Abstract
Growing evidence indicates that cyclin dependent kinases regulatory subunit 2 (CKS2) serves an essential role in the regulation of multiple cellular processes in diverse human cancer types. The present study investigated the contribution of CKS2 to breast cancer (BC) progression. In the present study, CKS2 expression in BC was detected using Oncomine and The Cancer Genome Atlas database. The association between expression levels and clinical features was explored using Kaplan-Meier plotter and the Breast Cancer Gene-Expression Miner Version 4.0 (bc-GenExMiner) online database. In addition, the roles of CKS2 in BC progression were examined. It was identified that CKS2 expression was significantly increased in BC tissues at the mRNA and protein levels. Bc-GenExMiner demonstrated that high CKS2 expression was associated with a positive estrogen receptor status, progesterone receptor status, nodal status and basal-like status. High CKS2 expression was markedly associated with poor overall survival, relapse-free survival, and distant metastasis-free survival in patients with BC. Moreover, functional assays revealed that CKS2 inhibition suppressed cell proliferation and invasion ability in vitro and reduced tumor growth in vivo. Thus, the present findings suggested that CKS2 may act as a potential biomarker and therapeutic target for the treatment of BC.
Keywords: breast cancer, cyclin dependent kinases regulatory subunit 2, prognosis
Introduction
Breast cancer (BC) is one of the most common malignancies in women worldwide and the incidence has increased continuously in recent years (1,2). Every year, there are ~1.7 million new cases of breast cancer diagnosed globally, and >30% of patients succumb to BC (3). In spite of great advances in diagnosis and therapy for the disease, the overall 5-year survival rate for patients with BC remains low (4,5). Thus, it is important to explore effective diagnostic biomarkers and therapeutic targets to improve the prognosis of patients with BC.
Cyclin-dependent kinases regulatory subunit 2 (CKS2) was identified in 1990, and the gene is located at chromosome 9q22 (6). CKS2 is a member of the cell cycle dependent protein kinase subunits family, which is involved in cell cycle regulation (7,8). A previous study demonstrated that CKS2 may serve critical roles in early embryonic development and somatic cell division (9). Increasing numbers of studies demonstrate that CKS2 may exert functions in tumor progression. For example, Chen et al (10) reported that CKS2 expression was increased and acted as a biomarker for predicting superficial bladder cancer progression to muscle-invasive cancer. Lin et al (11) demonstrated that depletion of CKS2 expression compromised cell proliferation and enhanced chemotherapy-induced apoptosis in HepG2 cells. Wang et al (12) reported that CKS2 was upregulated and correlated with poor overall survival in patients with BC.
The present study explored CKS2 expression in patients with BC based on a number of public databases, to illustrate its prognostic and potential therapeutic value. Moreover, the functions of CKS2 in BC progression were determined. The present results demonstrated that CKS2 may serve as a prognostic biomarker and potential therapeutic target in BC treatment.
Materials and methods
Oncomine database analysis
The Oncomine gene expression array database (www.oncomine.org) was used to assess the CKS2 mRNA expression levels in four BC datasets, including Sørlie Breast (13), Curtis Breast (14), Sørlie Breast 2 (15) and The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) Breast. In the present study, CKS2 mRNA expression in BC samples and normal individuals was compared using a Student's t-test.
Human Protein Atlas
CKS2 protein expression levels in BC tissues and normal tissues were reviewed in the Human Tissue Atlas (http://www.proteinatlas.org/) (16).
bcGenExMiner v4.0
Breast Cancer Gene-Expression Miner v4.0 (bcGenExMiner v4.0), a statistical mining tool of published annotated genomic data including 36 annotated genomic datasets and three classical mining functions: Expression, prognosis, and correlation (17,18). The correlation between CKS2 expression and the risk of any event of relapse (AE) or metastatic relapse (MR) in patients with BC was determined by univariate Cox analysis.
Kaplan-Meier analysis
Kaplan-Meier Plotter (www.kmplot.com) is an online database including gene expression data and clinical data (19). In the present study, Kaplan-Meier Plotter was used to evaluate the overall survival (OS), relapse-free survival (RFS) and distant metastasis-free survival (DMFS) of patients with BC. Individuals were separated into two groups based on median gene expression; high (≥median expression) and low expression (<median expression).
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from BC cells was isolated using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocols. The complementary DNA (cDNA) was generated from 100 ng of total RNA using the PrimeScript™ Reverse Transcription kit (Takara Bio, Inc., Otsu, Japan) according to the manufacturer's protocols. The relative expression of CKS2 was detected using SYBR® Green Master Mix (Takara Biotechnology Co., Ltd., Dalian, China) using a Step One Plus Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR was performed as follows: 94°C for 30 sec, then 30 cycles of 56°C for 30 sec and 72°C for 90 sec, and final extension at 72°C for 5 min. GAPDH was used as an endogenous control. The relative expression level of CKS2 was calculated via the 2ΔΔCq method (20). The primers used were as follows: CKS2; forward, 5′-CTTCGCGCTCTCGTTTCATT-3′; and reverse, 5′-CACCAAGTCTCCTCCACTCC-3′; and GAPDH; forward, 5′-GTCGATGGCTAGTCGTAGCATCGAT-3′ and reverse, 5′-TGCTAGCTGGCATGCCCGATCGATC-3′.
Cell culture and transfection
Human BC cell lines (MCF-7, BT-474, HCC1937 and MDA-MB-231) and a normal human breast epithelial cell line (MCF-10A) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were maintained in Dulbecco's modified Eagle's medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences) at 37°C with 5% CO2.
Small interfering RNA (siRNA) for CKS2 and the corresponding negative controls were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China) and transfected into the cells to a final oligonucleotide concentration of 10 nmol/l. Transfection was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's protocol. The siRNA sequences were as follows: si-CKS2-1, 5′-GCUGGGUUCAUUACAUGAUdTdT-3′; si-CKS2-2, 5′-CAGAACCACAUAUUCUUCUdTdT-3′, the transfection was performed 24 h prior to subsequent experiments.
Cell proliferation assay
A Cell Counting Kit-8 (CCK8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to analyze the proliferation of BC cells. In brief, cells were seeded into 96-well plates and cultured for the indicated times. Subsequently, 10 µl CCK8 solution was added and incubated for 2 h at 37°C. The absorbance at 450 nm was determined using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Cell invasion assay
For the cell invasion assay, transfected cells (1×105) in 200 µl serum-free DMEM were seeded into the upper invasion chambers (8 µm pore size; EMD Millipore, Billerica, MA, USA) coated with Matrigel, while 600 µl medium supplemented with 10% FBS was added to the lower chamber. After 48 h incubation at 37°C, cells invading the bottom of the membrane were fixed using 4% paraformaldehyde for 5 min at room temperature and stained with 0.3% crystal violet dye for 5 min at room temperature. The images were captured using a light microscope (magnification, ×100; Nikon Corporation, Tokyo, Japan).
Western blot analysis
Total protein was from cells using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) including 1% phenylmethanesulfonyl fluoride. The concentration of total protein was determined using a bicinchoninic acid protein assay kit (Beyotime Institute of Technology). Proteins (40 µg/lane) were separated via 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (EMD Millipore). Following blocking with 5% non-fat milk overnight at room temperature, membranes were incubated with primary antibodies against CKS2 (1:1,000; cat. no. ab240129, Abcam, Cambridge, UK), and GAPDH (1:10,000; cat. no. ab181602, Abcam) overnight at 4°C. Membranes were then incubated with a horseradish peroxidase-conjugated secondary anti-rabbit immunoglobulin G antibody (1:20,000; cat. no. sc-2004, Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Bands were visualized using enhanced chemiluminescence reagent (7Sea Biotech, Shanghai, China).
Xenograft assays in nude mice
All animal experiments were performed according to the approved protocols of the Animal Care and Use Committee at Tongji University School of Medicine (Shanghai, China). A total of 6 female BALB/c nude mice (10 weeks of age; 20–22 g; n=3 mice/group) were obtained from the Beijing Experimental Animal Research Center (Beijing, China) and were housed under specific pathogen-free conditions at 20–26°C, 40–70% humidity and a 12/12 h light/dark cycle. The mice had free access to food and water. Transfected MCF-7 cells (2×106) were injected into the dorsal flanks of the animals to form a single tumor. The mice were sacrificed by dislocation of the neck following anesthetization by CO2 inhalation (air displacement rate, 10–30%/min) at 6 weeks following cell injection, and the tumor weights and volumes were determined. The tumor volume (V) was calculated using the following formula: V=π/6 × L × W × H; where W is width, L is length and H is height. The animal experiments performed in the present study were approved by the Animal Ethics Committee Review Board at Tongji University School of Medicine (no. T-2018-01-1197).
Statistical analysis
All data are presented as the mean ± standard deviation from three independent experiments and the statistical analyses were performed using SPSS 18.0 (SPSS, Inc., Chicago, IL, USA). Student's t-tests were used for the analysis of statistical significance between two groups, and one-way analysis of variance followed by Dunnett's post hoc test was applied to analyze the statistical significance among three groups or more. P<0.05 was considered as to indicate a statistically significant difference.
Results
Analysis of CKS2 expression based on Oncomine and the Human Protein Atlas database
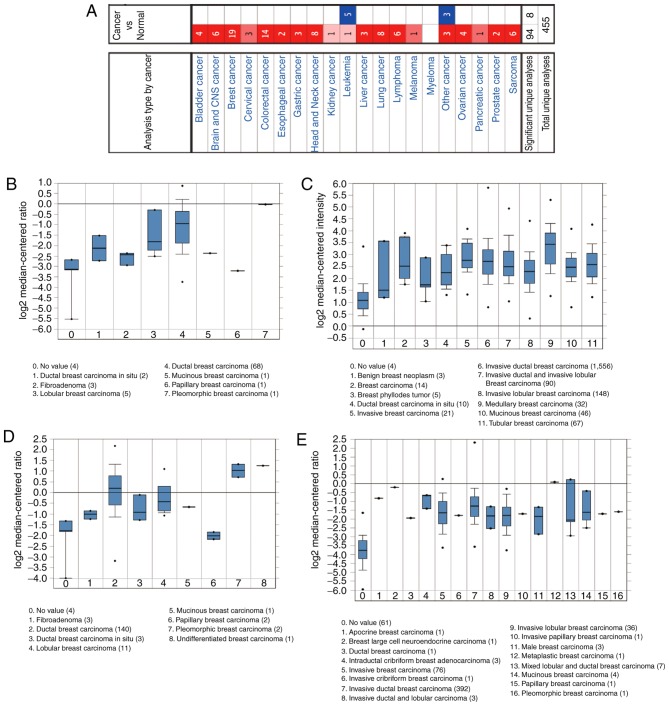
First, CKS2 expression in 20 types of cancer was examined using the Oncomine database. The results demonstrated that CKS2 mRNA expression was notably increased in BC samples (Fig. 1A). According to the Sørlie Breast (13), Curtis Breast (14), Sørlie Breast 2 (15) and TCGA Breast datasets, it was observed that CKS2 expression was notably increased in BC tissues (Fig. 1B-E). Moreover, the Human Protein Atlas database demonstrated that CKS2 protein expression was decreased in normal breast samples (Fig. 2A), and increased in BC tissues (Fig. 2B).
Figure 1.
CKS2 mRNA expression in malignant tumors based on the Oncomine database. (A) The mRNA expression of CKS2 in different types of cancers. The mRNA expression of CKS2 was upregulated across the whole set of BC tissues compared with normal controls in four datasets from the Oncomine database: (B) Sorlie Breast; (C) Curtis Breast; (D) Sorlie Breast 2; and (E) The Cancer Genome Atlas Breast. CKS2, cyclin dependent kinases regulatory subunit 2.
Figure 2.

CKS2 protein expression in BC tissues from the Human Protein Atlas database. (A) IHC staining for CKS2 in 2 cases of normal breast tissues (a and b). (B) IHC staining for CKS2 in 6 cases of BC (a-f). Magnification, ×20. BC, breast cancer; IHC, immunohistochemistry; CKS2, cyclin dependent kinases regulatory subunit 2.
Correlation between CKS2 expression and clinical features in patients with BC
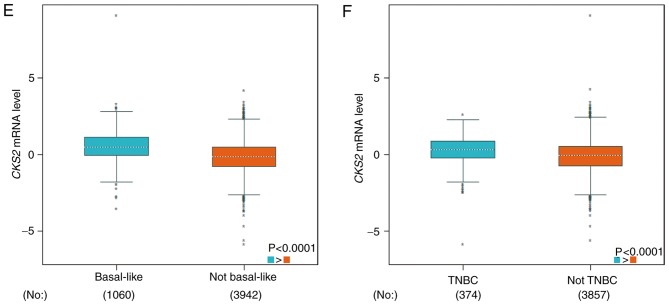
Next, the bc-GenExMiner database was used to determine the association between CKS2 expression and clinical features in patients with BC. The results demonstrated that CKS2 mRNA expression was significantly decreased in estrogen receptor (ER)-positive and progesterone receptor (PR)-positive BC patients (Fig. 3A and B). However, there was no significant difference between the erb-b2 receptor tyrosine kinase 2 (HER2)-positive group and the HER2-negative group (Fig. 3C). In addition, it was demonstrated that CKS2 mRNA expression was significantly increased patients with BC with positive nodal status, basal-like status and triple-negative status (Fig. 3D-F).
Figure 3.
Association between CKS2 mRNA expression and clinical features in patients with breast cancer. (A) ER status; (B) PR status; (C) HER2 status; (D) nodal status; (E) basal-like status; and (F) TNBC. ER, estrogen receptor; PR, progesterone receptor; HER2, erb-b2 receptor tyrosine kinase 2; TNBC, triple negative breast cancer; N, node; CKS2, cyclin dependent kinases regulatory subunit 2.
Correlation between CKS2 expression and prognosis in patients with BC
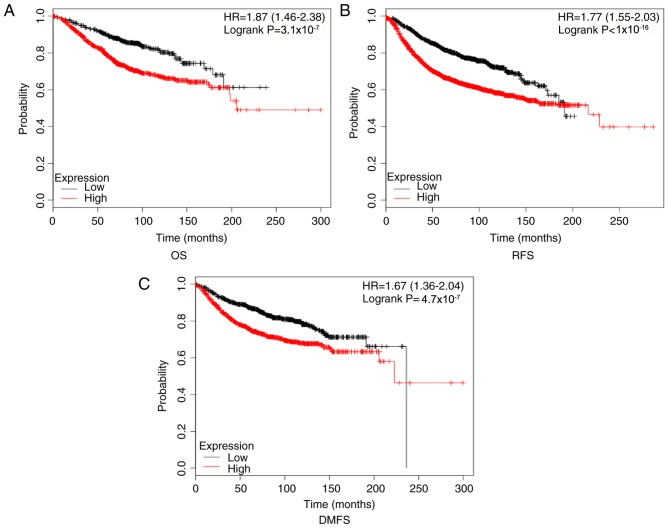
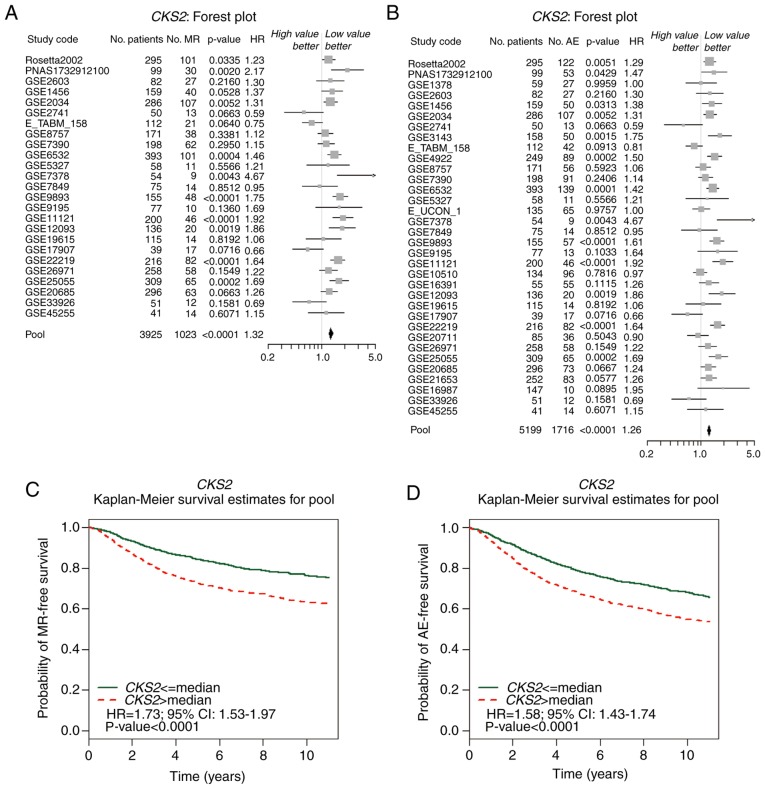
Kaplan-Meier Plotter results illustrated that high CKS2 mRNA expression was correlated with a poor OS, RFS, and DMFS in patients with BC (Fig. 4). The bc-GenExMiner results indicated that high CKS2 expression was associated with an increased risk of MR [Fig. 5A; hazard ratio (HR)=1.32, 95% confidence interval (CI): 1.25–1.40; P<0.0001] and AE (Fig. 5B; HR=1.26; 95% CI: 1.21–1.32; P<0.0001). The survival curve demonstrated that high CKS2 expression was associated with poor MR-free survival (Fig. 5C; HR=1.73; 95% CI: 1.53–1.97; P<0.0001) and AE-free survival (Fig. 5D; HR=1.58; 95% CI: 1.43–1.74; P<0.0001).
Figure 4.
Kaplan-Meier plotter reveals the prognostic value of CKS2 mRNA expression in patients with BC. High CKS2 expression was associated with (A) poor OS, (B) poor RFS, and (C) poor DMFS in patients with BC. OS, overall survival; RFS, recurrence-free survival; DMFS, distant metastasis-free survival; BC, breast cancer; CKS2, cyclin dependent kinases regulatory subunit 2; HR, hazard ratio.
Figure 5.
High CKS2 expression is associated with improved AE-free and MR-free survival in patients with BC based on Gene-Expression Miner v4.0. The forest plots illustrate univariate Cox analysis of CKS2 expression and the risk of (A) AE or the risk of (B) MR in patients with BC. Kaplan-Meier survival analysis revealed the association between CKS2 expression and (C) AE-free survival and (D) MR-free survival in patients with BC. AE, any event of relapse; MR, metastatic relapse; CKS2, cyclin dependent kinases regulatory subunit 2; BC, breast cancer; HR, hazard ratio; CI, confidence interval.
CKS2 inhibition decreased BC cells progression
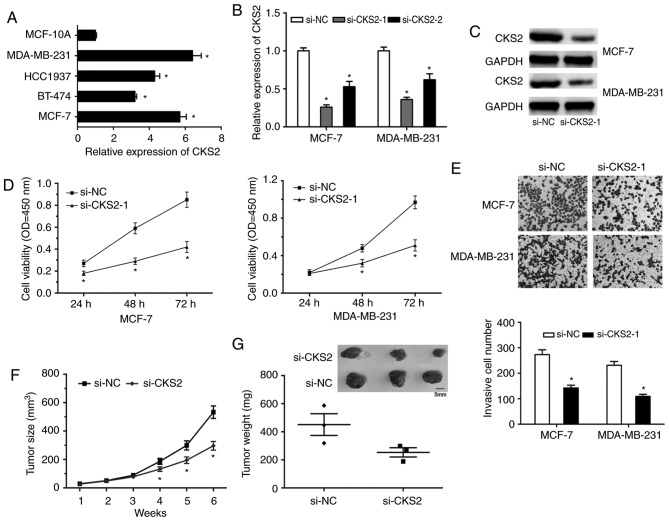
To examine the effects of CKS2 on BC progression, CKS2 expression was first determined in BC cells (Fig. 6A; P<0.05). MCF-7 and MDA-MB-231 cells were selected for the functional experiments. Following si-CKS2 transfection, the expression of CKS2 was determined by RT-qPCR and western blot analyses (Fig. 6B and C; P<0.05). The CCK-8 assay revealed that CKS2 inhibition significantly decreased BC cell proliferation capacity (Fig. 6D; P<0.05). The Transwell invasion assay demonstrated that the invasive capability of BC cells transfected with si-CKS2 was significantly decreased (Fig. 6E; P<0.05). Furthermore, the influence of CKS2 on BC cell growth was assessed in vivo. The results indicated that the average weights and volumes of the tumors were significantly decreased in the CKS2 knockdown group (Fig. 6F and G; P<0.05).
Figure 6.
CKS2 inhibition reduces BC progression. (A) RT-qPCR was used to determine CKS2 expression in BC cell lines (MCF-7, BT-474, HCC1937 and MDA-MB-231) and the normal breast epithelial cell line MCF-10. (B) RT-qPCR and (C) western blotting were used to test the transfection efficiency of si-CKS2. (D) CCK-8 and (E) Transwell invasion assays were used to explore the impacts of si-CKS2 on cell viability and invasion ability (magnification, ×100). The effects of CKS2 knockdown on (F) tumor size and (G) weight were examined in vivo. *P<0.05 vs. respective control. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; si, small interfering; CKS2, cyclin dependent kinases regulatory subunit 2; BC, breast cancer; NC, negative control; OD, optical density.
Discussion
CKS2 belongs to the mammalian cyclin kinase subunit family, which has two members: CKS1 and CKS2 (21). Previous studies indicated that CKS1 was increased in various cancer types, including prostate cancer, esophageal squamous cell carcinoma, nasopharyngeal carcinoma and glioma (22–25). The effects of CKS1 have been studied thoroughly; however, the roles of CKS2 remain unclear.
A previous study reported that the expression of CKS2 was increased in bladder cancer and associated with cancer progression (9). In the present study, the Oncomine database revealed that CKS2 mRNA expression was markedly increased in invasive ductal breast carcinoma, apocrine breast carcinoma, invasive breast carcinoma, and mixed lobular and ductal breast carcinoma. The Human Protein Atlas database indicated that CKS2 protein expression was significantly increased in BC tissues. Additionally, the bc-GenExMiner database revealed that high CKS2 expression was correlated with ER-positive, PR-positive, positive nodal and positive basal-like status, indicative of fast-growing invasive tumors. These data indicated that CKS2 may serve critical roles in BC tumorigenesis.
Kaplan-Meier Plotter analysis demonstrated that high CKS2 mRNA expression was associated with poor OS, RFS and DMFS in patients with BC. In addition, bc-GenExMiner analysis revealed that high CKS2 expression was associated with a higher risk of MR and AE. Survival curve analysis revealed that high CKS2 expression was associated with poor MR-free survival and AE-free survival in patients with BC. These data suggested that CKS2 may act as a potential prognostic biomarker in BC.
Next, the roles of CKS2 in BC tumorigenesis were investigated. In vitro functional assays revealed that CKS2 inhibition significantly decreased BC cell proliferation and invasion capacity compared with the si-NC group. Moreover, the in vivo assay revealed that the average weights and volumes of the tumors were significantly decreased in the CKS2 knockdown group. Therefore, the present data suggested that CKS2 may act as an oncogene in BC. Kang et al (26) reported that CKS2 was significantly upregulated in gastric cancers and promoted cell growth by decreasing p53 and p21cip1 expression. Additionally, Ji et al (27) revealed that increased expression of CKS2 was associated with hepatocellular carcinoma cell proliferation and downregulated the expression of phosphatase and tensin homolog. Future experiments should investigate whether CKS2 promotes the proliferation of BC cells via similar mechanisms.
In conclusion, the present study identified that CKS2 may act as a novel potential oncogene in BC, serving important roles in cell proliferation and invasion; however, the research had the following limitations. Firstly, the data on the expression of CKS2 in BC and its association with clinical features were obtained from bioinformatics, and require further analysis in BC samples. Secondly, the underlying mechanisms of the association between CKS2 and cell proliferation and invasion remain to be investigated. Future studies will aim to determine these underlying mechanisms to identify approaches by which CKS2 may be targeted in the treatment of BC.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Youth Science Foundation (grant no. 81502426).
Availability of data and materials
All data generated or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
NQH, ZLW and HGL performed the majority of the experiments, analyzed the data and drafted the manuscript. HH, XMW, and FQY contributed to the acquisition of data and revised the manuscript. HGL designed the study and revised the manuscript.
Ethics approval and consent to participate
The animal experiments performed in the present study were approved by the Animal Ethics Committee Review Board at Tongji University School of Medicine (Shanghai, Chin; no. T-2018-01-1197).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Tao Z, Shi A, Lu C, Song T, Zhang Z, Zhao J. Breast cancer: Epidemiology and etiology. Cell Biochem Biophys. 2015;72:333–338. doi: 10.1007/s12013-014-0459-6. [DOI] [PubMed] [Google Scholar]
- 3.DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66:31–42. doi: 10.3322/caac.21320. [DOI] [PubMed] [Google Scholar]
- 4.Qian C, Guan M, Si C, Shen H, Jin T, Zhang T. Identification of differentially expressed profiles of lncRNAs and mRNAs in ER-negative and HER-2 positive breast cancer. Arch Med Sci Civil Dis. 2017;2:148–160. [Google Scholar]
- 5.Hortobagyi GN, Stemmer SM, Burris HA, Yap YS, Sonke GS, Paluch-Shimon S, Campone M, Blackwell KL, André F, Winer EP, et al. Ribociclib as first-line therapy for HR-positive, advanced breast cancer. N Engl J Med. 2016;375:1738–1748. doi: 10.1056/NEJMoa1609709. [DOI] [PubMed] [Google Scholar]
- 6.You H, Lin H, Zhang Z. CKS2 in human cancers: Clinical roles and current perspectives. Mol Clin Oncol. 2015;3:459–463. doi: 10.3892/mco.2015.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liberal V, Martinsson-Ahlzén HS, Liberal J, Spruck CH, Widschwendter M, McGowan CH, Reed SI. Cyclin-dependent kinase subunit (Cks) 1 or Cks2 overexpression overrides the DNA damage response barrier triggered by activated oncoproteins. Proc Natl Acad Sci USA. 2012;109:2754–2759. doi: 10.1073/pnas.1102434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litchfield DW. Protein kinase CK2: Structure, regulation and role in cellular decisions of life and death. Biochem J. 2003;369:1–15. doi: 10.1042/bj20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinsson-Ahlzén HS, Liberal V, Grünenfelder B, Chaves SR, Spruck CH, Reed SI. Cyclin-dependent kinase-associated proteins Cks1 and Cks2 are essential during early embryogenesis and for cell cycle progression in somatic cells. Mol Cell Biol. 2008;28:5698–5709. doi: 10.1128/MCB.01833-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen R, Feng C, Xu Y. Cyclin-dependent kinase-associated protein Cks2 is associated with bladder cancer progression. J Int Med Res. 2011;39:533–540. doi: 10.1177/147323001103900222. [DOI] [PubMed] [Google Scholar]
- 11.Lin L, Fang Z, Lin H, You H, Wang J, Su Y, Wang F, Zhang ZY. Depletion of Cks1 and Cks2 expression compromises cell proliferation and enhance chemotherapy-induced apoptosis in HepG2 cells. Oncol Rep. 2016;35:26–32. doi: 10.3892/or.2015.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Xu L, Liu Y, Chen J, Jiang H, Yang S, Tan H. Expression of cyclin kinase subunit 2 in human breast cancer and its prognostic significance. Int J Clin Exp Pathol. 2014;7:8593–8601. [PMC free article] [PubMed] [Google Scholar]
- 13.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sørlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pontén F, Jirström K, Uhlen M. The Human Protein Atlas-a tool for pathology. J Pathol. 2008;216:387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 17.Lu XF, Zeng D, Liang WQ, Chen CF, Sun SM, Lin HY. FoxM1 is a promising candidate target in the treatment of breast cancer. Oncotarget. 2018;9:842–852. doi: 10.18632/oncotarget.23182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jézéquel P, Frénel JS, Campion L, Guérin-Charbonnel C, Gouraud W, Ricolleau G, Campone M. bc-GenExMiner 3.0: New mining module computes breast cancer gene expression correlation analyses. Database (Oxford) 2013;2013:bas060. doi: 10.1093/database/bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Zhao H, Lu Z, Bauzon F, Fu H, Cui J, Locker J, Zhu L. p27T187A knockin identifies Skp2/Cks1 pocket inhibitors for advanced prostate cancer. Oncogene. 2017;36:60–70. doi: 10.1038/onc.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Zhou Y, Tu B, Bu Y, Liu A, Kong J. Long noncoding RNA MALAT1 affects the efficacy of radiotherapy for esophageal squamous cell carcinoma by regulating Cks1 expression. J Oral Pathol Med. 2017;46:583–590. doi: 10.1111/jop.12538. [DOI] [PubMed] [Google Scholar]
- 24.Xu L, Fan S, Zhao J, Zhou P, Chu S, Luo J, Wen Q, Chen L, Wen S, Wang L, Shi L. Increased expression of Cks1 protein is associated with lymph node metastasis and poor prognosis in nasopharyngeal carcinoma. Diagn Pathol. 2017;12:2. doi: 10.1186/s13000-016-0589-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D, Zhi T, Xu X, Bao Z, Fan L, Li Z, Ji J, Liu N. MicroRNA-936 induces cell cycle arrest and inhibits glioma cell proliferation by targeting CKS1. Am J Cancer Res. 2017;7:2131–2143. [PMC free article] [PubMed] [Google Scholar]
- 26.Kang MA, Kim JT, Kim JH, Kim SY, Kim YH, Yeom YI, Lee Y, Lee HG. Upregulation of the cycline kinase subunit CKS2 increases cell proliferation rate in gastric cancer. J Cancer Res Clin Oncol. 2009;135:761–769. doi: 10.1007/s00432-008-0510-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji X, Xue Y, Wu Y, Feng F, Gao X. High-expressed CKS2 is associated with hepatocellular carcinoma cell proliferation through down-regulating PTEN. Pathol Res Pract. 2018;214:436–441. doi: 10.1016/j.prp.2017.12.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are available from the corresponding author upon reasonable request.