Abstract
Fatty acid binding protein 4 (FABP4), a subtype of fatty acid-binding protein family, shows critical roles in metabolism and inflammation. However, its roles on regulating renal interstitial fibrosis (RIF) remain unclear. In this work, LPS-stimulated in vitro models on NRK-52E and NRK-49F cells, and in vivo UUO models in rats and mice were established. The results showed that comparing with control groups or sham groups, the expression levels of α-SMA, COL1A, COL3A, IL-1β, IL-6, and TNF-α in LPS-stimulated cells or UUO animals were significantly increased. Meanwhile, the levels of TC, TG, and free fatty acid were also significantly increased as well as the obvious lipid droplets, and the serum levels of BUN, Cr were significantly increased with large amounts of collagen deposition in renal tissues. Further investigation showed that compared with control groups or sham groups, the expression levels of FABP4 in LPS-stimulated cells and UUO animals were significantly increased, resulting in down- regulating the expression levels of PPARγ, upregulating the levels of p65 and ICAM-1, and decreasing the expression levels of ACADM, ACADL, SCP-2, CPT1, EHHADH, and ACOX1. To deeply explore the mechanism of FABP4 in RIF, FABP4 siRNA and inhibitor interfered cell models, and UUO model on FABP4 knockout (KO) mice were used. The results showed that the expression levels of α-SMA, COL1A, and COL3A were significantly decreased, the deposition of lipid droplets decreased, and the contents of TC, TG, and free fatty acids were significantly decreased after gene silencing. Meanwhile, the expression levels of PPAR-γ, ACADM, ACADL, SCP-2, CPT1, EHHADH, and ACOX1 were upregulated, the levels of p65 and ICAM-1 were downregulated, and the mRNA levels of IL-1β, IL-6, and TNF-α were decreased. Our results supported that FABP4 contributed to RIF via promoting inflammation and lipid metabolism, which should be considered as one new drug target to treat RIF.
Subject terms: Target validation, End-stage renal disease
Introduction
Renal fibrosis, a common pathological process during the progression of chronic kidney disease (CKD) to the end stage renal disease (ESRD), includes glomerular sclerosis (GS) and renal interstitial fibrosis (RIF), in which renal interstitial lesions are more important than glomerular lesions to demonstrate the severity of renal function decline and prognostic prediction1. RIF with the accumulation of collagen components in renal interstitium can be caused by various pathogenic factors including glomerulonephritis, chronic pyelonephritis, obstructive nephropathy, diabetic nephropathy, hypertensive nephropathy, and kidney transplantation2. Thus, RIF, an important global public health issue, can seriously threaten human health and bring great economic burden to families and society.
At present, the detailed molecular mechanisms of RIF are not completely clarified. Multiple pathophysiological changes including inflammation, apoptosis and oxidative stress can cause fibrosis. Recent studies have found that massive proteinuria can cause the overload of free fatty acids (FFAs) in renal interstitial cells and lipid hydroperoxides after oxidization in mitochondria and lysosomes3. The molecules associated with reactive oxygen species (ROS) can destroy cell membrane, and cause severe renal interstitial damage and fibrosis. It has also been found that increased absorption of FFAs can result in apoptosis and damage of renal interstitial cells4. Under normal conditions, fatty acid oxidation can produce energy for renal tubular epithelial cells. However, reduced fatty acid metabolism can cause ATP depletion, cell death, lipid accumulation, and ultimately lead to RIF. On the other hand, transforming growth factor-β (TGF-β) can reduce fatty acid oxidation in renal tubular epithelial cells to promote renal fibrosis5. In addition, macrophages, the predominant infiltrating immune cells, can produce various proinflammatory cytokines, which are closely associated with renal fibrosis6. Monocyte chemoattractant protein-1 (MCP-1), an important proinflammatory cytokine, has important role in the progression of tubulointerstitial fibrosis7. Thus, regulating lipid metabolism and inflammation should be one effective method to control RIF.
Fatty acid-binding protein 4 (FABP4), a subtype of fatty acid-binding protein family, is a key transmitter of lipid metabolism and inflammatory reaction8. FABP4 is positively correlated with FFAs, and high level of FABP4 can directly damage endothelial cells, while the injured endothelial cells can promote FABP4 level, followed by deposition of triglyceride and cholesterol, together with lipid metabolism disorders9. It has been reported that peroxisome proliferator activated receptorγ (PPARγ), one target gene of FABP4, can be negative feedback controlled by FABP410. However, PPARγ is able to decrease NF-κB activity, which can also effectively inhibit the expression levels of intercellular cell adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule 1 (VCAM-1)11. Activated PPARγ can suppress the production of tumor necrosis factor-α (TNF-α), Interleukin-1 (IL-1), IL-4, and IL-6 with anti-inflammatory effect12. Meanwhile, PPARγ can regulate the processes of fatty acid transport, oxidation and decomposition by regulating the expression levels of fatty acid transporter, fatty acid binding protein, and Carnitine palmitoyltransferase-1 (CPT1)13. In addition, FABP4 can adjust the eicosanoid balance by regulating the activities of cyclooxygenase 2 (COX2) and leukotriene A4 (LTA4), and ultimately affect the functions of macrophages and inflammation14. Moreover, FABP4 can regulate obesity-induced neuroinflammation through FABP4-uncoupling protein 2 (UCP2) axis15. In diabetic nephropathy, FABP4 can regulate apoptosis of renal interstitial cells via adjusting endoplasmic reticulum stress, which may also serve as a marker of renal injury16. In addition, inhibition of FABP4 can reduce hepatic liver ischemia- reperfusion injury17. Hence, FABP4 plays critical roles in regulating inflammation and lipid metabolism. However, there have no studies concerning the roles of FABP4 in RIF. Therefore, the aim of the present work was to investigate the function of FABP4 in regulating RIF.
Materials and methods
Chemicals and materials
Dulbecco’s Modified Eagle’s Medium (DMEM) was purchased from KeyGEN (Jiangsu, China). The assay kits of blood urea nitrogen (BUN), creatinine (Cr), total cholesterol (TC), triglyceride (TG), and free fatty acids (FFAs) were obtained from Nanjing Jiancheng Institute of Biotechnology (Nanjing, China). Tissue Protein Extraction Kit and the Bicinchoninic acid (BCA) Protein Assay Kit were purchased from Beyotime Institute of Biotechnology (Jiangsu, China). Tris, sodium dodecyl sulfate (SDS) and 4,6-diamidino-2-phenylindole (DAPI) were purchased from Sigma (St. Louis, MO, USA). Lipopolysaccharide (LPS) and Oil red O staining solution were purchased from Solarbio Technology Co., Ltd (Beijing, China) and KeyGEN (Jiangsu, China). CCK-8 was purchased from Selleck Chemicals (Houston, USA). Lipofectamine 2000 was obtained from GenePharma (Shanghai, China). RNAisoPlus, PrimeScript®RT reagent Kit with DNA Eraser (Perfect Real Time) and SYBR®Premix Ex TaqTMII (Tli RNaseH Plus) were purchased from TaKaRa Biotechnology Co., Ltd. (Dalian, China). FABP4 siRNA was purchased from RIBOBIO Co., Ltd. (Guangzhou, China). Mouse and Rat FABP4 ELISA Kits were purchased from Boster Biological Technology Co., Ltd. (California, USA). FABP4 inhibitor BMS309403 was purchased from MedChemExpress (Shanghai, China).
Cell culture
NRK-52E cell line (a rat proximal tubular epithelial cell) and NRK-49F cell line (a kind of rat fibroblast) were purchased from the Institute of Biochemistry Cell Biology (Shanghai, China) and BeNa Culture Collection (Beijing, China), which were maintained in DMEM or MEM NEAA supplemented with 10% FBS and antibiotics (100 IU/mL penicillin and 100 mg/mL streptomycin) in a humidified atmosphere of 5% CO2 and 95% O2 at 37 °C.
LPS-induced cell proliferation
NRK-52E and NRK-49F cells were plated in 96-well plates at a density of 1 × 105 cells/mL for 24 h before challenge with various concentrations of LPS (0, 25, 50, 100, 200, and 400 ng/mL) for different times (3, 6, 12, and 24 h). Then, CCK-8 solution was added to the plates for 4 h incubation at 37 °C, and the absorbance was measured at 450 nm with a microplate reader (Thermo, Waltham, MA, USA).
Unilateral ureteral obstruction (UUO) model on rats and mice
Male Sprague Dawley rats (180–220 g) and Male C57BL/6J mice (18–22 g) were purchased from the Experimental Animal Center of Dalian Medical University (Dalian, China) (SCXK (Liao): 2013-0003). FABP4 knockout (KO) and WT mice (20–22 g) were purchased from Nanjing Biomedical Research Institute of Nanjing University (Nanjing, China) (SCXK (Su): 2015-0001). The animals were maintained in a controlled environment under standard conditions with temperature at 21–25 °C and relative humidity at 55–70%. The animals were provided with ample food and water and maintained on a 12 h/12 h light-dark cycle. The rats and mice were randomly divided into sham group and UUO model group (n = 7). FABP4 KO and WT mice were randomly divided into sham group of WT mice, UUO model group of WT mice, sham group of FABP4-KO mice, and UUO model group of FABP4-KO mice (n = 5). The animals in UUO groups were achieved by ligating the left ureter with 3-0 silk through a left lateral incision. The animals in sham-operated groups were used as the control. After 4 weeks of rats and 7 days of mice18,19, the animals were sacrificed. The blood samples were obtained to produce serum after centrifugation (1200 × g, 4 °C), which were stored at −20 °C. The kidney samples were harvested and stored at −80 °C.
Assessment of biochemical parameters
The protein samples from NRK-52E cells and NRK-49F cells were extracted using cold lysis buffer containing 1 mM phenylmethyl sulfonyl fluoride according to the protocol, and the contents were determined using BCA Protein Assay Kit. The levels of TC, TG, and FFAs in cell lysates were measured using the commercial kits20. The serum levels of BUN, Cr, TC, TG, and FFAs of rats and mice were also detected using the commercial kits according to the manufacturer’s instructions.
ELISA assay
The serum protein levels of FABP4 in rats and mice were measured using the assay kits according to the manufacturer’s instructions21.
Histological assay
Formalin-fixed renal tissue was embedded in paraffin. The portion of renal cortex was separated and fixed in formaldehyde (10%), and then the section with 5 μm thick was stained with H&E, Masson, Sirius red staining assays. Images were acquired and the histological evaluations were performed using a light microscope (Nikon Eclipse TE2000-U, NIKON, Japan) with 400× magnification. In addition, the images of Sirius red polarization assay were captured using a polarized light microscope (NIKON Eclipse Ci, NIKON, Japan) with 400× magnification.
Oil Red O staining of cells and renal tissues
The frozen tissue slices or formal in-fixed cells were washing with PBS, and then Oil red O solution was added and incubated at room temperature in the dark for 30 min, which were finally washed with 60% isopropanol and PBS. Then, the images were captured using a microscope (Olympus, Tokyo, Japan) with 400× magnification.
Immunofluorescence assay
For immunofluorescence staining of FABP4, α-SMA, and COL1A, the tissue slices or formal in-fixed cells were incubated with anti-FABP4, anti-α-SMA, and anti- COL1A antibodies, respectively, in one humidified chamber at 4 °C overnight, followed by incubation with an alexa fluorescein-labeled secondary antibody at 37 °C for 1 h. Eventually, cell nuclei were stained with DAPI (5 mg/mL) for 15 min. Then, the immune stained images were captured using a fluorescence microscope (Olympus, Tokyo, Japan) with 400× magnification.
Western blot assay
Total protein samples from kidney tissues of rats and mice, NRK-52E and NRK- 49F cells were extracted using cold lysis buffer containing 1 mM phenylmethyl sulfonyl fluoride according to the manufacturer’s protocol, and the protein content was determined using the BCA protein assay kit. The protein samples were loaded onto SDS-PAGE gel (8–12%), electrophoretically separated and transferred onto a PVDF membrane (Millipore, USA). After blocking nonspecific binding sites for 1 h with 5% dried skim milk in TTBS at 37 °C, the membrane was individually incubated for overnight at 4 °C with the primary antibodies listed in Supplemental Table S1. Then, the membrane was incubated at room temperature for 2 h with horseradish peroxidase-conjugated antibodies at a 1:2000 dilution. Protein expression was detected by the enhanced chemiluminescence (ECL) method. Protein bands were imaged using a Bio-Spectrum Gel Imaging System (UVP, Upland, CA, USA) and normalized with GAPDH as an internal control (IOD of objective protein versus IOD of GAPDH protein).
Quantitative real-time PCR assay
Total RNA samples were obtained from kidney tissues, NRK-52E, and NRK-49F cells using RNAiso Plus reagent following the manufacturer’s protocol. Each RNA sample was reverse transcribed into cDNA using the PrimeScript1 RT reagent Kit. The forward (F) and reverse (R) primers used in the present study are given in Supplemental Table S2. Among the data from each sample, the Ct value of the target genes was normalized to that of GAPDH. The unknown template in our study was calculated using the standard curve for quantitative analysis.
FABP4 siRNA treatment
FABP4-targeted siRNA and control siRNA were dissolved in DMEM or MEM NEAA and then equilibrated for 5 min at room temperature. NRK-52E and NRK-49F cells were transfected with FABP4 siRNA or non-binding control siRNA using Lipofectamine 2000 reagent according to the protocol. Then, the levels of TC, TG, and FFAs were detected. Oil Red O staining and immunofluorescence assay were carried out for detecting lipid droplets and the expression levels of α-SMA, COL1A, and FABP4 after transfection. In addition, the protein levels of FABP4, PPARγ, SCP2, ACADM, ACADL, EHHADH, CPT1, ACOX1, p65, ICAM-1, and the mRNA levels of α-SMA, COL1A, COL3A, IL-1β, IL-6, and TNF-α were measured after transfection.
Statistical analysis
The data and statistical analysis in this study comply with the recommendations on experimental design and analysis in pharmacology. The data are expressed as the mean ± SD. GraphPad Prism 6.01 software (Paragraph Software, Inc, La Jolla, CA, USA) was used to handle these data and only when a minimum of n = 5 independent samples was acquired. Statistical significance was determined by one-way ANOVA. Analysis between two individual groups was determined by Student t-test. The results were considered to be statistically significant at p < 0.05.
Results
Fibrosis and lipid metabolism disorders in cells caused by LPS
As shown in Fig. 1a, the expression levels of α-SMA, COL1A1, and COL3A1 were significantly increased (p < 0.01) in NRK-52E and NRK-49F cells after LPS treatment compared with control groups. (The survival rates of NRK-52E and NRK-49F treated by LPS are shown in Supplemental Fig. S1). Based on these results, LPS at the concentration of 200 ng/mL for NRK52E cells, and 25 ng/mL for NRK49F cells under 3 h treatment were selected in the rest of the experiments. The results of immunofluorescence assay also showed the high expression levels of α-SMA (green light) and COL1A (red light) in LPS-stimulated cells. In addition, as shown in Fig. 1b, Oil red O staining revealed that lipid droplets were accumulated in NRK-52E and NRK-49F cells after LPS treatment compared with control groups. Furthermore, the levels of TC, TG and FFAs were significantly increased by 38.76, 79.57, and 85.73% in LPS-induced NRK-52E cells compared with un-treated cells, and increased by 30.97, 70.07, and 85.16% in LPS-induced NRK-49F cells compared with control group.
Fig. 1. Renal fibrosis and lipid metabolism disorders in vitro and in vivo.
a The mRNA levels of α-SMA, COL1A, COL3A, and the protein levels of α-SMA and COL1A based on immunofluorescence staining (400× original magnification) in NRK-52E and NRK-49F cells (n = 3). b The lipid droplets based on Oil Red O staining (400× original magnification), and the levels of TG, TC, and FFAs in NRK-52E and NRK-49F cells (n = 5). c The serum Cr and BUN levels, and H&E staining (400× original magnification) of the kidney tissues in mice and rats (n = 7). d Masson and Sirius red staining (400× original magnification), and Sirius red polarized light observation (400× original magnification) of the kidney tissues in mice and rats. e The mRNA levels of α-SMA, COL1A1, COL3A1, and the protein levels of α-SMA and COL1A in the kidney tissues of mice and rats based on immunofluorescence staining (400× original magnification). f The lipid droplets based on Oil O Red staining (400× original magnification), and the levels of TG, TC, and FFAs in mice and rats (n = 7). Data are presented as the mean ± SD. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with sham groups or control groups
Renal injury and histopathological changes in UUO rats and mice
As shown in Fig. 1c, compared with sham groups, the levels of serum Cr (mice 25.506 ± 4.596 μmol/L; rats 53.344 ± 16.458 μmol/L) and BUN (mice 10.833 ± 1.803 mmol/L; rats 8.333 ± 1.883 mmol/L) in UUO animals were significantly increased. The results of H&E staining showed that the kidneys of the animals in sham groups exhibited integral tubular cell structure. However, the histopathological changes including swelling in renal tubular epithelial cells, vacuoles degeneration, disappearing of brush border, coagulation necrosis, and massive inflammatory cells infiltration in UUO animals were obviously found compared with sham groups.
UUO induces RIF and lipid metabolism disorders
As shown in Fig. 1d, compared with sham groups, the interstital and perivascular collagen depositions were obviously found in UUO animals, and the expression levels of COL1A (green) and COL3A (orange red) were heavily increased compared with sham groups, as well as the results of polarized light observation. As shown in Fig. 1e, the mRNA levels of α-SMA, COL1A1, and COL3A1 in UUO animals were markedly increased compared with sham group. In addition, immunofluorescence assay showed that the expression levels of α-SMA (green) and COL1A (red) in UUO animals were markedly increased compared with sham groups. As shown in Fig. 1f, Oil Red O staining indicated that the lipid droplets in renal tissue were significantly increased in UUO animals. At the same time, compared with sham groups, the levels of TC, TG, and FFAs were significantly increased by 21.19, 29.43, and 40.38% in UUO mice, and increased by 44.74, 61.81, and 34.38% in UUO rats.
Expression levels of FABP4 in RIF
As shown in Fig. 2a, b, in LPS-induced cells, the expression levels of FABP4 were significantly increased compared with control groups. Similarly, in UUO rats and mice, the expression levels of FABP4 were significantly increased compared with sham groups (Fig. 2c, d). In addition, the serum protein levels of FABP4 in UUO rats and mice were significantly increased with p < 0.01 compared with sham groups (Fig. 2e).
Fig. 2. Increased protein levels of FABP4 in RIF models.
a, b The protein levels of FABP4 in NRK-52E and NRK-49F cells based on immunofluorescence staining and western blotting assay (n = 3). c, d The protein levels of FABP4 in mice and rats based on immunofluorescence staining and western blotting assay (n = 3). e Serum protein levels of FABP4 in mice and rats based on Elisa assay (n = 7). Data are presented as the mean ± SD. **p < 0.01 compared with sham groups or control groups
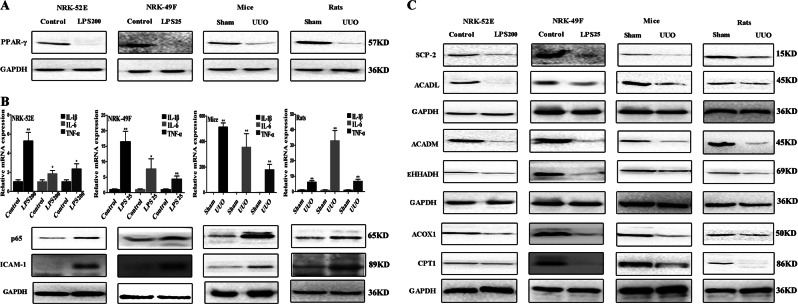
FABP4 adjusts PPARγ to regulate inflammation and fatty acid oxidation
As shown in Fig. 3a, the protein levels of PPARγ in LPS-stimulated cells or UUO animals were significantly reduced compared with control groups or sham groups in vitro and in vivo. As shown in Fig. 3b, the mRNA levels of IL-1β, IL-6, and TNF-α in LPS -stimulated cells or UUO animals were significantly increased, and the protein levels of p65 and ICAM-1 were also significantly increased compared with control groups or sham groups. As shown in Fig. 3c, the expression levels of some proteins associated with fatty acid oxidation including ACADL, ACADM, CPT1, ACOX1, SCP-2, and EHHADH were significantly decreased in LPS-treated cells or UUO animals compared with control groups or sham groups (Details of fold changes and significances of the proteins in western blot assay are shown in Supplemental Fig. S2).
Fig. 3. FABP4 adjusts PPARγ to regulate inflammation in vitro and in vivo.
a The protein levels of PPARγ in vitro and in vivo. b The mRNA levels of IL-1β, IL-6, and TNF-α, and the protein levels of p65 and ICAM-1 in vitro and in vivo. c The protein levels of EHHADH, ACOX1, SCP-2, CPT1, ACADM, and ACADL in vitro and in vivo. Data are presented as the mean ± SD (n = 3). *p < 0.05 and **p < 0.01 compared with control or sham groups
FABP4 siRNA inhibits inflammation and reinforces PPARγ signal in vitro
As shown in Fig. 4a, the expression levels of FABP4 were massively reduced in FABP4 siRNA groups compared with LPS-stimulated cells. Besides, western blotting assay illustrated that the expression levels of FABP4 were markedly decreased in FABP4 siRNA groups compared with LPS-stimulated cells. In contrast, the expression levels of PPARγ in FABP4 siRNA-treated NRK-52E and NRK-49F cells were significantly increased compared with LPS-stimulated cells. In addition, compared with LPS- stimulated cells, FABP4 siRNA suppressed inflammation by decreasing the mRNA levels of IL-1β, IL-6, and TNF-α in NRK-52E and NRK-49F cells, and the protein levels p65 and ICAM-1 were also obviously reduced (Fig. 4b). Besides, the data in Fig. 4c showed that, compared with LPS-stimulated cells, FABP4 inhibition improved fatty acid oxidation via reinforcing PPARγ signal by affecting the protein levels ACADL, ACADM, CPT1, ACOX1, SCP-2, and EHHADH (Details of fold changes and significances of these proteins in western blot assay are shown in Supplemental Fig. S3).
Fig. 4. FABP4 siRNA attenuates fibrosis and lipid metabolism disorders, and reinforces PPARγ signal in cells.
a The protein levels of FABP4 and PPARγ based on immunefluorescence staining (400× magnification) and western blotting assay in FABP4 siRNA transfected NRK-52E and NRK-49F cells. b The mRNA levels of IL-1β, IL-6, and TNF-α, and the protein levels of p65 and ICAM-1 in NRK-52E and NRK-49F cells after FABP4 siRNA transfection. c The protein levels of EHHADH, ACOX1, SCP-2, CPT1, ACADM, and ACADL in NRK-52E and NRK-49F cells after FABP4 siRNA transfection. d The mRNA levels of α-SMA, COL1A1 and COL3A1 in NRK-52E and NRK-49F cells after FABP4 siRNA transfection and the protein levels of α-SMA and COL1A based on immunofluorescence assay (400× magnification) in NRK-52E and NRK-49F cells after FABP4 siRNA transfection. e The levels of TC, TG, and FFAs in NRK-52E and NRK-49F cells after FABP4 siRNA transfection and the lipid droplets based on Oil O Red staining (400× magnification) in NRK-52E and NRK-49F cells after FABP4 siRNA transfection. Data are presented as the mean ± SD (n = 3). **p < 0.01 control groups compared with LPS-groups; ## p < 0.01 and # p < 0.05 LPS groups compared with FABP4 siRNA groups
FABP4 siRNA attenuates fibrosis and lipid metabolism disorders in cells
As shown in Fig. 4d, compared with LPS-stimulated cells, the mRNA levels of α-SMA, COL1A1, and COL3A1 were significantly decreased in FABP4 siRNA groups. Meanwhile, FABP4 siRNA attenuated LPS-induced fibrosis in vitro by reducing the expression levels of α-SMA (green) and COL1A (red). As shown in Fig. 4e, the levels of TC, TG, and FFAs in FABP4 siRNA-treated NRK-52E cells were reduced by 55.27, 33.33, and 42.13%, and decreased by 51.01, 33.41, and 53.65% in NRK-49F cells compared with LPS-stimulated cells. Furthermore, the results of Oil Red O staining showed that the depositions of lipid droplets in FABP4 siRNA-treated cells were obviously reduced compared with LPS-stimulated cells.
FABP4 inhibitor decreases FABP4 expression, reinforces PPARγ signal, and reverses fibrosis and lipid metabolism disorders in cells
As shown in Fig. 5a, the expression levels of FABP4, p65, and ICAM were decreased in FABP4 inhibitor groups, while the expression levels of PPARγ in FABP4 inhibitor-treated NRK-52E and NRK-49F cells were increased compared with LPS stimulated cells. The data in Fig. 5b showed that the expression levels of ACADL, ACADM, CPT1, ACOX1, SCP-2, and EHHADH in NRK-52E and NRK-49F cells were increased by FABP4 inhibitor compared with LPS-stimulated cells. In addition, as shown in Fig. 5c, the mRNA levels of α-SMA, COL1A1 and COL3A1 were significantly decreased in FABP4 inhibitor groups and the expression levels of α-SMA (green) and COL1A (red) were reduced compared with LPS groups (p < 0.05). Moreover, the results of Oil Red O staining in Fig. 5d showed that the depositions of lipid droplets, and the levels of TC, TG, and FFAs in FABP4 inhibitor-treated cells were significantly decreased compared with LPS-stimulated cells (p < 0.05).
Fig. 5. FABP4 inhibitor attenuates fibrosis and lipid metabolism disorders, and reinforces PPARγ signal in cells.
a The protein levels of FABP4, PPARγ, p65, and ICAM based on immunefluorescence staining (400× magnification) and western blotting assay after treatment by FABP4 inhibitor in NRK-52E and NRK-49F cells. b The protein levels of EHHADH, ACOX1, SCP-2, CPT1, ACADM, and ACADL in NRK-52E and NRK-49F cells treated by FABP4 inhibitor. c The protein levels of α-SMA and COL1A based on immunofluorescence assay (400× magnification), and the mRNA levels of α-SMA, COL1A1, and COL3A1 in NRK-52E and NRK-49F cells after treatment by FABP4 inhibitor in NRK-52E and NRK-49F cells. d The lipid droplets based on Oil O Red staining (400× magnification) and the levels of TC, TG, and FFAs in NRK-52E and NRK-49F cells after FABP4 inhibitor treatment. Data are presented as the mean ± SD (n = 3). **p < 0.01 control groups compared with LPS groups, ## p < 0.01 and # p < 0.05 LPS groups compared with FABP4 inhibitor groups
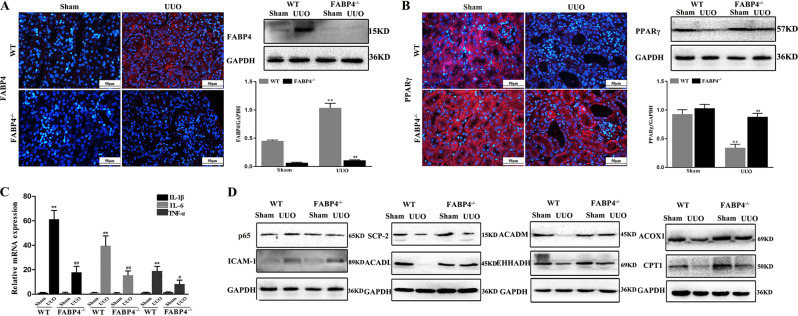
FABP4 KO downregulates FABP4 expression and reinforces PPARγ signal in mice
As expected, the expression level of FABP4 in FABP4 KO mice was not detected (Fig. 6a). As shown in Fig. 6b, the expression level of PPARγ was significantly increased in UUO FABP4 KO mice compared with WT mice. In addition, compared with UUO WT animal, the mRNA levels of IL-1β, IL-6, and TNF-α in UUO FABP4 KO mice were reduced, and the protein levels of p65 and ICAM-1 were also decreased (Fig. 6c, d). In addition, the data in Fig. 6d showed that, compared with UUO WT mice, FABP4 knockdown attenuated lipid metabolism disorders via inhibiting PPARγ signal by affecting the protein levels of ACADL, ACADM, CPT1, ACOX1, SCP-2, and EHHADH in UUO FABP4 KO mice (Details of fold changes and significances of these proteins in Western blot assay are shown in Supplemental Fig. S4).
Fig. 6. FABP4 KO downregulates FABP4 expression and reinforces PPARγ signal in mice.
a The protein levels of FABP4 based on immunofluorescence staining (400× magnification) and western blotting assay in FABP4 KO mice. b The protein levels of PPARγ based on immunofluorescence staining (400× magnification) and western blotting assay in FABP4 KO mice. c The mRNA levels of IL-1β, IL-6, and TNF-α in FABP4 KO mice. d The protein levels of p65, ICAM-1, EHHADH, ACOX1, SCP-2, CPT1, ACADM, and ACADL in FABP4 KO mice. Data are presented as the mean ± SD (n = 3). **p < 0.01 Sham group compared with UUO mice; ## p < 0.01 and # p < 0.05 UUO WT mice compared with UUO FABP4−/−mice
FABP4 KO protects renal function in mice
As shown in Fig. 7a, the levels of Cr (59.712 ± 8.991 μmol/L) and BUN (13.720 ± 1.297 mmol/L) were significantly increased in UUO WT mice. However, in FABP4 KO mice after UUO operation, the levels of Cr (18.975 ± 4.959 μmol/L) and BUN (9.002 ± 1.316 mmol/L) were significantly reduced. The results of H&E staining showed that in UUO FABP4 KO mice, the extent of tubular dilation or atrophy and the infiltration of inflammatory cells were markedly improved compared with UUO WT animal.
Fig. 7. FABP4 KO protects renal function, attenuates RIF, and reverses lipid metabolism disorders in mice.
a The serum levels of Cr, BUN, and H&E staining (400× original magnification) of the kidney tissue in FABP4 KO mice (n = 5). b Degree of fibrosis based on Masson and Sirius Red staining (400× original magnification), and Sirius Red polarized light observation (400× original magnification) in FABP4 KO mice. c The mRNA levels of α-SMA, COL1A1, COL3A1, and the protein levels of α-SMA and COL1A based on immunofluorescence staining (400× magnification) in FABP4 KO mice (n = 3). d The lipid droplets based on Oil O Red staining (400× original magnification), and the levels of TG, TC, and FFA in FABP4 KO mice. Data are presented as the mean ± SD. **p < 0.01 and *p < 0.05 Sham group compared with UUO FABP4 KO mice; ## p < 0.01 and # p < 0.05 UUO WT mice group compared with UUO FABP4−/−mice
FABP4 KO attenuates RIF and lipid metabolism disorders in mice
Compared with UUO WT mice, collagen deposition was reduced and fibrosis degree was significantly improved in UUO FABP4 KO mice (Fig. 7b). Under polarized light observation, the expression levels of COL1A (green) and COL3A (orange red) were heavily decreased in UUO FABP4 KO mice compared with UUO WT mice. At the same time, as shown in Fig. 7c, the mRNA levels of α-SMA, COL1A1, and COL3A1 were significantly decreased in UUO FABP4 KO mice compared with UUO WT mice. In addition, Oil Red O staining observed that the deposition of lipid droplets was significantly reduced in FABP4 KO UUO mice compared with UUO WT mice (Fig. 7d). Furthermore, the levels of TC, TG, and FFAs were also significantly reduced by 18.74, 48.28, and 44.30%.
Discussion
RIF is the pathological basis or pathological feature that leads to chronic renal failure, which is also the best histologic predictor of renal functional decline in CKD22. Therefore, it is necessary to explore effective drug targets for research and development of innovative drugs to treat RIF.
Recently, some works have reported that dyslipidemia associated with lipid metabolism disorder plays an important role in the pathogenesis of RIF23. Lemos et al. have observed that inhibiting Interleukin-1R (IL-1R) signal transducer kinase IRAK4 (Interleukin-1 receptor-associated kinase 4) can abrogate fibrosis and reduce tubular injury24. In our study, the survival rates of NRK-52E and NRK-49F after challenge with LPS were detected and UUO models in mice for 7 days and rats for 4 weeks were established25,26. The results showed that in vivo experiments, compared with sham groups, the levels of serum Cr and BUN in UUO animals were significantly increased (p < 0.05), as well as the histopathological changes. At the same time, the levels of inflammation-related factors including IL-1β, IL-6, and TNF-α were increased in UUO mice and rats. In addition, we also found that the levels of TC, TG, and FFAs were increased, and lipid depositions were obvious in UUO animals. The same results were verified in vitro experiments. Therefore, inflammation and lipid metabolism disorders played important roles in the process of RIF.
Therefore, it is important to find drug targets that can regulate inflammation and lipid metabolism to treat RIF. FABP4, a member of the fatty acid-binding protein family, is involved in lipid metabolism and inflammation27,28. Recent studies have shown that FABP4 inhibitor can reduce lipid-induced ER stress-associated inflammation, ameliorate lipid deposits and suppress ROS and nuclear factor-kappaB (NF-κB) nuclear translocation29. In other hand, FABP4 deficiency can alter adipocyte biology and fatty acid metabolism to regulate systemic insulin resistance, dyslipidemia, and lipotoxicity30. However, the mechanisms of FABP4 in RIF via regulating inflammation and dyslipidemia remain poorly defined.
Some reports have shown that FABPs can correlate with PPARγ, in which FABP4 can specifically connect with PPARγ31. Besides, in FABP4-deficient macrophages, the cholesterol accumulation and alterations in pro-inflammatory responsiveness can be suppressed32. At the same time, the shortage of FABP4 alters lipid composition in macrophages and enhances PPARγ activity, leading to the elevated expression of CD36 and enhanced uptake of modified low density lipoprotein33. In our present study, the results showed that the occurrences of RIF were accompanied with the increased expression of FABP4 and the decreased expression of PPARγ in vivo and in vitro experiments. Then, FABP4 siRNA and inhibitor interfered NRK-52E and NRK-49F cells, and FABP4 KO mice were used. We found that knocking down and inhibition of FABP4 promoted PPARγ expression. These data proved that downregulation of FABP4 increased PPARγ expression in feedback, as a result of reversing RIF.
Inflammation is a driver of RIF. PPARγ can regulate NF-κB activation, and activated PPARγ can be induced by FABP4, and then PPARγ binds to p65 in the nucleus to prevent the activation of NF-κB-RE (Nuclear factor kappa beta response element) during inflammation34,35. In the present study, FABP4 inhibition or knockdown increased PPAR-γ expression, and decreased the protein levels of p65 and ICAM-1 to suppress inflammation. These data indicated that suppression of FABP4 inhibited NF-κB activation to improve inflammation against RIF.
In recent years, a number of animal experiments and clinical studies have shown that lipid metabolism disorders can also promote glomerulosclerosis and tubulointerstitial injury36. Some researchers have reported that FABP4 KO can prevent obesity-induced insulin resistance and reduce the rate of lipolysis with normal diet of mice37. In the present study, we found that FABP4 KO in mice or FABP4 siRNA and inhibitor in cells decreased lipid deposition, and reversed the levels of TC, TG, FFAs. By contrast, the expression levels of the proteins associated with fatty acid oxidation including SCP2, ACADL, ACADM, CPT1, ACOX1, and EHHADH were increased, suggesting that the mechanism of FABP4 in regulating RIF might result from controlling lipid metabolism disorders. However, FABP4 KO is full KO, not specific for kidney tissues. Since the main source of FABP4 is adipose tissue, it is possible that the effects observed in FABP4 KO mice would be consequence of the reduction of FABP4 circulating levels found in the animals, in addition to the lack of FABP4 specifically in kidney.
In conclusion, FABP4 deteriorated RIF via promoting inflammation and lipid metabolism disorders (Fig. 8), which should be considered as one new drug target against RIF.
Fig. 8. Schematic diagram of the mechanisms of FABP4 on RIF.
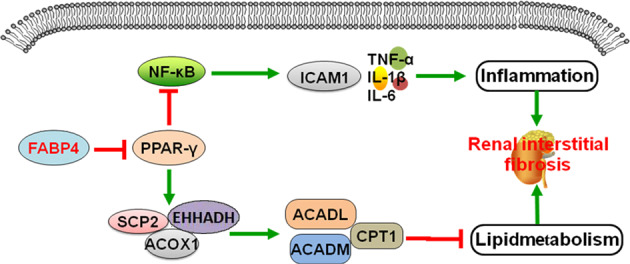
FABP4 downregulated PPAR-γ expression, up-regulated the protein levels of p65 and ICAM-1, and decreased the protein levels of ACADM, ACADL, SCP-2, CPT1, EHHADH, and ACOX1 to promote inflammation and lipid metabolism
Supplementary information
Acknowledgements
This work was supported by the Key Research and Development Project of Liaoning Province (2017225090), the Special Grant for Translational Medicine, Dalian Medical University (2015004), the Basic Scientific Research Projects of Liaoning University (No. LF2017010), the Natural Science Foundation of Liaoning Province (201602223) and the Project of Leading Talents of Dalian, China.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
This article has been retracted. Please see the retraction notice for more detail: https://dx.doi.org/10.1038/s41419-021-04084-x
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yujie Qiao, Liping Liu
Edited by R A Knight
Change history
8/20/2021
A Correction to this paper has been published: 10.1038/s41419-021-04084-x
Supplementary information
Supplementary information accompanies this paper at (10.1038/s41419-019-1610-5).
References
- 1.Gewin LS. Renal fibrosis: primacy of the proximal tubule. Matrix Biol. 2018;68:248–262. doi: 10.1016/j.matbio.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deelman L, Sharma K. Mechanisms of kidney fibrosis and the role of antifibrotic therapies. Curr. Opin. Nephrol. Hypertens. 2009;18:85–90. doi: 10.1097/MNH.0b013e32831c50a1. [DOI] [PubMed] [Google Scholar]
- 3.Howard CR, Stadler K. Albumin-bound fatty acids, but not albumin itself, alter mitochondrial bioenergetics in renal proximal tubular cells. Free Radic. Biol. Med. 2011;51:S66–S66. doi: 10.1016/j.freeradbiomed.2011.10.172. [DOI] [Google Scholar]
- 4.Allison SJ. Fibrosis: dysfunctional fatty acid oxidation in renal fibrosis. Nat. Rev. Nephrol. 2015;11:64. doi: 10.1038/nrneph.2014.244. [DOI] [PubMed] [Google Scholar]
- 5.Kang HM, et al. Defective fatty acid oxidation in renal tubular epithelial cells plays a key role in kidney fibrosis development. Na.t Med. 2015;21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eardley KS, Cockwell P. Macrophages and progressive tubulointerstitial disease. Kidney Int. 2005;68:437–455. doi: 10.1111/j.1523-1755.2005.00422.x. [DOI] [PubMed] [Google Scholar]
- 7.Taniguchi H, et al. Involvement of mcp-1 in tubulointerstitial fibrosis through massive proteinuria in anti-gbm nephritis induced in wky rats. J. Clin. Immunol. 2007;27:409–429. doi: 10.1007/s10875-007-9085-z. [DOI] [PubMed] [Google Scholar]
- 8.Hotamisligil GS, Bernlohr DA. Metabolic functions of FABPs-mechanisms and therapeutic implications. Nat. Rev. Endocrinol. 2015;11:592–605. doi: 10.1038/nrendo.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H, et al. Uncoupling lipid metabolism from inflammation through fatty acid binding protein-dependent expression of UCP2. Mol Cell. Biol. 2015;35:1055–1065. doi: 10.1128/MCB.01122-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grygiel-Górniak B. Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications-a review. Nutr. J. 2014;13:17. doi: 10.1186/1475-2891-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shrestha UK, Xia B. Role of peroxisome proliferator activated receptor-gamma and its ligands in inflammatory bowel disease. J. Adv. Int. Med. 2012;1:33–38. doi: 10.3126/jaim.v1i1.5838. [DOI] [Google Scholar]
- 12.Boss M, Kemmerer M, Brüne B, Namgaladze D. Fabp4 inhibition suppresses pparγ activity and vldl-induced foam cell formation in il-4-polarized human macrophages. Atherosclerosis. 2015;240:424–430. doi: 10.1016/j.atherosclerosis.2015.03.042. [DOI] [PubMed] [Google Scholar]
- 13.Hsiao PJ, et al. Pioglitazone enhances cytosolic lipolysis, β-oxidation and autophagy to ameliorate hepatic steatosis. Sci. Rep. 2017;7:9030. doi: 10.1038/s41598-017-09702-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto T, et al. Transcriptome and metabolome analyses in exogenous FABP4- and FABP5-treated adipose-derived stem cells. PLoS ONE. 2016;11:e0167825. doi: 10.1371/journal.pone.0167825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duffy CM, Xu H, Nixon JP, Bernlohr DA, Butterick TA. Identification of a fatty acid binding protein4-ucp2 axis regulating microglial mediated neuroinfla- mmation. Mol. Cell. Neurosci. 2017;80:52–57. doi: 10.1016/j.mcn.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu LE, et al. Identification of fatty acid bind-ing protein 4 as an adipokine that regulates insulin secretion during obesity. Mol. Metab. 2014;4:465–473. doi: 10.1016/j.molmet.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu B, et al. Fatty acid binding protein-4 (fabp4) is a hypoxia inducible gene that sensitizes mice to liver ischemia/reperfusion injury. J. Hepatol. 2015;63:855–862. doi: 10.1016/j.jhep.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eddy AA, Lopez-Guisa JM, Okamura DM, Yamaguchi I. Investigating mechan- isms of chronic kidney disease in mouse models. Pediatr. Nephrol. 2012;27:1233–1247. doi: 10.1007/s00467-011-1938-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chevalier RL, Forbes MS, Thornhill BA. Ureteral obstruction as a model of renal interstitial fibrosis and obstructive nephropathy. Kidney Int. 2009;75:1145–1152. doi: 10.1038/ki.2009.86. [DOI] [PubMed] [Google Scholar]
- 20.Xu L, et al. MiR-125a-5p ameliorates hepatic glycolipid metabolism disorder in type 2 diabetes mellitus through targeting of STAT3. Theranostics. 2018;8:5593–5609. doi: 10.7150/thno.27425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elie A, et al. Local enrichment of fatty acid-binding protein 4 in the pericardial cavity of cardiovascular disease patients. PLoS ONE. 2018;13:e0206802. doi: 10.1371/journal.pone.0206802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphreys BD. Mechanisms of renal fibrosis. Annu. Rev. Physiol. 2012;7:10–17. doi: 10.1146/annurev-physiol-022516-034227. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, et al. The metabolic syndrome and chronic kidney disease in U.S. adults. Ann. Int. Med. 2004;140:167–174. doi: 10.7326/0003-4819-140-3-200402030-00007. [DOI] [PubMed] [Google Scholar]
- 24.Lemos DR, et al. Interleukin-1β activates a MYC-dependent metabolic switch in kidney stromal cells necessary for progressive tubulointerstitial fibrosis. J. Am. Soc. Nephrol. 2018;29:1690–1705. doi: 10.1681/ASN.2017121283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang C, et al. Chitosan/sirna nanoparticles targeting cyclooxygenase type 2 attenuate unilateral ureteral obstruction-induced kidney injury in mice. Theranostics. 2015;5:110–123. doi: 10.7150/thno.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee ES, et al. Sarpogrelate hydrochloride ameliorates diabetic nephropathy associated with inhibition of macrophage activity and inflammatory reaction in db/db mice. PLoS ONE. 2017;12:e0179221. doi: 10.1371/journal.pone.0179221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong CQ, et al. FABP4 suppresses proliferation and invasion of hepatocellular carcinoma cells and predicts a poor prognosis for hepatocellular carcinoma. Cancer Med. 2018;7:2629–2640. doi: 10.1002/cam4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge XN, et al. FABP4 regulates eosinophil recruitment and activation in allergic airway inflammation. Am. J. Physiol. Lung Cell Mol. Physiol. 2018;315:L227–L240. doi: 10.1152/ajplung.00429.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosquet A, et al. FABP4 inhibitor BMS309403 decreases saturated-fatty-acid- induced endoplasmic reticulum stress-associated inflammation in skeletal muscle by reducing p38 mapk activation. Biochim. Biophys. Acta. 2018;1863:604–613. doi: 10.1016/j.bbalip.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan NS, et al. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol. Cell. Biol. 2002;22:5114–5127. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makowski L, Brittingham KC, Reynolds JM, Suttles J, Hotamisligil GS. The fatty acid-binding protein, aP2, coordinates macrophage cholesterol trafficking and inflammatory activity. Macrophage expression of aP2 impacts peroxisome proliferator-activated receptor gamma and ikappab kinase activitie. J. Biol. Chem. 2005;280:12888–12895. doi: 10.1074/jbc.M413788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosen ED, Spiegelman BM. Ppargamma: a nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 34.Lefterova MI, et al. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen F, et al. Phosphorylation of ppargamma via active erk1/2 leads to its physical association with p65 and inhibition of NF-kappabeta. J. Cell. Biochem. 2003;90:732–744. doi: 10.1002/jcb.10668. [DOI] [PubMed] [Google Scholar]
- 36.Abrass CK. Lipid metabolism and renal disease. Contrib. Nephrol. 2006;151:106. doi: 10.1159/000095323. [DOI] [PubMed] [Google Scholar]
- 37.Uysal KT, Scheja L, Wiesbrock SM, Bonnerweir S, Hotamisligil GS. Improved glucose and lipid metabolism in genetically obese mice lacking aP2. Endocrinology. 2000;141:3388–3396. doi: 10.1210/endo.141.9.7637. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.