Abstract
Even with increasing evidence for roles of glycolytic enzymes in controlling cancerous characteristics, the best target of candidate metabolic enzymes for lessening malignancy remains under debate. Pyruvate is a main glycolytic metabolite that could be mainly converted into either lactate by Lactate Dehydrogenase A (LDHA) or acetyl-CoA by Pyruvate Dehydrogenase E1 component α subunit (PDHA1) catalytic complex. In tumor cells, accumulating lactate is produced whereas the conversion of pyruvate into mitochondrial acetyl-CoA is less active compared with their normal counterparts. This reciprocal molecular association makes pyruvate metabolism a potential choice of anti-cancer target. Cellular and molecular changes were herein assayed in Head and Neck Squamous Cell Carcinoma (HNSCC) cells in response to LDHA and PDHA1 loss in vitro, in vivo and in clinic. By using various human cancer databases and clinical samples, LDHA and PDHA1 levels exhibit reversed prognostic roles. In vitro analysis demonstrated that decreased cell growth and motility accompanied by an increased sensitivity to chemotherapeutic agents was found in cells with LDHA loss whereas PDHA1-silencing exhibited opposite phenotypes. At the molecular level, it was found that oncogenic Protein kinase B (PKB/Akt) and Extracellular signal-regulated kinase (ERK) singling pathways contribute to pyruvate metabolism mediated HNSCC cell growth. Furthermore, LDHA/PDHA1 changes in HNSCC cells resulted in a broad metabolic reprogramming while intracellular molecules including polyunsaturated fatty acids and nitrogen metabolism related metabolites underlie the malignant changes. Collectively, our findings reveal the significance of pyruvate metabolic fates in modulating HNSCC tumorigenesis and highlight the impact of metabolic plasticity in HNSCC cells.
Abbreviations: 4-NQO, 4-nitroquinoline 1-oxide; 5-FU, 5-fluouracil; ABC, ATP-binding cassette; ACACB, Acetyl-CoA carboxylase beta; ALDH, Aldehyde dehydrogenase; CDDP, Cisplatin; DCA, Dicholoroacetate; DLAT, Dihydrolipoamide S-acetyltransferase; DLD, Dihydrolipoamide dehydrogenase; DON, 6-diazo-5-oxo-L-norlucine; ECM, Extracellular matrix; EGCG, Epigallocatechin gallate; EMT, Epithelial-mesenchymal transition; ENO, Enolase; ERK, Extracellular signal-regulated kinase; FASN, Fatty acid synthase; hOSCC, human oral squamous cell carcinoma; GC-FID, Gas chromatograph-flame ionization detector; GLS1, Glutaminase 1; Gluts, Glucose transporters; GLUD1/2, Glutamine dehydrogenase 1/2; G3PDH, Glyceraldehyde-3-phosphate dehydrogenase; HNSCC, Head and neck squamous cell carcinoma; IC50, Half maximal inhibitory concentration; INN, Silibinin; LDHA, Lactate dehydrogenase A; LC–MS, Liquid chromatography-mass spectrophotometry; MUFAs, Monounsaturated fatty acids; MTT, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; NHOK, Normal human oral keratinocytes; OCR, Oxygen consumption rate; OS, Overall survival; OxPhos, Oxidative phosphorylation; PDC, Pyruvate dehydrogenase complex; PDHA1, Pyruvate dehydrogenase E1 component α subunit; PDK1, Pyruvate dehydrogenase kinase 1; PDP1, Pyruvate dehydrogenase phosphatase 1; PEP, Phosphoenolpyruvate; PFK1, Phosphofructokinase 1; PGAM1, Phosphoglycerate mutase 1; PKB/Akt, Protein kinase B; PKM2, Pyruvate kinase M2; PPP, Pentose phosphate pathway; PUFAs, Polyunsaturated fatty acids; ROS, Reactive oxygen species; SCD1, Stearoyl-CoA desaturase 1; SFAs, Saturated fatty acids; shRNA, short-hairpin RNA; SREBF1/2, Sterol regulatory element-binding transcription factor 1/2; TAXOL, Paclitaxel; TCA, Tricarboxylic acid; TCGA, The Cancer Genomic Atlas
Introduction
In the physiological aspect, glucose and oxygen availability are key factors for determination of cellular metabolism. A single glucose molecule could be transported into cells via membranous glucose transporters (Gluts) and metabolized by a multi-step glycolysis to generate pyruvate. Normal cells in non-malignant tissues are exposed to various levels of oxygen with respect to their distance from the closest blood vessel developing an evolutionary choice of “Pasteur effect” as a system to fine-tune cell metabolism. Many rapidly growing cells, on the contrary, rely primarily on glucose fermentation during proliferation regardless of oxygen availability, known as aerobic glycolysis or the “Warburg effect” [1], [2]. Despite being less efficient for energy production, aerobic glycolysis is a metabolic hallmark uniquely observed in cancer cells compared to its normal counterparts, and the detection of up-regulated expression and activity of Gluts in cancer cells partly explains that cancer cells are highly dependent on glucose uptake for their survival [3]. The reversal of the Warburg phenotype had therefore been considered as one of the targets to develop anti-cancer drugs [4].
Recent studies indeed showed down-regulated malignancy in various tumors deficient for glycolytic molecules or its metabolites [5]. For example, loss of glyceraldehyde-3-phosphate dehydrogenase (G3PDH), Enolase (ENO), Phosphoglycerate Mutase 1 (PGAM1) and Pyruvate kinase M2 (PKM2) attenuates Warburg phenotype and down-regulated cell malignancy in different human cancer cells including Head and Neck Squamous Cell Carcinoma (HNSCC), leukemia as well as gastric and lung cancers, through the regulation of anti-apoptotic protein and pro-inflammatory chemokine [6], [7], [8]. At the molecular level, it was found that a dynamic post-translational modification of proteins by O-linked β-N-acetylglucosamine (O-GlcNAcylation) on phosphofructokinase 1 (PFK1) inhibited PFK1 activity and redirected glucose flux through Pentose Phosphate Pathway (PPP) conferring a selective growth advantage on cancer cells revealing a novel regulatory mechanism of metabolic pathways for therapeutic intervention [9]. As for metabolites, a recent study demonstrated that phosphoenolpyruvate (PEP) serves as a metabolic checkpoint molecule of tumor-reactive T cells and could modulate anti-tumor T cell responses [10]. On the other hand, although some cancers exhibited mutations in the nuclear encoded mitochondrial TriCarboxylic Acid (TCA) cycle enzymes that produce oncogenic metabolites, the impacts of Oxidative Phosphorylation (OxPhos) related factors in regulating cancer malignancy, however, are largely unknown.
Among all metabolic molecules, the enzymatic catalysis to define pyruvate metabolism could be a good target to drive metabolic forces away from aerobic glycolysis towards mitochondrial OxPhos, thereby lessening neoplastic properties in cancer cells. Pyruvate metabolism and carbon flux is altered in many human diseases including cancers [11]. Pyruvate could either be oxidatively metabolized in mitochondrion to form acetyl-CoA or oxaloacetate (OAA) or be reductively converted into organic acids/alcohols (e.g., lactate, acetate, or ethanol) and alanine via the Cahill cycle in cytosol [12]. Two key factors defining the by-products of pyruvate catabolism, Lactate dehydrogenase A (LDHA) and Pyruvate dehydrogenase complex (PDC), have drawn increasing attention for controlling tumorous phenotypes. LDHA respectively catalyzes the conversion of NADH and pyruvate into NAD+ and lactate, which could be transported outside of cancer cells resulting in a more acidic microenvironment that triggers cell malignancy [13]. Previous studies indicated that LDHA expression is enriched in many human cancers including gastric, colon, lung cancers and leukemia [14], while LDHA suppression modulates cell growth and motility, possibly via metabolic changes towards to OxPhos in order to re-oxidize NADH and produce ATP [15].
On the contrary, the other arm of pyruvate utilization is to convert pyruvate into acetyl-CoA controlled by PDC that is comprised of multiple copies of three distinct enzymes including pyruvate dehydrogenase (PDH, E1), dihydrolipoamide S-acetyltransferase, (DLAT, E2) and dihydrolipoamide dehydrogenase (DLD, E3) in humans [16]. Among them, PDHE1 alpha subunit named pyruvate dehydrogenase A1 (PDHA1) serves as a rate-limiting subunit of PDC [17]. PDHA1 activity is mainly regulated by balance between Pyruvate Dehydrogenase Kinase 1 (PDK1) and Pyruvate Dehydrogenase Phosphatase 1 (PDP1). PDK1 mediated PDHA1 phosphorylation inactivates PDC as PDP1 enhances PDC activity by down-regulating PDHA1 phosphorylation. Former studies indicated that PDC activation led to an elevated Warburg phenotype resulting in an increased cell growth in different cancers implying that PDC-mediated pyruvate metabolism plays a critical role in controlling cancerous identity [18]. Although a PDK1 inhibitor such as Dicholoroacetate (DCA) was emphasized for its role in suppressing tumor growth, DCA exhibited low-potency, required high effective dose [19] and routinely caused peripheral neurological condition, cataract formation and testicular degeneration in vivo limiting its anti-cancer application [20]. It therefore becomes imperative to clarify the anti-cancer impacts by targeting the other PDC-associated molecules, such as PDHA1.
Current prognosis of HNSCC remains poor due to (i) a failure to diagnose disease at an early stage (ii) recurrent primary or secondary tumors, and (iii) minimal effectiveness of conventional treatments [21]. It was widely accepted that viral infection (e.g., human papillomavirus, HPV), betel nut chewing, smoking, alcohol consumption, and imbalanced metabolism could trigger gene mutations and epigenetic modulations during HN tumorigenesis [22]. While our previous finding demonstrated that hyperglycemic conditions facilitated HNSCC malignancy [23] and a prospective study reported that elevated lactate levels in HNSCC tumor tissues could serve as a predictive marker for poorer Overall Survival (OS) and Recurrence Free Survival (RFS) [24], the present study was conducted to examine multifaceted cellular, molecular and metabolic changes of HNSCCs in response to genetic or pharmaceutical modulations for LDHA and PDHA1 in order to delineate the role of pyruvate metabolism in regulating HNSCC malignancy.
Materials and Methods
Chemicals, HNSCC Cells, Animal and Human Tissues
Puromycin, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), DCA, linolenic acid (LA), spermine, cisplatin (CDDP), 5-fluouracil (5-FU), Paclitaxel (TAXOL), curcumin, epigallocatechin gallate (EGCG), silibinin (INN), metformin, ERK1/2 inhibitor PD98059 and glutaminase inhibitor 6-diazo-5-oxo-L-norlucine (DON) were purchased from Sigma. PKB/Akt inhibitor MK-2206 was obtained from Selleckchem. HNSCC cell lines including SAS and HSC3 (tongue cancer), FaDu (hypopharyngeal cancer) and OECM-1 (Oral squamous cell carcinoma, OSCC) were obtained elsewhere [23]. Animal procedures were approved in accordance with the Institutional Animal Care and Use Committee (IACUC), National Yang-Ming University (NYMU) [23], [25]. For xenografic tumor growth assay, cells were subcutaneously injected at the back of nude mice from National Laboratory Animal Centre and tumor size/mass was recorded. Clinical human HNSCC tissues (Supplementary Table 1) were obtained under approval of the Institutional Review Boards (IRB) of MacKay Memorial Hospital (IRB approval number: 12MMHIS178) and National Taiwan University Hospital (IRB approval number: 203106005RINC), Taiwan.
Establishment of LDHA/PDHA1 Deficient/Reporter HNSCC Cells
Human HNSCC cells were grown and maintained in indicated conditions [23]. The plasmids encoding small hairpin RNA (shRNA) targeting LDHA/PDHA1 genes were purchased from National RNAi Core Facility (NRCF), Academic Sinica, Taiwan (Supplementary Table 2). Lentiviral vectors containing shLDHA/shPDHA1/shLuc/shLacZ were generated in 293 T cells. LDHA and PDHA1 deficient HNSCC cells were cultured in medium containing 4 μg/ml puromycin. For establishment of reporter HNSCC cells, the plasmids containing Gaussia Luciferase (GLuc) reporter protein driven by LDHA (HPRM21172-PG02) and PDHA1 (HPRM22843-PG02) promoters were purchased from GeneCopoeia™. pLDHA-GLuc and pPDHA1-GLuc transfected cells were selected with 5 μg/ml puromycin and survived colonies were hand-picked using cloning ring.
Cellular and Molecular Assays
Cells growth, cell cycle, Annexin V-FITC based cell apoptosis, transwell-based cell migration/invasion, quantitative real-time PCR (qRT-PCR), Western blot, immunostaining and Human antibody array analysis were performed followed by protocols previously described [23]. Measurements were carried out using ELISA reader (BIO-TEK instruments, USA) and Beckman Coulter Cytomics FC500 Flow Cytometry at Instrumentation Resource Center (IRC), NYMU. Primers for qRT-PCR analysis (Supplementary Table 3) and antibodies used for Western blot and immunostaining analysis (Supplementary Table 4) are listed. Image J was used to quantify protein expression.
XF Metabolic Assay
Measurements were made with a Seahorse XF instrument. Cells were seeded in 24-well Seahorse tissue culture microplates. Approximately 45 minutes prior to the assay, the culture medium was exchanged with a low-buffered and serum-free medium to ensure accurate ECAR readings. For detection of acute drug responses, Oxygen Consumption Rates (OCRs) were measured followed by a sequential additions of compounds (Oligomycin, FCCP and Rotenone/Antimycin A).
Measurement of Extracellular Lactate, Intracellular Pyruvate/ATP, Glucose Uptake and PDH Activity
Lactate colorimetric/fluorometric assay kit, ATP colorimetric/fluorometric Assay kit, Pyruvate assay kit and Pyruvate Dehydrogenase Activity Colorimetric Assay kit (Biovision) and Glucose Uptake Cell-based Assay kit (Cayman Chemical) were carried out following manufacturer's instructions. Cell number or isolated genomic DNA content (Bioman Scientific Co. Ltd.) was used to normalize detections.
Luciferase Assay
Promoter activity of pLDHA-GLuc and pPDHA1-GLuc containing HNSCC cells were assayed using Secrete-Pair™ Dual Luminescence Assay Kit (GeneCopeia™) normalized by DNA content. In brief, cell culture medium was collected after indicated treatments, mixed with GLuc Assay Working Solution and measured by ELISA reader at IRC, NYMU.
H2DCF-DA Assay for ROS Detection
For intracellular ROS analysis, HNSCC cells were resuspended in PBS containing 2% FBS and loaded with 10 μM DCF-DA (Thermo Fisher Scientific) or 2.5 μM CellROX Deep Red (Thermo Fisher Scientific) and incubated at 37 °C for 30 minutes. An oxidation insensitive analog of Carboxy-DCFDA (Thermo Fisher Scientific) staining was used as a control to detect the probe uptake, probe efflux, and nonspecific probe activation. Cells were washed and resuspended in PBS containing 10 μM propidium iodide (PI) solution for analysis on a Beckman Coulter Cytomics FC500 Flow Cytometry at IRC, NYMU.
Metabolomics/Lipidomic Analysis
Cellular metabolites were analyzed using Liquid Chromatography-Mass Spectrophotometry (LC–MS) at Metabolomics Core laboratory at Genomics Centre of National Taiwan University following the previous described protocol [26]. In short, cells were washed, pelleted and treated with methanol and iced LC–MS grade water followed by vigorous vortex and centrifuged to remove cell debris. The supernatants were collected for LC–MS metabolic profiling analysis. For lipid profiling, cellular lipids were extracted in methanol/chloroform (v/v = 1:2) mixture and analyzed using Gas chromatograph-flame ionization detector (GC-FID) at Yuanpei University of Medical Technology following the procedure previously described [27].
Statistical Analysis
For Kaplan–Meier survival analysis, based on FPKM value of each gene, patients were classified into two expression groups and the correlation between expression level and patient survival was examined. Genes with a median expression less than FPKM 1 were excluded. The prognosis of each group of patients was examined by Kaplan–Meier survival estimators, and the survival outcomes of the two groups were compared by log-rank tests. The maximally separated Kaplan–Meier plots are presented in the Human Protein Atlas. All in vitro analyses were performed using Microsoft Excel and statistical software Prism 5 (GraphPad). All quantitative results were presented as mean ± SEM and significant difference was defined as the P-value <.05.
Results
LDHA and PDHA1 Expression Determines HNSCC Clinic Outcomes
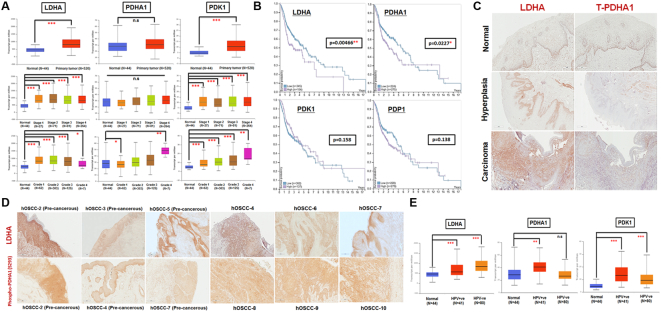
The clinical impacts of pyruvate metabolic molecules LDHA, PDHA1 as well as PDHA1 associated regulators PDK1 and PDP1 in HNSCCs was first examined using The Cancer Genomic Atlas (TCGA) based UALCAN [28] and The Human Protein Atlas (www.proteinatlas.org) [29] databases. LDHA and PDK1 transcripts, in agreement with previous findings in other cancers [14], are enriched in HNSCCs in a stage- and grade-dependent manner; PDHA1, on the contrary, exhibited no significant changes between normal and primary HNSCC tumor tissues when the analysis was stratified by stages (Figure 1A). Further analysis to define the role of differential LDHA/PDHA1/PDK1/PDP1 expression level for OS probability indicated that higher LDHA and PDHA1 expression, but not for PDK1 and PDP1 expression, seems to associate with poorer OS in HNSCC patients within 5 years while, in long-term (> 6 years) setting, patients with higher PDHA1 expressing HNSCC tumors exhibited a steady survival trend (Figure 1B). LDHA/PDHA1 expression in HNSCCs were further determined in human and mouse head and neck cancer tissues. Immunohistochemical (IHC) analysis demonstrated that, in contrast to normal oral epithelium, LDHA (Figure 1, C and D) and phosphorylated PDHA1 (S293; inactive form) (Figure 1D) expression was strongly detected in human Oral Squamous Cell Carcinoma (hOSCC) as well as in human pre-cancerous hyperplastic oral epithelial lesions; total PDHA1 protein, nevertheless, is weakly expressed in oral cancerous tissues (Figure 1C). Similar results were also observed in 4-nitroquinoline 1-oxide (4-NQO) induced mouse tongue cancers (Supplementary Fig. 1).
Figure 1.
LDHA Level and PDC Activity Reversely Correlate HNSCC Pathology in Clinic. (A) Box-whisker plots showed the analysis for LDHA, PHDA1 and PDK1 mRNA levels in normal and primary HNSCC tissues as well as tumor tissues, stratified by clinical stages and grades, from UALCAN database. Level 3 TCGA RNA-seq data corresponding to the primary tumor and normal samples for each gene is represented as Box-whisker plots. Highcharts was used to generate the visualization representing interquartile range (IQR) [28]. The boundary of the box closest to zero indicates the 25th percentile, a black line within the box marks the median and the boundary of the box farthest from 0 indicates the 75th percentile. Whiskers above and below the box indicate the maximum and minimal values, respectively. (B) Kaplan–Meier analysis for cancer-specific survival rates in HNSCC patients in The Human Protein Atlas classified by LDHA, PDHA, PDK1 and PDP1 expression. The expression cut-off values are 226.3, 13.4, 2.7 and 9.3 FPKM (Fragments Per Kilobase of transcript per Million mapped reads) for LDHA, PDHA1, PDK1 and PDP1, respectively. IHC analysis for (C, D) LDHA, (C) total PDHA1 and (D) phosphorylated PDHA1 (S293) expression in human oral normal, hyperplastic and cancerous tissues. (E) Statistic analysis for LDHA, PHDA1 and PDK1 mRNA expression in normal, HPV+ and HPV− HNSCC tissues from UALCAN database. Data are presented as mean ± SEM. ***P < .001; **P < .01; *P < .05; n.s. = non-significant.
It was noteworthy that LDHA and PDK1 are greatly expressed in both HPV+ and HPV− HNSCC patients whereas PDHA1 expression is only significantly increased in HPV+, but not in HPV−, HNSCC population compared with normal counterparts (Figure 1E). Numbers of epidemiological studies have established a causal role for HPV, particularly type 16, in HNSCCs [30]. While patients with HPV+ HNSCC tumors exhibited higher response rates after chemo/radiation treatments and improved OS; a recent large international study, nevertheless, found that HPV DNA is only positively stained in 18–22%, 3–4%, and 1.5–3% of cancers originated from oropharynx, oral cavity and larynx, respectively implying that most HNSCC tumors are HPV−, despite substantial HPV contribution to HNSCCs [31]. Collectively, these analyses suggest that the combinational expression of LDHA and PDHA1 could potentially serve as an important indicator for clinical outcomes in the majority of HNSCC population.
LDHA and PDHA1 Loss Display Opposite Effects in Regulating HNSCC Cell Growth and Cell Motility
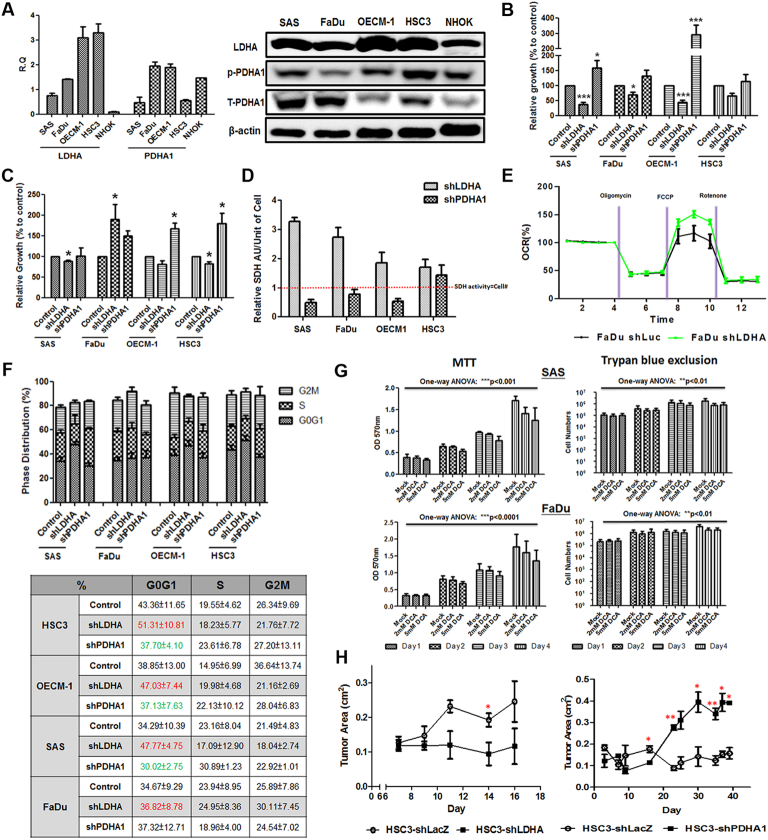
In different HNSCC cells, LDHA mRNA and protein expression is enriched compared to Normal Human Oral Keratinocytes (NHOK) while PDHA1 mRNA/protein is abundantly detected (Figure 2A). Multifaceted phenotypes were evaluated in LDHA and PDHA1 deficient HNSCC cells (Supplementary Fig. 2 and 3) to gain a systemic overview of pyruvate metabolism mediated malignant changes. Trypan blue exclusion (Figure 2B) assays demonstrated that LDHA-silencing led to decreased cell growth in HNSCC cells whereas upregulated cell growth was detected in response to PDHA1 loss. Interestingly, it was found that mitochondrial succinate dehydrogenase (SDH) activity, measured by MTT assay (Figure 2C), per unit of cell was upregulated in LDHA deficient cells while PDHA1 downregulation led to decreased SDH activity revealing a potential metabolic shift in response to LDHA/PDHA1 knockdown (Figure 2D). Indeed, Seahorse analysis showed an enhanced mitochondrial Oxygen Consumption Rate (OCR) in LDHA-silencing FaDu cells compared with control cells (Figure 2E) whilst PDHA1 knockdown only showed decreased cellular respiration (Supplementary Fig. 4). Further analysis confirmed that differential cell cycling change (Figure 2F) and deregulated cell apoptosis (Supplementary Fig. 5) underlie LDHA/PDHA1 mediated HNSCC cell growth changes. In addition to genetic manipulations, an effective DCA treatment (Supplementary Fig. 6) significantly suppressed HNSCC cell growth in a dose- and time-dependent manner (Figure 2G). In vivo xenografic tumor growth in response to LDHA/PDHA1 loss was next examined. It was found that LDHA downregulation led to significantly smaller HNSCC-bearing tumors while PDHA1 silencing resulted in greater tumors compared with control cells (Figure 2H).
Figure 2.
Differential Cell Growth in Response to LDHA/PDHA1 Knockdown in HNSCC Cells in vitro and Xenografic Tumor Growth in vivo. (A) Respective real-time RT-PCR and Western blot analysis for LDHA/total PDHA1 (T-PDHA1)/Phosphorylated PDHA1(S293, p-PDHA1) mRNA and protein expression in HNSCC and NHOK cells. Cell growth was assayed in LDHA- and PDHA1-silencing HNSCC cells using (B) trypan exclusion and (C) MTT assays. (D) Mitochondrial SDH activity in single unit of LDHA- and PDHA1-silencing HNSCC cells was determined by the values of MTT levels divided by corresponding cell number. Red dotted line indicated the condition that SDH activity represents compatible cell number as in control cells. (E) Seahorse metabolic analysis showed an elevated OCR in LDHA deficient FaDu cells. (F) Flow cytometry based cell cycle analysis was performed in LDHA/PDHA1 deficient HNSCC cells. LDHA loss leads to a greater percentage of G0/G1 cells (red) whereas PDHA1 deficiency results in lower G0/G1 cell proportion (green). (G) MTT assay showed suppressed cell growth in response to treatments of PDK1 inhibitor DCA in HNSCC SAS and FaDu cells. (H) In vivo HNSCC-bearing xenografic tumor growth analysis showed that LDHA loss downregulated tumor growth while PDHA1-silencing results in bigger and heavier tumor mass. shLuc vector is used as a control plasmid. R.Q.: Relative Quantification. Data are presented as mean ± SEM (N≥3). ***P < .001; **P < .01; *P < .05.
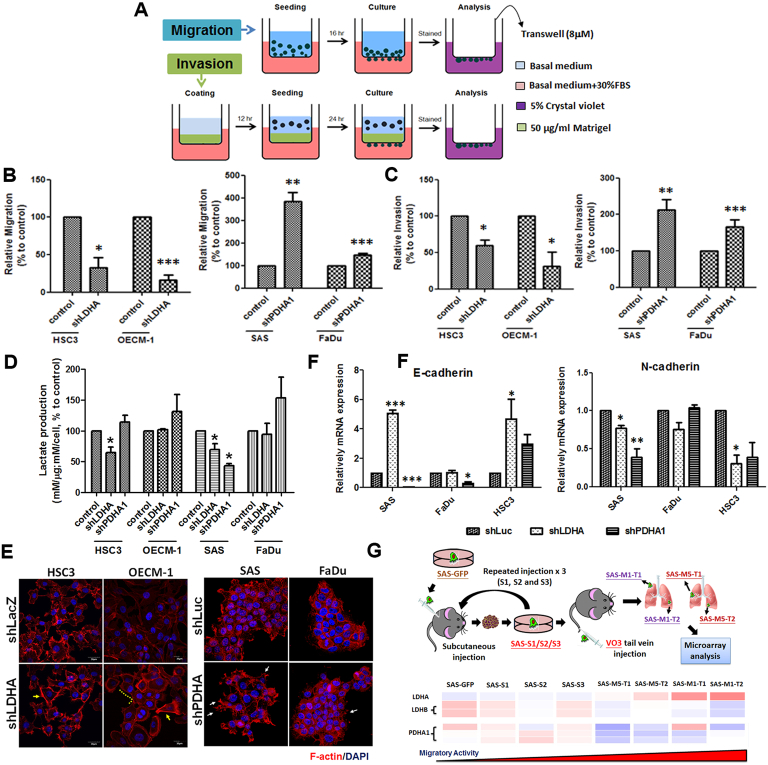
Potential impact of LDHA/PDHA1 loss in controlling cell motility was next determined using transwell-based migration and invasion assays (Figure 3A). The results demonstrated that LDHA deficiency could lead to decreased cell migration and invasion; on the contrary, PDHA1-silencing led to greater cell motility in HNSCC cells (Figure 3, B and C). On a cellular and molecular basis, cancer cell movement could be regulated by multiple factors including lactate levels, cytoskeletal organization and epithelial-Mesenchymal Transition (EMT)-associated mechanisms [32], [33], [34]. In response to LDHA loss, a decreased extracellular lactate production (Figure 3D) as well as changes of EMT markers E-cadherin and N-cadherin (Figure 3E) was detected predominantly in SAS and HSC3 cells; in contrast, PDHA1 knockdown seems to lead to an inconsistent trend in different HNSCC cells, implying that lactate and EMT may partly contribute to LDHA/PDHA1 mediated cell motility. Moreover, immunofluorescence staining analysis (IFA) showed that lamellipodia-like F-actin structures were predominantly detected in LDHA deficient cells as filopodia-like protrusions were more evident in PDHA1-silencing HNSCC cells (Figure 3F). Roles of LDHA and PDHA1 in controlling in vivo tumor metastasis was further defined in primary and metastatic xenografic tumors established previously [25]. Microarray analysis showed that LDHA is enriched in highly migrating cells while PDHA1 and LDHB, another LDH protein that catalyze lactate to pyruvate, was downregulated in this model (Figure 3G). In short, multiple molecular cues, though in various degrees, play essential roles in regulating cell migration upon LDHA/PDHA1 loss.
Figure 3.
Cell Motility Altered in LDHA/PDHA1 Deficient HNSCC Cells. (A) Trans-well based cell migration and invasion was assayed in LDHA/PDHA1 deficient HNSCC cells. LDHA loss led to decreased (B) migration and (C) invasion where PDHA1 decrease results in the opposite effect in HNSCC cells. The changes of cell motility could result from (D) different extracellular lactate production; (E) rearrangement of lamellipodia-like (yellow lined and arrows) and filopodia-like (white arrows) F-actin derived structures; and (F) altered EMT-associated mRNA expression. (G) Microarray analysis showed a reverse correlation between LDHA and LDHB/PDHA1 mRNA expression in serial subcutaneous injected in situ (S1/2/3) and tail vein injected metastatic (M1/M5) SAS derived tumors. Data are presented as mean ± SEM (N≥3). ***P < .001; **P < .01; *P < .05.
LDHA/PDHA1 Modulates Drug Sensitivity via Regulations of Stemness and Differentiation in HNSCCs
The therapeutic efficacy of chemotherapeutic agents, CDDP, 5-FU and TAXOL, in LDHA/PDHA1-silencing HNSCC cells was next defined by detection of half maximal inhibitory concentration (IC50). The results showed a common pattern of decreased IC50 level for tested chemotherapy drugs in LDHA deficient HNSCC cells compared with control cells whereas PDHA1 downregulation resulted in greater IC50 concentration (Figure 4A). On the cellular level, a number of studies reported that stemness, EMT activation and ATP-binding cassette (ABC) complex contribute to chemoresistance in HNSCCs [35], [36]. Stemness markers Aldehyde dehydrogenase (ALDH) activity and ABCC family mRNA expression was decreased in response to LDHA knockdown (Figure 4, B and C) whereas Nanog and Oct4 mRNA were upregulated in PDHA1-silencing HNSCC cells (Figure 4D). On the other hand, the protein expression of epithelial differentiation marker involucrin in LDHA/PDHA1-silencing HNSCC cells negatively correlated with stemness (Figure 4E), further supporting that alterations of LDHA and PDHA1 expression modulated stemness and differentiation status, which differentially regulate drug sensitivity in HNSCC cells.
Figure 4.
LDHA/PDHA1 Loss Modulate Therapeutic Sensitivity in HNSCC Cells. (A) Decreased IC50 for clinical chemotherapeutic agent CDDP, 5-FU and paclitaxel in LDHA deficient HNSCC cells as higher IC50 was detected in response to PDHA1 loss. This effect is probably regulated via (B) alterations of ALDH activity, (C) ABCC family and (D) Nanog/Oct4 mRNA expression or/and (E) differential changes of cellular differentiation marker involucrin protein by Western blot and immunofluorescence staining analysis in LDHA/PDHA1-silencing HNSCC cells. (F) LDHA/PDHA1 reporter systems were established to explore metabolism-mediated anti-cancer molecules. Dietary anti-neoplastic agents curcumin, EGCG, AMPK activator metformin and herbal extract compound silibinin, but not CDDP, suppressed LDHA promoter activity and activated PDHA1 promoter activity in FaDu and OECM-1 cells. Data are presented as mean ± SEM (N≥3). ***P < .001; **P < .01; *P < .05.
Overall phenotypic changes suggest LDHA and PDHA1 act opposite in determination of HNSCC malignancy, the potential to explore pyruvate metabolism mediated anti-cancer molecule(s) was therefore examined. HNSCC cells containing vectors with secreted luciferase reporter protein driven by LDHA and PDHA1 promoters were successfully established. Numbers of dietary anti-neoplastic compounds as well as CDDP were applied to clarify the efficacy of anti-cancer effects of these compounds in LDAH/PDHA1 reporter cells. The results interestingly showed that LDHA promoter activity was suppressed and PDHA1 promoter was activated under the treatment of dietary anti-cancer drugs, but not by CDDP, implying that CDDP exhibits its anti-cancer effect unlikely through metabolism based machinery (Figure 4F).
PKB/Akt and ERK Signaling Pathway Underlie LDHA/PDHA1 Mediated HNSCC Cell Growth
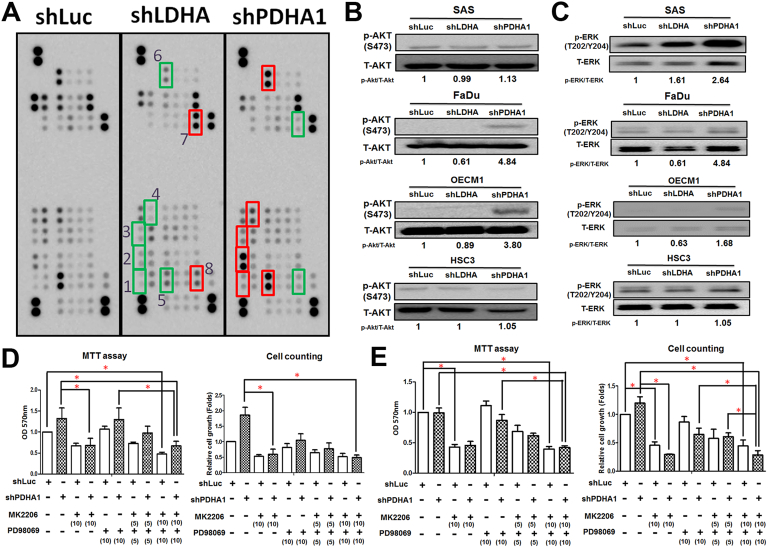
LDHA/PDHA1 mediated molecular regulatory players for HNSCC malignancy were next explored. It was widely accepted that tumor cells co-opt numbers of signaling pathways to allow them to proliferate, survive and invade other tissues [37]. Based on our data showing opposing cellular changes in LDHA- and PDHA1-silencing HNSCC cells, the candidate regulators should exhibit reversed expression levels, in comparison with control cells, in response to LDHA and PDHA1 loss. By using a large-throughput phosphorylation protein array (Figure 5A), it was found that p38alpha kinase (T180/Y182), ERK1/2 (T202/Y204, T185/Y187), JNK1/2/3 (T183/Y185, T221/Y223), Akt (S473), CREB (S133) and C-Jun (S63) proteins (higher expression in response to PDHA1 knockdown) as well as STAT3 (Y727) and Chk2 (T68) proteins (higher expression in response to LDHA knockdown) fulfilled these criteria. Ras–ERK and PI3K-Akt signaling pathways that play central roles in multiple cancerous processes were next emphasized for their roles in controlling LDHA/PDHA1 mediated HNSCC cell growth. Increased PKB/Akt (Figure 5B) and ERK1/2 (Figure 5C) phosphorylation was detected in PDHA1-silencing HNSCC cells. With effective treatments of ERK1/2 inhibitor PD98059 and PKB/Akt inhibitor MK2206 in HNSCC cells (Supplementary Fig. 7), PDHA1 mediated cell growth enhancement was abolished in OECM-1 (Figure 5D) and FaDu (Figure 5E) cells. Interestingly, a combinational treatment of PD98059 and MK2206 seems to display a dose-dependent synergetic effect in controlling PDHA1 mediated HNSCC cell growth.
Figure 5.
PKB/Akt and Erk Signaling Pathways Underlie LDHA/PDHA1 Mediated Malignant Changes in HNSCC Cells. (A) Human Protein array analysis for phosphorylated proteins differentially expressed in LDHA and PDHA1-silencing OECM-1 cells were performed. Green boxes indicated oncogenic phosphorylated molecules: (1) p38 alpha (T180/Y182), (2) Erk1/2 (T202/Y204, T185/Y187), (3) JNK1/2/3 (T183/Y185, T221/Y223), (4) Akt (S473), (5) CREB (S133) and (6) c-Jun (S63). Red boxes indicate tumor suppressor phosphorylated molecules: (7) STAT3 (Y727) and (8) Chk2 (T68). Western blot analysis showed opposite changes of phosphorylated (B) PKB/Akt and (C) Erk proteins in LDHA- and PDHA1-silencing HNSCC cells. MTT and trypan blue exclusion assays for PDHA1-silencing (D) OECM-1 and (E) FaDu cells treated with PKB/Akt inhibitor MK2206 and Erk1/2 inhibitor PD98059 suggested PKB/Akt and Erk activities contribute to PDHA1-mediated cell growth upregulation in a dose-dependent manner. Doses (μM) of inhibitors are shown in parentheses. Data are presented as mean ± SEM (N≥3). *P < .05.
LDHA/PDHA1 Loss Leads to Distinct Metabolic Reprogramming in HNSCC Cells
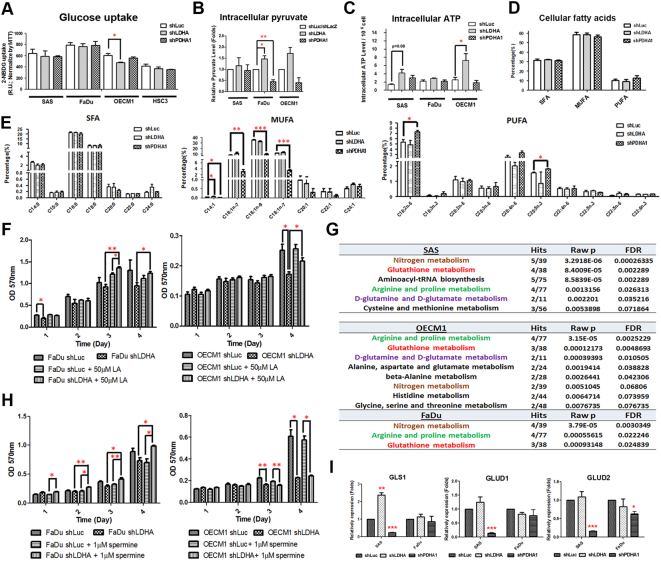
Metabolic plasticity was recently proposed by several groups suggesting that cancer cells might have a complex machinery to adapt to unfavorable metabolic stresses [38]. We therefore sought to determine metabolic changes in HNSCC cells with LDHA/PDHA1-silencing. Multiple metabolic indices including glucose uptake activity, intracellular pyruvate level and cellular ATP level, were examined. While Seahorse analysis (Figure 2E) and lactate assay (Figure 3D) suggested potential metabolic shift in response to LDHA/PDHA1 loss, no significant changes of glucose uptake activity was observed in LDHA- and PDHA1-silencing HNSCC cells compared with control cells (Figure 6A). Interestingly, accumulated intracellular pyruvate (Figure 6B) and increased ATP levels (Figure 6C) were found in response to LDHA loss despite the LDHA-silencing HNSCC cells displaying decreased cell malignancy, implying that glycolytic energetics could not compensate for LDHA mediated HNSCC cellular changes. In addition to energy, further analysis for intracellular amino acid, nucleotide and lipid abundance was also carried out to define whether LDHA/PDHA1 deficiency could affect cellular biomass in HNSCC cells, thereby leading to corresponding phenotypic changes. The results showed that similar amounts of polypeptides and nucleotides per unit of cells were detected between control and LDHA/PDHA deficient cells (Supplementary Fig. 8). As for cellular lipid content, a GC-FID based lipidomic analysis was performed to determine the total lipid amount and differential lipid compositions in LDHA/PDHA1-silencing HNSCC cells. The data showed an increasing amount of saturated fatty acids (SFAs) and monounsaturated fatty acids (MUFAs), lipids produced from de novo lipogenic pathways, as well as exogenously preferential polyunsaturated fatty acids (PUFAs) [39] in both LDHA and PDHA1 deficient HNSCC cells (Supplementary Fig. 9), suggesting that increased lipid levels are important in response to LDHA/PDHA1 loss but may not contribute to the corresponding phenotypic alterations. As a very recent study showed that tumor suppressor retinoblastoma (Rb) gene alteration regulated cellular lipid composition, leading to suppression of malignant progression in rat neurinoma cells [40], the analysis for fatty acid (FA) compositional changes in LDHA/PDHA-silencing HNSCC cells was therefore performed. Although there were no obvious changing trends of SFAs, MUFAs and PUFAs in LDHA/PDHA1-silencing cells compared with control cells (Figure 6D), in highly malignant PDHA1-silencing cells, MUFAs were significantly decreased at the expense of higher PUFA proportion, revealing a potential regulatory mechanism that HNSCC cells gain growth advantages by increasing external PUFA uptake (Figure 6E). The importance of PUFAs in controlling LDHA/PDHA-mediated HNSCC cell growth was further confirmed by the detection of rescued cell growth of LDHA-silencing HNSCC cells cultivated in medium with addition of LA, the most abundant PUFAs in cells, in comparison with the non-treated group (Figure 6F).
Figure 6.
Differential Metabolic Changes in LDHA/PDHA1 Deficient HNSCC Cells. (A) Glucose uptake, (B) intracellular pyruvate and (C) ATP levels were determined in LDHA and PDHA1-silencing HNSCC cells. GC-FID based analysis for percentages of (D) total and (E) fractional cellular saturated fatty acids (SFAs), monounsaturated fatty acids (MUFAs) and increased polyunsaturated fatty acids (PUFAs) in LDHA/PDHA1-deficient SAS cells. Decreased MUFAs in accompany with increased PUFAs was detected in highly-proliferating PDHA1-silencing cells. (F) MTT assay showed rescued effects of PDHA1-silencing FaDu and OECM-1 cells supplemented with 50 μM linolenic acid (LA) compared with control cells. (G) Candidate pyruvate metabolism mediated regulatory metabolites, highlighted in red, in HNSCC cells were determined based on LC–MS analysis. Boxes represent fold changes of metabolites in LDHA (red) and PDHA1 (green) deficient HNSCC cells compared with control cells. (H) MTT assay showed rescued effects of LDHA-silencing FaDu and OECM-1cells supplemented with 1 μM spermine compared with control cells. (I) Real-time RT-PCR analysis for glutaminase (GLS1) and glutamine dehydrogenase 1/2 (GLUD1/2) in LDHA/PDHA1-silencing SAS and FaDu cells. Data are presented as mean ± SEM (N≥3). ***P < .001; **P < .01; *P < .05.
In line with the “metabolic reprogramming” concept during neoplastic transformation, recent studies attempt to link mitochondrial dysfunction to carcinogenesis. Indeed, deregulated mitochondrial metabolites, known as oncometabolites, including 2-hydroxyglutarate, succinate, and fumarate all contribute to tumorigenesis [41]. To define potential cellular metabolites associated with LDHA/PDHA1-mediated phenotypes, LC–MS based analysis for metabolites in LDHA-/PDHA1-silencing HNSCC cells was performed (Supplementary Fig. 10). The results showed similar but distinct metabolite changes in LDHA/PDHA1-silencing HNSCC cells from different origins (Supplementary Fig. 11). By using the “Pathway analysis” module on the MetaboAnalyst website, it was found that metabolites relevant to LDHA/PDHA1 changes in HNSCC cells, defined by the metabolic pathways with P < .01 and False Discovery Rate (FDR) <0.1, are involved in nitrogen metabolism, glutathione metabolism, arginine and proline metabolism and D-glutamine/D-glutamate metabolism (Figure 6G). The specific metabolites that correlate with related phenotypes in response to LDHA or PDHA1 loss were further determined by analyzing metabolite changes between shLDHA or shPDHA1 treated, regardless of cell origins, and control HNSCC cells (N = 9 in each experimental group). The candidate metabolites in response to LDHA/PDHA1 loss required to fulfill the criteria showing a reversed changing trend in LDHA and PDHA1-silencing cells. Based on these criteria, the results uncovered that AMP, aspartic acid, L-acetylcarnitine, spermine and Uridine 5′-monophosphate could be potential LDAH/PDHA1-mediated regulatory metabolites (Supplementary Fig. 12 and 13).
To validate the importance of these metabolites in regulating HNSCC cell malignancy in response to LDHA/PDHA1 knockdown, molecules involved in metabolism of arginine and glutamine were focused based on previous recognition of their roles in controlling tumorous phenotype [42]. While cancer cells often exhibit great intracellular polyamine content, previous studies demonstrated that the end product of arginine metabolism spermine, a positively charged alkyl polyamines, is involved in cancer development [43]. At the molecular level, it was found that spermine could promote initiation of protein translation and safeguard DNA from reactive oxygen species (ROS)-induced damages by serving as a ROS scavenger in mammalian cells [44]. While an elevated ROS level in HNSCC cells was detected in response to LDHA loss (Supplementary Fig. 14), we hypothesized that spermine could potentially modulate LDHA mediated cell growth. Indeed, with the administration of spermine in slow-growing LDHA-silencing cells, decreased cell growth could be ameliorated (Figure 6H). Glutamine/glutamate metabolism is another pathway drawing our attention for further examination in LDHA/PDHA1-mediated malignant changes. The impacts of glutamine metabolism in different cancers for tumor-associated phenotypes have been extensively studied [45]. As a recent investigation reported that glutamine/glutamate pathway serves as a driver for compensatory anaplerotic flux in human cells [46], glutamine/glutamate metabolism could therefore be a potential mediator to trigger glycolysis/OxPhos shift in LDHA/PDHA1-silencing HNSCC cells. In our experimental setting, glutamine metabolism-related molecules glutaminase (GLS1) and glutamine dehydrogenase 1/2 (GLUD1/2) mRNA expression is enriched in LDHA-silencing cells but decreased in PDHA1 deficient SAS cells (Figure 6I) indicating that LDHA loss reprogrammed HNSCC cell metabolism preferentially towards the mitochondrial pathway.
Discussion
Cancers exhibit unique metabolic features different from their normal counterparts making it possible to target tumor-favorable metabolic cues for development of an anti-cancer strategy. One way to practice this concept is to reverse cancer-specific metabolic characteristics back to a normal cell-like state, thereby lessening cell malignancy [4]. While numbers of studies defined regulatory roles of LDHA and PDC in controlling different cancer development [14], [47] (Supplementary Table 5), a simultaneous evaluation of two factors in individual cancer types with the same experimental settings has not been reported. In the present study, it was demonstrated that LDHA loss suppresses whereas PDHA1 loss triggers HNSCC neoplastic progression in vitro, in vivo and in clinic. Based on UALCAN and The Human Protein Atlas databases, in addition to HNSCCs, differential LDHA/PDHA1 mRNA expression in identifying cancer-specific survival rates was also found in patients with adrenocortical carcinoma, brain lower-grade glioma, glioblastoma multiforme, pancreatic adenocarcinoma, kidney renal papillary cell carcinoma and cervical cancer, suggesting that manipulation for pyruvate metabolic factors could be an good prognostic indicator for a wide range of cancers. Furthermore, according to rare detection of deletions/mutations for LDHA and PDHA1 genes in HNSCC cells (Supplementary Table 6) as well as in clinical HNSCC tissues (N = 1476) using cBioPortal cancer genomic database (http://www.cbioportal.org) (LDHA:0.7%; PDHA1:2.6%], the shRNA mediated genetic manipulation could be an ideal design to faithfully reflect the responses in most HNSCC tumor cells upon changes of LDHA/PDHA1 expression by treatments of potential therapeutic drugs. Indeed, the established reporter system described here for screening neoplastic inhibitors supported the idea that compounds could inhibit LDHA and facilitate PDHA1 transcriptional activity are likely metabolism-based anti-cancer candidates. By taking advantage of this system, future work is currently on the way to determine traditional herbal compounds for their anti-tumor potential.
One of the striking findings from current study is the detection of differential metabolic alteration in response to LDHA/PDHA1 loss. With analysis for both cellular energy and biomass, less-malignant LDHA-silencing HNSCC cells possess higher ATP levels while more aggressive PDHA1 deficient HNSCC cells showed no significant energetic changes. As no obvious change in glucose uptake activity was found under a condition of LDHA/PDHA1 loss, the detection of an accumulating intracellular pyruvate level in LDHA-silencing cells indicated that HNSCC cells less preferentially utilize mitochondrial metabolic pathways for growth, in agreement with the concept that mitochondrial dysfunction serves as a disease predisposition in most cancers [1], [2], [12], [48]. The rationale could also be supported by the data showing a growth arrest in LDHA deficient HNSCC cells, despite an increased glutamine metabolic molecules and intracellular glutamine/glutamate levels being detected. Collectively, the results revealed an alternative metabolism based machinery to aid HNSCC cellular survival in an unfavorable environment (e.g., LDHA loss), even though the compensatory effects may not sufficiently meet the demands to rescue cancer cell growth.
Another interesting discovery from our results is the impact of cellular FA composition and HNSCC-specific oncometabolites in regulating HNSCC cell malignancy. In comparison with glucose metabolism, alterations in FA metabolism in cancer cells have received less attention. FAs, both saturated and unsaturated kinds, are required for energy storage, membranous organelle homeostasis, and generation of signaling molecules. Although the total amount of SFAs, MUFAs and PUFAs increased in both LDHA and PDHA1-deficient HNSCC cells, the increased FAs in LDHA and PDHA1-deficient cells seem to come from different sources. Significant increased exogenous-preferential PUFAs were detected in PDHA1-silencing cells, implying that PUFAs may play a dominant role in modulating HNSCC cell growth. This finding is consistent with previous studies showing that supplementation of PUFAs Linoleate (C18:2), Arachidonate (C20:4) and Docosahexaenoic acid (DHA; C22:6) in breast cancer cells suppressed caspase3 mediated cell apoptosis thereby facilitating cell growth [49]. At the molecular level, it was unexpectedly found that mRNA expression of most de novo lipogenic enzymes, including sterol regulatory element-binding transcription factor 1/2 (SREBF1/2), acetyl-CoA carboxylase beta (ACACB), fatty acid synthase (FASN) and stearoyl-CoA desaturase 1 (SCD1) are enhanced in PDHA1-silencing HNSCC cells (Supplementary Fig. 15). While PDHA1 loss would likely lead to a decrease of acetyl-CoA, the precursor molecule of de novo lipogenesis, intracellular SFAs and MUFAs may be still downregulated compared with control cells. As for cellular metabolites, in addition to spermine, the metabolomics analysis also uncovered several potential LDHA/PDHA1-mediated metabolites in controlling HNSCC malignancy. For instance, in agreement with a very recent study reporting that aspartate is a limiting metabolite for tumor cell growth under hypoxic condition [50], aspartate levels were also altered in response to LDHA/PDHA1 manipulation and positively correlated with cellular malignancy in HNSCC cells. In summary, our results confirm that modulation of pyruvate metabolic fates in HNSCC cells could be a great target to reverse cell malignancy or to develop anti-tumor drugs. It also highlights the impact of metabolic plasticity in HNSCC cells and the importance to examine molecular/metabolic cues systemically upon manipulation of the individual metabolic pathway.
The following are the supplementary data related to this article.
Summary for impacts of LDHA/PDH in controlling tumorigenesis
Supplementary material
Acknowledgments
Acknowledgements
This work is supported by grants from Ministry of Science and Technology, Taiwan (Most-103-2314-B-010-024-MY3 and 106-2314-B-010-005-MY3); Yen Tjing Ling Medical Foundation (CI-105-8) and a grant from Ministry of Education, Aiming for the Top University Plan as well as the Higher Education Sprout Project by the Ministry of Education (MOD) in Taiwan. Authors would like to thank Professor Kuo-Wei Chang (National Yang-Ming University) for providing experimental materials. We thank the National RNAi Core Facility at Academia Sinica in Taiwan for providing shRNA reagents and The Metabolomics Core Laboratory, National Taiwan University, for Metabolomics services. We also thank Dr. Jude Clapper (Taipei American School) and Ms. Courtney Anne Curtis for critical review and English corrections for the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Funding sources: This work is supported by grants from Ministry of Science and Technology, Taiwan (Most-103-2314-B-010-024-MY3 and 106–2314-B-010-005-MY3); Yen Tjing Ling Medical Foundation (CI-105-8) and a grant from Ministry of Education, Aiming for the Top University Plan as well as the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 3.Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol. 2005;202:654–662. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- 4.Wilde L, Roche M, Domingo-Vidal M, Tanson K, Philp N, Curry J, Martinez-Outschoorn U. Metabolic coupling and the Reverse Warburg Effect in cancer: Implications for novel biomarker and anticancer agent development. Semin Oncol. 2017;44:198–203. doi: 10.1053/j.seminoncol.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granchi C, Minutolo F. Anticancer agents that counteract tumor glycolysis. ChemMedChem. 2012;7:1318–1350. doi: 10.1002/cmdc.201200176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, Seo JH, Shan C, Dai Q, Zhang L, Xie J. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22:585–600. doi: 10.1016/j.ccr.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller FL, Colla S, Aquilanti E, Manzo VE, Genovese G, Lee J, Eisenson D, Narurkar R, Deng P, Nezi L. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012;488:337–342. doi: 10.1038/nature11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwon OH, Kang TW, Kim JH, Kim M, Noh SM, Song KS, Yoo HS, Kim WH, Xie Z, Pocalyko D. Pyruvate kinase M2 promotes the growth of gastric cancer cells via regulation of Bcl-xL expression at transcriptional level. Biochem Biophys Res Commun. 2012;423:38–44. doi: 10.1016/j.bbrc.2012.05.063. [DOI] [PubMed] [Google Scholar]
- 9.Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA, 3rd, Peters EC, Driggers EM, Hsieh-Wilson LC. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337:975–980. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray LR, Tompkins SC, Taylor EB. Regulation of pyruvate metabolism and human disease. Cell Mol Life Sci. 2014;71:2577–2604. doi: 10.1007/s00018-013-1539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbet C, Feron O. Tumour acidosis: from the passenger to the driver's seat. Nat Rev Cancer. 2017;17:577–593. doi: 10.1038/nrc.2017.77. [DOI] [PubMed] [Google Scholar]
- 14.Miao P, Sheng S, Sun X, Liu J, Huang G. Lactate dehydrogenase A in cancer: a promising target for diagnosis and therapy. IUBMB Life. 2013;65:904–910. doi: 10.1002/iub.1216. [DOI] [PubMed] [Google Scholar]
- 15.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 16.Yu X, Hiromasa Y, Tsen H, Stoops JK, Roche TE, Zhou ZH. Structures of the human pyruvate dehydrogenase complex cores: a highly conserved catalytic center with flexible N-terminal domains. Structure. 2008;16:104–114. doi: 10.1016/j.str.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahl HH, Brown GK, Brown RM, Hansen LL, Kerr DS, Wexler ID, Patel MS, De Meirleir L, Lissens W, Chun K. Mutations and polymorphisms in the pyruvate dehydrogenase E1 alpha gene. Hum Mutat. 1992;1:97–102. doi: 10.1002/humu.1380010203. [DOI] [PubMed] [Google Scholar]
- 18.Fan J, Kang HB, Shan C, Elf S, Lin R, Xie J, Gu TL, Aguiar M, Lonning S, Chung TW. Tyr-301 phosphorylation inhibits pyruvate dehydrogenase by blocking substrate binding and promotes the Warburg effect. J Biol Chem. 2014;289:26533–26541. doi: 10.1074/jbc.M114.593970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stacpoole PW, Kurtz TL, Han Z, Langaee T. Role of dichloroacetate in the treatment of genetic mitochondrial diseases. Adv Drug Deliv Rev. 2008;60:1478–1487. doi: 10.1016/j.addr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stacpoole PW, Moore GW, Kornhauser DM. Toxicity of chronic dichloroacetate. N Engl J Med. 1979;300:372. doi: 10.1056/NEJM197902153000726. [DOI] [PubMed] [Google Scholar]
- 21.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 22.Li WC, Lee PL, Chou IC, Chang WJ, Lin SC, Chang KW. Molecular and cellular cues of diet-associated oral carcinogenesis--with an emphasis on areca-nut-induced oral cancer development. J Oral Pathol Med. 2015;44:167–177. doi: 10.1111/jop.12171. [DOI] [PubMed] [Google Scholar]
- 23.Liu CJ, Chang WJ, Chen CY, Sun FJ, Cheng HW, Chen TY, Lin SC, Li WC. Dynamic cellular and molecular modulations of diabetes mediated head and neck carcinogenesis. Oncotarget. 2015;6:29268–29284. doi: 10.18632/oncotarget.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blatt S, Voelxen N, Sagheb K, Pabst AM, Walenta S, Schroeder T, Mueller-Klieser W, Ziebart T. Lactate as a predictive marker for tumor recurrence in patients with head and neck squamous cell carcinoma (HNSCC) post radiation: a prospective study over 15 years. Clin Oral Investig. 2016;20:2097–2104. doi: 10.1007/s00784-015-1699-6. [DOI] [PubMed] [Google Scholar]
- 25.Chen YS, Huang WL, Chang SH, Chang KW, Kao SY, Lo JF, Su PF. Enhanced filopodium formation and stem-like phenotypes in a novel metastatic head and neck cancer cell model. Oncol Rep. 2013;30:2829–2837. doi: 10.3892/or.2013.2772. [DOI] [PubMed] [Google Scholar]
- 26.Lai YS, Chen WC, Kuo TC, Ho CT, Kuo CH, Tseng YJ, Lu KH, Lin SH, Panyod S, Sheen LY. Mass-Spectrometry-Based Serum Metabolomics of a C57BL/6J Mouse Model of High-Fat-Diet-Induced Non-alcoholic Fatty Liver Disease Development. J Agric Food Chem. 2015;63:7873–7884. doi: 10.1021/acs.jafc.5b02830. [DOI] [PubMed] [Google Scholar]
- 27.Chuang LT, Glew RH, Li CC, VanderJagt DJ, Broyles JS, Ray GM, Shah VO. Comparison of the fatty acid composition of the serum phospholipids of controls, prediabetics and adults with type 2 diabetes. J Diabetes Mellitus. 2012;2:393–401. doi: 10.4236/jdm.2012.24061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 30.Vokes EE, Agrawal N, Seiwert TY. HPV-Associated Head and Neck Cancer. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv344. [DOI] [PubMed] [Google Scholar]
- 31.Castellsague X, Alemany L, Quer M, Halec G, Quiros B, Tous S, Clavero O, Alos L, Biegner T, Szafarowski T. HPV Involvement in Head and Neck Cancers: Comprehensive Assessment of Biomarkers in 3680 Patients. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djv403. [DOI] [PubMed] [Google Scholar]
- 32.San-Millan I, Brooks GA. Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis. 2017;38:119–133. doi: 10.1093/carcin/bgw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fife CM, McCarroll JA, Kavallaris M. Movers and shakers: cell cytoskeleton in cancer metastasis. Br J Pharmacol. 2014;171:5507–5523. doi: 10.1111/bph.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 35.Guan GF, Zhang DJ, Zheng Y, Wen LJ, Yu DJ, Lu YQ, Zhao Y. Significance of ATP-binding cassette transporter proteins in multidrug resistance of head and neck squamous cell carcinoma. Oncol Lett. 2015;10:631–636. doi: 10.3892/ol.2015.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulsum S, Sudheendra HV, Pandian R, Ravindra DR, Siddappa G, R N, Chevour P, Ramachandran B, Sagar M, Jayaprakash A. Cancer stem cell mediated acquired chemoresistance in head and neck cancer can be abrogated by aldehyde dehydrogenase 1 A1 inhibition. Mol Carcinog. 2017;56:694–711. doi: 10.1002/mc.22526. [DOI] [PubMed] [Google Scholar]
- 37.Sever R, Brugge JS. Signal transduction in cancer. Cold Spring Harb Perspect Med. 2015;5 doi: 10.1101/cshperspect.a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keibler MA, Wasylenko TM, Kelleher JK, Iliopoulos O, Vander Heiden MG, Stephanopoulos G. Metabolic requirements for cancer cell proliferation. Cancer Metab. 2016;4:16. doi: 10.1186/s40170-016-0156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Currie E, Schulze A, Zechner R, Walther TC, Farese RV., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muranaka H, Hayashi A, Minami K, Kitajima S, Kohno S, Nishimoto Y, Nagatani N, Suzuki M, Kulathunga LAN, Sasaki N. A distinct function of the retinoblastoma protein in the control of lipid composition identified by lipidomic profiling. Oncogene. 2017;6:e350. doi: 10.1038/oncsis.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Z, Ibekwe E, Chornenkyy Y. Metabolic Alterations in Cancer Cells and the Emerging Role of Oncometabolites as Drivers of Neoplastic Change. Antioxidants (Basel) 2018;7:E16. doi: 10.3390/antiox7010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris CR, Hamilton-Reeves J, Martindale RG, Sarav M, Ochoa Gautier JB. Acquired amino acid deficiencies: a focus on arginine and glutamine. Nutr Clin Pract. 2017;32:30S–47S. doi: 10.1177/0884533617691250. [DOI] [PubMed] [Google Scholar]
- 43.Bae DH, Lane DJR, Jansson PJ, Richardson DR. The old and new biochemistry of polyamines. Biochim Biophys Acta. 2018;1862:2053–2068. doi: 10.1016/j.bbagen.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 44.Mandal S, Mandal A, Johansson HE, Orjalo AV, Park MH. Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc Natl Acad Sci U S A. 2013;110:2169–2174. doi: 10.1073/pnas.1219002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Pavlova NN, Thompson CB. Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. EMBO J. 2017;36:1302–1315. doi: 10.15252/embj.201696151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Q, Kirk K, Shurubor YI, Zhao D, Arreguin AJ, Shahi I, Valsecchi F, Primiano G, Calder EL, Carelli V. Rewiring of glutamine metabolism is a bioenergetic adaptation of human cells with mitochondrial DNA mutations. Cell Metab. 2018;27:1007–1025 e1005. doi: 10.1016/j.cmet.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stacpoole PW. Therapeutic targeting of the pyruvate dehydrogenase complex/pyruvate dehydrogenase kinase (PDC/PDK) axis in cancer. J Natl Cancer Inst. 2017;109 doi: 10.1093/jnci/djx071. [DOI] [PubMed] [Google Scholar]
- 48.Lleonart ME, Grodzicki R, Graifer DM, Lyakhovich A. Mitochondrial dysfunction and potential anticancer therapy. Med Res Rev. 2017;37:1275–1298. doi: 10.1002/med.21459. [DOI] [PubMed] [Google Scholar]
- 49.Hardy S, El-Assaad W, Przybytkowski E, Joly E, Prentki M, Langelier Y. Saturated fatty acid-induced apoptosis in MDA-MB-231 breast cancer cells. A role for cardiolipin. J Biol Chem. 2003;278:31861–31870. doi: 10.1074/jbc.M300190200. [DOI] [PubMed] [Google Scholar]
- 50.Garcia-Bermudez J, Baudrier L, La K, Zhu XG, Fidelin J, Sviderskiy VO, Papagiannakopoulos T, Molina H, Snuderl M, Lewis CA. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat Cell Biol. 2018;20:775–781. doi: 10.1038/s41556-018-0118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary for impacts of LDHA/PDH in controlling tumorigenesis
Supplementary material