Abstract
MicroRNAs (miRNAs) have been proven to play a crucial role in postmenopausal osteoporosis (PMO), and studies on their diagnostic value have been increasing. In our study, we aim to identify the key miRNAs in the PMO that might be potential biomarkers. A comprehensive systematic literature search was conducted by searching PubMed, Web of Science, Embase and Cochrane Library databases. In the total of 16 independent miRNA expression studies which contained 327 PMO patients and 328 postmenopausal (PM) healthy control samples, miRNAs were evaluated by using robust rank aggregation (RRA) method. A statistically significant meta-signature of up-regulated hsa-miR-133a-3p (P = 1.38e−03) was determined. Then bioinformatics analysis to recruit putative target genes prediction of hsa-miR-133a-3p and pathway enrichment analysis to reveal what biological processes this miRNA may affect were conducted. It was indicated that pathways were commonly associated with adrenergic signaling in cardiomyocytes, adherens junction, PI3K-Akt signaling pathway and AMPK signaling pathway. Furthermore, STRING and Cytoscape tools were used to visualize the interactions between target genes of hsa-miR-133a-3p. Six genes were detected as hub genes among 576 targets which were CDC42, RHOA, EGFR, VAMP2, PIK3R2 and FN1. After Kyoto Encyclopedia of Genes and Genomes pathway analysis, it was detected that these hub genes were mostly enriched in signaling pathways and cancer. In this meta-analysis, it is stated that circulating hsa-miR-133a-3p may serve as a potential non-invasive biomarker and therapeutic target in PMO.
Keywords: biomarker, circulating miRNAs, meta-analysis, Postmenopausal osteoporosis, Robust rank aggregation
Introduction
Osteoporosis is a systemic skeletal disorder, which is common in postmenopausal (PM) women, characterized by an increased risk of bone fragility and a decrease in bone mass [1]. Bone homeostasis requires a balance between bone-forming osteoblast cells and bone-resorbing osteoclast cells [2]. When this balance is impaired, normal bone remodeling cannot keep bone mass stable and leads to develop osteopenia and osteoporosis [3]. Traditionally, dual-X-ray absorptiometry (DEXA) which measures bone mineral density (BMD) is routinely used to assess the risk of fracture in osteoporosis [4].
MicroRNAs (miRNA) are short noncoding and single-stranded molecules in 18–24 nucleotides length which play key roles in translation and expression of genes via binding to the 3′untranslated region (3′UTR) of target messenger RNAs (mRNA) [5,6]. miRNAs have been identified as playing critical roles in biological processes (BPs) like differentiation and development; thus abundant studies associated with dysregulated miRNA expression in bone tissue and circulatory biofluids in osteoporosis [3]. According to these studies, miRNAs contribute to the pathogenesis of osteoporosis and therapeutic potential of them should be considered as they regulate several kinds of cells in bone homeostasis [3].
miRNA profiling datasets were arising rapidly with the improvement of high-throughput technologies. Using of different technological platforms and small sample size in studies results miRNA data sets show inconsistent results between various studies. In our study, we aimed to identify the key circulating miRNAs in the postmenopausal osteoporosis (PMO) that might be non-invasive potential biomarkers. Considering our aim and these problems, we found a substantial way to increase the statistical power of profiling data by combining the results of several studies. We combined these results via conducting a meta-analysis applying the robust rank aggregation (RRA) method [7], followed by pathway analysis, to identify miRNA dysregulation in PMO and the pathways that key miRNAs may affect [8]. The RRA strategy has been generated for comparison of different ranked gene lists and identification of commonly overlapping genes. This is a proper and efficient method for discrimination of statistically significant miRNA meta-signature and is particularly beneficial when experiments are performed by distinct technological platforms include different gene sets and full rankings of miRNAs are not available [9]. miRNA meta-signature investigation and identification of involved pathways would present potential targets for additional experimental studies of PMO development.
Materials and methods
Search strategies
We conducted a systematic literature search to identify miRNA expression profiling studies in PMO published up to 9 October 2018. The databases searched included PubMed, Web of Science, Embase and Cochrane Library through the MESH search headings ‘(miRNA OR microRNA OR miR) AND (osteoporosis)’. The searches were limited to English language studies and only full text published studies were included. We evaluated potentially relevant studies according to their titles and abstracts by using Rayyan which is a free application that dramatically speeds up the process of screening and selecting studies [10]. Moreover, each article was controlled to evaluate the publication type through its existed database manually.
Study selection
The inclusion criteria for the selection of eligible studies were defined as: (1) circulating miRNA expression profiling on PMO patients; (2) have to compare PMO samples with PM healthy control samples; (3) miRNA expressions were reported to be profiled by using miRNA microarray, next-generation sequencing (NGS) or qRT-PCR; (4) have to report cut-off criteria of dysregulated miRNAs; (5) they reported fold changes (even if non-explicit); (6) have to report sample sizes. The exclusion criteria were defined as: (1) studies on tissues, cell lines and animals; (2) review literature and case reports.
Relevant papers were selected by two authors (E.P. and T. D.) and decided the list of studies be included carefully.
Data extraction
From the full text and supplementary data of each study, the subsequent eligible information was collected and documented as first author, date of publication, country of study, sample sizes, tissue types, miRNA expression profiling assay type, number of probes, the list of dysregulated miRNAs and their related fold changes if given and cut-off criteria of dysregulated miRNAs. The authors directly contacted the correspondence of the studies to reach the gene lists which were not given in the full text and supplementary information. All miRNA names were standardized according to miRBase v22 by using miRNAmeConverter available in Bioconductor R package [11].
Robust rank aggregation analysis
RRA method was used in our meta-analysis which is a free package in R software (http://cran.R-project.org/). This approach is utilized for comparison of different ranked gene lists, and is a definite and powerful approach for identification of differentially expressed miRNA integrated signature where a P-value would be assigned for each miRNA in the ranked lists to rerank these miRNAs and determine their significance. This method applies a probabilistic model for aggregation. The RRA method is robust to noise, and it facilitates the computation of significance probabilities to each of the elements in the final ranking [8,12]. All miRNAs from included studies were ranked with this method by using their P-values (P<0.05).
Prediction of target gene differentially expressed miRNAs
In the present study MultiMiR package (http://multimir.ucdenver.edu/) was constructed to predict targets of miRNAs, which cover 14 databases. MultiMiR package was used to predict targets of miRNAs by DIANA-microT, ElMMo, MicroCosm, miRanda, miRDB, PicTar, PITA and TargetScan databases and also, validated targets of miRNAs were acquired from miRecords, miRTarBase and TarBase databases with the criterion of primary score listed in top 35 [13]. Only targets predicted by at least three algorithms or those validated by one database were kept for analyses [14].
Functional enrichment analysis
The Database Annotation for Visualization and Integrated Discovery (DAVID) is an online program which allows functional annotation of the huge number of genes obtained from several genomic resources [15]. We used the DAVID database to perform gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis tools to implement the enrichment analysis. The consensus targets of miRNAs were used as input in screening GO and KEGG pathway analyses. P-values≤0.05 after Benjamini correction were considered enriched for the genes targeted by the selected miRNA [14].
Protein–protein interaction network construction of the target genes
The STRING database (http://string-db.org/) is online software that aims to provide a crucial evaluation and integration of protein–protein interaction (PPI), including direct and indirect connections [16]. Cytoscape is one of the most favorite open-source software tools for the visual exploration of biomedical networks composed of protein, gene and other types of interactions [17]. The target genes were mapped to STRING with a confidence score>0.9 as a cut-off criterion to evaluate the PPI information and then interactions were visualized with Cytoscape. Node degree≥15 was set as the cut-off criterion to screen the hub genes. Furthermore, KEGG pathway analysis was applied to these hub genes by using DAVID with a P-values≤0.05 after Benjamini correction cut-off criterion.
Results
Literature search and included studies
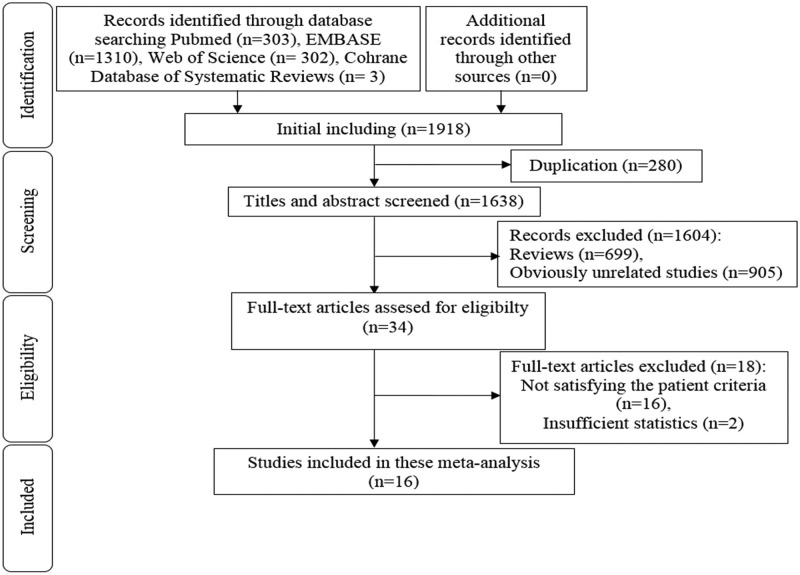
The flow diagram demonstrating the strategy used to include in this meta-analysis is shown in Figure 1. 1918 possible relevant studies were detected in Pubmed, Web of Science, EMBASE and Cochrane databases considering our criteria. After eliminating duplicated publications, reviews and unrelated studies according to our inclusion/exclusion criteria, 34 articles met the eligibility. Finally following full-text analysis 16 articles included in this meta-analysis. The main information about the included studies was given in Table 1 [18–33].
Figure 1. The flow diagram for study selection.
Table 1. Characteristics of included miRNA profiling studies.
| No | Author | Date | Country | Sample type | # of samples (P/C) | Assay type | # of probes | Total | Up- regulate | Down- regulate | Cut-off criteria |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Li Z | 2018 | China | Blood (serum) | 20 (10/10) | qRT-PCR | 1 | 1 | 1 | 0 | P<0.05 |
| 2 | Jiménez-Ortega RF | 2017 | Mexico | Blood (monocyte) | 12 (6/6) | Affimetrix GeneChip miRNA 4.0 Array | 2578 | 35 | 3 | 3 | FC>0.5, P<0.05 |
| 3 | Bedene A | 2016 | Slovenia | Blood (plasma) | 74 (17/57) | qRT-PCR | 9 | 1 | 1 | 0 | P<0.05 |
| 4 | Yavropoulou MP | 2017 | Greece | Blood (serum) | 100 (70/30) | qRT-PCR | 14 | 5 | 2 | 3 | P<0.05 |
| 5 | Meng J | 2015 | China | Blood | 56 (32/24) | qRT-PCR | 1 | 1 | 1 | 0 | P<0.05 |
| 6 | Li H | 2014 | China | Blood (serum) | 80 (40/40) | qRT-PCR | 3 | 2 | 1 | 1 | P<0.01 |
| 7 | Cao Z | 2014 | America | Blood (monocyte) | 20 (10/10) | qRT-PCR | 4 | 1 | 1 | 0 | P<0.05 |
| 8 | Wang Y | 2012 | America | Blood (monocyte) | 20 (10/10) | TaqMan Human MicroRNA Array v1.0 | 365 | 2 | 2 | 0 | P<0.05 |
| 9 | Chen J | 2016 | China | Blood (serum) | 29 (10/19) | qRT-PCR | 15 | 4 | 0 | 4 | P<0.05 |
| 10 | Liu H | 2017 | China | Blood (plasma) | 40 (20/20) | qRT-PCR | 2 | 2 | 2 | 0 | P<0.05 |
| 11 | Chen H | 2017 | China | Blood (serum) | 60 (30/30) | qRT-PCR | 5 | 3 | 3 | 0 | P<0.05 |
| 12 | Chen C | 2013 | China | Blood | 20 (10/10) | miRNA microarray, LC Sciences | 721 | 7 | 3 | 4 | P<0.05 |
| 13 | Seeliger C | 2014 | Germany | Blood (serum) | 60 (30/30) | qRT-PCR | 13 | 9 | 9 | 0 | P<0.05 |
| 14 | Chen R | 2018 | China | Blood | 18 (9/9) | qRT-PCR | 150 | 14 | 4 | 7 | P<0.05 |
| 15 | Ramírez-Salazar EG | 2018 | China | Blood (serum) | 40 (20/20) | TaqMan Array Human MicroRNA A+B Cards Set v3.0 | 754 | 7 | 7 | 0 | FC≥2, P≤0.05 |
| 16 | Jin D | 2018 | China | Blood | 6 (3/3) | Sequencing (Illumina HiSeq platform) | NR | 13 | 3 | 10 | P<0.05 |
Abbreviations: C, Control; FC, fold change; P, patient.
Differentially expressed miRNAs in postmenopausal osteoporosis vs. healthy postmenopausal controls
In the included 16 miRNA expression profiling studies, 75 miRNAs were reported to be differentially expressed in groups of 327 PMO patients and 328 PM healthy controls. Among these miRNAs 32 were down-regulated, 43 were up-regulated and 10 miRNAs (hsa-miR-133a-3p, hsa- miR-148a-3p, hsa-miR-21-5p, hsa-miR-124-3p, hsa-miR-2861, hsa-miR-23a-3p, hsa-miR-328-3p, hsa-miR-107, hsa-miR-125b-5p and hsa-miR-100-5p) were reported in at least two studies. Furthermore four miRNAs (hsa-miR-21-5p, hsa-miR-2861, hsa-miR-23a-3p and hsa-miR-100-5p) were reported to be dysregulated in both directions (Figure 2).
Figure 2. Up- and down-regulated miRNAs of PMO vs. PM healthy control samples in 16 included studies.
* indicates miRNAs reported in more than one study, # indicates miRNAs reported to be dysregulated in both directions.
Afterward, using the RRA method, we identified a statistically significant meta-signature of one up-regulated miRNA; hsa-miR-133a-3p considering the permutation P-value (1.38e−03). Only this miRNA was detected in two datasets of included studies by using RRA.
Target prediction of differentially expressed hsa-miR-133a-3p
Target genes of hsa-miR-133a-3p were evaluated via bioinformatics analysis. The combination of eight different in silico predicted and three validated databases was systematically screened by using MultiMiR for miRNA–target interactions. According to our criterion, a total of 3153 target genes were predicted initially, and only 576 genes were essential for further studies.
The enrichment analysis for predicted targets of hsa-miR-133a-3p
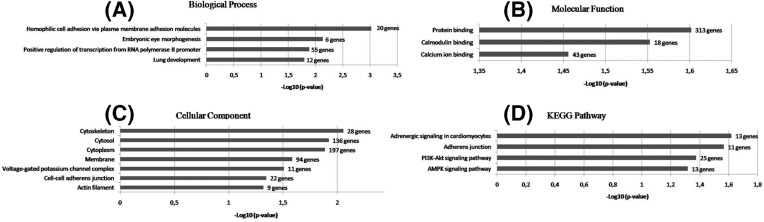
Using consensus target genes of hsa-miR-133a-3p enrichment analysis were performed to highlight the biological function of our PMO-miR meta-signature. Following the enrichment analysis by DAVID software, it was detected the most significantly enriched GO terms on BPs are homophilic cell adhesion via plasma membrane adhesion molecules, embryonic eye morphogenesis, positive regulation of transcription from RNA polymerase II promoter and lung development (Figure 3A). We listed all the BP, cellular components (CCs) (Figure 3B), molecular functions (MFs) (Figure 3C), and KEGG pathways (P<0.05 after Benjamini correction). Also, the KEGG pathway analysis showed that target genes were enriched in adrenergic signaling in cardiomyocytes, adherens junction, PI3K-Akt signaling pathway and AMPK signaling pathway (Figure 3D).
Figure 3. Enriched GO terms of 576 target genes obtained from the database for annotation (A) BP; (B) CC; (C) MF; (D) Enriched KEGG pathway.
PPI network and identification of hub genes
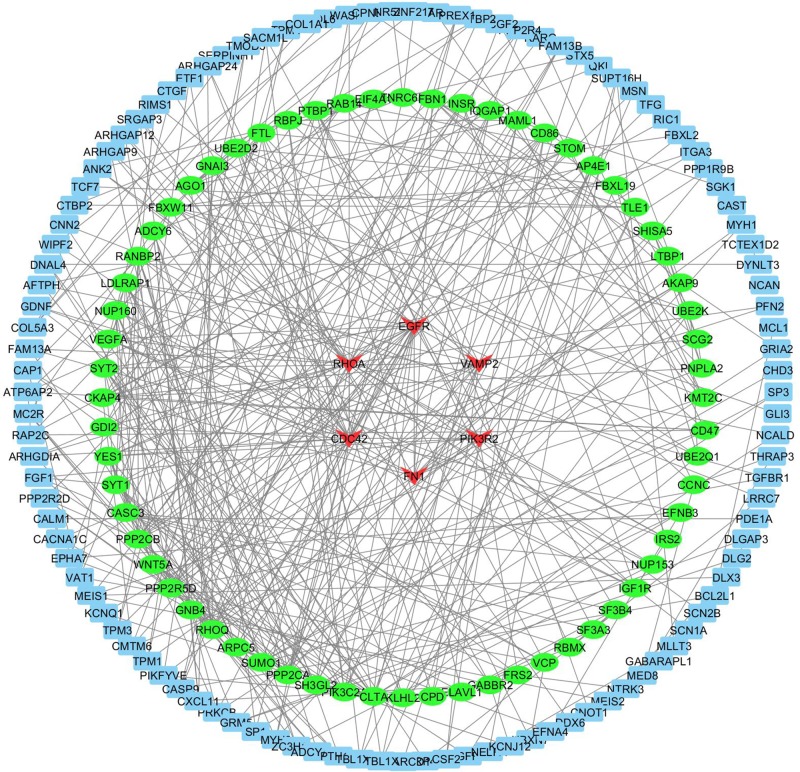
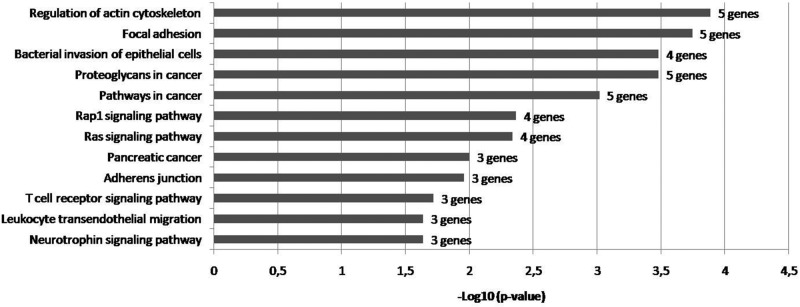
Target genes were used to establish the PPI network by STRING, which composed of 572 nodes and 509 edges. Subsequently, we analyzed the STRING results using Cytoscape (Figure 4.) and six genes in the PPI network were identified as hub genes (node degree≥15). These included CDC42 (degree = 33), RHOA (degree = 25), EGFR (degree = 24), VAMP2 (degree = 19), PIK3R2 (degree = 16), and FN1 (degree = 15). KEGG pathway enrichment analysis of these hub genes was performed by DAVID. The pathway enrichment analysis revealed that hub genes were mostly enriched in signaling pathways and cancer (P-values≤0.05 after Benjamini correction) (Figure 5).
Figure 4. The PPI network of the target genes of miR–133a–3p from which six hub genes colored in red were identified according to the value of degrees.
Figure 5. Enriched KEGG pathway of hub genes.
Discussion
According to the World Health Organization, osteoporosis is one of the most common diseases, and 30–50% of all women in the world suffer fractures due to osteoporosis throughout their lives [34]. Circulating miRNAs are ideal non-invasive biomarkers as they are stable in body fluids and can easily be detected by using confirmed techniques for quantification [35]. miRNAs can regulate the expression of multiple mRNAs so that they are involved in almost all key BPs like proliferation, differentiation, migration and apoptosis [36]. There has been a growing interest in the regulation of miRNA studies for the development and progression of various diseases in recent years.
DEXA is a gold standard method for the measurement of BMD, used in the diagnosis of osteoporosis. To our knowledge, the present study is the first meta-analysis concerning PMO circulating miRNA expression profiles and identifying specific miRNA as a potential biomarker in PMO. Serum biomarker analyzing is beneficial as dysregulation of specific circulating miRNAs in the serum might present valuable potential diagnostic predictors for PMO.
Although there are some studies in the literature on PMO and circulating miRNAs, no meta-analysis is available, and consistent and precise data cannot be reached. In the present study, we aimed to use meta-analysis to recognize consistently dysregulated miRNAs, which were mentioned in profiling results to be candidate biomarkers for PMO. To overcome inconsistent results of miRNA dysregulation in PMO patients reported in the literature, RRA method was performed in the present study. Ranked list of miRNAs from each included study was aggregated into a single gene ranking and analyzed using the RRA method, which approved hsa-miR-133a-3p as the only statistically significant miRNA associated with PMO (P = 0.00138).
Dysregulation of miR-133a-3p has been identified in several types of cancers, such as bladder [37], colorectal [38], osteosarcoma [39], non-small cell lung [40] and esophageal cancers [41]. Two genes encode human mature miR-133a: MIR133A1 for miR-133a1 at 18q11.2 and MIR133A2 for miR-133a2 at 20q13.33. These two genes encode different premature miRNAs but produce the identical mature miR-133a sequence. Genetic researches also discovered the association of 18q11.2 to osteoporosis-related traits [42] and the linkage of 20q13 to bone phenotypes [43,44]. Wang et al. aimed to identify important miRNAs in human circulating monocytes of 20 high and low BMD PM women and found significant up-regulation of miR-133a in the low BMD group by both array and qRT-PCR analyses. They suggest that miR-133a in circulating monocytes is a potential biomarker for PMO [25]. Besides, Li et al. searched the levels of miR-133a in the plasma of 120 Chinese PM women who were divided into three groups of normal, osteopenia and osteoporosis according to the T-scores. Up-regulation of miR-133a was validated in the plasma of osteoporosis and osteopenia cases versus the normal group and suggested a potential use of miR-133a as sensitive and plasma biomarker for PMO [18].
Also, there are studies available concerning circulating miRNAs in PMO based on different inclusion patient criteria in the literature. Studies on the circulating miRNAs in PMO patients that were compared with the PM women without osteoporosis were included in this meta-analysis. As noted in Figure 1, studies comparing PMO patients with osteoarthritic controls [45] and PMO women with and without fracture (control) [46,47] were excluded as they did not meet patient criteria.
MiR-133a was initially examined as a muscle-specific miRNA involved in the regulation of muscle-cell differentiation and pathogenesis of heart disease [48]. Afterward, it was determined that miR-133a plays a crucial role in bone morphogenesis and fracture healing [49]. RUNX2 is involved in the differentiation of mesenchymal stem cells into osteoblast and bone formation [50,51]. MiR-133a can inhibit the secretion and expression of bone formation-promoting substances and other molecules related to osteoblast differentiation via binding to RUNX2 [52]. It was shown that the expression level of miR-133a was significantly increased and RUNX2 protein expression level was significantly decreased in bone tissues of patients with fracture nonunion, suggesting that the miR-133a expression level was closely related to fracture healing [53].
In an included study to our meta-analysis, in vitro experiments demonstrated that miR-133a was up-regulated during osteoclastogenesis and overexpression of this miRNA promoted RANKL induced differentiation of RAW264.7 and THP-1 cells into osteoclasts. And also in vivo experiments carried out in the present study showed that osteoclastogenesis related factors levels changed in serum, lumbar spine BMD increased and bone histomorphology changed in miR-133a knockdown ovariectomized rats. These results indicated that miRNA-133a is involved in the regulation of PMO by promoting osteoclast differentiation [18].
Target gene prediction for miR-133a-3p was performed by using MultiMiR. 576 genes were detected as the target of this miRNA and KEGG pathway analysis has shown that these genes were mostly enriched in adrenergic signaling in cardiomyocytes and adherens junction. Some other reports in the literature that show the association of these two pathways with osteoporosis [54,55]. Furthermore, target genes of miR-133a-3p were used to establish the PPI network by using Cytoscape and six genes in the PPI network were identified as hub genes (degree≥15). These included CDC42, RHOA, EGFR, VAMP2, PIK3R2 and FN1. Increased activity of osteoclasts is related with several bone diseases, including PMO. After adherence to the bone, the osteoclasts become polarized and reorganize their cytoskeleton and membrane to form unique domains including the sealing zone. This zone is a dense ring of F-actin-rich podosomes delimiting the ruffled border, where protons and proteases are secreted to demineralize and degrade the bone matrix, respectively. These processes are dependent on the activity of small GTPases. CDC42 and RHOA are members of Rho GTPases that regulate podosome assembly and their organization into the sealing zone [56]. Epidermal growth factor receptor (EGFR) is one member of the transmembrane growth factor receptor proteins [57]. It has been shown that the EGFR network was closely associated with bone biology and pathology; for example, it affected osteoblastic bone formation and bone mass in different ways. Recently, Zhu et al. found that the EGFR system suppressed osteoblast differentiation and have crucial functions in skeletal homeostasis [58]. In compliance with all this information, the KEGG pathway analysis of the hub genes showed that these genes are mostly enriched in regulation of actin cytoskeleton and focal adhesion.
Conclusions
In conclusion, hsa-miR-133a-3p seems to be a potential biomarker for PMO, since its levels are up-regulated in patients suffering from PMO. This miRNA would enable the analysis of osteoporosis without requiring DEXA so that radiation exposure will be reduced in these patients. Thus, a deeper examining of the role of miRNAs in osteoporosis can inspire critical implications for the early diagnosis and prevention of osteoporosis. It can also provide unique opportunities to develop novel pharmacologic approaches for osteoporosis.
Abbreviations
- BMD
bone mineral density
- BP
biological process
- CC
cellular component
- DAVID
Database Annotation for Visualization and Integrated Discovery
- DEXA
dual-X-ray absorptiometry
- EGFR
epidermal growth factor receptor
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MF
molecular function
- miRNA
microRNA
- mRNA
messenger RNA
- PM
postmenopausal
- PMO
postmenopausal osteoporosis
- PPI
protein–protein interaction
- RRA
robust rank aggregation
Author Contribution
Elif Pala and Tuba Denkçeken designed the meta-analysis, searched aimed studies, extracted corresponding data, prepared the manuscript and approved the manuscript for submission.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
References
- 1.Wade S.W. et al. (2014) Estimating prevalence of osteoporosis: examples from industrialized countries. Arch. Osteoporos. 9, 182 10.1007/s11657-014-0182-3 [DOI] [PubMed] [Google Scholar]
- 2.Kanis J.A. et al. (2019) European guidance for the diagnosis and management of osteoporosis in postmenopausal women. Osteoporos. Int. 30, 3–44 10.1007/s00198-018-4704-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao W. et al. (2018) Therapeutic potential of microRNAs in osteoporosis function by regulating the biology of cells related to bone homeostasis. J. Cell. Physiol. 233, 9191–9208 10.1002/jcp.26939 [DOI] [PubMed] [Google Scholar]
- 4.Siris E.S. et al. (2014) The clinical diagnosis of osteoporosis: a position statement from the National Bone Health Alliance Working Group. Osteoporos. Int. 25, 1439–1443 10.1007/s00198-014-2655-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 6.Lee H.K. et al. (2013) Mesenchymal stem cells deliver synthetic microRNA mimics to glioma cells and glioma stem cells and inhibit their cell migration and self-renewal. Oncotarget 4, 346–361 10.18632/oncotarget.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sempere L.F. (2011) Integrating contextual miRNA and protein signatures for diagnostic and treatment decisions in cancer. Expert Rev. Mol. Diagn. 11, 813–827 10.1586/erm.11.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolde R. et al. (2012) Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 28, 573–580 10.1093/bioinformatics/btr709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez M.L. and Nourbakhsh M. (2011, RNA mapping protocols: northern blot and amplification of cDNA ends. In Disease Gene Identification, pp. 199–220, Springer; [DOI] [PubMed] [Google Scholar]
- 10.Ouzzani M. et al. (2016) Rayyan – a web and mobile app for systematic reviews. Syst. Rev. 5, 210 10.1186/s13643-016-0384-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haunsberger S.J., Connolly N.M.C. and Prehn J.H.M. (2017) miRNAmeConverter: an R/bioconductor package for translating mature miRNA names to different miRBase versions. Bioinformatics 33, 592–593 [DOI] [PubMed] [Google Scholar]
- 12.Zang S. et al. (2014) Identification of differentially-expressed genes in intestinal gastric cancer by microarray analysis. Genomics Proteomics Bioinformatics 12, 276–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ru Y. et al. (2014) The multiMiR R package and database: integration of microRNA–target interactions along with their disease and drug associations. Nucleic Acids Res. 42, e133–e133 10.1093/nar/gku631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almeida R.S. et al. (2019) MicroRNA expression profiles discriminate childhood T‐from B‐ acute lymphoblastic leukemia. Hematol. Oncol. 37, 103–112 10.1002/hon.2567 [DOI] [PubMed] [Google Scholar]
- 15.Huang D.W., Sherman B.T. and Lempicki R.A. (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 16.Szklarczyk D. et al. (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon P. et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z., Zhang W. and Huang Y. (2018) MiRNA-133a is involved in the regulation of postmenopausal osteoporosis through promoting osteoclast differentiation. Acta Biochim. Biophys. Sin. (Shanghai) 50, 273–280 10.1093/abbs/gmy006 [DOI] [PubMed] [Google Scholar]
- 19.Jiménez-Ortega R.F. et al. (2017) Identification of microRNAs in human circulating monocytes of postmenopausal osteoporotic Mexican-Mestizo women: A pilot study. Exp. Therapeutic Med. 14, 5464–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bedene A. et al. (2016) MiR-148a the epigenetic regulator of bone homeostasis is increased in plasma of osteoporotic postmenopausal women. Wien. Klin. Wochenschr. 128, 519–526 10.1007/s00508-016-1141-3 [DOI] [PubMed] [Google Scholar]
- 21.Yavropoulou M.P. et al. (2017) Expression of microRNAs that regulate bone turnover in the serum of postmenopausal women with low bone mass and vertebral fractures. Eur. J. Endocrinol. 176, 169–176 10.1530/EJE-16-0583 [DOI] [PubMed] [Google Scholar]
- 22.Meng J. et al. (2015) Identification of miR-194-5p as a potential biomarker for postmenopausal osteoporosis. Peer J. 3, e971 10.7717/peerj.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H. et al. (2014) Plasma miRNA levels correlate with sensitivity to bone mineral density in postmenopausal osteoporosis patients. Biomarkers 19, 553–556 10.3109/1354750X.2014.935957 [DOI] [PubMed] [Google Scholar]
- 24.Cao Z. et al. (2014) MiR-422a as a potential cellular microRNA biomarker for postmenopausal osteoporosis. PLoS One 9, e97098 10.1371/journal.pone.0097098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y. et al. (2012) MiR-133a in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. PLoS One 7, e34641 10.1371/journal.pone.0034641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J. et al. (2016) Identification of suitable reference gene and biomarkers of serum miRNAs for osteoporosis. Sci. Rep. 6, 36347 10.1038/srep36347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H. et al. (2018) MiR‐96 regulates bone metabolism by targeting osterix. Clin. Exp. Pharmacol. Physiol. 45, 602–613 10.1111/1440-1681.12912 [DOI] [PubMed] [Google Scholar]
- 28.Chen H. et al. (2017) Evaluation of MicroRNA 125b as a potential biomarker for postmenopausal osteoporosis. Tropical J. Pharmaceutical Res. 16, 641–647 10.4314/tjpr.v16i3.20 [DOI] [Google Scholar]
- 29.Chen C. et al. (2014) MiR‐503 regulates osteoclastogenesis via targeting RANK. J. Bone Miner. Res. 29, 338–347 10.1002/jbmr.2032 [DOI] [PubMed] [Google Scholar]
- 30.Seeliger C. et al. (2014) Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J. Bone Miner. Res. 29, 1718–1728 10.1002/jbmr.2175 [DOI] [PubMed] [Google Scholar]
- 31.Chen R. et al. (2018) Circulating microRNAs, miR-10b-5p, miR-328-3p, miR-100 and let-7, are associated with osteoblast differentiation in osteoporosis. Int. J. Clin. Exp. Pathol. 11, 1383–1390 [PMC free article] [PubMed] [Google Scholar]
- 32.Ramírez-Salazar E.G. et al. (2018) Serum miRNAs miR-140-3p and miR-23b-3p as potential biomarkers for osteoporosis and osteoporotic fracture in postmenopausal Mexican- Mestizo women. Gene 679, 19–27 10.1016/j.gene.2018.08.074 [DOI] [PubMed] [Google Scholar]
- 33.Jin D. et al. (2018) Systematic analysis of lncRNAs, mRNAs, circRNAs and miRNAs in patients with postmenopausal osteoporosis. Am. J. Translational Res. 10, 1498. [PMC free article] [PubMed] [Google Scholar]
- 34.Rachner T.D., Khosla S. and Hofbauer L.C. (2011) Osteoporosis: now and the future. Lancet North Am. Ed. 377, 1276–1287 10.1016/S0140-6736(10)62349-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Assmann T.S. et al. (2018) MicroRNAs and diabetic kidney disease: systematic review and bioinformatic analysis. Mol. Cell. Endocrinol. 477, 90–102 10.1016/j.mce.2018.06.005 [DOI] [PubMed] [Google Scholar]
- 36.Fabris L. et al. (2016) The potential of microRNAs as prostate cancer biomarkers. Eur. Urol. 70, 312–322 10.1016/j.eururo.2015.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshino H. et al. (2011) The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br. J. Cancer 104, 808 10.1038/bjc.2011.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W. et al. (2015) MicroRNA-133a functions as a tumor suppressor by targeting IGF-1R in hepatocellular carcinoma. Tumor Biol. 36, 9779–9788 10.1007/s13277-015-3749-8 [DOI] [PubMed] [Google Scholar]
- 39.Fujiwara T. et al. (2014) Clinical relevance and therapeutic significance of microRNA‐133a expression profiles and functions in malignant osteosarcoma‐initiating cells. Stem Cells 32, 959–973 10.1002/stem.1618 [DOI] [PubMed] [Google Scholar]
- 40.Lan D. et al. (2015) MiR-133a is downregulated in non-small cell lung cancer: a study of clinical significance. Eur. J. Med. Res. 20, 50 10.1186/s40001-015-0139-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akanuma N. et al. (2014) MicroRNA-133a regulates the mRNAs of two invadopodia-related proteins, FSCN1 and MMP14, in esophageal cancer. Br. J. Cancer 110, 189 10.1038/bjc.2013.676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu Y.-H. et al. (2010) An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLos Genet. 6, e1000977 10.1371/journal.pgen.1000977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell B.D. et al. (2000) Genes influencing variation in serum osteocalcin concentrations are linked to markers on chromosomes 16q and 20q. J. Clin. Endocrinol. Metab. 85, 1362–1366 [DOI] [PubMed] [Google Scholar]
- 44.Ralston S.H. et al. (2005) Loci for regulation of bone mineral density in men and women identified by genome wide linkage scan: the FAMOS study. Hum. Mol. Genet. 14, 943–951 10.1093/hmg/ddi088 [DOI] [PubMed] [Google Scholar]
- 45.Panach L. et al. (2015) Serum circulating microRNAs as biomarkers of osteoporotic fracture. Calcif. Tissue Int. 97, 495–505 10.1007/s00223-015-0036-z [DOI] [PubMed] [Google Scholar]
- 46.Zhu Y. et al. (2016) Six miRNAs identified serving as prognostic and predictive markers for osteoporosis by miRNA high-throughput method. Int. J. Clin. Exp. Med. 9, 15226–15234 [Google Scholar]
- 47.Heilmeier U. et al. (2016) Serum miRNA signatures are indicative of skeletal fractures in postmenopausal women with and without type 2 diabetes and influence osteogenic and adipogenic differentiation of adipose tissue-derived mesenchymal stem cells in vitro. J. Bone Miner. Res. 31, 2173–2192 10.1002/jbmr.2897 [DOI] [PubMed] [Google Scholar]
- 48.Rao P.K. et al. (2006) Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. 103, 8721–8726 10.1073/pnas.0602831103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koutsoulidou A. et al. (2011) Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev. Biol 11, 34 10.1186/1471-213X-11-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito T. et al. (2004) Acceleration effect of human recombinant bone morphogenetic protein-2 on differentiation of human pulp cells into odontoblasts. J. Endod. 30, 205–208 10.1097/00004770-200404000-00005 [DOI] [PubMed] [Google Scholar]
- 51.Okazaki K. and Sandell L.J. (2004) Extracellular matrix gene regulation. Clin. Orthop. Relat. Res. 427, S123–S128 10.1097/01.blo.0000144478.51284.f3 [DOI] [PubMed] [Google Scholar]
- 52.Li Z. et al. (2008) A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc. Natl. Acad. Sci. 105, 13906–13911 10.1073/pnas.0804438105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng H. et al. (2018) MiR-133a inhibits fracture healing via targeting RUNX2/BMP2. Eur. Rev. Med. Pharmacol. Sci. 22, 2519–2526 [DOI] [PubMed] [Google Scholar]
- 54.Wei J. et al. (2016) Multiple analyses of large-scale genome-wide association study highlight new risk pathways in lumbar spine bone mineral density. Oncotarget 7, 31429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chai Y. et al. (2019) Identification of core genes and prediction of miRNAs associated with osteoporosis using a bioinformatics approach. Oncol. Lett. 17, 468–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Itzstein C., Coxon F.P. and Rogers M.J. (2011) The regulation of osteoclast function and bone resorption by small GTPases. Small GTPases 2, 117–130 10.4161/sgtp.2.3.16453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herbst R.S. (2004) Review of epidermal growth factor receptor biology. Int. J. Radiat. Oncol. Biol. Phys. 59, S21–S26 10.1016/j.ijrobp.2003.11.041 [DOI] [PubMed] [Google Scholar]
- 58.Zhu J. et al. (2011) EGFR signaling suppresses osteoblast differentiation and inhibits expression of master osteoblastic transcription factors Runx2 and Osterix. J. Cell. Biochem. 112, 1749–1760 10.1002/jcb.23094 [DOI] [PMC free article] [PubMed] [Google Scholar]