Abstract
Radiotherapy is the primary treatment option for nasopharyngeal carcinoma (NPC). Local recurrence and metastasis caused by radioresistance become a bottleneck of curative effect for patients with NPC. Currently, serum predictive biomarkers of radioresistance are scare. We enrolled NPC patients, who underwent radiotherapy in the Department of Oncology, Xiangya Hospital, Central Southern University, and analyzed the serum proteins profiles in NPC patients using with quantitative label-free proteomics using ultra-definition MS. Patients were divided into those who were radioresistant and radiosensitive by the overall reduction (≤50% or >50%, respectively) in tumor extent. The MS/MS spectrum database search identified 911 proteins and 809 proteins are quantitatable. Eight proteins significantly up-regulated and 12 serum proteins were significantly down-regulated in the radioresistance group compared with radiosensitivity group (P<0.05). Finally, five proteins entered the optimal models, including secreted protein acidic and cysteine rich (SPARC) (P =0.032), serpin family D member 1S (ERPIND1) (P =0.040), complement C4B (C4B) (P =0.017), peptidylprolyl Isomerase B (PPIB) (P =0.042), and family with sequence similarity 173 member A (FAM173A) (P =0.017). In all patient, the area under the curves (AUC) for SPARC, SERPIND, C4B, PPIB, and FAM173A were 0.716 (95% CI: 0.574–0.881), 0.697 (95% CI: 0.837–0.858), 0.686 (95% CI: 0.522–0.850), 0.668 (95% CI: 0.502–0.834) and 0.657 (95% CI: 0.512–0.825), respectively. The AUC of five selected proteins was 0.968 (95% CI: 0.918–1.000) with the sensitivity of 0.941 and the specificity of 0.926. Our result indicated that a panel including five serum protein (SPARC SERPIND1 C4B PPIB FAM173A) based on serum proteomics provided a high discrimination ability for radiotherapy effects in NPC patients. Studies with larger sample size and longer follow-up outcome are required.
Keywords: biomarkers, Nasopharyngeal carcinoma, radioresistance, serum proteomics
Introduction
Nasopharyngeal carcinoma (NPC) is an Epstein–Barr (EB) virus-associated epithelial malignancy exhibiting unusual ethnic and geographical distributions [1,2]. NPC is a typical regional disease; most patients live in Southern Asia (particularly Southeast China) [3]. The incidence ranges from 30 to 50 cases per 10,000 of the population [4]. NPC is highly malignant; both lymph node and distant metastases may develop early, without obvious symptoms. Most patients have middle- and late-stage disease when diagnosed [5]. Radiation therapy is the major therapeutic modality used to treat NPC, and most NPC patients can be cured if the disease is diagnosed and treated at an early stage. The 5-year survival rate of patients with stages I–IVB NPC is approximately 85% [6]. However, some patients do not benefit from radiotherapy due to radioresistance caused by local recurrence and distant metastasis [7]. Radio resistance and radiation-induced cell damage are affected by the treatment methods employed and genetic differences in cell cycle regulation, apoptotic/anti-apoptotic mechanisms, and DNA repair pathways [8,9]. However, the details remain unclear. A previous study used a microarray to identify certain NPC radioresistance genes, but the data were not confirmed in other studies [10–12]. It is likely that the data reflect tissue specificities rather than radioresistance mechanisms. New techniques enable the exploration of tumor genetic profiles. To date, the molecular mechanism of radioresistance remains unclear; NPC is caused by many different factors, and few serum biomarkers predictive of radioresistance are known [13]. Besides, the present treatment guidelines have certain limitations, and treatments do not vary by tumor stage or lymph node metastasis status. However, histological staging and molecular subtyping have shown that treatment sensitivity is affected by stage [14,15]. Thus, treatment should be patient specific, and biomarkers of radiotherapeutic sensitivity are urgently required. Finding biomarkers of radiation sensitivity would not only benefit the individual but also reduce healthcare costs. Previous study showed that occurrence and development process of NPC involved many tumor-associated proteins [16]. Protein overexpression led by related gene abnormality in serum cannot only reflect the degree of malignancy but also lead to dynamic changes of protein expression after the excision of primary lesion and radiotherapy, indicating that abnormal expression of certain proteins was associated with NPC treatment and prognosis [17]. Surface-enhanced laser desorption/ionization time-of-flight mass spectrometry renders serum proteomics possible. Here, we analyzed the serum proteins of NPC patients who were either radiosensitive or radioresistant to build a diagnostic panel of prognosis after radiotherapy.
Materials and methods
Patients and blood samples
We enrolled NPC patients who underwent radiotherapy in the Department of Oncology, Xiangya Hospital, Central Southern University, China, from April to June 2018. The selection criteria were pathologically confirmed NPC; no prior radiotherapy, chemotherapy, surgery, adjuvant therapy, or anticarcinoma drug therapy; and stage III or IV disease. Serum samples were collected prior to treatment; all patients were over 18 years of age. The exclusion criteria were any severe infection; any other tumor; any severe liver, kidney, immune system, or hematological disease; and/or any prior cancer treatment. Blood samples were collected into tubes containing EDTA and an antithrombotic agent and stored at –80°C. The study was approved by the ethics committee of Xiangya Hospital, Central Southern University. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki, and that all subjects provided written informed consent.
Clinical data
We extracted clinical data from electronic medical records. We recorded gender, age, cancer type, American Joint Committee on Cancer stage, nasopharyngeal gross tumor volume (GTV), lymph node GTV prior to treatment, lymph node remission rate, overall tumor reduction rate, routine blood data (the numbers of red and white blood cells, neutrophils, and lymphocytes), and the levels of hemoglobin, α-fetoprotein, carcinogenic embryonic antigen, CA125, CA242, C19F, EB virus DNA, EAIgA, and EBVCA IgA. Tumor biomarkers were quantitated using a chemiluminescence method (Immulite; DPC, U.S.A.). The EB virus DNA was quantitated via fluorescence-PCR. Treatment included induction chemotherapy (docetaxel 120–164 m−2 and cisplatin 60–120 m−2 on days 1–2) and concurrent chemotherapy (cisplatin 65–120 m−2 on days 1–2) and radiotherapy (70.4 Gy/2.2 Gy over 32 fractions [PGTVnx], 70.4 Gy/2.2Gy over 32 fractions [PGTVnd], 60.8 Gy/1.9 Gy over 32 fractions [PTV1], and 54 Gy/1.8 Gy over 30 fractions [PTV2]). Radiotherapy sensitivity was then evaluated as the reduction in the maximum cross-sectional area of the primary lesion, calculated as the maximum transverse diameter × the maximum vertical length.
Experimental
The experimental procedures are shown in Figure 1.
Figure 1. Technical route of quantitative serum proteomics in NPC patients.
Protein extraction
Serum samples were collected. Protein concentration was determined using a BCA kit according to the manufacturer’s instructions. First, cellular debris was removed by centrifugation at 12,000 g at 4°C for 10 min and the supernatants transferred to new tubes. High-abundance proteins were removed with the aid of a Pierce Top 12 Abundant Protein Depletion Spin Column Kit (Thermo Fisher Scientific).
Trypsin digestion
Prior to digestion, protein solutions were reduced with 5 mM dithiothreitol for 30 min at 56°C, alkylated with 11 mM iodoacetamide for 15 min at room temperature in the dark, and subjected to ultrafiltration three times. Trypsin (Promega, U.S.A.) was added to a final concentration of 15 ng/μl followed by overnight digestion (37°C), and each peptide solution was centrifuged at 12,000 gat 4°C for 15 min and the peptide concentration then determined using the BCA kit according to the manufacturer’s instructions.
Tandem mass tag labeling
After trypsin digestion, the peptides were desalted on a Strata X C18 SPE column (Phenomenex), vacuum-dried, reconstituted in 0.5 M triethylammonium bicarbonate buffer (TEAB), and processed according to the manufacturer’s protocol. Briefly, one unit of TMT reagent was thawed and reconstituted in acetonitrile. The peptide mixtures were incubated with the reagent for 2 h at room temperature, then pooled, desalted, and dried via vacuum centrifugation.
High performance liquid chromatography fractionation
Tryptic peptides were fractionated by high-pH reverse-phase high-performance liquid chromatography using an Agilent 300 Extend C18 column (5 μm particle diameter, 4.6 mm internal diameter, 250 mm in length). Briefly, peptides were first separated into 60 fractions using a gradient of 8–32% acetonitrile (pH 9.0) over 60 min, then combined into nine fractions, and dried by vacuum centrifugation.
LC-MS/MS
Tryptic peptides were dissolved in 0.1% (v/v) formic acid (solvent A) and directly loaded onto an in-house reverse-phase analytical column (15 cm in length and 75 μm in internal diameter). The gradient featured a rise from 6 to 18% solvent B (0.1% [v/v] formic acid in 98% [v/v] acetonitrile) over 40 min and a further rise from 18 to 28% over 12 min, followed by a rise to 80% (v/v) acetonitrile over 4 min and a hold at 80% (v/v) acetonitrile for the last 4 min, at a constant flow rate of 300 nl/min, using an EASY-nLC 1000 UPLC system.
Database search
MS/MS data were processed using the Maxquant search engine (version 1.5.2.8). Tandem mass spectra were searched against a human database concentrated with a reverse decoy database (https://web.expasy.org/docs/swiss-prot_guideline.html). Trypsin/P was the specified cleavage enzyme; up to two missed cleavages were allowed. The precursor ion mass tolerances were set to 20 and 5 ppm in the first and main searches, respectively, and the mass tolerance to 0.02 Da. Carbamidomethylation of Cys was specified as the fixed modification, and Met oxidation as a variable modification. The false detection rate was adjusted to <1% and the minimum peptide score to >40.
Statistical analysis
Patients were divided into those who were radioresistant and radiosensitive by the overall reduction (≤50% or >50%, respectively) in tumor extent [18]; we compared their clinical data and serum protein expression levels. Data that were normally distributed are expressed as means ± S.D. and categorical data as counts with percentages. All identified proteins were divided into two groups by the median expression level. Univariate logistic regression was used to explore the relationships amongst clinical parameters, protein expression levels, and overall tumor reductions. All proteins were evaluated in terms of their differential diagnostic utilities. We drew receiver operating characteristic curves and calculated the areas under the curves (AUCs), sensitivities, specificities, Youden indices, and accuracies. Proteins expressed at significant levels were subjected to logistic regression. We created a diagnostic protein panel and evaluated the diagnostic utility thereof as follows: 0.5 < AUC < 0.7, low utility; 0.7 ≤ AUC < 0.9, moderate utility; and AUC ≥0.9, high Utility [19]. A P-value <0.05 was considered to reflect significance.
Results
General patient characteristics
We enrolled 44 NPC patients, 29 males (65.9%), and 15 females (34.1%). Their general characteristics are listed in Table 1. The mean patient age was 45.6 ± 11.2 years. Of all patients, 47.7% had stage III and 52.3% stage IV disease. Eight patients exhibited differentiated and 36 exhibited undifferentiated pathologies. The radiosensitive group contained 27 (61.4%) patients and the radioresistant group 17 patients (38.6%). Univariate logistic regression identified no significant between-group differences between any clinical parameter or the overall tumor reduction rate (Table 1 and Supplementary Material 1).
Table 1. Clinicopathological parameters of included patient with NPC.
| Parameters | Number (%) |
|---|---|
| Age | |
| <40 | 20 (45.5%) |
| ≥40 | 24 (54.5%) |
| Gender | |
| Male | 29 (65.9%) |
| Female | 15 (34.1%) |
| TNM stage | |
| III | 21 (47.7%) |
| IV | 23 (52.3%) |
| Pathological type | |
| Differentiated | 8 (18.2%) |
| Undifferentiated | 36 (81.8%) |
| Prior treatment | |
| Nasopharynx GTVnx | 41.9 ± 25.6 |
| Lymph GTVnx | 23.9 ± 38.4 |
| Lymph reduction rate | 0.57 ± 0.29 |
| >0.50 | 26 (59.1%) |
| ≤0.5 | 18 (40.9%) |
| Overall reduction rate (%) | 0.57 ± 0.23 |
| >0.50 | 27 (61.4%) |
| ≤0.5 | 17 (38.6%) |
Differential protein screening
The MS/MS spectral search identified 911 proteins, of which 809 were quantitatable (Supplementary Material 2). Proteins were divided into those expressed at high and low levels by reference to the median expression level. Univariate logistic regression identified 20 candidate predictive proteins (Table 2). The serum expression levels of ‘secreted protein acidic and cysteine-rich’ (SPARC), ‘serpin family D member 1’ (SERPIND1), ‘complement C4B’ (C4B), ‘pro-platelet basic protein’ (PPBP), ‘podocalyxin-like protein’ (PODXL), ‘serglycin’ (SRGN), ‘peptidylprolyl isomerase B’ (PPIB), ‘S100 calcium-binding Protein A4’ (S100A4), and ‘cathepsin F’ (CTSF) were significantly higher in the radioresistant than in the radiosensitive group (all P<0.05). The ‘endoplasmic reticulum aminopeptidase 1’ (ERAP1), ‘vitamin D-binding protein’ (GC), ‘inter-α-trypsin inhibitor heavy chain 1’ (ITIH1), ‘neuropilin 1’ (NRP1 or C1R), ‘multiple inositol-polyphosphate phosphatase 1’ (MINPP1), ‘coagulation factor XIII A chain’ (F13A1), ‘complement C1qB chain’ (C1QB), ‘inter-α-trypsin inhibitor heavy chain 2’ (ITIH2), ‘insulin-like growth factor binding protein 6’ (IGFBP6), and ‘family with sequence similarity 173 member A’ (FAM173A) proteins were significantly down-regulated in the radioresistant compared with the radiosensitive group (all P<0.05). We subjected the levels of the 20 candidate proteins to stepwise multivariate logistic regression. Finally, five proteins were used to create the optimal model (Table 3): SPARC (P=0.032), SERPIND1 (P=0.040), C4B (P=0.017), PPIB (P=0.042), and FAM173A (P=0.017).
Table 2. Univariate logistic regression of groups of proteins for overall reduction rate.
| Protein | Expression level | β | SE | Waldχ2 | P | OR | 95% CI | |
|---|---|---|---|---|---|---|---|---|
| SPARC | Up-regulation | 1.872 | 0.703 | 7.098 | 0.008 | 6.50 | 1.64 | 25.76 |
| ERAP1 | Down-regulation | −1.741 | 0.679 | 6.572 | 0.010 | 0.18 | 0.05 | 0.66 |
| SERPIND1 | Up-regulation | 1.709 | 0.697 | 6.015 | 0.014 | 5.53 | 1.41 | 21.66 |
| GC | Down-regulation | −1.569 | 0.671 | 5.468 | 0.019 | 0.21 | 0.06 | 0.78 |
| C4B | Up-regulation | 1.569 | 0.671 | 5.468 | 0.019 | 4.80 | 1.29 | 17.88 |
| ITIH1 | Down-regulation | −1.569 | 0.671 | 5.468 | 0.019 | 0.21 | 0.06 | 0.78 |
| PPBP | Up-regulation | 1.553 | 0.693 | 5.023 | 0.025 | 4.73 | 1.22 | 18.39 |
| PODXL | Up-regulation | 1.406 | 0.665 | 4.472 | 0.035 | 4.08 | 1.11 | 15.02 |
| NRP1 | Down-regulation | −1.406 | 0.665 | 4.472 | 0.035 | 0.25 | 0.07 | 0.90 |
| C1R | Down-regulation | −1.406 | 0.665 | 4.472 | 0.035 | 0.25 | 0.07 | 0.90 |
| SRGN | Up-regulation | 1.406 | 0.665 | 4.472 | 0.035 | 4.08 | 1.11 | 15.02 |
| PPIB | Up-regulation | 1.406 | 0.665 | 4.472 | 0.035 | 4.08 | 1.11 | 15.02 |
| CTSF | Up-regulation | 1.406 | 0.665 | 4.472 | 0.035 | 4.08 | 1.11 | 15.02 |
| MINPP1 | Down-regulation | −1.406 | 0.665 | 4.472 | 0.035 | 0.25 | 0.07 | 0.90 |
| F13A1 | Down-regulation | −1.299 | 0.651 | 3.979 | 0.046 | 0.27 | 0.08 | 0.98 |
| C1QB | Down-regulation | −1.299 | 0.651 | 3.979 | 0.046 | 0.27 | 0.08 | 0.98 |
| ITIH2 | Down-regulation | −1.299 | 0.651 | 3.979 | 0.046 | 0.27 | 0.08 | 0.98 |
| IGFBP6 | Down-regulation | −1.299 | 0.651 | 3.979 | 0.046 | 0.27 | 0.08 | 0.98 |
| S100A4 | Up-regulation | 1.299 | 0.651 | 3.979 | 0.046 | 3.67 | 1.02 | 13.14 |
| FAM173A | Down-regulation | −1.299 | 0.651 | 3.979 | 0.046 | 0.27 | 0.08 | 0.98 |
CTSF, cathepsin F; S100A4, S100 calcium binding protein A4.
Table 3. Multivariate logistic regression by stepwise for overall reduction rate.
| Gene | β | SE | Wald χ2 | P | OR | 95% CI | |
|---|---|---|---|---|---|---|---|
| SPARC | 3.025 | 1.406 | 4.627 | 0.032 | 20.6 | 1.308 | 324.4 |
| SERPIND1 | 2.582 | 1.258 | 4.215 | 0.040 | 13.22 | 1.124 | 155.4 |
| C4B | 3.723 | 1.561 | 5.692 | 0.017 | 41.39 | 1.943 | 881.7 |
| PPIB | 3.103 | 1.523 | 4.150 | 0.042 | 22.27 | 1.125 | 440.8 |
| FAM173A | −4.248 | 1.778 | 5.707 | 0.017 | 0.014 | <0.001 | 0.466 |
| Intercept | −4.798 | 1.574 | 9.291 | 0.002 | |||
PPIB, peptidylprolyl isomerase B.
The NPC diagnostic utilities of serum proteins
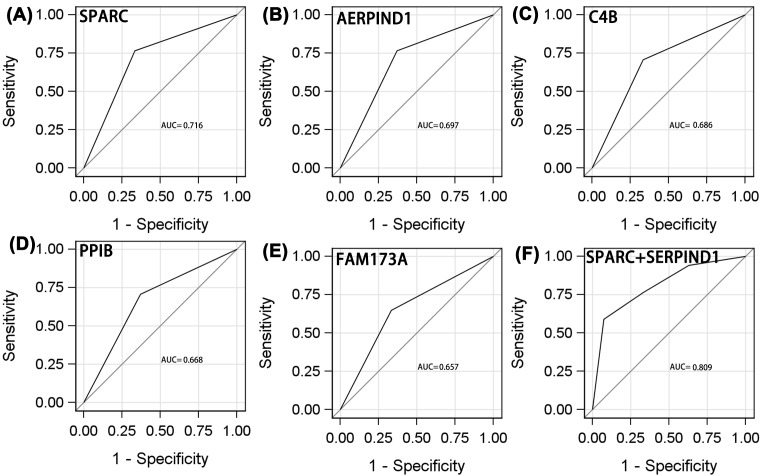
We assessed the differential diagnostic utilities of each protein and combinations of two, three, four, and five proteins with the largest AUCs. For all patients, the AUCs for SPARC, SERPIND, C4B, PPIB, and FAM173A were 0.716 (95% CI: 0.574–0.881), 0.697 (95% CI: 0.837–0.858), 0.686 (95% CI: 0.522–0.850), 0.668 (95% CI: 0.502–0.834), and 0.657 (95% CI: 0.512–0.825), respectively (Figure 2A–E and Table 4). The respective sensitivities and specificities were 0.765 and 0.667, 0.765 and 0.630, 0.706 and 0.667, 0.706 and 0.630, and 0.647 and 0.667. Of the individual proteins, SPARC exhibited the highest diagnostic utility.
Figure 2. ROC analyses of protein to distinguish adverse efficacy from NPC patients who receive radiotherapy.
((A) SPARC; (B) AERPIND1; (C) C4B; (D) PPIB; (E) FAM173A; (F) SPARC+ SERPIND1).
Table 4. Diagnostic sensitivity and specificity for groups of serum proteins in NPC efficacy.
| Variables in Model | Sensitivity | Specificity | Youden index | AUC | Correct rate |
|---|---|---|---|---|---|
| SPARC | 0.765 | 0.667 | 0.431 | 0.716 | 0.705 |
| SERPIND1 | 0.765 | 0.630 | 0.394 | 0.697 | 0.681 |
| C4B | 0.706 | 0.667 | 0.372 | 0.686 | 0.681 |
| PPIB | 0.706 | 0.630 | 0.336 | 0.668 | 0.659 |
| FAM173A | 0.647 | 0.667 | 0.313 | 0.657 | 0.659 |
| SPARC SERPIND1 | 0.588 | 0.926 | 0.514 | 0.809 | 0.795 |
| SPARC FAM173A | 0.765 | 0.667 | 0.431 | 0.804 | 0.727 |
| SERPIND1 C4B | 0.529 | 0.926 | 0.455 | 0.792 | 0.772 |
| SPARC C4B | 0.765 | 0.667 | 0.431 | 0.794 | 0.750 |
| SERPIND1 PPIB | 0.765 | 0.630 | 0.394 | 0.785 | 0.727 |
| C4B FAM173A | 0.412 | 0.963 | 0.374 | 0.777 | 0.750 |
| C4B PPIB | 0.787 | 0.588 | 0.588 | 0.769 | 0.841 |
| SPARC PPIB | 0.418 | 0.765 | 0.431 | 0.770 | 0.705 |
| PPIB FAM173A | 1.00 | 0.407 | 0.407 | 0.754 | 0.681 |
| SERPIND1 FAM173A | 0.765 | 0.630 | 0.394 | 0.761 | 0.704 |
| SPARC C4B FAM173A | 0.882 | 0.778 | 0.660 | 0.878 | 0.818 |
| SERPIND1 C4B PPIB | 0.765 | 0.815 | 0.579 | 0.862 | 0.795 |
| C4B PPIB FAM173A | 0.706 | 0.963 | 0.669 | 0.842 | 0.864 |
| SPARC SERPIND1 C4B | 0.941 | 0.741 | 0.682 | 0.883 | 0.818 |
| SPARC SERPIND1 FAM173A | 0.882 | 0.778 | 0.660 | 0.877 | 0.818 |
| SERPIND1 C4B FAM173A | 0.824 | 0.741 | 0.564 | 0.852 | 0.795 |
| SPARC SERPIND1 PPIB | 0.882 | 0.778 | 0.660 | 0.867 | 0.818 |
| SPARC PPIB FAM173A | 0.941 | 0.704 | 0.645 | 0.795 | 0.862 |
| SERPIND1 PPIB FAM173A | 0.941 | 0.704 | 0.645 | 0.795 | 0.855 |
| SPARC C4B PPIB | 0.647 | 0.889 | 0.535 | 0.815 | 0.795 |
| SERPIND1 C4B PPIB FAM173A | 0.706 | 0.963 | 0.669 | 0.914 | 0.863 |
| SPARC SERPIND1 C4B FAM173A | 0.824 | 0.963 | 0.786 | 0.928 | 0.909 |
| SPARC C4B PPIB FAM173A | 0.882 | 0.889 | 0.771 | 0.939 | 0.886 |
| SPARC SERPIND1 C4B PPIB | 0.941 | 0.852 | 0.793 | 0.886 | 0.902 |
| SPARC SERPIND1 PPIB FAM173A | 0.824 | 0.926 | 0.749 | 0.886 | 0.922 |
| SPARC SERPIND1 C4B PPIB FAM173A | 0.941 | 0.926 | 0.867 | 0.968 | 0.932 |
PPIB, peptidylprolyl isomerase B.
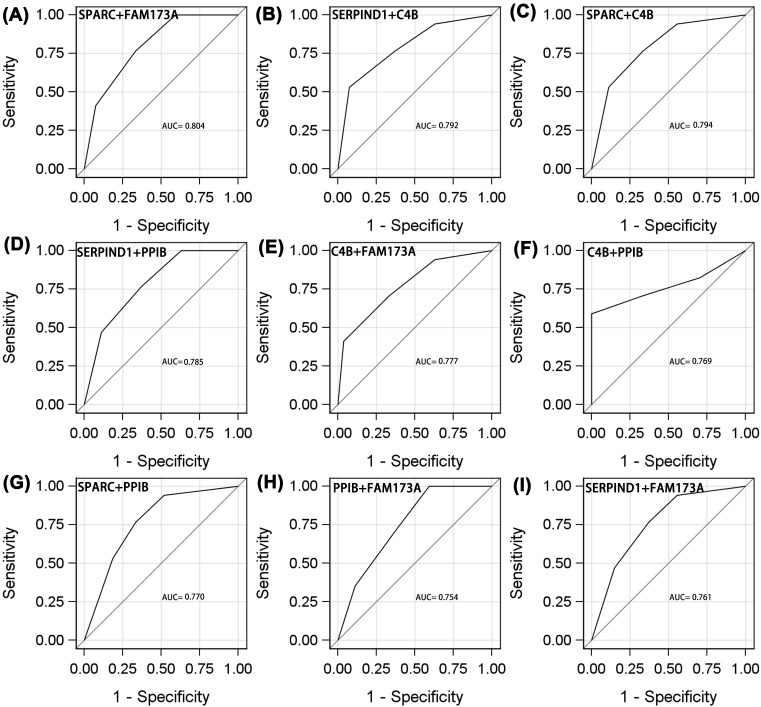
Of the combinations of two proteins (Figures 2F and 3A–I), SPARC/SERIND1 exhibited the highest AUC (0.809, 95% CI: 0.674–0.945), sensitivity (0.588), and specificity (0.926). PPIB/FAM173A was of lower diagnostic utility (AUC = 0.754, 95% CI: 0.613–0.894, sensitivity = 1.00, specificity = 0.407).
Figure 3. ROC analyses of protein to distinguish adverse efficacy from NPC patients who receive radiotherapy.
((A) SPARC+FAM173A; (B) SERPIND1+C4B; (C) SPARC+C4B; (D) SERPIND1+PPIB; (E) C4B+FAM173A; (F) C4B+PPIB; (G) SPARC+PPIB; (H) PPIB+FAM173A; (I) SERPIND1+FAM173A).
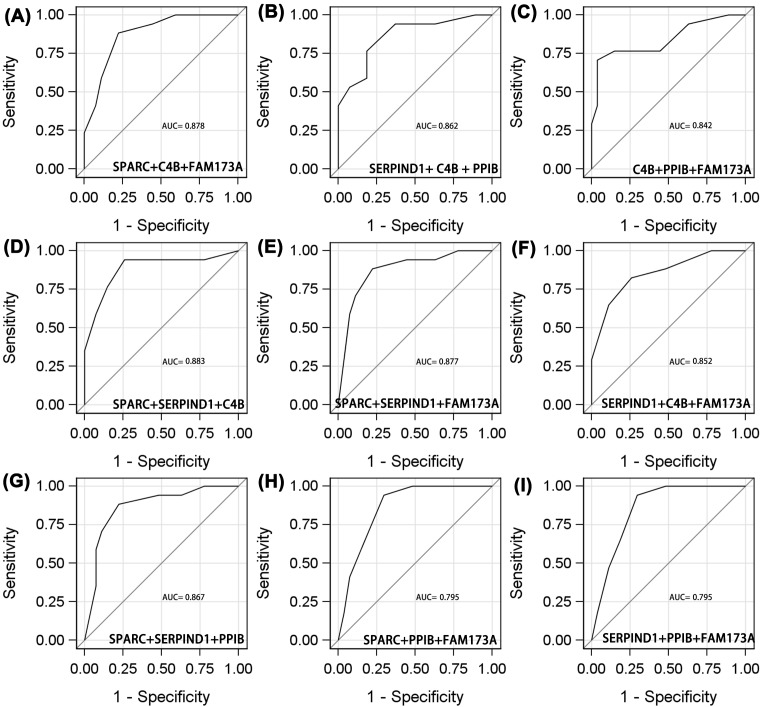
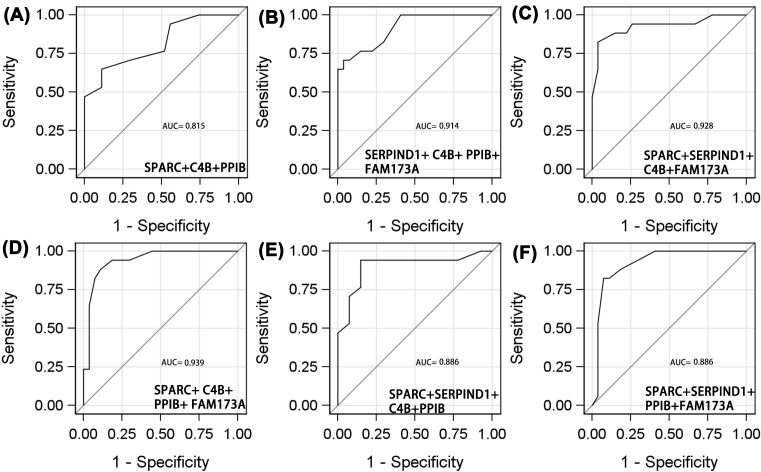
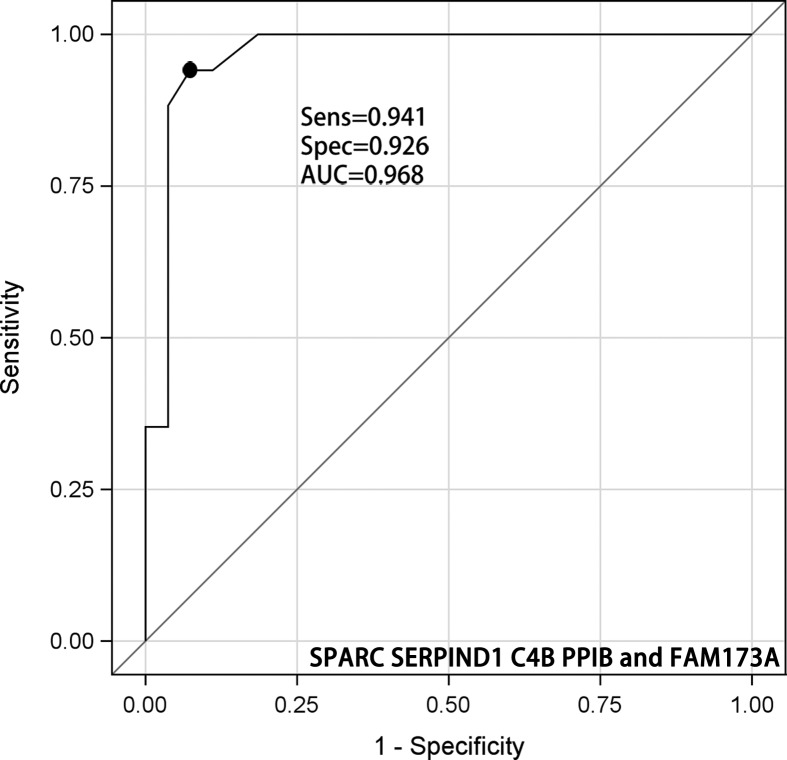
Of the three-protein combinations (Figure 4A–I), the diagnostic utility of SPARC/C4B/FAM173A was the same as those of SPARC/SERPIND1/FAM173A and SPARC/SERPIND1/PPIB. The AUCs, sensitivities, and specificities were 0.878, 0.882, and 0.778; 0.877, 0.882, and 0.778; and 0.867, 0.882, and 0.778, respectively. Of the four-protein combinations (Figure 5A–F), SPARC/C4B/PPIB/FAM173A was optimal, with an AUC of 0.939 (95% CI: 0.876–1.000), sensitivity 0.882, and specificity 0.889. The five-protein AUC was 0.968 (95% CI: 0.918–1.000), sensitivity 0.941, and specificity 0.926 (Figure 6). The diagnostic utilities of other protein combinations are shown in Table 4 and Figures 3–5.
Figure 4. ROC analyses of protein to distinguish adverse efficacy from NPC patients who receive radiotherapy.
((A) SPARC+C4B+FAM173A; (B) SERPIND1+ C4B + PPIB; (C) C4B+PPIB+FAM173A; (D) SPARC+SERPIND1+C4B; (E) SPARC+SERPIND1+FAM173A; (F) SERPIND1+C4B+FAM173A; (G) SPARC+SERPIND1+PPIB; (H) SPARC+PPIB+FAM173A; (I) SERPIND1+PPIB+FAM173A).
Figure 5. ROC analyses of protein to distinguish adverse efficacy from NPC patients who receive radiotherapy.
(A) SPARC+C4B+PPIB; (B) SERPIND1+ C4B+ PPIB+ FAM173A; (C) SPARC+SERPIND1+C4B+FAM173A; (D) SPARC+ C4B+ PPIB+ FAM173A; (E) SPARC+SERPIND1+C4B+PPIB; (F) SPARC+SERPIND1+PPIB+FAM173A.
Figure 6. ROC curve for selected model including SPARC SERPIND1 C4B PPIB and FAM173A.
Discussion
In the present study, we identified 874 proteins, 20 of which were differentially expressed in radioresistant and radiosensitive NPC patients. We used a SPARC/SERPIND1/C4B/ PPIB/FAM173A panel to predict the response of NPC to radiotherapy. The panel afforded a sensitivity of 0.941 and a specificity of 0.926 with an AUC of 0.968 and thus effective. To our knowledge, this is the first study to explore the differential diagnostic utilities of serum proteins in terms of NPC radiosensitivity.
NPC is more sensitive to ionizing radiation than other head-and-neck cancers. Radiotherapy is the primary treatment option. Recently, improvements in radiotherapy have enhanced the survival of even patients with advanced disease [20]. However, some NPC patients do not respond to radiotherapy. Local recurrence and distant metastasis attributable to radioresistance impede treatment. Serum biomarkers of NPC radioresistance would be useful in predicting outcomes, facilitating exploration of the underlying molecular mechanisms, and potentially identifying novel therapeutic targets. Many studies have explored the pathogenesis of NPC radioresistance. However, no study has explored changes in serum protein levels. The present results had some differences from previous reports in other cancers [21,22]. Several reasons may explain the differences: (1) different diseases: Li performed analysis in esophageal cancer, Skiöld performed in breast cancer; (2) samples size difference: Li had ten samples, Skiöld had eight radiation-sensitive and nine normo-sensitive, and we have 44 samples; and (3) different definition: Skiöld compared cancer patients after chemoradiotherapy treatment with the healthy control group. Both Li and our study focussed on radiotherapy sensitivity in cancer patients. Li defined radiotherapy sensitivity according to the radiotherapy oncology group. Our study refers to the overall remission rate of tumor volume.
Inflammation is a critical part of a broad biological response that is initiated after cell injury due to infection or sterile damage, such as cell death. These processes are activated by the immune system to terminate or neutralize destructive stimuli and initiate regeneration. Growing evidence has suggested that IR modulates the immune system through up-regulation of inflammatory mediators [23]. The present study also found some differences proteins related to immune system and inflammation such as C1QB, C4B, ITIH1, ITIH2, SERPIND1. These proteins were also reported by Jeonek et al. who explored the changes of the proteome and metabolome levels in the blood of cancer patients treated with radiotherapy [21]. The overall inflammatory factors were increased for cancer patients who received radiotherapy. The equilibrium between pro and anti-inflammatory cytokines just prior to radiotherapy is important, as this status may affect the rumor resistance to radiotherapy [24]. Moreover, radiotherapy also induces pro-inflammatory mechanism in tumor and normal cells after receiving sub-lethal does, primarily mediated through some transcription factors, which was related to carcinogenesis, inflammation and radio resistance [25]. Usually, most inflammatory factors are up-regulated after radiotherapy, such as SERPIND1 and C4B in the present study. However, some inflammatory and immune-related protein were down-regulated such as ITIH1, ITIH2, C1QB. These differential proteins indicated that blood biomarkers are extensive in monitoring the response to radiotherapy.
We found that the expression levels of SPARC, SERPIND1, C4B, PPIB, and FAM173A were significantly higher in radioresistant than in radiosensitive patients. SPARC encodes a cysteine-rich acidic matrix-associated protein located principally in the extracellular space and nucleus. The protein is involved in extracellular matrix synthesis and changes in cell shape. Some studies indicated that SPARC was a tumor suppressor, but others found that the protein gene was associated with tumor metastasis and invasion by changing the cell shape. Three transcriptional variants of the gene encode different protein isoforms. SPARC expression was positively correlated with NPC radioresistance and may affect NPC cell proliferation and growth. SERPIND1 expression was also associated with radioresistance. SERPIND1 belongs to the serpin gene superfamily. The protein is located both extracellularly and in the endoplasmic reticulum. SERPIND1 was reported to be a thrombin inhibitor that interacts with heparin [26]. To date, the role played by SERPIND1 in the context of cancer development remains largely unknown, particularly in NPC. Zhu et al. reported that SERPIND1 was a potential oncogene, triggering ovarian cancer, but the molecular mechanism involved was unclear [27]. High-level SERPIND1 expression was associated with poor prognosis and recurrence in patients with non-small cell lung cancer (NSCLC). The gene promoted the activity of, invasion by, and filopodial dynamics within NSCLC cells by activating the PI3K signaling pathway [28]. High-level serum SERPIND1 expression was observed in patients with B-cell acute lymphoblastic leukemia [29]. In our present study, SERPIND1 was up-regulated in the serum of radioresistant NPC patients. This finding is novel, suggesting a mechanism of radioresistance in some NPC patients, and perhaps serving as a very relevant marker of radioresistance. Further research is required.
The C4B gene lies in the major histocompatibility complex class III region of chromosome 6. The encoded protein locates to the plasma membrane and the extracellular matrix, and (to a minor extent) the nucleus. C4B is a component of the classical activation pathway, and is expressed in a precursor form that is then proteolytically cleaved, prior to secretion, into a trimer composed of α, β, and γ chains. Hepatitis B virus X protein up-regulated C4b-binding protein α synthesis via activation of transcription factor Sp1 to protect hepatoma cells from attacks by complement [30]. The level of the C4B α-chain was significantly higher in patients with pancreatic ductal adenocarcinoma than in healthy controls. The C4B level was of moderate diagnostic utility in terms of pancreatic ductal adenocarcinoma diagnosis (AUC = 0.860), serving as a novel serum biomarker for detecting early stage disease [31]. In the present study, C4B was of moderate utility in terms of diagnosing NPC radioresistance (AUC = 0.686).
PPIB, also termed cyclophilin B, is located principally in the endoplasmic reticulum, and is a cyclosporine-binding protein regulating cyclosporine A-mediated immunosuppression. Recently, it was shown that cyclophilin B induced chemoresistance, interacting with MDM2 to degrade wild-type p53 in colorectal cancer patients [32]. Paul reported that PPIB expression was associated with radioresistance in head-and-neck squamous cell carcinoma, affecting outcomes after radiotherapy. Inhibition of PPIB expression by siRNA increased the radiosensitivity of cancer cells and inhibited DNA repair [33]. We confirmed that these findings using NPC serum samples; the gene may be a useful radiotherapeutic target. FAM173A is located principally in the cytosol but has been little studied. Serum levels of FAM173A were down-regulated in NPC patients; thus, it was negatively associated with radioresistance in NPC patients. FAM173A is found throughout the cytoplasm, and radiotherapy may affect protein synthesis. Further study is needed.
We created a panel of five serum proteins predicting the short-term efficacy of radiotherapy (radiosensitivity) in NPC patients. Compared with other materials, such as miRNAs and lncRNAs, serum proteins are easy to obtain and handle, and our panel, can be simply applied. However, several limitations of the work should be addressed. Liu et al. also identified some serum proteins related with radiosensitive [34]. The present identified proteins were different previous study. The main reasons may be that there are some differences between our study and this article. In our study, patients were divided into those who were radioresistant and radiosensitive by the overall remission (≤50% or >50%, respectively) in tumor extent. We tend to focus on recent effect. The published paper focussed on recurrence and complete remission. The definition of radioresistance may vary if long-term efficacy is considered; more work is needed. Our sample size was small due to the expense of serum proteomics. A larger sample size is required to confirm our findings. Additionally, unexplored factors potentially affecting the results may have been in play.
In summary, we used proteomics to create a panel of five serum proteins (SPARC, SERPIND1, C4B, PPIB, and FAM173A) predicting the radiotherapeutic response in NPC patients. The panel was highly discriminatory. Future studies should confirm our findings in larger patient numbers with longer follow-up, and further explore the mechanism of induced radioresistance.
Supporting information
Supplementary Material 1.
Supplementary Material 2. Univariate logistic regression of clinical parameters for overall reduction rate.
Acknowledgments
We thanks for the help from Department of Oncology, Xiangya Hospital, Central South University.
Abbreviations
- AUC
area under the curve
- C4B
complement C4B
- C1QB
complement C1q B chain
- ERAP1
endoplasmic reticulum aminopeptidase 1
- F13A1
coagulation factor XIII A chain
- FAM173A
family with sequence similarity 173 member A
- GC
vitamin D binding protein
- GTV
gross tumor volume
- IGFBP6
insulin like growth factor binding protein 6
- ITIH1
inter-α-trypsin inhibitor heavy chain 1
- ITIH2
inter-α-trypsin inhibitor heavy chain 2
- MINPP1
multiple inositol-polyphosphate phosphatase 1
- NPC
nasopharyngeal carcinoma
- NRP1
neuropilin 1
- NSCLC
non-small cell lung cancer
- PODXL
podocalyxin like
- PPBP
pro-platelet basic protein
- PPIB
peptidylprolyl isomerase B
- SERPIND1
serpin family D member 1
- SPARC
secreted protein acidic and cysteine rich
- SRGN
serglycin
Funding
The present study was supported by the Youth Foundation of Xiangya Hospital Central South University [grant number 2018Q012], China Postdoctoral Science Foundation [grant number 2017M612597], Hunan Department of Science and Technology Foundation China [grant number 2015JC3026] and National Key Clinical Program (Department of Oncology, Xiangya Hospital Central South).
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
ZGY and SLF participated in this study design. LZZ wrote the manuscript and analyzed the data. ZQ and LN provided the blood samples. ZK and LC collected the clinical data. LYY revised the manuscript. All authors approved the final version manuscript.
References
- 1.Han B.L., Xu X.Y., Zhang C.Z., Wu J.J., Han C.F., Wang H.. et al. (2012) Systematic review on Epstein-Barr virus (EBV) DNA in diagnosis of nasopharyngeal carcinoma in Asian populations. Asian Pac. J. Cancer Prev. 13, 2577–2581 10.7314/APJCP.2012.13.6.2577 [DOI] [PubMed] [Google Scholar]
- 2.Fu J., Li Z. and Li N. (2018) The association between COX-2 gene rs5275 polymorphism and nasopharyngeal carcinoma risk. Pathol. Res. Pract. 10.1016/j.prp.2018.07.028 [DOI] [PubMed] [Google Scholar]
- 3.Chang E.T. and Adami H.O. (2006) The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol. Biomarkers Prev. 15, 1765–1777 10.1158/1055-9965.EPI-06-0353 [DOI] [PubMed] [Google Scholar]
- 4.Chua M., Wee J., Hui E.P. and Chan A. (2016) Nasopharyngeal carcinoma. Lancet 387, 1012–1024 10.1016/S0140-6736(15)00055-0 [DOI] [PubMed] [Google Scholar]
- 5.Lee A.W., Ma B.B., Ng W.T. and Chan A.T. (2015) Management of nasopharyngeal carcinoma: current practice and future perspective. J. Clin. Oncol. 33, 3356–3364 10.1200/JCO.2015.60.9347 [DOI] [PubMed] [Google Scholar]
- 6.Su S.F., Han F., Zhao C., Chen C.Y., Xiao W.W., Li J.X.. et al. (2012) Long-term outcomes of early-stage nasopharyngeal carcinoma patients treated with intensity-modulated radiotherapy alone. Int. J. Radiat. Oncol. Biol. Phys. 82, 327–333 10.1016/j.ijrobp.2010.09.011 [DOI] [PubMed] [Google Scholar]
- 7.Chen Z.T., Liang Z.G. and Zhu X.D. (2015) A review: proteomics in nasopharyngeal carcinoma. Int. J. Mol. Sci. 16, 15497–15530 10.3390/ijms160715497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu C., Liang Z., Huang J., Zhao R. and Su C. Wang S.. et al. (2012) MiR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle 11, 785–796 10.4161/cc.11.4.19228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Q., He Y., Zhao Y., Wang Y., Kuang W. and Shen L. (2017) A study of 358 cases of locally advanced nasopharyngeal carcinoma receiving intensity-modulated radiation therapy: improving the seventh edition of the American Joint Committee on Cancer T-Staging System. Biomed. Res. Int 2017, 1419676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogawa K., Utsunomiya T., Mimori K., Tanaka F., Haraguchi N., Inoue H.. et al. (2006) Differential gene expression profiles of radioresistant pancreatic cancer cell lines established by fractionated irradiation. Int. J. Oncol. 28, 705–713 [PubMed] [Google Scholar]
- 11.Fukuda K., Sakakura C., Miyagawa K., Kuriu Y., Kin S., Nakase Y.. et al. (2004) Differential gene expression profiles of radioresistant oesophageal cancer cell lines established by continuous fractionated irradiation. Br. J. Cancer 91, 1543–1550 10.1038/sj.bjc.6602187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang J.T., Chan S.H., Lin C.Y., Lin T.Y., Wang H.M., Liao C.T.. et al. (2007) Differentially expressed genes in radioresistant nasopharyngeal cancer cells: gp96 and GDF15. Mol. Cancer Ther. 6, 2271–2279 10.1158/1535-7163.MCT-06-0801 [DOI] [PubMed] [Google Scholar]
- 13.Ahmad P., Sana J., Slavik M., Slampa P., Smilek P. and Slaby O. (2017) MicroRNAs Involvement in Radioresistance of Head and Neck Cancer. Dis. Markers 2017, 8245345 10.1155/2017/8245345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q., Yin Z., Wang T., Chen L. and Li Z. (2018) Gemcitabine and cisplatin regimen facilitates prognosis of advanced nasopharyngeal carcinoma. Cancer Med. 7, 2985–2992 10.1002/cam4.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z., Li Y., Yan S., Fu J., Zhou Q., Huang X.. et al. (2017) Nimotuzumab combined with concurrent chemoradiotherapy benefits patients with advanced nasopharyngeal carcinoma. Onco. Targets Ther. 10, 5445–5458 10.2147/OTT.S141538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu X.D., Su F., Liang Z.G., Li L., Qu S., Liang X.. et al. (2014) Identification of patients with nasopharyngeal carcinoma by serum protein profiling using surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. Int. J. Clin. Oncol. 19, 579–585 10.1007/s10147-013-0621-y [DOI] [PubMed] [Google Scholar]
- 17.Ho D.W., Yang Z.F., Wong B.Y., Kwong D.L., Sham J.S., Wei W.I.. et al. (2006) Surface-enhanced laser desorption/ionization time-of-flight mass spectrometry serum protein profiling to identify nasopharyngeal carcinoma. Cancer 107, 99–107 [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y., Shen L., Huang X., Jing D., Huang D., Fu J.. et al. (2017) High expression of Ki-67 acts a poor prognosis indicator in locally advanced nasopharyngeal carcinoma. Biochem. Biophys. Res. Commun. 494, 390–396 10.1016/j.bbrc.2017.09.118 [DOI] [PubMed] [Google Scholar]
- 19.Fu J., Li Y., Li N. and Li Z. (2018) Comprehensive analysis of clinical utility of three-dimensional ultrasound for benign and malignant breast masses. Cancer Manag. Res. 10, 3295–3303 10.2147/CMAR.S176494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Zhao J., Zhao Y., Yang Z., Lei M., Li Z.. et al. (2018) Impact of paranasal sinus invasion on advanced nasopharyngeal carcinoma treated with intensity-modulated radiation therapy: the validity of advanced T stage of AJCC/UICC eighth edition staging system. Cancer Med. 7, 2826–2836 10.1002/cam4.1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skiold S., Azimzadeh O., Merl-Pham J., Naslund I., Wersall P., Lidbrink E.. et al. (2015) Unique proteomic signature for radiation sensitive patients; a comparative study between normo-sensitive and radiation sensitive breast cancer patients. Mutat. Res. 776, 128–135 10.1016/j.mrfmmm.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 22.Li C., Xia G., Jianqing Z., Mei Y., Ge B. and Li Z. (2014) Serum differential protein identification of Xinjiang Kazakh esophageal cancer patients based on the two-dimensional liquid-phase chromatography and LTQ MS. Mol. Biol. Rep. 41, 2893–2905 10.1007/s11033-014-3145-2 [DOI] [PubMed] [Google Scholar]
- 23.Di Maggio F.M., Minafra L., Forte G.I., Cammarata F.P., Lio D., Messa C.. et al. (2015) Portrait of inflammatory response to ionizing radiation treatment. J. Inflamm. 12, 14 10.1186/s12950-015-0058-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunther S., Ostheimer C., Stangl S., Specht H.M., Mozes P., Jesinghaus M.. et al. (2015) Correlation of Hsp70 serum levels with gross tumor volume and composition of lymphocyte subpopulations in patients with squamous cell and adeno non-small cell lung cancer. Front. Immunol. 6, 556 10.3389/fimmu.2015.00556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubner Y., Muth C., Strnad A., Derer A., Sieber R., Buslei R.. et al. (2014) Fractionated radiotherapy is the main stimulus for the induction of cell death and of Hsp70 release of p53 mutated glioblastoma cell lines. Radiat. Oncol. 9, 89 10.1186/1748-717X-9-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rau J.C., Deans C., Hoffman M.R., Thomas D.B., Malcom G.T., Zieske A.W.. et al. (2009) Heparin cofactor II in atherosclerotic lesions from the pathobiological determinants of atherosclerosis in youth (PDAY) study. Exp. Mol. Pathol. 87, 178–183 10.1016/j.yexmp.2009.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu L., Guo Q., Jin S., Feng H., Zhuang H., Liu C.. et al. (2016) Analysis of the gene expression profile in response to human epididymis protein 4 in epithelial ovarian cancer cells. Oncol. Rep. 36, 1592–1604 10.3892/or.2016.4926 [DOI] [PubMed] [Google Scholar]
- 28.Liao W.Y., Ho C.C., Hou H.H., Hsu T.H., Tsai M.F., Chen K.Y.. et al. (2015) Heparin co-factor II enhances cell motility and promotes metastasis in non-small cell lung cancer. J. Pathol. 235, 50–64 10.1002/path.4421 [DOI] [PubMed] [Google Scholar]
- 29.Cavalcante M.S., Torres-Romero J.C., Lobo M.D., Moreno F.B., Bezerra L.P., Lima D.S.. et al. (2016) A panel of glycoproteins as candidate biomarkers for early diagnosis and treatment evaluation of B-cell acute lymphoblastic leukemia. Biomark. Res. 4, 1 10.1186/s40364-016-0055-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng G., Li J., Zheng M., Yang Z., Liu Y., Zhang S.. et al. (2016) Hepatitis B virus X protein up-regulates C4b-binding protein alpha through activating transcription factor Sp1 in protection of hepatoma cells from complement attack. Oncotarget 7, 28013–28026 10.18632/oncotarget.8472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sogawa K., Takano S., Iida F., Satoh M., Tsuchida S., Kawashima Y.. et al. (2016) Identification of a novel serum biomarker for pancreatic cancer, C4b-binding protein alpha-chain (C4BPA) by quantitative proteomic analysis using tandem mass tags. Br. J. Cancer. 115, 949–956 10.1038/bjc.2016.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi T.G., Nguyen M.N., Kim J., Jo Y.H., Jang M., Nguyen N.. et al. (2018) Cyclophilin B induces chemoresistance by degrading wild-type p53 via interaction with MDM2 in colorectal cancer. J. Pathol. 10.1002/path.5107 [DOI] [PubMed] [Google Scholar]
- 33.Williams P.D., Owens C.R., Dziegielewski J., Moskaluk C.A., Read P.W., Larner J.M.. et al. (2011) Cyclophilin B expression is associated with in vitro radioresistance and clinical outcome after radiotherapy. Neoplasia 13, 1122–1131 10.1593/neo.111398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S.C., Tsang N.M., Chiang W.C., Chang K.P., Hsueh C., Liang Y.. et al. (2013) Leukemia inhibitory factor promotes nasopharyngeal carcinoma progression and radioresistance. J. Clin. Invest. 123, 10.1172/JCI63428 [DOI] [PMC free article] [PubMed] [Google Scholar]