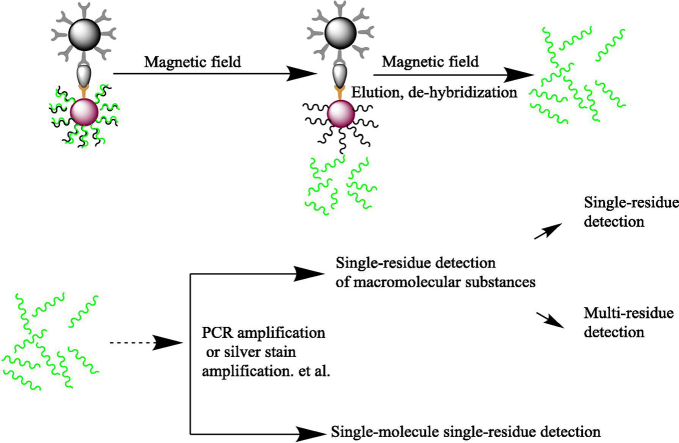
Graphical abstract
Keywords: Bio-barcode assay, Protein, Application, Multi-residue detection of macromolecules, Single-molecule single-residue detection
Highlights
-
•
This review describes the principle of the bio-barcode assay (BCA) and provides a comparison of method applications.
-
•
The review summarizes the application of BCA for the detection of macromolecules.
-
•
It summarizes the applications of BCA technology for small–molecule detection in recent years.
-
•
BCA technology makes up for the shortcomings of other technologies.
-
•
It summarizes the feasibility, deficiencies and expectations of BCA technology.
Abstract
With the rapid development of nanotechnology, the bio-barcode assay (BCA), as a new diagnostic tool, has been gradually applied to the detection of protein and nucleic acid targets and small-molecule compounds. BCA has the advantages of high sensitivity, short detection time, simple operation, low cost, good repeatability and good linear relationship between detection results. However, bio-barcode technology is not yet fully formed as a complete detection system, and the detection process in all aspects and stages is unstable. Therefore, studying the optimal reaction conditions, optimizing the experimental steps, exploring the multi-residue detection of small-molecule substances, and preparing immuno-bio-barcode kits are important research directions for the standardization and commercialization of BCA. The main theme of this review was to describe the principle of BCA, provide a comparison of its application, and introduce the single-residue and multi-residue detection of macromolecules and single-residue detection of small molecules. We also compared it with other detection methods, summarized its feasibility and limitations, expecting that with further improvement and development, the technique can be more widely used in the field of stable small-molecule and multi-residue detection.
Introduction
In recent years, there has been continuous development and exploration in the fields of medicine, clinical detection, molecular biology, immunology, and nanotechnology, among others [1], [2], [3], [4], [5], [6]. There are now higher requirements for the trace analysis of macromolecules and small molecules, such as proteins and nucleic acids, as well as agricultural and veterinary drugs and environmental pollutants. Traditional immunoassay methods have difficulty in the ultrasensitive detection of proteins, mainly due to the lack of direct amplification techniques, such as PCR. Recently, BCA has become a new technology in various fields, such as clinical diagnosis and trace analysis, and has the advantages of high sensitivity, simple operation and low cost. This technology can be used to realize indirect amplification of a probe and is widely used for the highly sensitive detection of DNA and protein [7], [8], [9], [10].
This ultrasensitive nanoparticle (NP) amplification detection system was originally proposed in 2003 by Mirkin et al. [11] of Northwestern University in the United States for the detection of prostate-specific antigen (PSA). The traditional BCA technique uses double-stranded DNA as a bio-barcode, with one strand connected to a gold nanoparticle (AuNP) via a Au-S chain and the other indicating the analyte. The disadvantage of this method is related with poor hybridization, which affects the experimental results to some extent. The method was improved by Thaxton et al. [12] in 2005 via the use of thiol-modified single-stranded DNA instead of double-stranded DNA. The addition of dithiothreitol (DTT) allows the Au-S chain to covalently bind to the DNA on the AuNP, which greatly simplifies the experimental procedure and enhances its quantitative ability.
After more than ten years of exploration and research, BCA technology now has high specificity and ultrahigh sensitivity that is 5–6 orders of magnitude higher than that of ELISA [13], [14], [15]. Compared to PCR, BCA compensates for its complexity, cost, time-consuming nature, labouriousness, and disadvantage of providing only a narrow quantitative range of target DNA after amplification; thus, BCA technology can achieve rapid and efficient trace detection and provide new ideas and platforms for detection in fields related to clinical medicine, food toxins, environmental analysis, and drug detection, among others.
This review searched articles from 2003 to 2019 through the Web of Science in English, and the keywords are Bio-barcode assay; Protein; Nucleic acid; Pesticide residues, Biotoxin, etc.
The principle of bio-barcode technology and a method application comparison
BCA technology is a classic example of nanogold diagnostic technology. AuNP have a high electron density, dielectric and catalytic properties, and may be combined with a variety of biological macromolecules without affecting their biological activity [16], [17].
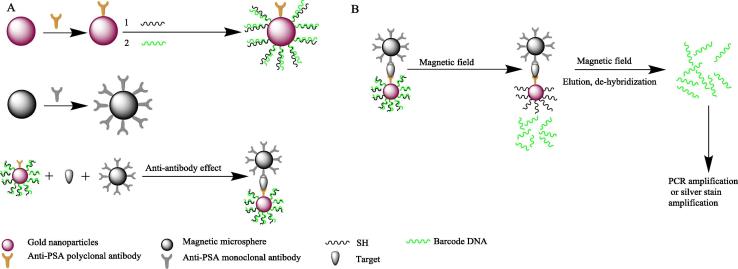
The method involves the use of two probes: magnetic beads coated with monoclonal antibodies for the protein, thereby producing a magnetic probe that can adsorb onto the target protein, and AuNP prepared by the trisodium citrate reduction method and coated with anti-target protein antibody and thiol-modified barcode DNA. Then, a magnetic field can be used to form a sandwich-like complex of the two probes with a test sample containing a target protein (e.g., sera, pathogen culture, body fluid) to form “magnetic microsphere-target protein-AuNP”. After dissociation of the labelled DNA barcode strands on the gold nanoprobes via de-hybridization elution release, the target protein content can be determined by the selected colorimetric, fluorescence labelling, biochip or other detection method [18], [19], [20], [21], [22], [23]. The main schematic diagram is shown in Fig. 1.
Fig. 1.
Schematic diagram of the biological barcode assay. (A). Probe preparation. (B). Generation of a sandwich structure with the target and separate detection.
Signal amplification detection methods commonly used for the BCA mainly include chip methods [24], [25], [26], [27], fluorescence labelling methods [28], [29], [30], [31], colorimetric methods [32], [33], [34], biosensor methods [35], [36], [37], [38], and immuno-PCR methods [39], [40], [41], [42]. Occasionally, DNA barcodes are amplified by a combination of techniques, such as real-time PCR [43], [44], [45]. BCA technology achieves high sensitivity and simple detection due to multiple signal amplification and the lack of a need for enzyme amplification. This specificity of detection is achieved via monoclonal and polyclonal antibodies corresponding to the target protein. An overview of common signal amplification methods for BCA is shown in Table 1.
Table 1.
Comparison of common BCA signal amplification methods.
| Method | Target | Year | Country | Sensitivity | Time | Principle | Advantages | Disadvantages | References |
|---|---|---|---|---|---|---|---|---|---|
| Chip methods | Anthrax | 2004 | USA | 5 × 10−15 mol/L | 3–4 h | A chip with a surface-fixed capture probe is hybridized with AuNP labelled with the complementary DNA sequence barcode, and silver staining is performed for scanning analysis. | Small, portable, fast. The barcode DNA is highly concentrated on the surface of the chip, and the silver staining method can further amplify the detection signal with high sensitivity. |
It is sometimes inconvenient to amplify and detect non-nucleic acid macromolecules. | [25] |
| Human Immunodeficiency Type 1 Capsid (p24) Antigen | 2007 | USA | 100 fg/mL | 2–3 h | [27] | ||||
| Hepatitis B Virus Deoxyribonucleic Acid | 2010 | China | 10−15 mol/L | 1.5 h | [26] | ||||
| Human IgG | 2013 | China | 1 pg/mL | 14 h | [24] | ||||
| Fluorescent labelling | Salmonella enterica serovar Enteritidis | 2009 | USA | 1 ng/mL | 4 h | Barcode DNA is labelled with fluorescent dyes and detected by collecting signals using a fluorescence scanner. | Wide application range, improved sensitivity, short detection time. |
This combination of technologies is not yet mature, and it is necessary to further optimize the experimental steps and conditions to reduce the cost of testing. | [30] |
| Bluetongue virus | 2012 | China | 10−2 fg/mL | 3 h | [28] | ||||
| Ricin toxin | 2012 | China | 1 fg/mL | 3 h | [29] | ||||
| Multiple DNAs (HCV and HIV) | 2017 | China | 5 × 10−12 mol/L | 2 h | [31] | ||||
| Colorimetric methods | Cytokines-IL-2 | 2005 | USA | 30 × 10−18 mol/L | 3 h | The sequence is hybridized with the labelled AuNP, and the experimental results are determined according to the change in the colour of the solution. | Simple, portable, low cost. The experimental results can be observed more intuitively through colour changes. |
The experimental steps are complicated. Reproducibility and sensitivity need to be further improved. |
[32] |
| Cytokines-IL-2 | 2007 | USA | 10−18 mol/L | 3 h | [33] | ||||
| Pesticide triazophos | 2017 | China | 14 ng/L | 1 h | [34] | ||||
| Biosensor methods | HTLV-I and HTLV-II | 2009 | China | 1.71 × 10−12 mol/L; 1.5 × 10−12 mol/L | 1.5 h | The AuNP is used as a signal amplifier, and the magnetic probe is used as a splitter. The signal amplification and silver staining form a complex structure, and other techniques are used for detection. |
Variety of sensors, simple operation, good portability, short response time, high sensitivity, low background signal. | The detection sensitivity cannot meet the practical requirements for large-scale applications. Experimental steps are complicated. |
[38] |
| Human platelet antigen | 2010 | Germany | 2 × 10−12 mol/L | 150 s | [35] | ||||
| The protective antigen A (pagA) gene of Bacillus anthracis and the insertion element (Iel) gene of Salmonella enteritidis | 2010 | USA | 0.5 ng/mL; 50 pg/mL | 1 h | [36] | ||||
| Escherichia coli O157:H7 | 2018 | China | 50 CFU/mL | 1 h | [37] | ||||
| IPCR | Hantaan virus nucleocapsid protein HCV core antigen | 2009 | China | 10 fg/mL | 1.5 h | Detection of target by antigen-antibody specificity and PCR amplification technology. Sometimes used in conjunction with fluorescence PCR technology. |
High sensitivity and specificity and rapid detection Can be applied in the diagnosis of cancer, Salmonella, and animal diseases and in food safety testing. |
Separate IPCR technology, quantitative uncertainty. The instrument is expensive, and the results cannot be stored for a long time. |
[39] |
| Polychlorinated biphenyls (PCBs) 77 | 2014 | China | 1.72 pg/L | 10 h | [40] | ||||
| PCBs-Aroclor 1248 | 2015 | China | 2.55 pg/L | 10 h | [41] | ||||
| Staphylococcal enterotoxin B | 2019 | China | 0.269 pg/mL | 3 h | [42] | ||||
Development and application of bio-barcode detection technology
Single-residue detection of macromolecules
Protein detection
Protein is the material basis of human cells and tissues [46], and the detection of protein biomarkers is of great significance for clinical diagnosis and treatment. However, in many early stages of disease, the concentrations of protein markers are considerably low, and conventional ELISA-based methods are not helpful in the diagnosis of various diseases. Traditionally, ELISA-based methods for detecting trace amounts of proteins have been unable to detect protein label concentrations, and their sensitivity has not yet met clinical requirements [47], [48], [49], [50]. Bio-barcode detection technology is 5–6 orders of magnitude more sensitive than conventional ELISA, thus making it a highly sensitive and highly specific detection method.
Since 2003, BCA technology has been applied to detect several protein targets. Georganopoulou et al. [51] used BCA technology for the detection of amyloid-derived diffusible ligands (ADDL) in cerebrospinal fluid (CSF) and detected 50 ADDL. Thus, BCA technology can provide a high-sensitivity, high-throughput, rapid, and reliable detection method for the clinical diagnosis of Alzheimer's disease. In 2007, Nam et al. [33] used SiO2 microspheres instead of AuNP to modify barcode DNA and antibodies and then detected interleukin 2 (IL-2) by colorimetry. IL-2 is a secreted cytokine that plays a key role in a variety of infectious, inflammatory, and immune diseases. This method detects IL-2 targets as low as 30 aM but does not guarantee reproducibility. Although BCA technology has increased sensitivity and speed, for the maturation of bio-barcode technology, it is also necessary to optimize the relevant experimental parameters. Yin et al. [28] established a method for the specific detection of the blue-tongue virus (BTV) outer nuclear protein VP7 by ultrasensitive BCA technology, which relied on real-time PCR. The limit of detection (LOD) was 0.1 fg/mL, which is 7 orders of magnitude lower than that of conventional ELISA. During the course of the experiment, due to false positives caused by laboratory contamination, multiple washing and cleaning steps are required, and real-time PCR reduces the possibility of DNA contamination to some extent. Zheng et al. [37] used a colorimetric biosensor to rapidly aggregate AuNP and then used smart phone imaging for the detection of E. coli O157:H7. The amount of E. coli was determined by the conversion of the colour of AuNP from blue to red. The LOD was 50 CFU/mL and the method had good specificity and sensitivity but could not meet the LOD of 1 CFU/mL required for food testing. As such, there is a need to further optimize experimental conditions and increase sensitivity. Li et al. [52] performed the rapid detection and sensitive determination of E. coli O157:H7 bacteria via AuNP labelling and inductively coupled plasma mass spectrometry (ICP-MS). Because of the signal amplification characteristics of AuNP and the high sensitivity of ICP-MS, the assay was capable of detecting at least 500 E. coli O157:H7 cells in a 1 mL sample. Cui et al. [53] established a sensitive and selective detection method for the H. pylori DNA sequence using a novel bio-barcode-based DNA sensing approach. The DNA sensor exhibited a detection limit of 1 × 10−15 M. Yin et al. [29] reported the use of real-time quantitative PCR for BCA technology in the specific detection of ricin in water. The method showed a coefficient of variation ranging from 3.39 to 6.84% and an LOD of 1 fg/mL, which was 6 orders of magnitude greater than that of conventional ELISA. Later, the technology was improved: the sandwich structure was directly subjected to real-time PCR bio-barcode detection reaching an LOD of 0.01 fg/mL, representing a 100-fold increase in sensitivity. Furthermore, in 2018, Zhang et al. [54] reported a new bio-barcode-based split-type photoelectrochemical (PEC) immunoassay for the sensitive detection of PSA using polymerase-triggered rolling circle amplification, accompanied by the enzymatic biocatalytic precipitation reaction, and the LOD reached as low as 1.8 pg/mL.
In the clinical diagnosis of specific protein markers, chemiluminescence, ELISA, ELISA electrophoresis, and radioimmunoassays are commonly used. However, because early markers of tumours and other disease markers are typically present at trace amounts, the specificity and sensitivity of classic detection methods are insufficient for such clinical tests. In recent years, bio-barcode technology has demonstrated strong sensitivity and specificity through continuous improvements, relying on the combination with various technologies [55], [56], [20], [57], [58], [59], [60], [61], [62], [63]. Zhang et al. [64] developed a highly sensitive and selective electrochemical DNA biosensor for detecting Ag+ based on DNA-Au bio-barcode and silver-enhanced amplification. Since the sandwich hybridization assay format and the silver enhancement have different electrochemical signal amplification linear ranges from 5 pM to 50 μM and an LOD as low as 2 pM, the method showed good analytical performance for the sensitive transduction of Ag+ recognition. Broto et al. [65] first reported the quantitative analysis of C-reactive protein (CRP) in plasma samples. The method can quantify the biomarker in a plasma sample in the range of 900–12500 ng/mL with excellent accuracy. The assay can also be used to monitor biomarkers in patients suspected of having or at risk of cerebrovascular disease (CVD) or related inflammatory diseases. Xing et al. [66] established a method for the synthesis of novel carcinoembryonic antigen (CEA) probes based on hollow quenched gold nanoparticles (HPGNP) and fluorescence quenching. By optimizing the experimental conditions, the LOD reached 1.5 pg/mL and the linear range of the probe was 2–100 pg/mL. Narmani et al. [67] established a fluorescence DNA biosensor method based on MMP and NP for the detection of the Vibrio cholera O1 OmpW gene. The results showed that the linear range was from 5 to 250 ng/mL and that the LOD was 2.34 ng/mL. Amini et al. [68] established a method based on two probes and bio-barcode DNA for the detection of Staphylococcus aureus protein A. The results showed that the standard curve was linear from 102 to 107 CFU/mL and that the LOD for both PBS and real samples was 86 CFU/mL. Li et al. [69] developed an immunoassay based on tyramine signal amplification (TSA) and AuNP labelling for the highly sensitive detection of alpha-fetoprotein (AFP) by ICP-MS. The LOD was 1.85 pg/mL, and the linear range was 0.005–2 ng/mL. Moreover, the assay showed good repeatability and can be applied to detect other macromolecules in human sera.
Nucleic acid detection
Nucleic acids are the macromolecules of DNA and RNA and one of the most basic substances required for life [70]. As a standard technique for nucleic acid detection, PCR technology is a highly sensitive method for amplifying specific DNA fragments. However, it relies on enzymatic amplification, requires expensive reagents and is time-consuming [71], [72], [73], [74], [75], [76], [77]. On the other hand, compared to PCR, BCA technology has achieves high sensitivity, and simple detection while being less time consuming and labour intensive. BCA technology is capable of rapid, low-cost detection and can be applied clinically for the rapid and joint detection of DNA and viruses of epidemic diseases under different conditions.
Wang et al. [26] used chip BCA technology for the detection of trace amounts of hepatitis B virus (HBV) DNA with a sensitivity of 10–15 mol/L. The detection time was less than 1.5 h, and the test results showed a good linear relationship with the HBV DNA levels and no false-positive results. This method can be used for the rapid screening of HBV DNA and other microbial genes in the serum of hepatitis B patients. Based on high-sensitivity BCA technology, Tang et al. [27] used the chip scanning method to detect HIV-1. The linear range was determined to be from 0.1 to 500 pg/mL, demonstrating a sensitivity approximately 150 times greater than that of conventional ELISA. Hill et al. [78] first applied bio-barcode technology to detect genomic double-stranded DNA isolated from Bacillus subtilis cells. The core of this approach was the use of blocking oligonucleotides during heat denaturation of the double-stranded DNA. The LOD was 2.5 fM; thus, this method can provide a technical platform for the improvement and development of biological material detection systems. Zhang et al. [30] used BCA technology to detect Salmonella via a fluorescence-based method. The fluorescence signal of the released barcode DNA was exponentially related to the target DNA concentration, and the LOD was 1 ng/mL. Breaking through the traditional microbial culture method used to detect Salmonella will further ensure better food safety control.
Chen et al. [39] developed a functionalized nanogold-enhanced hypersensitivity immuno-PCR method to detect Hantavirus nucleocapsid protein (HNP); the method, based on PCR/gel electrophoresis and SYBR-Green real-time fluorescence PCR, achieved a LOD of 10 fg/mL. Li et al. [79] established a triple amplification system based on ICP-MS for the detection of HBV through a combination of nicking-displacement, rolling circle amplification (RCA) and bio-barcode probes. This assay exhibited a LOD of 3.2 × 10−17 M. As the level of fluorescent PCR increases, ICP-MS technology and experimental methods are also optimized continuously [80], [81], [82], [83], [84], [85], [86], [87]. In 2017, Yin et al. [88] established a real-time PCR method with a LOD of 1 fg/mL for the detection of hepatitis C virus (HCV) core antibodies using a TaqMan probe. By improving the method, a 100-fold increase in sensitivity for the detection of HCV was achieved, and the false positives caused by the interference of other DNA sequences were reduced. Zhang et al. [89] developed a hybridized chain reaction (HCR) amplification method combined with AuNP labelling for the ICP-MS-based detection of H9N2 virions. The LOD achieved was 0.12 ng/mL, and the method revealed high specificity and sensitivity. Through optimization of the experimental steps and the technological combinations, the sensitivity of bio-barcode detection technology has been continuously improved, allowing its application to the detection of nucleic acids in various fields.
Multi-residue detection of macromolecules
There is a one-to-one correspondence between the bio-barcode DNA strand and the target protein, and the multi-residue analysis of a target substance can be achieved by designing a corresponding DNA barcode based on the capture probe of the target. In other words, BCA technology is capable of the simultaneous detection of multiple targets in one sample.
In 2006, Stoeva et al. [90] reported a method using bio-barcoded NP probes for the simultaneous detection of three protein cancer markers: PSA, (a prostate cancer marker); human chorionic gonadotropin (HCG, a testicular cancer marker); and alpha-fetoprotein (AFP, a liver cancer marker). The method was performed in a 96-well plate format in a high-throughput manner on buffer or serum samples. The barcodes were detected with the chip-based scanometric method, and with a sensitivity up to fmol/L. Thus, the technical breakthrough of bio-barcode detection in the field of the multi-residue detection of macromolecular substances has been realized.
Li et al. [91] labelled DNA with different types of fluorescent dyes and simultaneously detected five sequences and sources of DNA using fluorescently labelled NP-DNA bio-barcodes; the detection limit was 620 aM. The final detection step was completed within 30 s. In 2008, He et al. [92] used a 3730 capillary DNA analyser to detect four viral DNA sequences at concentrations of 5 pmol/L using bio-barcode detection technology within 40 min. Lin et al. [31] developed a novel nanoenzyme-based bio-barcode fluorescence amplification assay that can simultaneously detect HIV and HCV DNA. The method mainly used bimetallic (PtAu) NP and showed excellent characteristics, including peroxidase activity for the simultaneous oxidation of non-fluorescent substrates into fluorescent reagents for the simultaneous detection of HIV and HCV genes under both enzyme-free and label-free conditions. Within the range from 10 pM to 500 pM with a regression coefficient of 0.9945, the LOD reached 5 pM. The measured results showed high sensitivity and accuracy. Thus, this approach will also be an important research direction for the simultaneous detection of biological macromolecules.
The establishment of multi-residue detection of biological macromolecules is of great significance for clinical disease diagnosis, drug analysis and detection of DNA from pathogens. Currently, technology for the simultaneous detection of biological macromolecules continues to be explored and developed.
Single-molecule single-residue detection
Agricultural and veterinary drug testing
Semicarbazide is a hydrazine small-molecule compound that is a metabolite of the veterinary drug nitrofurazone. It is often considered a marker for judging the abuse of nitrofurazone in animal-derived foods. Tang et al. [93] proposed a functional AuNP bio-barcode detection technology for detecting the hapten CPSEM (a nitrofurazone derivative). PCR was combined with indirect competition ELISA to convert the enzyme signal into a DNA signal. The sensitivity reached up to 8 pg/mL, which is approximately 25 times that of conventional ELISA.
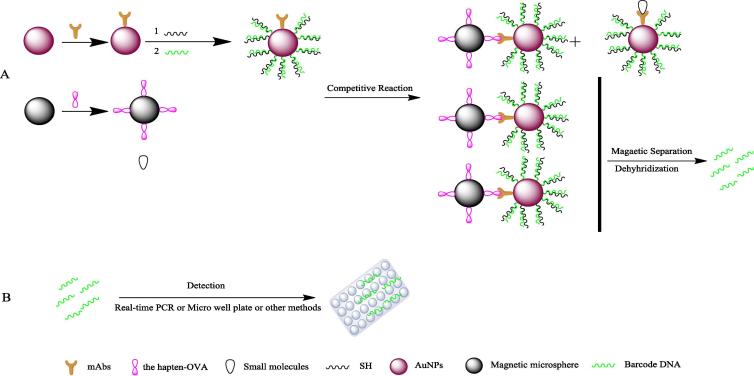
Nanomaterials are less commonly used in the detection of small-substances, such as additives in food and pesticide residues, than macromolecules; one of the main reasons is the structure of small molecules, which cannot bind to two antibodies due to steric hindrance. To solve this problem, the double sandwich structure can be replaced with a competition model [94], [95], [4], [96]. The main schematic diagram is shown in Fig. 2. Sun et al. [97] developed competitive AuNP that improved real-time immuno-PCR techniques (GNP-rt-IPCR) to detect diethyl phthalate (DEP) in foodstuff samples. By optimizing the experimental conditions, a rather low linearity was achieved within a range from 4 pg/L to 40 ng/L, and the LOD was 1.06 pg/L. Zhang et al. [98] detected the small molecule triazophos in water, rice, cucumber, cabbage and apple samples based on a competitive immunoassay with BCA. The method showed a linear range of 0.01–20 μg/L, and the LOD was 6 ng/L. Du et al. [99], [100] established a bio-barcode competitive immunoassay method based on a microplate platform. By designing different DNA strands, the detection of triazophos pesticides was realized. The detection range of this method was from 2.5 × 10−2 to 40.0 ng/mL, and the sensitivity was 1.96 × 10−2 ng/mL. As such, this method provides a new direction for the rapid detection and screening of pesticide residues. Competitive colorimetric immunoassays are a new method that makes up for the shortcoming of long-term analysis and expensive equipment for single-residue detection of single-molecule substances in gas chromatography, liquid chromatography and other common detection methods [101], [102], [103], [104], [105], [106], [107], [108]. These new methods have promising accuracy and sensitivity and provide broad prospects for the rapid detection of pesticide residues in the environment, agricultural products and foods, as well as the screening of proteins.
Fig. 2.
Schematic diagram of the biological barcode competition model. (A). Probe preparation. (B). Generation of a competition structure with the target and separate detection.
Biotoxin detection
Yu et al. [109] established a BCA technology for the detection of aflatoxin B1 (AFB1) with a sensitivity of approximately 10−8 ng/mL, which is much higher than the sensitivity of ELISA. This work is also the first use of BCA technology for the analysis of AFB1 in herbal medicine. This method can be used for the trace detection of AFB1 in peanuts, cashew nuts and other nut-based foods.
Zhang et al. [110] developed a new high-sensitivity method based on BCA and RCA technology to detect T-2 toxin in food. This method exhibited a LOD of 0.26 pg/mL, a linear range of 0.002–200 ng/mL, a good recovery and a relative standard deviation of 88.65% to 10.04% and 0.6% to 13.1%, respectively. This method showed potential for the ultrasensitive detection of various small molecules in complex matrices.
Environmental pollutant detection
PCB are a class of typical persistent organochlorine compounds that are widely found in the environment, are highly toxic, and bioaccumulate, as they are difficult to degrade; thus, PCB present a major threat to the ecosystem and to human health [111].
Yang et al. [40] established a sensitive immunosorbent bio-barcode detection method based on real-time immuno-PCR to detect 3,4,3′,4′-tetrachlorobiphenyl. The linear range was 5 pg/L-10 ng/L, and the LOD was 1.72 pg/L. The coefficient of variation was within the specified range; therefore, this method could be used for rapid semi-quantitative PCB detection.
Yang et al. [41] established a bio-barcode method based on real-time immuno PCR for the analysis and detection of PCBs in environmental samples. The lower LOD of this method was 2.55 pg/L, and the method could successfully detect Aroclor 1248 in seaweed samples collected from the East China Sea in Zhejiang Province. The recovery range was from 84% to 104%. Thus, this method provides a new detection technology for applications in the detection of PCBs.
BCA versus other techniques
Compared with PCR technology, BCA has simple operation and high specificity. It replaces the PCR step with barcode amplification technology, which reduces PCR-related equipment cost and possible pollution. In addition, it does not require enzyme participation, which decreases reagent and transportation costs. The detection range is wide, and the test substance can be detected as long as their corresponding monoclonal antibody and polyclonal antibody can be obtained. Thus, this technology has great advantages in detecting some pathogenic microorganisms that are not suitable for detection by PCR technology. Compared with ELISA technology, BCA has high sensitivity and can be used in combination with the chip method, colorimetric method and fluorescence method. Its sensitivity is 5–6 orders of magnitude higher than that of conventional ELISA. Moreover, the detection time is at least 3 h less than that of ELISA. Due to insufficient detection sensitivity, ELISA cannot detect certain low-concentration substances and cannot be applied to all samples. In comparison, BCA has a wider detection range. For small-molecule substances, chromatography, mass spectrometry, spectroscopy, biosensors and other methods are commonly used for detection. However, the price of instruments and equipment for these methods is relatively high. Compared with other methods, the bio-barcode method is simple, inexpensive, specific and sensitive.
Limitations
Nevertheless, the BCA technology also has some drawbacks. First, in terms of nucleic acid detection, while BCA technology does not require enzymes for amplification, the sensitivity is not superior to that of PCR technology. While the results are reproducible, they can be difficult to accurately quantify, and false positives can be a problem. For example, there are certain false positives in the silver staining reaction, and other methods, such as the colorimetric method and fluorescent labelling method that can be used for quantitative detection. Further exploration is needed to optimize the experimental conditions and operating procedures to further improve the detection sensitivity. In some detection methods, barcode DNA needs to be amplified by PCR, or the amplified barcode is subjected to electrophoresis or chip detection by combined technology. In these cases, detection relies on expensive equipment, which limits the widespread application of these methods in practice. Second, BCA technology has been applied for the multi-residue detection of macromolecules but not small molecules, which could be an important direction for future exploration. Third, the reagents and materials required for bio-barcode-based experiments are easier to prepare than those needed for other detection methods, and their specificity also depends on the specificity of the monoclonal antibodies used in the detection system; however, the relatively high cost of commercial monoclonal antibodies and polyclonal antibodies will affect the application of this technology in actual detection. The preparation of test kit products and promotion of their use are also an important research direction. Fourth, the preparation of probes takes a long time, as the experimental preparation of some probes require 72 h or more; therefore, the reaction needs to be optimized to further shorten the preparation and detection time.
Conclusions and future perspectives
In this review, we introduced the directions and applications of several bio-barcode detection assays. After more than ten years of development and exploration, BCA technology has enabled the establishment of a simple and reliable high-efficiency system for the single-residue or multi-residue detection of macromolecular substances, such as proteins, and the single-residue detection of small molecules. As they have high sensitivity and specificity, these methods have demonstrated strong advantages for applications in clinical disease diagnosis, food safety testing, and chemical contaminant testing.
First, the nanomaterials used in bio-barcode detection technology are safe and not easily denatured by binding to target molecules; in addition, the high specificity and high sensitivity allow broad application prospects. Second, BCA technology can design barcode DNA of different lengths and sequences according to different targets to complete multi-residue detection. Third, compared with chromatographic methods and other detection methods, it is cost-effective, fast and simple in the single-residue detection of small molecules.
BCA technology is not yet fully mature, and each component of a method may affect its sensitivity and specificity. Therefore, exploring the optimal reaction conditions, reducing the testing costs, further simplifying the operation steps, improving the detection sensitivity, shortening the time of preparation and testing, realizing the detection of multiple substances in the same reaction system, and developing and commercializing BCA technology-based immunization kits are important directions for future research.
Conflict of interest
The authors have declared no conflict of interest.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Acknowledgements
The authors gratefully acknowledge the support of the National Key Research Program of China (2017YFF0210201), the National Natural Science Foundation (31671938), and the Central Public-Interest Scientific Institution Basal Research Fund for the Chinese Academy of Agricultural Sciences (Y2017JC13).
Biographies

Yuanshang Wang, is a M.S. candidate at Institute of Quality Standards and Testing Technology for Agro-products, Chinese Academy of Agricultural Sciences. Her research focused on quality safety of agricultural product.

Maojun Jin received his BSc degree in plant protection in 2004, and received his Doctor degree majored in pesticide science in Zhejiang University in 2009. He is currently the associate professor of Institute of Quality Standards & Testing Technology for Agro-Products, Chinese Academy of Agricultural Sciences. He mainly focuses on the development of immunoassay for the trace detection of pollutants in food. Until now, he published more than 40 papers, 20 of which were cited by SCI.

Ge Chen received her M.S. degree from Institute of Quality Standards & Testing Technology for Agro-Products, Chinese Academy of Agricultural Sciences in 2017. She is a Doctoral student at Institute of Quality Standards and Testing Technology for Agro-products, Chinese Academy of Agricultural Sciences. Her research focused on quality safety of agricultural product.

Xueyan Cui is a M.S. candidate at Institute of Quality Standards and Testing Technology for Agro-products, Chinese Academy of Agricultural Sciences. Her research focused on quality safety of agricultural product.

Yudan Zhang is woking at Institute of Quality Standards & Testing Technology for Agro-Products, Chinese Academy of Agricultural Sciences. Her research focused on quality safety of agricultural product.

Mingjie Li, is a M.S. candidate at Institute of Quality Standards and Testing Technology for Agro-products, Chinese Academy of Agricultural Sciences. Her research focused on quality safety of agricultural product.

Yun Liao, is a M.S. candidate at Institute of Quality Standards and Testing Technology for Agro-products, Chinese Academy of Agricultural Sciences. Her research focused on quality safety of agricultural product.

Xiuyuan Zhang, is a M.S. candidate at Institute of Quality Standards and Testing Technology for Agro-products, Chinese Academy of Agricultural Sciences. Her research focused on quality safety of agricultural product.

Guoxin Qin is working at Agro-products Quality Safety and Testing Technology Research Institute, Guangxi Academy of Agricultural Sciences. His research focused on quality safety of agricultural product.

Feiyan Yan, is working at Agro-products Quality Safety and Testing Technology Research Institute, Guangxi Academy of Agricultural Sciences. Her research focused on quality safety of agricultural product.

A. M. Abd El-Aty is a Professor of Pharmacology, Cairo University, Egypt and currently (from Jan 2018 to date) appointed as a Foreign Professor at Pharmacology Department, Faculty of Medicine, Ataturk University, Erzurum, Turkey. From 2013 till Jan 2018 he was a Brain Pool fellow in Chonnam National University, Kwangju and a Foreign Professor in Konkuk University, Seoul, Republic of Korea. His era of interest is “Food Science and Technology”; in particular, xenobiotic analysis using various extractions as well as analytical methods. He published more than 270 articles in prestigious journals, with current h-index= 27 (Scopus database). At the Editorial level, he is acting as a Managing as well as an Associate Editor of Journal of Advanced Research; Associate Editor of Lipids in Health and Disease; Advisory board member of Biomedical Chromatography and Separation Science Plus. He is also a member of 2017–2021 Expert Roster of the Joint (FAO/WHO) Expert Committee on Food Additives

Jing Wang received B.S. degree from Heilongjiang University in 1985. she was working in the Northeast Agriculture University between 1985 and 1996.After obtained her PhD, she was appointed to be an associate professor, professor of the Northeast Agriculture University and of Harbin Institute of Technology from 1996 to 2005. She is currently a professor and director of residues research department of the Institute of Quality Standards & Testing Technology for Agro-products, CAAS. She has engaged in studies of food safety and testing technology/screening novel products with bioactivities.
Footnotes
Peer review under responsibility of Cairo University.
Contributor Information
Maojun Jin, Email: jinmaojun@caas.cn.
Jing Wang, Email: wangjing05@caas.cn.
References
- 1.Trévisan M., Schawaller M., Quapil G., Souteyrand E., Mérieux Y., Cloarec J.P. Evanescent wave fluorescence biosensor combined with DNA bio-barcode assay for platelet genotyping. Biosens Bioelectron. 2010;26(4):1631–1637. doi: 10.1016/j.bios.2010.08.038. [DOI] [PubMed] [Google Scholar]
- 2.Perez J.W., Vargis E.A., Russ P.K., Haselton F.R., Wright D.W. Detection of respiratory syncytial virus using nanoparticle amplified immuno-polymerase chain reaction. Anal Biochem. 2011;410(1):141–148. doi: 10.1016/j.ab.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broto M., Salvador J.P., Galve R., Marco M.P. Biobarcode assay for the oral anticoagulant acenocoumarol. Talanta. 2018;178:308–314. doi: 10.1016/j.talanta.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Lee H., Lee D., Park J.H., Song S.H., Jeong I.G., Kim C.S. High throughput differential identification of TMPRSS2-ERG fusion genes in prostate cancer patient urine. Biomaterials. 2017;135:23–29. doi: 10.1016/j.biomaterials.2017.04.049. [DOI] [PubMed] [Google Scholar]
- 5.Hee A.J., Lee K.J., Choi J.W. Gold nanoparticles-based barcode analysis for detection of norepinephrine. J Biomed Nanotechnol. 2016;12(2):357–365. doi: 10.1166/jbn.2016.2185. [DOI] [PubMed] [Google Scholar]
- 6.An J.H., Kim T.H., Oh B.K., Choi J.W. Detection of dopamine in dopaminergic cell using nanoparticles-based barcode DNA analysis. J Nanosci Nanotechnol. 2012;12(1):764–768. doi: 10.1166/jnn.2012.5403. [DOI] [PubMed] [Google Scholar]
- 7.Amini B., Kamali M., Salouti M., Yaghmaei P. Fluorescence bio-barcode DNA assay based on gold and magnetic nanoparticles for detection of Exotoxin A gene sequence. Biosens Bioelectron. 2017;92:679–686. doi: 10.1016/j.bios.2016.10.030. [DOI] [PubMed] [Google Scholar]
- 8.Tang S., Gu Y., Lu H., Dong H., Zhang K., Dai W. Highly-sensitive microRNA detection based on bio-bar-code assay and catalytic hairpin assembly two-stage amplification. Anal Chim Acta. 2018;1004:1–9. doi: 10.1016/j.aca.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Sanvicens N., Pastells C., Pascual N., Marco M.P. Nanoparticle-based biosensors for detection of pathogenic bacteria. TrAC, Trends Anal Chem. 2009;28(11):1243–1252. [Google Scholar]
- 10.Brakmann S. DNA-based barcodes, nanoparticles, and nanostructures for the ultrasensitive detection and quantification of proteins. Angew Chem Int Ed. 2010;36(3):5730–5734. doi: 10.1002/anie.200461112. [DOI] [PubMed] [Google Scholar]
- 11.Nam J.M., Thaxton C.S., Mirkin C.A. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 12.Thaxton C.S., Hill H.D., Georganopoulou D.G., Stoeva S.I., Mirkin C.A. A bio-barcode assay based upon dithiothreitol induced oligonucleotide release. Anal Chem. 2005;77(24):8174–8178. doi: 10.1021/ac0514265. [DOI] [PubMed] [Google Scholar]
- 13.Bao Y.P., Wei T.F., Lefebvre P.A., An H., He L., Kunkel G.T. Detection of protein analytes via nanoparticle-based bio bar code technology. Anal Chem. 2006;78(6):2055–2059. doi: 10.1021/ac051798d. [DOI] [PubMed] [Google Scholar]
- 14.Lee C.Y., Shiau R.J., Chou H.W., Hsieh Y.Z. Combining aptamer-modified gold nanoparticles with barcode DNA sequence amplification for indirect analysis of ethanolamine. Sens Actuators, B. 2018;254:189–196. [Google Scholar]
- 15.Loo J.F., Yang C., Tsang H.L., Lau P.M., Yong K.T., Ho H.P. An aptamer Bio-barCode (ABC) assay using SPR, RNase H, and probes with RNA and gold-nanorods for anti-cancer drug screening. Analyst. 2017;142(19):3579–3587. doi: 10.1039/c7an01026e. [DOI] [PubMed] [Google Scholar]
- 16.An J.H., Oh B.K., Choi J.W. Detection of tyrosine hydroxylase in dopaminergic neuron cell using gold nanoparticles-based barcode DNA. J Biomed Nanotechnol. 2013;9(4):639–643. doi: 10.1166/jbn.2013.1525. [DOI] [PubMed] [Google Scholar]
- 17.Azzazy H.M., Mansour M.M., Samir T.M., Franco R. Gold nanoparticles in the clinical laboratory:principles of preparation and applications. Clin Chem Lab Med. 2012;50(2):193–209. doi: 10.1515/CCLM.2011.732. [DOI] [PubMed] [Google Scholar]
- 18.Wei Y., Zhou C.Y., Li Y.L. A novel approach to establish bio-barcode in cells by CRISPR/Cas9. Biotechnol Bull. 2017 [Google Scholar]
- 19.Ye S., Mao Y., Guo Y., Zhang S. Enzyme-based signal amplification of surface-enhanced Raman scattering in cancer-biomarker detection. TrAC, Trends Anal Chem. 2014;55:43–54. [Google Scholar]
- 20.Zhou Z., Li T., Huang H., Chen Y., Liu F., Huang C. A dual amplification strategy for DNA detection combining bio-barcode assay and metal-enhanced fluorescence modality. Chem Commun. 2014;50(87):13373–13376. doi: 10.1039/c4cc05554c. [DOI] [PubMed] [Google Scholar]
- 21.Mishra A., Kumar J., Melo J.S. An optical microplate biosensor for the detection of methyl parathion pesticide using a biohybrid of Sphingomonas sp. cells-silica nanoparticles. Biosens Bioelectron. 2017;87(6):332–338. doi: 10.1016/j.bios.2016.08.048. [DOI] [PubMed] [Google Scholar]
- 22.Tomioka K., Yamaguchi T., Inoue M., Kajiwara K. Liposome-linked immunosorbent assay enhanced by immuno-PCR using plasmid-encapsulated liposomes. Biochem Eng J. 2018;137:352–357. [Google Scholar]
- 23.Xu J., Jiang B., Su J., Xiang Y., Yuan R., Chai Y. Background current reduction and biobarcode amplification for label-free, highly sensitive electrochemical detection of pathogenic DNA. Chem Commun. 2012;48(27):3309–3311. doi: 10.1039/c2cc18107j. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z., Zhou B., Wang H., Lu F., Liu T., Song C. Highly sensitive detection of human IgG using a novel bio-barcode assay combined with DNA chip technology. J Nanopart Res. 2013;15(9):1964. [Google Scholar]
- 25.Nam J.M., Stoeva S.I., Mirkin C.A. Bio-bar-code-based DNA detection with PCR-like sensitivity. JACS. 2004;126(19):5932–5933. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y., Mao H.J., Zang G.Q., Zhang H.L., Jin Q.H., Zhao J.L. Detection of Hepatitis B virus deoxyribonucleic acid based on gold nanoparticle probe chip. Chinese J Anal. Chem. 2010;38(8):1133–1138. [Google Scholar]
- 27.Tang S., Zhao J., Storhoff J.J., Norris P.J., Little R.F., Yarchoan R. Nanoparticle-Based biobarcode amplification assay (BCA) for sensitive and early detection of human immunodeficiency type 1 capsid (p24) antigen. J Acquir Immune Defic Syndr. 2007;46(2):231–237. doi: 10.1097/QAI.0b013e31814a554b. [DOI] [PubMed] [Google Scholar]
- 28.Yin H.Q., Jia M.X., Yang S., Jing P.P., Wang R., Zhang J.G. Development of a highly sensitive gold nanoparticle probe-based assay for bluetongue virus detection. J Virol Methods. 2012;183(1):45–48. doi: 10.1016/j.jviromet.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 29.Yin H.Q., Jia M.X., Yang S., Wang S.Q., Zhang J.G. A nanoparticle-based bio-barcode assay for ultrasensitive detection of ricin toxin. Toxicon. 2012;59(1):12–16. doi: 10.1016/j.toxicon.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Zhang D., Carr D.J., Alocilja E.C. Fluorescent bio-barcode DNA assay for the detection of Salmonella enterica serovar Enteritidis. Biosens. Bioelectron. 2009;24(5):1377–1381. doi: 10.1016/j.bios.2008.07.081. [DOI] [PubMed] [Google Scholar]
- 31.Lin X., Liu Y., Tao Z., Gao J., Deng J., Yin J. Nanozyme-based bio-barcode assay for high sensitive and logic-controlled specific detection of multiple DNAs. Biosens Bioelectron. 2017;94:471–477. doi: 10.1016/j.bios.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Nam J.M., Wise A.R., Groves J.T. Colorimetric bio-barcode amplification assay for cytokines. Anal Chem. 2005;77(21):6985–6988. doi: 10.1021/ac0513764. [DOI] [PubMed] [Google Scholar]
- 33.Nam J.M., Jang K.J., Groves J.T. Detection of proteins using a colorimetric bio-barcode assay. Nat Protoc. 2007;2(6):1438. doi: 10.1038/nprot.2007.201. [DOI] [PubMed] [Google Scholar]
- 34.Du P., Jin M., Chen G., Zhang C., Cui X., Zhang Y. Competitive colorimetric triazophos immunoassay employing magnetic microspheres and multi-labeled gold nanoparticles along with enzymatic signal enhancement. Microchim Acta. 2017;184(10):3705–3712. [Google Scholar]
- 35.Trévisan Marie, Schawaller M., Quapil G., Souteyrand E., Mérieux Y., Cloarec J.P. Evanescent wave fluorescence biosensor combined with DNA bio-barcode assay for platelet genotyping. Biosens Bioelectron. 2011;26(4):1631–1637. doi: 10.1016/j.bios.2010.08.038. [DOI] [PubMed] [Google Scholar]
- 36.Zhang D., Huarng M.C., Alocilja E.C. A multiplex nanoparticle-based bio-barcoded DNA sensor for the simultaneous detection of multiple pathogens. Biosens Bioelectron. 2011;26(4):1736–1742. doi: 10.1016/j.bios.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Zheng L., Cai G., Wang S., Liao M., Li Y., Lin J. A microfluidic colorimetric biosensor for rapid detection of Escherichia coli O157:H7 using gold nanoparticle aggregation and smart phone imaging. Biosens Bioelectron. 2018;124:143–149. doi: 10.1016/j.bios.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X., Su H., Bi S., Li S., Zhang S. DNA-based amplified electrical bio-barcode assay for one-pot detection of two target DNAs. Biosens Bioelectron. 2009;24(8):2730–2734. doi: 10.1016/j.bios.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 39.Chen L., Wei H., Guo Y., Cui Z., Zhang Z., Zhang X.E. Gold nanoparticle enhanced immuno-PCR for ultrasensitive detection of Hantaan virus nucleocapsid protein. J Immunol Methods. 2009;346(1–2):64–70. doi: 10.1016/j.jim.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Yang G.X., Zhuang H.S., Chen H.Y. A sensitive immunosorbent bio-barcode assay based on real-time immuno-PCR for detecting 3, 4, 3', 4'-tetrachlorobiphenyl. Anal Bioanal Chem. 2014;406(6):1693–1700. doi: 10.1007/s00216-013-7583-9. [DOI] [PubMed] [Google Scholar]
- 41.Yang G.X., Zhuang H.S., Chen H.Y. A gold nanoparticle based immunosorbent bio-barcode assay combined with real-time immuno-PCR for the detection of polychlorinated biphenyls. Sens Actuators, B. 2015;214:152–158. [Google Scholar]
- 42.Xu Y., Huo B., Li C., Peng Y., Tian S., Fan L. Ultrasensitive detection of staphylococcal enterotoxin B in foodstuff through dual signal amplification by bio-barcode and real-time PCR. Food Chem. 2019;283:338–344. doi: 10.1016/j.foodchem.2018.12.128. [DOI] [PubMed] [Google Scholar]
- 43.Niemeyer C.M., Adler M., Wacker R. Immuno-PCR: high sensitivity detection of proteins by nucleic acid amplification. Trends Biotechnol. 2005;23(4):208–216. doi: 10.1016/j.tibtech.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 44.Duan R., Zhou X., Xing D. Electrochemiluminescence biobarcode method based on cysteamine-gold nanoparticle conjugates. Anal Chem. 2010;82(8):3099–3103. doi: 10.1021/ac100018z. [DOI] [PubMed] [Google Scholar]
- 45.Tang Y., Wang H., Xiang J. A sensitive immunosorbent bio-barcode assay combining PCR with icELISA for detection of gonyautoxin 2/3. Anal Chim Acta. 2010;657(2):210–214. doi: 10.1016/j.aca.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 46.Kim E.Y., Stanton J., Korber B.T., Krebs K., Bogdan D., Kunstman K. Detection of HIV-1 p24 Gag in plasma by a nanoparticle-based bio-barcode-amplification method. Nanomedicine. 2008;3(3):293–303. doi: 10.2217/17435889.3.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan J., Brennand D.M., Bradley N., Beirong G.A.O., Bruckdorfer R., Jacobs M. 3-Nitrotyrosine in the proteins of human plasma determined by an ELISA method. Biochem J. 1998;330(2):795–801. doi: 10.1042/bj3300795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Espana F., Griffin J.H. Determination of functional and antigenic protein C inhibitor and its complexes with activated protein C in plasma by ELISA's. Thromb Res. 1989;55(6):671–682. doi: 10.1016/0049-3848(89)90298-3. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Y., You F., Zhong J., Wang H., Ding H. Development of an ELISA for identification of immunodominant protein antigens of Mycoplasma hyopneumoniae. Chin J Biotechnol. 2018;34(1):44–53. doi: 10.13345/j.cjb.170220. [DOI] [PubMed] [Google Scholar]
- 50.Zhang L., Li H.Y., Li W., Shen Z.Y., Wang Y.D., Ji S.R. An ELISA assay for quantifying monomeric C-reactive protein in plasma. Front Immunol. 2018;9:511. doi: 10.3389/fimmu.2018.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Georganopoulou D.G., Chang L., Nam J.M., Thaxton C.S., Mufson E.J., Klein W.L. Nanoparticle based detection in cerebral spinal fluid of a solublepathogenic biomarker for Alzheimer’s disease. Proc Natl Acad Sci. 2005;102(7):2273–2276. doi: 10.1073/pnas.0409336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li F., Zhao Q., Wang C., Lu X., Li X.F., Le X.C. Detection of Escherichia coli O157:H7 using gold nanoparticle labeling and inductively coupled plasma mass spectrometry. Anal Chem. 2010;82(8):3399–3403. doi: 10.1021/ac100325f. [DOI] [PubMed] [Google Scholar]
- 53.Cui H.F., Xu T.B., Sun Y.L., Zhou A.W., Cui Y.H., Liu W. Hairpin DNA as a biobarcode modified on gold nanoparticles for electrochemical DNA detection. Anal Chem. 2015;87(2):1358–1365. doi: 10.1021/ac504206n. [DOI] [PubMed] [Google Scholar]
- 54.Zhang K., Lv S., Lin Z., Li M., Tang D. Bio-bar-code-based photoelectrochemical immunoassay for sensitive detection of prostate-specific antigen using rolling circle amplification and enzymatic biocatalytic precipitation. Biosens Bioelectron. 2018;101:159–166. doi: 10.1016/j.bios.2017.10.031. [DOI] [PubMed] [Google Scholar]
- 55.Goluch E.D., Stoeva S.I., Lee J.S., Shaikh K.A., Mirkin C.A., Liu C.A. Microfluidic detection system based upon a surface immobilized biobarcode assay. Biosens Bioelectron. 2009;24(8):2397–2403. doi: 10.1016/j.bios.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loo J.F.C., Lau P.M., Ho H.P., Kong S.K. An aptamer-based bio-barcode assay with isothermal recombinase polymerase amplification for cytochrome-c detection and anti-cancer drug screening. Talanta. 2013;115(115):159–165. doi: 10.1016/j.talanta.2013.04.051. [DOI] [PubMed] [Google Scholar]
- 57.Oh B.K., Nam J.M., Lee S.W., Mirkin C.A. A fluorophore-luorophorerem JM, Lee SW, earch combined with DNA c. Small. 2006;2(1):103–108. doi: 10.1002/smll.200500260. [DOI] [PubMed] [Google Scholar]
- 58.Pratiwi F.W., Rijiravanich P., Somasundrum M. Electrochemical immunoassay for Salmonella Typhimurium based on magnetically collected Ag-enhanced DNA biobarcode labels. Analyst. 2013;138(17):5011–5018. doi: 10.1039/c3an00606a. [DOI] [PubMed] [Google Scholar]
- 59.Bedin C., Crotti S., Tasciotti E., Agostini M. Diagnostic devices for circulating biomarkers detection and quantification. Curr. Med. Chem. 2017 doi: 10.2174/0929867324666171116124255. [DOI] [PubMed] [Google Scholar]
- 60.Huang L., Zheng L., Chen Y., Xue F., Cheng L., Adeloju S.B. A novel GMO biosensor for rapid ultrasensitive and simultaneous detection of multiple DNA components in GMO products. Biosens. Bioelectron. 2015;66:431–437. doi: 10.1016/j.bios.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 61.An J.H., Choi D.K., Lee K.J., Choi J.W. Surface-enhanced Raman spectroscopy detection of dopamine by DNA Targeting amplification assay in Parkisons's model. Biosens. Bioelectron. 2015;67:739–746. doi: 10.1016/j.bios.2014.10.049. [DOI] [PubMed] [Google Scholar]
- 62.Zhang X., Xu Y., Yang Y., Jin X., Ye S., Zhang S. A new signal-on photoelectrochemical biosensor based on a graphene/quantum-dot nanocomposite amplified by the dual-quenched effect of bipyridinium relay and AuNPs. Chem Eur J. 2012;18(51):16411–16418. doi: 10.1002/chem.201202213. [DOI] [PubMed] [Google Scholar]
- 63.Li Y., Liu B., Li X., Wei Q. Highly sensitive electrochemical detection of human telomerase activity based on bio-barcode method. Biosens Bioelectron. 2010;25(11):2543–2547. doi: 10.1016/j.bios.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y., Li H., Xie J., Chen M., Zhang D., Pang P. Electrochemical biosensor for silver ions based on amplification of DNA-Au bio-bar codes and silver enhancement. J Electroanal Chem. 2017;785:117–124. [Google Scholar]
- 65.Broto M., Galve R., Marco M.P. Sandwich NP-based biobarcode assay for quantification C-reactive protein in plasma samples. Anal Chim Acta. 2017;992:112–118. doi: 10.1016/j.aca.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 66.Xing T.Y., Zhao J., Weng G.J., Zhu J., Li J.J., Zhao J.W. Specific detection of carcinoembryonic antigen based on fluorescence quenching of hollow porous gold nanoshells with roughened surface. ACS Appl Mater Interf. 2017;9(42):36632–36641. doi: 10.1021/acsami.7b11310. [DOI] [PubMed] [Google Scholar]
- 67.Narmani A., Kamali M., Amini B., Kooshki H., Amini A., Hasani L. Highly sensitive and accurate detection of Vibrio cholera O1 OmpW gene by fluorescence DNA biosensor based on gold and magnetic nanoparticles. Process Biochem. 2018;65:46–54. [Google Scholar]
- 68.Amini A., Kamali M., Amini B., Najafi A., Narmani A., Hasani L. Bio-barcode technology for detection of staphylococcus aureus protein A based on gold and iron nanoparticles. Int J Biol Macromol. 2019;124:1256–1263. doi: 10.1016/j.ijbiomac.2018.11.123. [DOI] [PubMed] [Google Scholar]
- 69.Li X.T., Chen B.B., He M., Xiao G.Y., Hu B. Gold nanoparticle labeling with tyramide signal amplification for highly sensitive detection of alpha fetoprotein in human serum by ICP-MS. Talanta. 2018;176:40–46. doi: 10.1016/j.talanta.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 70.Clyde D. Technology: Nucleic acid detection-it's elementary with SHERLOCK! Nat. Rev. Genet. 2017;18(7):392. doi: 10.1038/nrg.2017.40. [DOI] [PubMed] [Google Scholar]
- 71.Cartwright C.P., Pherson A.J., Harris A.B., Clancey M.S., Nye M.B. Multicenter study establishing the clinical validity of a nucleic-acid amplification based assay for the diagnosis of bacterial vaginosis. Diagn Microbiol Infect Dis. 2018;92(3):173–178. doi: 10.1016/j.diagmicrobio.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 72.Yao D., Yu F., Kim J., Scholz J., Nielsen P.E., Sinner E.K. Surface plasmon field-enhanced fluorescence spectroscopy in PCR product analysis by peptide nucleic acid probes. Nucleic Acids Res. 2004;32(22) doi: 10.1093/nar/gnh175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barletta J., Bartolome A., Constantine N.T. Immunomagnetic quantitative immuno-PCR for detection of less than one HIV-1 virion. J Virol Methods. 2009;157(2):122–132. doi: 10.1016/j.jviromet.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 74.Pecchia S., Lio D.D. Development of a rapid PCR-Nucleic Acid Lateral Flow Immunoassay (PCR-NALFIA) based on rDNA IGS sequence analysis for the detection of Macrophomina phaseolina in soil. J Microbiol Methods. 2018;151:118–128. doi: 10.1016/j.mimet.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 75.Xu R., Wei S., Zhou G., Ren J., Liu Z., Tang S. Multiplex TaqMan locked nucleic acid real-time PCR for the differential identification of various meat and meat products. Meat Sci. 2018;137:41–46. doi: 10.1016/j.meatsci.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 76.Raigond B., Verma A., Kochhar T., Roach S., Sharma S., Chakrabarti S.K. Development of simplified and rapid nucleic acid release protocol for PCR based detection of Potato viruses. Phytoparasitica. 2018;46(2):255–262. [Google Scholar]
- 77.Faraji R., Behjati-Ardakani M., Moshtaghioun S.M., Kalantar S.M., Namayandeh S.M., Soltani M. The diagnosis of microorganism involved in infective endocarditis (IE) by polymerase chain reaction (PCR) and real-time PCR: a systematic review. Kaohsiung J Med Sci. 2018;34(2):71–78. doi: 10.1016/j.kjms.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hill H.D., Vega R.A., Mirkin C.A. Nonenzymatic detection of bacterial genomic DNA using the bio barcode assay. Anal Chem. 2007;79(23):9218–9223. doi: 10.1021/ac701626y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li X.M., Luo J., Zhang N.B., Wei Q.L. Nucleic acid quantification using nicking–displacement, rolling circle amplification and bio-bar-code mediated triple-amplification. Anal Chim Acta. 2015;881:117–123. doi: 10.1016/j.aca.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 80.Villa C., Costa J., Oliveira M.B.P.P., Mafra I. Novel quantitative real-time PCR approach to determine safflower (Carthamus tinctorius) adulteration in saffron (Crocus sativus) Food Chem. 2017;229:680–687. doi: 10.1016/j.foodchem.2017.02.136. [DOI] [PubMed] [Google Scholar]
- 81.Law I.L.G., Loo J.F.C., Kwok H.C., Yeung H.Y., Leung C.C.H., Hui M. Automated real-time detection of drug-resistant mycobacterium tuberculosis on a lab-on-a-disc by recombinase polymerase amplification. Anal Biochem. 2018;544:98–107. doi: 10.1016/j.ab.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 82.Zhang R., Lv S., Gong Y., Li Y., Ding C. Sensitive determination of Hg (II) based on a hybridization chain recycling amplification reaction and surface-enhanced Raman scattering on gold nanoparticles. Microchim Acta. 2018;185(8):363. doi: 10.1007/s00604-018-2907-2. [DOI] [PubMed] [Google Scholar]
- 83.Jeong A., Lim H.B. Magnetophoretic separation ICP-MS immunoassay using Cs-doped multicore magnetic nanoparticles for the determination of Salmonella typhimurium. Talanta. 2018;178:916–921. doi: 10.1016/j.talanta.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 84.Roper M.G., Guillo C. New technologies in affinity assays to explore biological communication. Anal Bioanal Chem. 2009;393(2):459. doi: 10.1007/s00216-008-2347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hsu I.H., Chen W.H., Wu T.K., Sun Y.C. Gold nanoparticle-based inductively coupled plasma mass spectrometry amplification and magnetic separation for the sensitive detection of a virus-specific RNA sequence. J Chromatogr A. 2011;1218(14):1795–1801. doi: 10.1016/j.chroma.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 86.He Q., Zhu Z., Jin L., Peng L., Guo W., Hu S. Detection of HIV-1 p24 antigen using streptavidin-biotin and gold nanoparticles based immunoassay by inductively coupled plasma mass spectrometry. J Anal At Spectrom. 2014;29(8):1477–1482. [Google Scholar]
- 87.Poturnayova A., Castillo G., Subjakova V., Tatarko M., Snejdarkova M., Hianik T. Optimization of cytochrome c detection by acoustic and electrochemical methods based on aptamer sensors. Sens Actuat, B. 2017;238:817–827. [Google Scholar]
- 88.Yin H.Q., Ji C.F., Yang X.Q., Wang R., Yang S., Zhang H.Q. An improved gold nanoparticle probe-based assay for HCV core antigen ultrasensitive detection. J Virol Meth. 2017;243:142–145. doi: 10.1016/j.jviromet.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 89.Zhang X., Xiao G., Chen B., He M., Hu B. Lectin affinity based elemental labeling with hybridization chain reaction for the sensitive determination of avian influenza A (H9N2) virions. Talanta. 2018;188:442–447. doi: 10.1016/j.talanta.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 90.Stoeva S.I., Lee J.S., Thaxton C.S., Mirkin C.A. Multiplexed DNA detection with biobarcoded nanoparticle probes. Angew Chem. 2006;45(20):3303–3306. doi: 10.1002/anie.200600124. [DOI] [PubMed] [Google Scholar]
- 91.Li Y., Cu Y.T., Luo D. Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes. Nat Biotechnol. 2005;23(7):885–889. doi: 10.1038/nbt1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He M., Li K., Xiao J., Zhou Y. Rapid bio-barcode assay for multiplex DNA detection based on capillary DNA Analyzer. J Virol Meth. 2008;151(1):126. doi: 10.1016/j.jviromet.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 93.Tang Y., Yan L., Xiang J.J., Wang W.Z., Yang H.Y. An immunoassay based on bio-barcode method for quantitative detection of semicarbazide. Eur Food Res Technol. 2011;232(6):963–969. [Google Scholar]
- 94.Le Lam T.N., Morvan C., Liu W., Bohn C., Jaszczyszyn Y., Bouloc P. Finding sRNA-associated phenotypes by competition assays: an example with Staphylococcus aureus. Methods. 2017;117:21–27. doi: 10.1016/j.ymeth.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 95.Ye S., Wang M., Wang Z., Zhang N., Luo X. A DNA-linker-DNA bifunctional probe for simultaneous SERS detection of miRNAs via symmetric signal amplification. Chem Commun. 2018;54(56):7786–7789. doi: 10.1039/c8cc02910e. [DOI] [PubMed] [Google Scholar]
- 96.Hong S., She Y., Cao X., Wang M., Zhang C., Zheng L. Biomimetic enzyme-linked immunoassay based on a molecularly imprinted 96-well plate for the determination of triazophos residues in real samples. RSC Adv. 2018;8(37):20549–20556. doi: 10.1039/c8ra03531h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun R., Zhuang H. An ultrasensitive gold nanoparticles improved real-time immuno-PCR assay for detecting diethyl phthalate in foodstuff samples. Anal Biochem. 2015;480:49–57. doi: 10.1016/j.ab.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 98.Zhang C., Du P., Jiang Z., Jin M., Chen G., Cao X. A simple and sensitive competitive bio-barcode immunoassay for triazophos based on multi-modified gold nanoparticles and fluorescent signal amplification. Anal Chim Acta. 2018;999:123–131. doi: 10.1016/j.aca.2017.10.032. [DOI] [PubMed] [Google Scholar]
- 99.Du P., Jin M., Zhang C., Chen G., Cui X., Zhang Y. Highly sensitive detection of triazophos pesticide using a novel bio-bar-code amplification competitive immunoassay in a micro well plate-based platform. Sens Actuat, B. 2018;256:457–464. [Google Scholar]
- 100.Du P., Jin M., Chen G., Zhang C., Jiang Z., Zhang Y. A competitive bio-barcode amplification immunoassay for small molecules based on nanoparticles. Sci Rep. 2016;6:38114. doi: 10.1038/srep38114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Akutsu K., Kitagawa Y., Yoshimitsu M., Takatori S., Fukui N., Osakada M. Problems and solutions of polyethylene glycol co-injection method in multiresidue pesticide analysis by gas chromatography-mass spectrometry: evaluation of instability phenomenon in type II pyrethroids and its suppression by novel analyte protectants. Anal Bioanal Chem. 2018;410(13):3145–3160. doi: 10.1007/s00216-018-1002-1. [DOI] [PubMed] [Google Scholar]
- 102.Tankiewicz M., Biziuk M. Fast, sensitive and reliable multi-residue method for routine determination of 34 pesticides from various chemical groups in water samples by using dispersive liquid-liquid microextraction coupled with gas chromatography-mass spectrometry. Anal Bioanal Chem. 2018;410(5):1533–1550. doi: 10.1007/s00216-017-0798-4. [DOI] [PubMed] [Google Scholar]
- 103.de Oliveira G.B., Vieira C.M.C.G., Orlando R.M., Faria A.F. Simultaneous determination of fumonisins B1 and B2 in different types of maize by matrix solid phase dispersion and HPLC-MS/MS. Food Chem. 2017;233:11–19. doi: 10.1016/j.foodchem.2017.04.091. [DOI] [PubMed] [Google Scholar]
- 104.Alemayehu Y., Tolcha T., Megersa N. Salting-out assisted liquid-liquid extraction combined with HPLC for quantitative extraction of trace multiclass pesticide residues from environmental waters. Am J Anal Chem. 2017;8(07):433. [Google Scholar]
- 105.Paal M., Zoller M., Schuster C., Vogeser M., Schütze G. Simultaneous quantification of cefepime, meropenem, ciprofloxacin, moxifloxacin, linezolid and piperacillin in human serum using an isotope-dilution HPLC–MS/MS method. J Pharm Biomed Anal. 2018;152:102–110. doi: 10.1016/j.jpba.2018.01.031. [DOI] [PubMed] [Google Scholar]
- 106.Huang Y., Shi T., Luo X., Xiong H., Min F., Chen Y. Determination of multi-pesticide residues in green tea with a modified QuEChERS protocol coupled to HPLC-MS/MS. Food Chem. 2019;275:255–264. doi: 10.1016/j.foodchem.2018.09.094. [DOI] [PubMed] [Google Scholar]
- 107.Timofeeva I., Shishov A., Kanashina D., Dzema D., Bulatov A. On-line in-syringe sugaring-out liquid-liquid extraction coupled with HPLC-MS/MS for the determination of pesticides in fruit and berry juices. Talanta. 2017;167:761–767. doi: 10.1016/j.talanta.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 108.Behniwal P.K., She J. Development of HPLC-MS/MS method for the simultaneous determination of metabolites of organophosphate pesticides, synthetic pyrethroids, herbicides and DEET in human urine. Int J Environ Anal Chem. 2017;97(6):548–562. [Google Scholar]
- 109.Yu Y.Y., Chen Y.Y., Gao X., Liu Y.Y., Zhang H.Y., Wang T.Y. Nanoparticle based bio-bar code technology for trace analysis of aflatoxin B1 in Chinese herbs. J Food Drug Anal. 2018;26(2):815–822. doi: 10.1016/j.jfda.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang M., Huo B.Y., Yuan S., Ning B.A., Bai J.L., Peng Y. Ultrasensitive detection of T-2 toxin in food based on bio-barcode and rolling circle amplification. Anal Chim Acta. 2018;1043:98–106. doi: 10.1016/j.aca.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 111.Tsutsumi T., Amakura Y., Nakamura M. Validation of the CALUX bioassay for the screening of PCDD/Fs and dioxin-like PCBs in retail fish. Analyst. 2003;128(5):486–492. doi: 10.1039/b300339f. [DOI] [PubMed] [Google Scholar]