 | |
| Homolog prediction | Alligator sinensis | 57,621 | 9,467.09 | 794.29 | 3.45 | 230.02 | 3,535.42 |
| Chelonia mydas | 37,629 | 15,048.25 | 1,093.96 | 4.9 | 223.12 | 3,575.20 | |
| Chrysemys picta bellii | 50,264 | 11,904.42 | 997.94 | 4.25 | 234.74 | 3,354.46 | |
| Deinag kistrodon acutus | 48,759 | 9,535.70 | 977.63 | 3.55 | 275.28 | 3,354.26 | |
| Gallus gallus | 45,081 | 11,902.30 | 950.46 | 3.91 | 243.04 | 3,762.56 | |
| Gekko japonicus | 34,511 | 14,745.27 | 1,136.85 | 4.8 | 236.7 | 3,578.43 | |
| Nanorana parkeri | 45,954 | 11,607.88 | 1,038.09 | 3.75 | 276.62 | 3,839.62 | |
| Parus major | 41,843 | 13,708.70 | 1,120.03 | 4.2 | 266.87 | 3,937.77 | |
| Pelodiscus sinensis | 46,338 | 11,270.86 | 981.39 | 4.05 | 242.04 | 3,368.40 | |
| Philomachus pugnax | 74,533 | 7,567.03 | 696.45 | 3.09 | 225.04 | 3,279.82 | |
| Xenopus leavis | 38,766 | 11,659.15 | 1,052.39 | 4.05 | 260 | 3,480.37 | |
| RNASeq | PASA | 106,250 | 23,132.29 | 1,040.89 | 5.91 | 176.10 | 4,498.48 |
| Cufflinks | 101,071 | 32,403.47 | 3,407.83 | 6.79 | 502.11 | 5,010.49 | |
| EVM | 396,95 | 17,966.58 | 987.68 | 5.19 | 190.45 | 4,056.02 | |
| Pasa-update* | 39,212 | 21,180.40 | 1,029.81 | 5.39 | 191.11 | 4,591.49 | |
| Final set* | 25,995 | 30,713.64 | 1,298.80 | 7.33 | 177.28 | 4,649.67 | |
*UTR regions were contained.
Functional annotation of genes
The functional annotation of protein-coding genes of the big-headed turtle genome were predicted by aligning protein sequences against public databases including SwissProt (http://www.gpmaw.com/html/swiss-prot.html), KEGG (http://www.genome.jp/kegg/) and TrEMBL (http://www.uniprot.org) using BLASTp with an E-value cutoff of 1 E−5. Protein motifs and domains were annotated using InterPro28, and Gene Ontology (GO)29 terms for each gene were retrieved from the corresponding InterPro results. Overall, 22,400 protein-coding genes (86.2%) were successfully annotated (Table 8).
Table 8.
Statistics of functional annotation.
| Type | Number | Percentage (%) |
|---|---|---|
| Total | 25,995 | — |
| NR | 22,357 | 86.0 |
| Swiss-Prot | 21,536 | 82.8 |
| KEGG | 19,560 | 75.2 |
| InterPro | 21,227 | 81.7 |
| Pfam | 19,277 | 74.2 |
| GO | 15,735 | 60.5 |
| Annotated | 22,400 | 86.2 |
| Unannotated | 3,595 | 13.8 |
Non-coding RNA annotation
The non-coding RNAs were predicted in the big-head genome based on four categories: ribosomal RNA (rRNA), transfer RNA (tRNA), microRNAs (miRNA) and small nuclear RNA (snRNA). tRNAscan-SE30 was applied to identify tRNA with eukaryotic parameters according to the characteristics of tRNA. Because of the highly conserved characteristics of rRNA, BLASTN31 was used to predict rRNA sequences by aligning with a human template with an E-value of 1 E−10. The miRNA and snRNA sequences were identified using INFERNAL32 by searching against the Rfam database. We identified a total of 409 rRNA, 2,089 tRNA, 16,050 miRNA, and 629 snRNA genes in the big-headed turtle genome (Table 9).
Table 9.
Summary of non-coding RNA.
| Type | Number | Average length (bp) | Total length (bp) | % of genome | |
|---|---|---|---|---|---|
| miRNA | 16,050 | 85.58 | 1,373,585 | 0.05921 | |
| tRNA | 2,089 | 74.90 | 156,473 | 0.00675 | |
| rRNA | rRNA | 409 | 160.37 | 65,591 | 0.00283 |
| 18S | 77 | 164.01 | 12,629 | 0.00054 | |
| 28S | 272 | 173.51 | 47,196 | 0.00204 | |
| 5.8S | 4 | 113.50 | 454 | 0.00002 | |
| 5S | 56 | 94.86 | 5,312 | 0.00023 | |
| snRNA | snRNA | 629 | 129.16 | 81,241 | 0.00350 |
| CD-box | 179 | 97.50 | 17,452 | 0.00075 | |
| HACA-box | 132 | 142.79 | 18,848 | 0.00081 | |
| splicing | 294 | 137.37 | 40,387 | 0.00174 | |
Data Records
This Whole Genome Shotgun project including the assembled genome sequence and the structural and functional annotation of genes has been deposited at DDBJ/ENA/GenBank under the accession QXTE0000000033. The version described in this paper is version QXTE01000000. Repeat annotation and Non-coding RNA annotation have been deposited at Figshare34. Raw read files have been deposited at NCBI Sequence Read Archive under the accession SRP15641935.
Technical Validation
Assessing the completeness of the genome assembly
To assess the quality of the genome assembly, we performed three independent evaluations as described below. First, the base content was counted with scaffolds longer than 100 bp and the results showed that the GC content for the big-headed turtle was 44.63% (Table 10), which was comparable to those of Chrysemys picta bellii, Chelonia mydas and Pelodiscus sinensis (Table 4). Second, the short-insert paired-end reads (250 bp and 500 bp) were mapped to the genome with BWA36. The mapping rate was 98.84% and the genome coverage was 99.57% (Table 11), indicating high reliability of genome assembly. Third, the SNPs (Single Nucleotide Polymorphisms) were counted to validate the uniformity of the genome using SAMtools37, and we found the ratio of homozygous SNPs was only 0.014% (Table 12), indicating that the assembly had a high base accuracy. Finally, CEGMA (http://korflab.ucdavis.edu/dataseda/cegma/) and BUSCO (http://busco.ezlab.org/) were used to evaluate the completeness of the assembly. CEGMA assessment showed that our assembly captured 226 (91.13%) of the 248 ultra-conserved core eukaryotic genes, of which 202 (81.45%) were complete (Table 13). BUSCO analysis showed that 95.2% and 2.6% of the expected vertebrate genes were identified as complete and fragmented, respectively, while 2.2% were considered missing in the assembly (Table 14).
Table 10.
Base content statistics of the genome.
| Type | Number (bp) | % of genome |
|---|---|---|
| A | 632,158,059 | 27.25 |
| T | 631,959,166 | 27.25 |
| C | 509,623,076 | 21.97 |
| G | 509,248,147 | 21.97 |
| N | 36,532,422 | 1.57 |
| Total | 2,319,520,870 | — |
| GC* | 1,018,871,223 | 44.63 |
*GC content of the genome without N.
Table 11.
Statistics of mapping ratio in genome.
| Type | Content | Value |
|---|---|---|
| Reads | Mapping rate (%) | 98.84 |
| Genome | Average sequencing depth (×) | 72.05 |
| Coverage (%) | 99.57 | |
| Coverage at least 4× (%) | 98.73 | |
| Coverage at least 10× (%) | 97.25 | |
| Coverage at least 20× (%) | 95.38 |
Table 12.
Number and density of SNPs in big-headed turtle genome.
| Type | Number | Proportion (%) |
|---|---|---|
| All SNPs | 5,319,363 | 0.233% |
| Heterozygous SNPs | 4,999,745 | 0.219% |
| Homozygous SNPs | 319,618 | 0.014% |
Table 13.
Assessment of CEGMA.
| Species | Complete | Complete + Partial | ||
|---|---|---|---|---|
| Proteins | Completeness (%) | Proteins | Completeness (%) | |
| Big-headed turtle | 202 | 81.45 | 226 | 91.13 |
Table 14.
Assessment of BUSCO.
| Species | Size | BUSCO notation assessment results* |
|---|---|---|
| Big-headed turtle | 2320 Mb | C: 95.2% [S: 94.2%, D: 1.0%], F: 2.6%, M: 2.2%, n: 2586 |
*C: Complete BUSCOs; S: Complete and single-copy BUSCOs; D: Complete and duplicated BUSCOs; F: Fragmented BUSCOs; M: Missing BUSCOs; n: Total BUSCO groups searched.
Annotation filtering and validation
The EVM software38 was used to merge the above results of gene annotation and 39,212 genes were obtained from the merged set. To further revise the genome annotation, we removed the following type of genes: (1) genes with overlap regions of TE ≥ 20%; (2) premature termination genes; (3) genes with only de novo predictive support. After filtering, 25,995 genes were retained. In addition, total RNA-seq reads of 7 tissues were mapped onto the big-headed turtle genome to further identify exon regions and splice positions using Tophat39 and Cufflinks40, and 20,028 (77.05%) genes had evidence supports of RNA data (RPKM value > 1) cross above 7 tissues.
ISA-Tab metadata file
Acknowledgements
The authors would like to thank Dr. Daniel Gaillard for checking the manuscript and Mr. Yufeng Wei for his help in sample collection. This work was supported by the National Natural Science Foundation of China (31471966), Guangdong Natural Science Foundation (2015A030313903), and the GDAS Special Project of Science and Technology Development (2017GDASCX-0107; 2018GDASCX-0107).
Author Contributions
S.P.G. and D.N.C. designed and supervised the project. M.W. performed bioinformatics analyses. Y.G. collected the samples. D.N.C., S.P.G. and M.W. were involved in the data analyses and wrote the manuscript. All authors have read and approved the final manuscript.
Code Availability
The execution of this work involved using many software tools, whose settings and parameters are described below.
(1) GCE: version1.0.0, parameters: -H 1; (2) SOAPdenovo: version2, k-mer size of 59; (3) GAPcloser: version1.12, parameters: -l 150 -p 31; (4) SSPACE: version3.0, default parameters; (5) RepeatMasker: Repeat Masker-open-4-0-6, parameters: -a -nolow -no_is -norna -parallel 1; (6) RepeatModeler: RepeatModeler-open-1.0.11, parameters: -database genome -engine ncbi -pa 15; (7) Tandem Repeats Finder: TRF-407b, parameters: 2 7 7 80 10 50 2000 -d -h; (8) TBLASTn: blast-2.2.26, parameters: -p tblastn -e 1e-05 -F T -m 8 -d; (9) GeneWise: version2.4.1, parameters: -tfor -genesf -gff; (10) Augustus: version3.2.3, parameters: –uniqueGeneId = true–noInFrameStop = true–gff3 = on–genemodel = complete–strand = both; (11) GlimmerHMM: version3.0.1, parameters: -g -f; (12) SNAP: snap-2013-11-29, default parameters; (13) Trinity: trinityrnaseq-2.1.1, parameters: –seqType fq-CPU 20–max_memory 200G–normalize_reads–full_cleanup–min_glue 2–min_kmer_cov 2–KMER_SIZE 25; (14) PASA: PASA_r20140417, default parameters; (15) InterPro: version29.0, perl-based version4.8, default parameters; (16) tRNAscan-SE: tRNAscan-SE-1.3.1, default parameters; (17) INFERNAL: version1.1rc4 (June 2013); (18) BLASTp: blast-2.2.26, parameters: -p blastn -e 1e-10 -v 10000 -b 10000; (19) BWA: bwa-0.7.8, parameters: mem -k 32 -w 10 -B 3 -O 11 -E 4 -t 10; (20) SAMtools: samtools-0.1.19, parameters: mpileup mpileup -m 2 -u; (21) EVM: VidenceModeler-1.1.1, parameters: –segmentSize 200000–overlapSize 20000; (22) Tophat: tophat-2.0.13, parameters: -p 6–max-intron-length 500000 -m 2–library-type fr-unstranded; (23) Cufflinks: cufflinks-2.1.1, parameters: -I 500000 -p 1–library-type fr-unstranded -L CUFF; (24) BUSCO: version3.0.2, OrthoDBv9_vertebrata.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
ISA-Tab metadata
is available for this paper at 10.1038/s41597-019-0067-9.
References
- 1.Rhodin, A. G. J. et al. Turtles of The World: Annotated Checklist And Atlas Of Taxonomy, Synonymy, Distribution, And Conservation Status 8th end (Chelonian Research Foundation and Turtle Conservancy, 2017).
- 2.Shen JW, Pike DA, Du WG. Movements and microhabitat use of translocated big-headed turtles (Platysternon megacephalum) in southern China. Chelonian Conserv. Bi. 2010;9:154–161. doi: 10.2744/CCB-0833.1. [DOI] [Google Scholar]
- 3.Sung YH, Hau BC, Karraker NE. Diet of the endangered big-headed turtle Platysternon megacephalum. PeerJ. 2016;4:e2784. doi: 10.7717/peerj.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang, M. W., Zong Y. & Ma J. F. Fauna Sinica (Reptilia 1): General Accounts Of Reptilia, Testudoformes And Crocodiliformes (Science Press, 1998).
- 5.Shen JW, Meng FW, Zhang YP, Du WG. Field body temperature and thermal preference of the big-headed turtle Platysternon megacephalum. Curr. Zool. 2013;59:626–632. doi: 10.1093/czoolo/59.5.626. [DOI] [Google Scholar]
- 6.Gong SP, et al. Disappearance of endangered turtles within China’s nature reserves. Curr. Biol. 2017;27:R170–R171. doi: 10.1016/j.cub.2017.01.039. [DOI] [PubMed] [Google Scholar]
- 7.Zheng C, et al. Recombination and evolution of duplicate control regions in the mitochondrial genome of the Asian big-headed turtle, Platysternon megacephalum. Plos One. 2013;8:e82854. doi: 10.1371/journal.pone.0082854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng QL, Nie LW, Pu YG. Complete mitochondrial genome of Chinese big-headed turtle, Platysternon megacephalum, with a novel gene organization in vertebrate mtDNA. Gene. 2006;380:14–20. doi: 10.1016/j.gene.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Parham JF, Feldman CR, Boore JL. The complete mitochondrial genome of the enigmatic bigheaded turtle (Platysternon): description of unusual genomic features and the reconciliation of phylogenetic hypotheses based on mitochondrial and nuclear DNA. BMC Evol. Biol. 2006;6:11. doi: 10.1186/1471-2148-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua L, et al. Isolation and characterization of 14 polymorphic microsatellite loci in the big-headed turtle (Platysternon megacephalum) Biochem. Genet. 2014;52:203–208. doi: 10.1007/s10528-014-9640-2. [DOI] [PubMed] [Google Scholar]
- 11.Peng J, et al. New microsatellite resources in Chinese big-headed turtle (Platysternon megacephalum) Conserv. Genet. Resour. 2010;2:55–57. doi: 10.1007/s12686-009-9162-0. [DOI] [Google Scholar]
- 12.Liu B, et al. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. Quant. Biol. 2013;35:62–67. [Google Scholar]
- 13.Li X, Waterman MS. Estimating the repeat structure and length of DNA sequences using L-tuples. Genome Res. 2003;13:1916–1922. doi: 10.1101/gr.1251803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boetzer M, Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014;15:211. doi: 10.1186/1471-2105-15-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaffer HB, et al. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 2013;14:R28. doi: 10.1186/gb-2013-14-3-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, et al. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat. Genet. 2013;45:701–706. doi: 10.1038/ng.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhagavan, N. V. & Ha, C. E. Essentials of Medical Biochemistry. (Academic Press, 2011).
- 19.Maja T-G, Nansheng C. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics. 2009;25:4.10.11–14.10.14. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 20.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birney E, Clamp M, Durbin R. Gene Wise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl 2):ii215–225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 24.Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 25.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell MA, Haas BJ, Hamilton JP, Mount SM, Buell CR. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulder N, Apweiler R. Inter Pro and Inter Pro Scan: tools for protein sequence classification and comparison. Methods Mol. Biol. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- 29.Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao, D. N., Wang, M., Ge, Y. & Gong, S. P. Platysternon megacephalum isolate DO16091913, whole genome shotgun sequencing project. GenBank, http://identifiers.org/ncbi/insdc:QXTE01000000 (2018).
- 34.Cao, D. N., Wang, M., Ge, Y. & Gong, S. P. Repeat annotation and Non-coding RNA annotation of Platysternon megacephalum. figshare, 10.6084/m9.figshare.7586408.v1 (2019).
- 35.NCBI Sequence Read Archive, http://identifiers.org/ncbi/insdc.sra:SRP156419 (2018).
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The execution of this work involved using many software tools, whose settings and parameters are described below.
(1) GCE: version1.0.0, parameters: -H 1; (2) SOAPdenovo: version2, k-mer size of 59; (3) GAPcloser: version1.12, parameters: -l 150 -p 31; (4) SSPACE: version3.0, default parameters; (5) RepeatMasker: Repeat Masker-open-4-0-6, parameters: -a -nolow -no_is -norna -parallel 1; (6) RepeatModeler: RepeatModeler-open-1.0.11, parameters: -database genome -engine ncbi -pa 15; (7) Tandem Repeats Finder: TRF-407b, parameters: 2 7 7 80 10 50 2000 -d -h; (8) TBLASTn: blast-2.2.26, parameters: -p tblastn -e 1e-05 -F T -m 8 -d; (9) GeneWise: version2.4.1, parameters: -tfor -genesf -gff; (10) Augustus: version3.2.3, parameters: –uniqueGeneId = true–noInFrameStop = true–gff3 = on–genemodel = complete–strand = both; (11) GlimmerHMM: version3.0.1, parameters: -g -f; (12) SNAP: snap-2013-11-29, default parameters; (13) Trinity: trinityrnaseq-2.1.1, parameters: –seqType fq-CPU 20–max_memory 200G–normalize_reads–full_cleanup–min_glue 2–min_kmer_cov 2–KMER_SIZE 25; (14) PASA: PASA_r20140417, default parameters; (15) InterPro: version29.0, perl-based version4.8, default parameters; (16) tRNAscan-SE: tRNAscan-SE-1.3.1, default parameters; (17) INFERNAL: version1.1rc4 (June 2013); (18) BLASTp: blast-2.2.26, parameters: -p blastn -e 1e-10 -v 10000 -b 10000; (19) BWA: bwa-0.7.8, parameters: mem -k 32 -w 10 -B 3 -O 11 -E 4 -t 10; (20) SAMtools: samtools-0.1.19, parameters: mpileup mpileup -m 2 -u; (21) EVM: VidenceModeler-1.1.1, parameters: –segmentSize 200000–overlapSize 20000; (22) Tophat: tophat-2.0.13, parameters: -p 6–max-intron-length 500000 -m 2–library-type fr-unstranded; (23) Cufflinks: cufflinks-2.1.1, parameters: -I 500000 -p 1–library-type fr-unstranded -L CUFF; (24) BUSCO: version3.0.2, OrthoDBv9_vertebrata.


