Abstract
Previous studies have suggested that long non-coding RNAs (lncRNAs) are closely associated with human diseases, particularly cancer, including cancer of the lung, breast and stomach. A variety of lncRNAs are abnormally expressed in cancer and participate in several pathways including cell proliferation and apoptosis; these elements are closely associated with the development of cancer. The Cancer Genome Atlas (TCGA) is an important cancer database. It consists of clinical data, genomic variation, mRNA, microRNA (miRNA) and lncRNAs expression, methylation and other data for various types of human cancer. In the present study, differential expression of RNA was identified using the edgeR package. A total 1,222 RNA sequencing profiles from patients with breast cancer were downloaded from TCGA. A competing endogenous RNA (ceRNA) network was constructed for breast cancer based on miRcode and miRTarBase. The top 10 lncRNAs were selected using Cox regression analysis. Survival analysis was performed using Kaplan-Meier analysis. A total of 1,028 breast cancer-associated lncRNAs and 89 miRNAs (fold change >2; P<0.05) were identified; among these, 93 lncRNAs and 19 miRNAs were included in the ceRNA network. Subsequently, 10 basic lncRNAs were selected and their associations with overall survival were identified. In addition, 5 lncRNAs (ADAM metallopeptidase with thrombospondin type 1 motif 9-antisense RNA 1, AL513123.1, chromosome 10 open reading frame 126, long intergenic non-protein coding RNA 536 and Wilms tumor 1 antisense RNA) were identified to be significantly associated with overall survival (P<0.05, log rank test). These results suggested that mRNAs, lncRNAs and miRNAs were involved in pathological mechanisms of breast cancer. The newly-identified ceRNA network included 93 breast cancer-specific lncRNAs, 19 miRNAs and 27 mRNAs. The results of the present study highlight the potential of lncRNAs in understanding the development and pathogenesis of breast cancer, and suggest novel concepts and an experimental basis for the identification of prognostic biomarkers and therapeutic targets for breast cancer.
Keywords: long non-coding RNA, microRNA, competing endogenous RNA network, breast cancer
Introduction
Breast cancer is one of the most widespread types of cancer in females, and its incidence is increasing yearly. It is expected that by 2018, the United States of America will have ~266,120 incident cases of invasive breast cancer and 63,960 cases of in situ breast cancer. Furthermore, it is estimated that 40,920 women will succumb to breast cancer (1). Similarly, in China, the incidence of breast cancer is the primary cause of mortality in women <45 years, followed by colorectal, liver and esophageal cancer (2). Following systemic treatment, patients with breast cancer continue to experience recurrence and metastasis, and the majority of patients eventually succumb to metastases. Therefore, it is important to identify diagnostic and prognostic markers, and potential therapeutic targets (3,4). The present study explored the mechanisms through which long non-coding RNAs (lncRNAs) act as competing endogenous RNAs (ceRNAs) to regulate target genes and participate in the prognosis and pathogenesis of breast cancer.
Over previous decades, 70–90% of the transcribed human genome has been identified. Relevant data indicate that the proportion of genes encoding proteins in the genome is >2%, and non-coding RNA accounts for the majority of the human transcriptome (5). Non-coding RNAs are a large class of RNA molecules that do not encode proteins, but which serve regulatory roles. Non-coding RNAs may be divided into three classes, by length: <50 nucleotides (nt), including microRNA (miRNA), small interfering RNA and Piwi-interacting RNA; 50–500 nt, including ribosomal RNA, transfer RNA, small nuclear RNA, small nucleolar RNA and lncRNA; >500 nt, including long mRNA-like non-coding RNAs and lncRNAs without polyA tails (6). lncRNA is a generic term for a class of RNA molecules with lengths of >200 nucleotides, and is one of the most active fields of study in molecular biology at present. lncRNAs regulate the gene expression of tumor cells through multiple modes of action, and therefore are widely involved in the occurrence and metastasis of tumors (7–9). lncRNAs are thought to serve an important role in the development of cancer; however, only a few have been well-characterized regarding their functional roles (10).
Differentially expressed ceRNAs have been identified in a number of diseases, particularly in cancer. Previous studies have demonstrated that ceRNAs affect the proliferation, growth, differentiation, apoptosis and other biological behaviors of cancer cells (11–13). miRNA response elements (MREs) are partially complementary sequences located on mRNAs that bind to miRNAs, and may inhibit the expression of miRNA target genes. miRNAs regulate hundreds of mRNAs, and one miRNA may be regulated by hundreds of mRNAs. Numerous types of RNA molecules constitute a ceRNA regulatory network, and the more MREs that are shared between them, the greater the potential for communication or co-regulation of a target gene (14). Theoretically, almost every RNA molecule possesses at least one MRE binding site that binds to an miRNA, thereby forming a ceRNA association with the corresponding miRNA (15). Among these, the interaction mechanisms between lncRNA and miRNA include: miRNA binding to and degrading lncRNA; lncRNA acting as miRNA sponge by binding to adsorbed miRNA; and truncation of lncRNA to generate miRNA (16). There is evidence that lncRNAs interact with miRNAs and act as ceRNAs to regulate the expression of miRNAs. CeRNAs serve important roles together with cancer-associated genes, and they have demonstrated potential in clinical tumor diagnosis and treatment (17,18).
As the largest database of cancer gene information currently available, The Cancer Genome Atlas (TCGA) includes 33 types of cancers in situ, advancing the understanding of the molecular basis of the onset of these diseases and improving the ability to diagnose, treat and prevent cancer. TCGA comprises multiple levels of tumor data, including genomic, transcriptomic, proteomic and epigenetic data (19). In the present study, according to the analysis of RNA expression profiles among 1,109 breast cancer tumor and 113 non-tumor tissue samples, a lncRNA-miRNA-mRNA regulatory network was successfully constructed. Furthermore, Cox regression analysis was used to select the top 10 differentially expressed lncRNAs (DElncRNAs), to understand the function of lncRNAs and their potential mechanisms in breast cancer. Survival analyses were also performed to identify prognostic genes.
Materials and methods
Study population
RNA sequencing data was downloaded from TCGA (https://www.cancergenome.nih.gov/). As of February 19, 2018, a total of 1,092 breast cancer cases were collected in the database. Univariate Cox analysis was performed using the library (survival) package in R software (version 3.4.3; R: A Language and Environment for Statistical Computing) (20) from the TCGA database. The present study adhered to the TCGA publication guidelines.
Screening differentially expressed genes
Breast cancer mRNA and miRNA sequencing data were derived from 1,222 samples, including 1,109 tumor samples (cohort Tumor) and 113 normal samples (cohort Normal). Normal sample and tumor sample data were then merged, and all expression values equal to zero were removed. Comparing the data from normal tissues with breast cancer samples, Perl language was used to extract the mRNA matrix file data from the RNA sequencing data (lncRNA, mRNA) downloaded by TCGA in the Windows Environment, running the program with cmd.exe, and then the data were converted into ensemble ID to obtain the gene symbol matrix file. The gene symbol matrix file contained the lncRNA and mRNA matrix, and gets-lncRNA symbol.pl and get-mRNA symbol.pl scripts were used to obtain the lncRNA symbol and mRNA symbol matrix data, respectively. DElncRNAs were measured using lncRNA symbol matrix and ‘edgeR’ package in R software (version 3.4.3), with thresholds of |log2 fold change (FC)| >2.0 and adjusted P<0.01. DEmiRNAs were defined as those with |log2FC| >2.0 and adjusted P<0.01.
Constructing the ceRNA network in breast cancer
DElncRNAs and DEmiRNAs were predicted using the miRcode (http://www.miRcode.org/), starBase (http://starbase.sysu.edu.cn/) (21), using miRTarBase online software to perform target gene predictions on the screened differential miRNAs (22). TargetScan (http://www.targetscan.org/vert_72/) and miRDB (http://mirdb.org/) databases were used to predict the targeting relationship between DEmiRNA and DEmRNA. The target DEmiRNA is entered into the database, which then shows all the target genes that may interact with the target microRNA, and then further screens the genes involved by the construction of a ceRNA network (23). To understand the functions of miRNA and lncRNA with the ceRNA network, gene co-expression network analysis was used, and the results were visualized with Cytoscape 3.6.0 (24).
Top 10 aberrantly expressed lncRNAs in breast cancer
The top 10 lncRNA were selected by receiver operating characteristics (ROC) curve and area under the curve (AUC) analysis. The prognostic roles of lncRNAs were examined with Kaplan-Meier curve analysis, and the log-rank test was conducted to distinguish survival time. P<0.05 was considered to indicate a statistically significant difference.
Analysis the lncRNAs with gene expression profiling interactive analysis (GEPIA)
Similarly, lncRNA expression levels in para-noncancerous tissues and cancer tissues were compared using the GEPIA database (http://gepia.cancer-pku.cn) (25), which was used to analyze the RNA sequencing data of 33 types of cancers from TCGA database.
Survival analysis
Using the survival time data for breast cancer in TCGA, the ‘Survival’ package in R software (version 3.4.3) was used to analyze the specific lncRNA, miRNA and mRNA associated with survival. Kaplan-Meier survival analysis was performed to analyze the correlation between DERNAs signature and breast cancer patient prognosis. P<0.05 was considered to indicate a statistically significant difference.
Results
DElncRNAs, DEmRNAs and DEmiRNAs
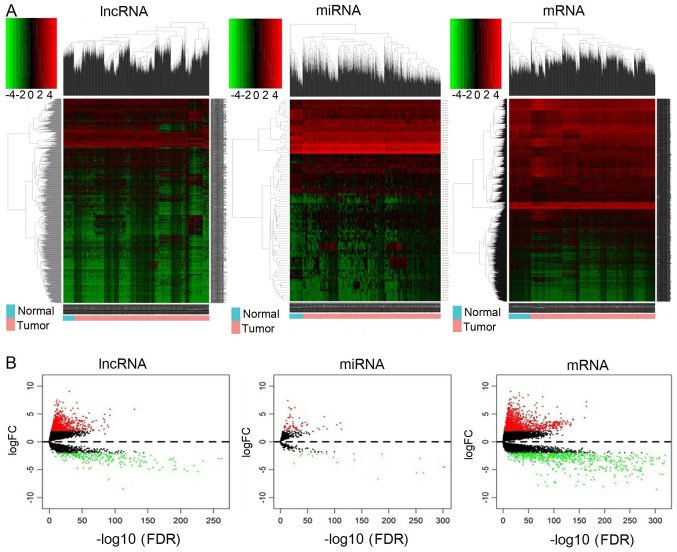
RNA expression levels in 1,109 breast cancer tumor and 113 normal tissue samples were investigated. Genes with |log2 FC| >1.5 and adjusted P<0.01 were considered as differentially expressed. The significant DEmiRNAs and DEmRNAs was identified in breast cancer samples. A total of 2,155 DEmRNAs, 89 DEmiRNAs and 1,028 lncRNAs were identified from TCGA data set using the ‘edgeR’ package in R. Of these, 1,434 mRNAs, 68 miRNAs and 777 lncRNAs were upregulated and 721 mRNAs, 21 miRNAs and 251 lncRNAs were downregulated in breast cancer tissues compared with normal tissues. The RNAs hierarchical clustering analyses are presented in Fig. 1A, and it was demonstrated that the expression levels of these three types of RNAs were significantly differentiated compared with the normal tissues. Finally, volcano plots were generated, and differences between the normal and tumor groups were identified (Fig. 1B).
Figure 1.
Heatmap and volcano plots demonstrating differential lncRNA, mRNA and miRNA expression between normal and cancer samples. (A) Heatmap of DElncRNAs, DEmiRNAs and DEmRNAs. Sample clusters are included above the heatmap. Clusters of DElncRNAs, DEmiRNAs and DEmRNAs are noted on the left of the heatmap. Red represents upregulated genes and green represents downregulated genes. Normal samples (blue) are on the left side of each heatmap, and tumor samples (pink) are on the right. (B) Volcano plots reflecting number, significance and reliability of differentially expressed lncRNAs, miRNAs and mRNAs. Red dots indicate upregulation and green dots indicate downregulation of lncRNAs, miRNAs and mRNAs. Black dots show the lncRNAs, miRNAs and mRNAs with expression of |log2FC|<2. The x-axis represents an adjusted FDR and the y-axis represents the value of log2 FC. Aberrantly expressed lncRNAs were calculated by DESeq R. In total, 777 upregulated and 251 downregulated lncRNAs, 68 upregulated and 21 downregulated miRNAs, and 1,434 upregulated and 721 downregulated mRNAs were identified. lncRNA, long non-coding RNA: miRNA, microRNA, DE, differentially expressed; FC, fold change; FDR, false discovery rate.
miRNA targeting lncRNAs predicted by miRcode and starBase
A total of 89 breast cancer-associated miRNAs were identified to be expressed in breast cancer and normal tissues. A total of 19 miRNAs were selected from 89 breast cancer-associated miRNAs in TCGA (|log2 FC| >3; P<0.001; Table I). It was identified that 19 miRNAs interacted with 93 lncRNAs (Table II).
Table I.
Breast cancer-specific miRNAs in the competing endogenous RNA network.
| miRNA | Expression change | log2 FC (T/N) | P-value | FDR |
|---|---|---|---|---|
| hsa-miR-141 | Upregulated | 2.31454976 | 2.06×10−88 | 6.36×10−87 |
| hsa-miR-200a | Upregulated | 2.197744863 | 5.86×10−74 | 1.32×10−72 |
| hsa-miR-145 | Downregulated | −2.240104939 | 4.89×10−190 | 3.59×10−188 |
| hsa-miR-182 | Upregulated | 2.45599701 | 2.74×10−71 | 5.75×10−70 |
| hsa-miR-206 | Downregulated | −3.892802244 | 1.7×10−26 | 9.3×10−26 |
| hsa-miR-204 | Downregulated | −2.548901757 | 2.02×10−61 | 3.29×10−60 |
| hsa-miR-21 | Upregulated | 2.26697984 | 5.18×10−115 | 2.54×10−113 |
| hsa-miR-375 | Upregulated | 2.657222381 | 8.83×10−30 | 5.36×10−29 |
| hsa-miR-183 | Upregulated | 3.040247685 | 3.28×10−106 | 1.28×10−104 |
| hsa-miR-122 | Upregulated | 7.377192392 | 8.45×10−15 | 2.46×10−14 |
| hsa-miR-96 | Upregulated | 3.37889678 | 5.04×10−113 | 2.28×10−111 |
| hsa-miR-187 | Upregulated | 3.256099283 | 1.11×10−24 | 5.49×10−24 |
| hsa-miR-301b | Upregulated | 2.966431803 | 7.76×10−40 | 7.59×10−39 |
| hsa-miR-429 | Upregulated | 2.767154505 | 9.75×10−82 | 2.49×10−80 |
| hsa-miR-210 | Upregulated | 3.146252439 | 5.59×10−52 | 7.62×10−51 |
| hsa-miR-144 | Downregulated | −2.790913655 | 4.88×10−100 | 1.59×10−98 |
| hsa-miR-137 | Upregulated | 2.533383379 | 6.50×10−11 | 1.48×10−10 |
| hsa-miR-184 | Upregulated | 4.310580766 | 1.02×10−23 | 4.84×10−23 |
| hsa-miR-100 | Downregulated | −2.005872118 | 3.19×10−80 | 7.8×10−79 |
hsa, Homo sapiens; miR, microRNA; FC, fold change; T/N, tumor/normal; FDR, false discovery rate.
Table II.
DElncRNAs interacting with the 19 DEmiRNAs retrieved from the miRcode database.
| lncRNA | miRNAs |
|---|---|
| AGAP11 | hsa-miR-141, hsa-miR-200a, hsa-miR-145, hsa-miR-182, hsa-miR-206, hsa-miR-204, hsa-miR-21, hsa-miR-375 |
| C2orf48 | hsa-miR-183, hsa-miR-204, hsa-miR-122 |
| SHANK2-AS3 | hsa-miR-96, hsa-miR-145, hsa-miR-187, hsa-miR-204, hsa-miR-122 |
| C20orf166-AS1 | hsa-miR-301b, hsa-miR-183, hsa-miR-429, hsa-miR-375 |
| C15orf54 | hsa-miR-301b, hsa-miR-182, hsa-miR-206, hsa-miR-429, hsa-miR-375 |
| AC127496.1 | hsa-miR-96, hsa-miR-182, hsa-miR-183, hsa-miR-204, hsa-miR-210, hsa-miR-122 |
| MIR7-3HG | hsa-miR-145, hsa-miR-204 |
| LINC00305 | hsa-miR-144, hsa-miR-204 |
| C10orf91 | hsa-miR-429, hsa-miR-204, hsa-miR-122 |
| WT1-AS | hsa-miR-96, hsa-miR-141, hsa-miR-200a, hsa-miR-145, hsa-miR-182, hsa-miR-206, hsa-miR-429, hsa-miR-375 |
| LINC00518 | hsa-miR-141, hsa-miR-200a, hsa-miR-145, hsa-miR-206, hsa-miR-204, hsa-miR-375 |
| LINC00221 | hsa-miR-301b, hsa-miR-96, hsa-miR-182, hsa-miR-204, hsa-miR-21 |
| TCL6 | hsa-miR-301b, hsa-miR-96, hsa-miR-137, hsa-miR-144, hsa-miR-145, hsa-miR-182, hsa-miR-183, hsa-miR-187, hsa-miR-206, hsa-miR-204, hsa-miR-210, hsa-miR-122, hsa-miR-375 |
| AC009093.1 | hsa-miR-183, hsa-miR-122 |
| C17orf102 | hsa-miR-96, hsa-miR-145, hsa-miR-206, hsa-miR-429, hsa-miR-21 |
| AC135178.1 | hsa-miR-122 |
| AF241725.1 | hsa-miR-145 |
| MUC2 | hsa-miR-145, hsa-miR-182, hsa-miR-183, hsa-miR-184, hsa-miR-429, hsa-miR-210, hsa-miR-122 |
| RMRP | hsa-miR-206, hsa-miR-122 |
| C1orf137 | hsa-miR-204 |
| AL391421.1 | hsa-miR-137, hsa-miR-144 |
| TDRG1 | hsa-miR-122 |
| C10orf126 | hsa-miR-141, hsa-miR-200a, hsa-miR-206, hsa-miR-375 |
| MUC19 | hsa-miR-301b, hsa-miR-96, hsa-miR-137, hsa-miR-144, hsa-miR-145, hsa-miR-182, hsa-miR-184, hsa-miR-187, hsa-miR-206, hsa-miR-429, hsa-miR-204, hsa-miR-122, hsa-miR-375 |
| UCA1 | hsa-miR-96, hsa-miR-182, hsa-miR-184, hsa-miR-206, hsa-miR-122 |
| LINC00488 | hsa-miR-96, hsa-miR-144, hsa-miR-206, hsa-miR-21, hsa-miR-122 |
| LINC00243 | hsa-miR-96, hsa-miR-145, hsa-miR-182, hsa-miR-206, hsa-miR-122, hsa-miR-375 |
| AL356479.1 | hsa-miR-429 |
| AL356310.1 | hsa-miR-206 |
| AC080037.1 | hsa-miR-429 |
| AL513123.1 | hsa-miR-141, hsa-miR-200a, hsa-miR-183 |
| LGALS8-AS1 | hsa-miR-122 |
| SMCR2 | hsa-miR-145, hsa-miR-183, hsa-miR-204 |
| PHEX-AS1 | hsa-miR-301b, hsa-miR-96, hsa-miR-145, hsa-miR-182, hsa-miR-204, hsa-miR-122 |
| LINC00466 | hsa-miR-96, hsa-miR-137, hsa-miR-141, hsa-miR-200a, hsa-miR-144, hsa-miR-183, hsa-miR-206, hsa-miR-429, hsa-miR-204, hsa-miR-21 |
| CHL1-AS2 | hsa-miR-183 |
| TSSC1-IT1 | hsa-miR-137, hsa-miR-141, hsa-miR-200a, hsa-miR-183 |
| LINC00337 | hsa-miR-145, hsa-miR-182, hsa-miR-375 |
| LINC00113 | hsa-miR-145 |
| LINC00351 | hsa-miR-21, hsa-miR-375 |
| ADIPOQ-AS1 | hsa-miR-144, hsa-miR-145, hsa-miR-182, hsa-miR-183, hsa-miR-184, hsa-miR-122, hsa-miR-375 |
| NAALADL2-AS2 | hsa-miR-183, hsa-miR-206 |
| LINC00355 | hsa-miR-141, hsa-miR-200a, hsa-miR-122 |
| LINC00392 | hsa-miR-183 |
| HOTAIR | hsa-miR-301b, hsa-miR-206, hsa-miR-204, hsa-miR-21, hsa-miR-375 |
| SRGAP3-AS2 | hsa-miR-145, hsa-miR-206 |
| HCG23 | hsa-miR-145 |
| LINC00200 | hsa-miR-183, hsa-miR-204 |
| LINC00404 | hsa-miR-141, hsa-miR-200a |
| SACS-AS1 | hsa-miR-144, hsa-miR-187 |
| EMX2OS | hsa-miR-182, hsa-miR-183, hsa-miR-184, hsa-miR-210 |
| ATXN8OS | hsa-miR-145, hsa-miR-183, hsa-miR-204, hsa-miR-210, hsa-miR-122hsa-miR-375 |
| LINC00210 | hsa-miR-96, hsa-miR-206, hsa-miR-204 |
| TLR8-AS1 | hsa-miR-182, hsa-miR-187, hsa-miR-206, hsa-miR-204 |
| LINC00460 | hsa-miR-206, hsa-miR-429 |
| POU6F2-AS2 | hsa-miR-137, hsa-miR-187, hsa-miR-375 |
| BOK-AS1 | hsa-miR-184 |
| MAGI2-AS3 | hsa-miR-137, hsa-miR-141, hsa-miR-200a, hsa-miR-144, hsa-miR-145, hsa-miR-429, hsa-miR-204, hsa-miR-210, hsa-miR-122 |
| CHL1-AS1 | hsa-miR-137, hsa-miR-187 |
| FNDC1-IT1 | hsa-miR-144 |
| DSCAM-AS1 | hsa-miR-137, hsa-miR-141, hsa-miR-200a, hsa-miR-204, hsa-miR-122 |
| LINC00484 | hsa-miR-141, hsa-miR-200a, hsa-miR-187, hsa-miR-206, hsa-miR-122 |
| ARHGEF7-AS2 | hsa-miR-187, hsa-miR-210, hsa-miR-122, hsa-miR-375 |
| RBMS3-AS3 | hsa-miR-96, hsa-miR-182, hsa-miR-204 |
| LINC00445 | hsa-miR-375 |
| NDP-AS1 | hsa-miR-145, hsa-miR-206hsa-miR-122 |
| AC009121.1 | hsa-miR-141, hsa-miR-200a |
| CLRN1-AS1 | hsa-miR-137, hsa-miR-206, hsa-miR-429, hsa-miR-204 |
| PDZRN3-AS1 | hsa-miR-141, hsa-miR-200a |
| AC080129.1 | hsa-miR-122 |
| AL109754.1 | hsa-miR-122 |
| MME-AS1 | hsa-miR-100, hsa-miR-182, hsa-miR-429 |
| LSAMP-AS1 | hsa-miR-183, hsa-miR-375 |
| ADAMTS9-AS1 | hsa-miR-301b, hsa-miR-96, hsa-miR-144, hsa-miR-145, hsa-miR-182, hsa-miR-21 |
| SYNPR-AS1 | hsa-miR-96, hsa-miR-182, hsa-miR-375 |
| ADAMTS9-AS2 | hsa-miR-301b, hsa-miR-96, hsa-miR-137, hsa-miR-141, hsa-miR-200a, hsa-miR-144, hsa-miR-145 |
| hsa-miR-182, hsa-miR-183, hsa-miR-184, hsa-miR-204, hsa-miR-122, hsa-miR-375 | |
| AC007731.1 | hsa-miR-183 |
| AL139002.1 | hsa-miR-301b, hsa-miR-210 |
| PEX5L-AS1 | hsa-miR-141, hsa-miR-200a |
| AC061992.1 | hsa-miR-301b, hsa-miR-204, hsa-miR-122 |
| AP000553.1 | hsa-miR-122 |
| LINC00461 | hsa-miR-96, hsa-miR-137, hsa-miR-141, hsa-miR-200a, hsa-miR-144, hsa-miR-145, hsa-miR-204, hsa-miR-210, hsa-miR-122 |
| ALDH1L1-AS2 | hsa-miR-301b, hsa-miR-145, hsa-miR-210 |
| MAST4-IT1 | hsa-miR-204 |
| LINC00536 | hsa-miR-96, hsa-miR-137, hsa-miR-182, hsa-miR-204 |
| OPCML-IT1 | hsa-miR-184, hsa-miR-375 |
| C8orf49 | hsa-miR-301b, hsa-miR-100, hsa-miR-184, hsa-miR-429, hsa-miR-122, hsa-miR-375 |
| AL589642.1 | hsa-miR-141, hsa-miR-200a, hsa-miR-145, hsa-miR-182, hsa-miR-183, hsa-miR-429, hsa-miR-204, hsa-miR-210, hsa-miR-122 |
| AC040173.1 | hsa-miR-96, hsa-miR-144, hsa-miR-182, hsa-miR-183, hsa-miR-429 |
| LINC00524 | hsa-miR-204 |
| LINC00052 | hsa-miR-145, hsa-miR-187 |
| PWRN1 | hsa-miR-137, hsa-miR-144, hsa-miR-145, hsa-miR-184, hsa-miR-21, hsa-miR-122 |
| LINC00261 | hsa-miR-301b, hsa-miR-144, hsa-miR-145, hsa-miR-182, hsa-miR-183, hsa-miR-206, hsa-miR-429, hsa-miR-204, hsa-miR-375 |
DE, differentially expressed; lncRNAs, long non-coding RNAs; hsa, Homo sapiens; miR, microRNA.
miRNA targets predicted by miRTarBase
Based on the miRNAs described in Table I, miRNA-targeted mRNA were identified by searching the TargetScan, miRDB and miRTarBase databases. A total of 27 mRNAs were identified including titin, chordin like 1, hydroxycarboxylic acid receptor 2, transforming growth factor β receptor 2, A-kinase anchoring protein 12, sprout RTK signaling antagonist 2, kelch like family member 40, sterile α motif domain containing 5, transcription elongation factor A like 7, KIT proto-oncogene receptor tyrosine kinase, cyclin B1, cell adhesion molecule L1 like, WAS protein family member 3, fibroblast growth factor (FGF) 2, SHC binding and spindle associated 1, serine rich and transmembrane domain containing 1, karyopherin α 2 (KPNA2), neurotrophic receptor tyrosine kinase 2 (NTRK2), cyclin E2, secreted frizzled related protein 1 (SFRP1), par-6 family cell polarity regulator β, ELAV like RNA binding protein 2, FGF receptor 3, cadherin 2, C-C motif chemokine ligand 20, solute carrier family 1 member 1 and homeobox B5 (Table III).
Table III.
Breast cancer-specific mRNAs in the competing endogenous RNA network.
| mRNA | Gene ID | Expression change | log2 FC (T/N) | P-value | FDR |
|---|---|---|---|---|---|
| TTN | ENSG00000155657 | Downregulated | −4.950085607 | 0.001 | 0.001 |
| CHRDL1 | ENSG00000101938 | Downregulated | −4.384645164 | 5.38×10−246 | 8.81×10−244 |
| HCAR2 | ENSG00000182782 | Downregulated | −3.359595565 | 2.72×10−233 | 3.83×10−231 |
| TGFBR2 | ENSG00000163513 | Downregulated | −2.148190475 | 4.53×10−182 | 4.04×10−180 |
| AKAP12 | ENSG00000131016 | Downregulated | −2.630546355 | 1.93×10−180 | 1.68×10−178 |
| SPRY2 | ENSG00000136158 | Downregulated | −2.373343848 | 3.08×10−179 | 2.63×10−177 |
| KLHL40 | ENSG00000157119 | Downregulated | −7.140824495 | 5.06×10−150 | 3.06×10−148 |
| SAMD5 | ENSG00000203727 | Downregulated | −2.921601554 | 1.42×10−134 | 5.06×10−150 |
| TCEAL7 | ENSG00000182916 | Downregulated | −2.087672875 | 1.82×10−121 | 7.64×10−120 |
| KIT | ENSG00000157404 | Downregulated | −2.904876101 | 5.47×10−120 | 2.24×10−118 |
| CCNB1 | ENSG00000134057 | Upregulated | 2.641558633 | 5.60×10−119 | 2.22×10−117 |
| CHL1 | ENSG00000134121 | Downregulated | −2.893484253 | 3.51×10−115 | 1.31×10−113 |
| WASF3 | ENSG00000132970 | Downregulated | −2.100371097 | 1.12×10−111 | 3.94×10−110 |
| FGF2 | ENSG00000138685 | Downregulated | −2.663380621 | 2.96×10−102 | 8.7×10−101 |
| SHCBP1 | ENSG00000171241 | Upregulated | 2.674370272 | 3.03×10−87 | 6.84×10−86 |
| SERTM1 | ENSG00000180440 | Downregulated | −3.596825432 | 3.8×10−81 | 7.75×10−80 |
| KPNA2 | ENSG00000182481 | Upregulated | 2.232771491 | 8.08×10−81 | 1.64×10−79 |
| NTRK2 | ENSG00000148053 | Downregulated | −2.532210134 | 3.89×10−70 | 6.46×10−69 |
| CCNE2 | ENSG00000175305 | Upregulated | 2.495744882 | 6.12×10−70 | 1.01×10−68 |
| SFRP1 | ENSG00000104332 | Downregulated | −2.49717591 | 1.76×10−46 | 1.59×10−45 |
| PARD6B | ENSG00000124171 | Upregulated | 2.263852741 | 8.63×10−35 | 5.35×10−34 |
| ELAVL2 | ENSG00000107105 | Upregulated | 2.693339772 | 2.05×10−34 | 1.25×10−33 |
| FGFR3 | ENSG00000068078 | Upregulated | 2.472470941 | 5.52×10−32 | 3.06×10−31 |
| CDH2 | ENSG00000170558 | Upregulated | 2.639030107 | 4.82×10−28 | 2.26×10−27 |
| CCL20 | ENSG00000115009 | Upregulated | 2.568683156 | 1.40×10−24 | 5.69×10−24 |
| SLC1A1 | ENSG00000106688 | Upregulated | 2.352249029 | 3.02×10−21 | 1.06×10−20 |
| HOXB5 | ENSG000001200755 | Upregulated | 2.001121261 | 1.91×10−15 | 5.15×10−15 |
FC, fold change; T/N, tumor/normal; FDR, false discovery rate.
Construction of a ceRNA network in breast cancer
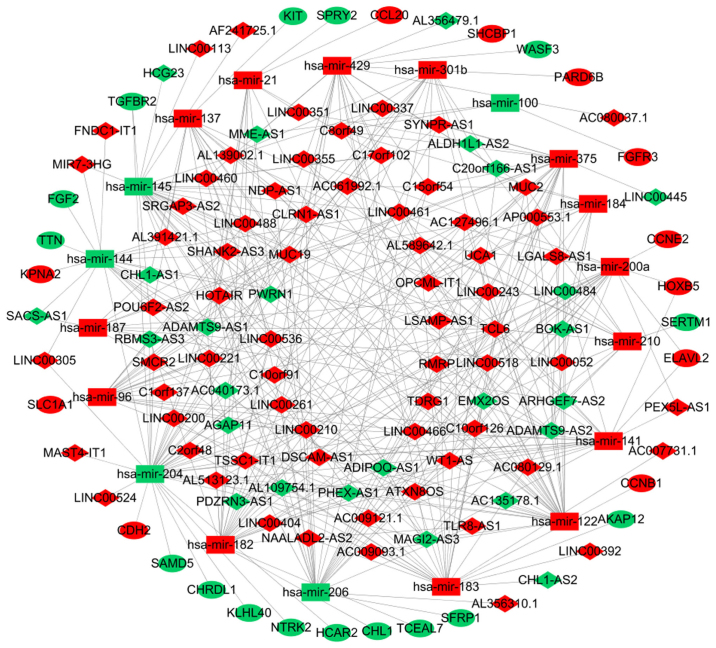
To improve the understanding of the role of DElncRNAs in breast cancer, a ceRNA network was constructed based on co-expressed lncRNAs-miRNAs and miRNAs-mRNAs. As demonstrated in Fig. 2, the ceRNA network was composed of 93 lncRNAs, 27 mRNAs and 19 miRNAs.
Figure 2.
Competing endogenous RNA network in breast cancer. The red nodes represent increased level of expression, while the green nodes represent decreased level of expression. Rectangles represent miRNAs, diamonds represent lncRNAs and ellipses represent mRNAs. The gray edges denote lncRNA-miRNA-mRNA interactions. lncRNA, long non-coding RNA; miR, microRNA.
ROC analysis of breast cancer-specific lncRNAs
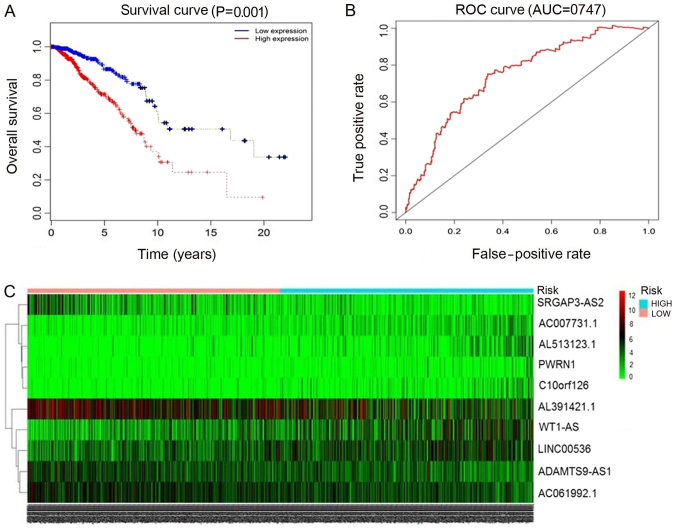
The 93 miRNAs targeting lncRNAs in breast cancer predicted by miRcode and starBase were selected. Then, Perl script was used to obtain the survival analysis data, and the clinicalexp file (lncRNA name, patient survival time, survival state) was combined with the survival data for univariate Cox analysis. In the univariate Cox analysis data, when the hazard ratio HR >1, this indicated that the higher the gene expression, the higher the risk of mortality, that is, the expression of the gene is contrary to survival. HR <1, indicated that the higher the expression of gene, the lower the risk of mortality, that is, the higher the survival rate. Survival-associated genes were then listed in order of ascending P-value, and the top 13 lncRNAs with P<0.05 were selected. lncRNAs were selected subsequent to entering the screening gene code in R software (version 3.4.3). Finally, the top 10 lncRNAs associated with breast cancer survival analysis were screened (Table IV; Fig. 3). These included: ADAM metallopeptidase with thrombospondin type 1 motif 9-antisense RNA 1 (ADAMTS9-AS1), AC061992.1, Wilms tumor 1 antisense RNA (WT1-AS), long intergenic non-protein coding RNA 536 (LINC00536), AL391421.1, SLIT-ROBO Rho GTPase activating protein 3 antisense RNA 2 (SRGAP3-AS2), Prader-Willi region non-protein coding RNA 1 (PWRN1), family with sequence similarity 230 member G (AC007731.1), chromosome 10 open reading frame 126 (C10orf126) and AL513123.1. Of these, the levels of ADAMTS9-AS1 and PWRN1 were downregulated in breast cancer tissues. The other 8 lncRNAs were upregulated in cancer tissues compared with in normal tissues. A total of 6 lncRNAs exhibited high prognostic values for distinguishing tumor tissues from normal tissues, with AUC values of >0.99.
Table IV.
Top 10 aberrantly expressed lncRNAs and AUC values in breast cancer.
| lncRNA | Gene ID | Expression change | log2 FC (T/N) | AUC | P-value |
|---|---|---|---|---|---|
| ADAMTS9-AS1 | ENSG00000241158 | Downregulated | −2.746216755 | 0.888 | <0.05 |
| AC061992.1 | ENSG00000266970 | Upregulated | 2.140133828 | 0.867 | <0.05 |
| WT1-AS | ENSG00000183242 | Upregulated | 4.901360082 | 1.102 | <0.05 |
| LINC00536 | ENSG00000249917 | Upregulated | 2.721025285 | 1.110 | <0.05 |
| AL391421.1 | ENSG00000204049 | Upregulated | 3.149701497 | 0.940 | <0.05 |
| SRGAP3-AS2 | ENSG00000228723 | Upregulated | 4.608297544 | 0.889 | <0.05 |
| PWRN1 | ENSG00000259905 | Downregulated | −2.274243394 | 1.243 | <0.05 |
| AC007731.1 | ENSG00000188280 | Upregulated | 2.840014701 | 1.182 | <0.05 |
| C10orf126 | ENSG000002043655 | Upregulated | 3.688082405 | 1.183 | <0.05 |
| AL513123.1 | ENSG00000236347 | Upregulated | 2.905568621 | 1.194 | <0.05 |
lncRNA; long-non-coding RNA; FC, fold change; T/N, tumor/normal; AUC, area under the curve.
Figure 3.
ROC curves and heatmap of the top 10 lncRNAs sorted by AUC in breast cancer. (A) The red line represents cases with high expression, and blue line represents cases with low expression. The x-axis indicates overall survival time (days), and the y-axis indicates the survival rate. Kaplan-Meier analysis was performed, and the curves were generated by R. (B) The red line represents the sensitivity curve, and the black line represents the identifying line. The x-axis indicates the false-positive rate. The y-axis indicates the true-positive rate. (C) Heatmap of the top 10 lncRNAs. Expression intensity increases from green to red, where green represents low expression, and red represents high expression. ROC, receiver operating characteristic; lncRNAs, long non-coding RNAs; AUC, area under the curve.
Additional analysis for the selected lncRNAs expression
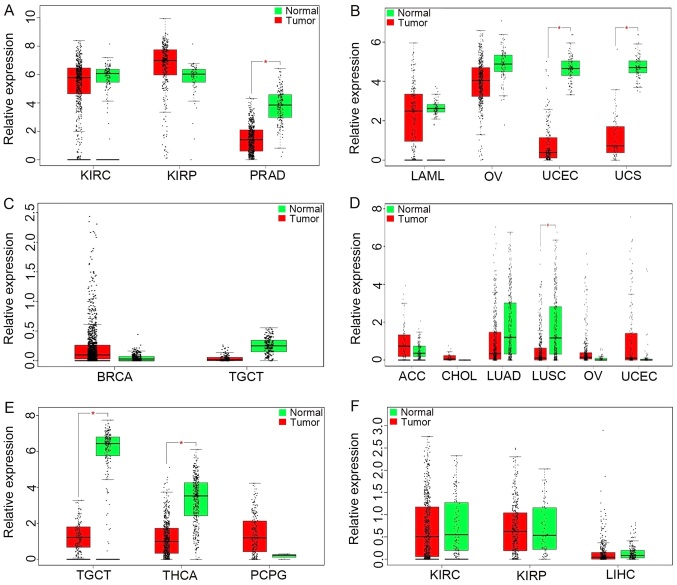
GEPIA results (Fig. 4A) revealed upregulation of ADAMTS9-AS1 in kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP) and prostate adenocarcinoma (PRAD). As demonstrated in Fig. 4B, the results also identified that WT1-AS levels were significantly increased in acute myeloid leukemia, ovarian cancer (OV), uterine corpus endometrial carcinoma (UCEC) and uterine carcinosarcomas compared with in normal tissues. Notably, the increased expression of LINC00536 was demonstrated in breast invasive carcinoma, while a decreased expression was demonstrated in testicular germ cell tumors (TGCT) (Fig. 4C). Furthermore, increased SRGAP3-AS2 expression was identified in adenoid cystic carcinoma, cholangiocarcinoma, OV and UCEC, while significant downregulation of SRGAP3-AS2 was observed in lung adenocarcinoma and lung squamous cell carcinoma (LUSC) (Fig. 4D). PWRN1 was observed to be downregulated in thyroid carcinoma and upregulated in TGCT, pheochromocytoma and paraganglioma (Fig. 4E). A decreased expression of C10orf126 was identified in KIRC, KIRP and liver hepatocellular carcinoma (Fig. 4F).
Figure 4.
lncRNAs expression among various types of cancer involved in The Cancer Genome Atlas based on GEPIA. (A) ADAMTS9-AS1, (B) WT1-AS, (C) LINC00536, (D) SRGAP3-AS2, (E) PWRN1 and (F) C10orf126. There is no description of the genes AC061992.1, AL391421.1, AC007731.1 and AL513123.1 in GEPIA. The y-axis indicates the log2 (transcripts per million + 1) for lncRNA expression. Red-colored bars represent the tumor tissue samples and the green-colored bars indicate the normal tissue samples. These charts were derived from GEPIA. lncRNAs; long non-coding RNAs; GEPIA, Gene Expression Profiling Interactive Analysis; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; PRAD, prostate adenocarcinoma; LAML, acute myeloid leukemia; OV, ovarian cancer; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcomas; BRCA, breast invasive carcinoma; TGCT, testicular germ cell tumors; ACC, adenoid cystic carcinoma; CHOL, cholangiocarcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; THCA, thyroid carcinoma; PCPG, pheochromocytoma and paraganglioma; LIHC, liver hepatocellular carcinoma. ADAMTS9-AS1, ADAM metallopeptidase with thrombospondin type 1 motif 9-antisense RNA 1; WT1-AS, Wilms tumor 1 antisense RNA; LINC00536, long intergenic non-protein coding RNA 536; SRGAP3-AS2, SLIT-ROBO Rho GTPase activating protein 3 antisense RNA 2; PWRN1, Prader-Willi region non-protein coding RNA 1; C10orf126, chromosome 10 open reading frame 126.
Survival analysis
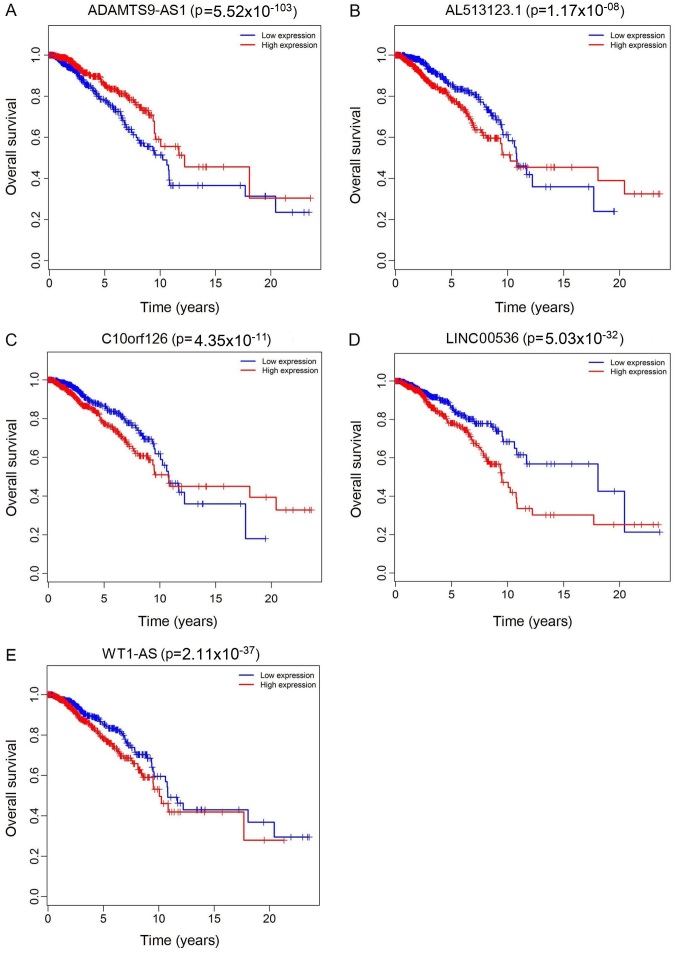
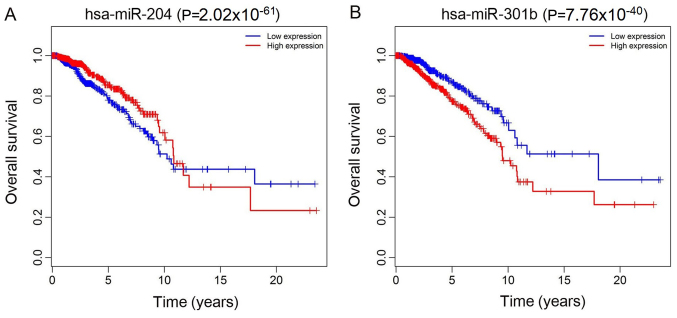
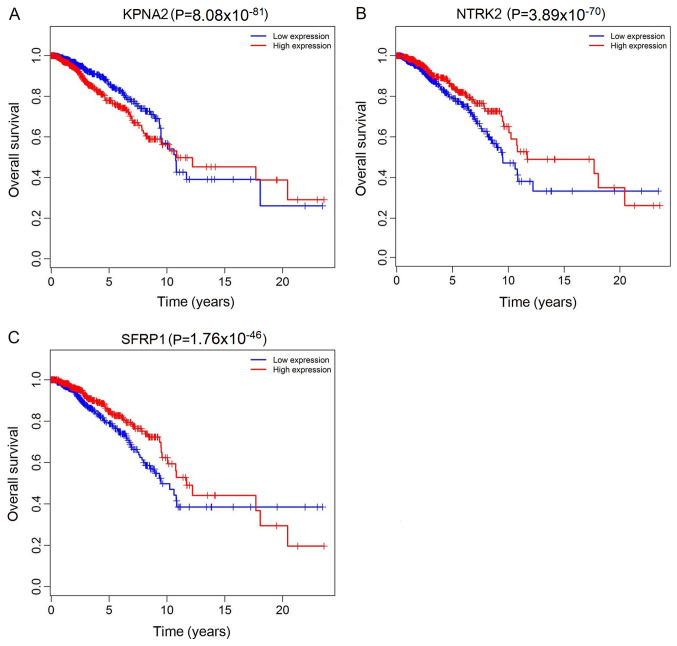
The top 10 lncRNAs were selected using ROC analysis. As demonstrated in Fig. 5, the expression levels of 5 DElncRNAs, including ADAMTS9-AS1, AL513123.1, C10orf126, LINC00536 and WT1-AS, were associated with overall survival (P<0.05). A total of 2 miRNAs (hsa-miR-204 and hsa-miR-301b) were associated with overall survival (P<0.05; Fig. 6). In addition, 3 mRNA (KPNA2, NTRK2 and SFRP1) were associated with overall survival (P<0.05; Fig. 7).
Figure 5.
Kaplan-Meier survival curves for 5 differentially expressed long non-coding RNAs associated with overall survival in breast cancer. (A) ADAMTS9-AS1 (P=5.52×10−103), (B) AL513123.1 (P=1.17×10−08), (C) C10orf126 (P=4.35×10−11), (D) LINC00536 (P=5.03×10−32) and (E) WT1-AS (P=2.11×10−37). The x-axis represents overall survival time (years) and the y-axis represents survival function. ADAMTS9-AS1, ADAM metallopeptidase with thrombospondin type 1 motif 9-antisense RNA 1; C10orf126, chromosome 10 open reading frame 126; LINC00536, long intergenic non-protein coding RNA 536; WT1-AS, Wilms tumor 1 antisense RNA.
Figure 6.
Kaplan-Meier survival curves for two differentially expressed miRNAs associated with overall survival in breast cancer. (A) hsa-miR-204 (P=2.02×10−61) and (B) hsa-miR-301b (P=7.76×10−40). The x-axis represents overall survival time (years) and the y-axis represents survival function. hsa, Homo sapiens; miRNA, microRNA.
Figure 7.
Kaplan-Meier survival curves for 3 mRNA associated with overall survival in breast cancer. (A) KPNA2 (P=8.08×10−81), (B) NTRK2 (P=3.89×10−70) and (C) SFRP1 (P=1.76×10−46). The x-axis represents overall survival time (years) and the y-axis represents survival function. KPNA2, karyopherin α 2; NTRK2, neurotrophic receptor tyrosine kinase 2; SFRP1, secreted frizzled related protein 1.
Discussion
In previous years, much attention has been paid to breast cancer pathogenesis. Nevertheless, clinical outcomes remain highly heterogeneous. Nik-Zainal et al (26) analyzed genome-wide data from 560 patients with breast cancer and identified that 93 oncogenes involved in the encoded proteins may cause breast cancer. Prognosis is an important indicator of disease treatment. Bioinformatics methods have been used to investigate the prognostic significance of lncRNAs in breast cancer. Several studies has also indicated that the aberrant expression of lncRNAs contributes to the development of different types of cancer: Xiao et al (27) revealed that MALAT1 may be used as a ceRNA to promote the expression of ZEB2 by expanding miR-200s, and may be a therapeutic target for kidney cancer; Wang et al (28) demonstrated that UCA1 and AATBC are not only included in the ceRNA network, but are also associated with overall survival in muscle-invasive bladder cancer; Chen et al (25) suggested that the top 10 aberrantly expressed lncRNAs identified in their study served important roles in LUSC through an lncRNA-mRNA network; and Li et al (29) described a novel gastric cancer-specific ceRNA network including 11 lncRNAs, 9 miRNAs and 41 mRNAs.
ceRNA transcripts may be used to competitively bind the same MRE with miRNAs, thereby relieving or mitigating the inhibitory effects of miRNAs on target genes. MREs are the basis for miRNA function, and other non-coding genes also interact with miRNAs through MREs. Recently, Zhou et al (30) used a Pearson's correlation analysis of miRNA-gene pairs to construct breast cancer-specific ceRNA networks. Regulation of phosphatase and tensin homolog expression in a 3′ untranslated region-and miRNA-dependent manner in breast cancer has been identified in another study (31). Furthermore, ceRNA crosstalk may be associated with regulatory mechanisms in breast cancer. Shen et al (32) and Yang et al (33) demonstrated that FAM83H-AS1 appeared to be a novel prognostic biomarker in breast cancer. Cheang et al (34) revealed that patients with breast cancer with high HER-2 protein expression levels and Ki67 index exhibited poor recurrence and disease-specific survival rates. Yuan et al (35) identified that the ceRNA crosstalk network in triple negative breast cancer included the differential expression of 22 hub mRNAs, 11 miRNAs and 14 lncRNAs. The present study obtained lncRNA, mRNA and miRNA data from TCGA to construct breast cancer-associated ceRNA networks, studied the regulatory mechanisms of lncRNA as ceRNAs in the progression of cancer, and also investigated their potential as prognostic biomarkers and therapeutic targets in breast cancer. A ceRNA network with differential expression of 27 mRNAs, 19 miRNAs and 93 lncRNAs was constructed.
The present study investigated the lncRNA expression profiles of a large cohort of patients in TCGA. A total of 93 DElncRNAs were identified in breast cancer samples compared with the normal samples. Among these, the top 10 lncRNA were significantly associated with overall survival. Furthermore, it was observed that the lncRNAs ADAMTS9-AS1, AL513123.1, C10orf126, LINC00536 and WT1-AS were included in the ceRNA network. Therefore, we hypothesized that these lncRNAs may serve significant roles in the pathogenesis and prognosis of breast cancer. Wang et al (36) revealed that antisense lncRNA ADAMTS9-AS1 was associated with the nearby coding gene ADAMTS9, which was involved in ovarian cancer progression. Li et al (37) identified that the lncRNA ADAMTS9-AS1 served as a novel prognostic biomarker for clinical application in esophageal squamous cell carcinoma. Crippa et al (38) hypothesized that LINC00536 disruption may have contributed to the onset of the clinical trichorhinophalangeal syndrome-like phenotype. Wang et al (39) suggested that WT1 was overexpressed in human hepatocellular carcinoma tissues, and was negatively correlated with overall survival in patients with hepatocellular carcinoma.
In the present study, a novel ceRNA network was constructed to identify the associations between miRNAs and lncRNAs. miRNAs also serve important roles in cell differentiation, biological development and disease progression. Several previous data have demonstrated that the mutual regulation between lncRNAs and miRNAs and their downstream target genes are closely associated with the occurrence and development of cancer (40–42). Zhang et al (43) used reverse transcription quantitative polymerase chain reaction and in situ hybridization for breast cancer tissue samples and cell lines. Cell phenotype experiments were performed to verify that lncRNA-GAS5 exon 4 bound miR-21 and inhibited the occurrence and development of breast cancer cells (43). The present study demonstrated that hsa-miR-204 was downregulated and hsa-miR-301b was upregulated in patients with breast cancer compared with healthy controls, and was associated with overall survival. Yuan et al (44) revealed that miR-204 suppressed the proliferation and metastasis of gastric cancer cells. Todorova et al (45) identified that miR-204 promoted prostate cancer-associated androgen-responsive genes and androgen receptor (AR) target genes and AR co-regulated molecules. Abmutalib et al (46) hypothesized that hsa-miR-301b may be involved in regulating lymph node metastasis in papillary thyroid carcinoma via interactions with hepatic leukemia factor, hypoxia-inducible factor and REL/nuclear factor kappa-light-chain-enhancer of activated B cells. Geng et al (47) demonstrated that overexpression of hsa-miR-301b significantly affected the cell cycle of human lung adenocarcinoma A549 cells. ceRNAs have been implicated in several biological processes, and abnormalities in the ceRNA network may lead to tumorigenesis.
In summary, the present study investigated lncRNA-mediated ceRNA interactions using miRNA, lncRNA and mRNA expression profiles in cancer and normal tissues. The results suggested that cancer-specific lncRNAs in breast cancer may be involved in the regulation of a complex ceRNA network. These data may provide novel insights into the clinical significance and regulatory mechanisms of lncRNA-mediated ceRNA networks, and identify novel lncRNAs as potential prognostic biomarkers and therapeutic targets for the diagnosis and treatment of breast cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from National Natural Science Foundation of China (grant no. 81660324) and the Key discipline Construction of the 13th Five-Year Plan in Xinjiang, China - Plateau Discipline Project.
Availability of data and materials
All data analyzed during this study are included in this published article.
Authors' contributions
SMF and LLL conceived and designed the manuscript. TT and ZA analyzed the data. TT wrote the paper. All authors read and approved the final manuscript
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Breastcancer.org. US Breast Cancer Statistics. http://www.breastcancer.org/symptoms/understand_bc/statistics. [Feb 18;2019 ];breast cancer org.
- 2.Linos E, Spanos D, Rosner BA, Linos K, Hesketh T, Qu JD, Gao YT, Zheng W, Colditz GA. Effects of reproductive and demographic changes on breast cancer incidence in China: A modeling analysis. J Natl Cancer Inst. 2008;100:1352–1360. doi: 10.1093/jnci/djn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgins MJ, Baselga J. Targeted therapies for breast cancer. J Clin Invest. 2011;121:3797–3803. doi: 10.1172/JCI57152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Győrffy B, Hatzis C, Sanft T, Hofstatter E, Aktas B, Pusztai L. Multigene prognostic tests in breast cancer: Past, present, future. Breast Cancer Res. 2015;17:11. doi: 10.1186/s13058-015-0514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- 8.Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng S, Zhang J, Su W, Bai S, Xiao L, Chen X, Lin J, Reddy RM, Chang AC, Beer DG, Chen G. Overexpression of LINC00152 correlates with poor patient survival and knockdown impairs cell proliferation in lung cancer. Sci Rep. 2017;7:2982. doi: 10.1038/s41598-017-03043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sang H, Liu H, Xiong P, Zhu M. Long non-coding RNA functions in lung cancer. Tumor Biol. 2015;36:4027–4037. doi: 10.1007/s13277-015-3449-4. [DOI] [PubMed] [Google Scholar]
- 11.Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Lu T, Wang Q, Liu J, Jiao W. Circular RNAs: Crucial regulators in the human body (Review) Oncol Rep. 2018;40:3119–3135. doi: 10.3892/or.2018.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S, Xiong F, Xiang B, Zhou M, Liao Q, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25. doi: 10.1186/s12943-017-0598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21:11680–11687. doi: 10.3748/wjg.v21.i41.11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Hou J, He D, Sun M, Zhang P, Yu Y, Chen Y. The emerging function and mechanism of ceRNAs in cancer. Trends Genet. 2016;32:211–224. doi: 10.1016/j.tig.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: Possible functions and clinical implications. J Med Genet. 2015;52:710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 18.Cheng DL, Xiang YY, Ji LJ, Lu XJ. Competing endogenous RNA interplay in cancer: Mechanism, methodology, and perspectives. Tumor Biol. 2015;36:479–488. doi: 10.1007/s13277-015-3093-z. [DOI] [PubMed] [Google Scholar]
- 19.Cline MS, Craft B, Swatloski T, Goldman M, Ma S, Haussler D, Zhu J. Exploring TCGA pan-cancer data at the UCSC cancer genomics browser. Sci Rep. 2013;3:2652. doi: 10.1038/srep02652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Core Team R, corp-author. R Foundation for Statistical Computing; Vienna, Austria: [Nov 30;2017 ]. A language and environment for statistical computing. version 3.4.3. [Google Scholar]
- 21.Jeggari A, Marks DS, Larsson E. miRcode: A map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015 Aug 12;4 doi: 10.7554/eLife.05005. (Epub ahead of print). doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen WJ, Tang RX, He RQ, Li DY, Liang L, Zeng JH, Hu XH, Ma J, Li SK, Chen G. Clinical roles of the aberrantly expressed lncRNAs in lung squamous cell carcinoma: A study based on RNA-sequencing and microarray data mining. Oncotarget. 2017;8:61282–61304. doi: 10.18632/oncotarget.18058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng J, Xiao W, Yu G, Yao W, Zhou H, et al. lncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6:38005–38015. doi: 10.18632/oncotarget.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Niu L, Jiang S, Zhai J, Wang P, Kong F, Jin X. Comprehensive analysis of aberrantly expressed profiles of lncRNAs and miRNAs with associated ceRNA network in muscle-invasive bladder cancer. Oncotarget. 2016;7:86174–86185. doi: 10.18632/oncotarget.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li F, Huang C, Li Q, Wu X. Construction and comprehensive analysis for dysregulated long non-coding RNA (lncRNA)-associated competing endogenous RNA (ceRNA) network in gastric cancer. Med Sci Monit. 2018;24:37–49. doi: 10.12659/MSM.905410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou S, Wang L, Yang Q, Liu H, Meng Q, Jiang L, Wang S, Jiang W. Systematical analysis of lncRNA-mRNA competing endogenous RNA network in breast cancer subtypes. Breast Cancer Res Treat. 2018;169:267–275. doi: 10.1007/s10549-018-4678-1. [DOI] [PubMed] [Google Scholar]
- 31.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen X, Xie B, Ma Z, Yu W, Wang W, Xu D, Yan X, Chen B, Yu L, Li J, et al. Identification of novel long non-coding RNAs in triple-negative breast cancer. Oncotarget. 2015;6:21730–21739. doi: 10.18632/oncotarget.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang F, Lv SX, Lv L, Liu YH, Dong SY, Yao ZH, Dai XX, Zhang XH, Wang OC. Identification of lncRNA FAM83H-AS1 as a novel prognostic marker in luminal subtype breast cancer. Onco Targets Ther. 2016;9:7039–7045. doi: 10.2147/OTT.S110055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan N, Zhang G, Bie F, Ma M, Ma Y, Jiang X, Wang Y, Hao X. Integrative analysis of lncRNAs and miRNAs with coding RNAs associated with ceRNA crosstalk network in triple negative breast cancer. Onco Targets Ther. 2017;10:5883–5897. doi: 10.2147/OTT.S149308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Fu Z, Dai C, Cao J, Liu X, Xu J, Lv M, Gu Y, Zhang J, Hua X, et al. lncRNAs expression profiling in normal ovary, benign ovarian cyst and malignant epithelial ovarian cancer. Sci Rep. 2016;6:38983. doi: 10.1038/srep38983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Yao Q, Zhao S, Wang Y, Li Y, Wang Z. Comprehensive analysis of differential co-expression patterns reveal transcriptional dysregulation mechanism and identify novel prognostic lncRNAs in esophageal squamous cell carcinoma. Onco Targets Ther. 2017;10:3095–3105. doi: 10.2147/OTT.S135312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crippa M, Bestetti I, Perotti M, Castronovo C, Tabano S, Picinelli C, Grassi G, Larizza L, Pincelli AI, Finelli P. New case of trichorinophalangeal syndrome-like phenotype with a de novo t(2;8)(p16.1;q23.3) translocation which does not disrupt the TRPS1 gene. BMC Med Genet. 2014;15:52. doi: 10.1186/1471-2350-15-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang N, Tan HY, Chan YT, Guo W, Li S, Feng Y. Identification of WT1 as determinant of heptatocellular carcinoma and its inhibition by Chinese herbal medicine Salvia chinensis benth and its active ingredient protocatechualdehyde. Oncotarget. 2017;8:105848–105859. doi: 10.18632/oncotarget.22406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai C, Huo Q, Wang X, Chen B, Yang Q. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun. 2017;485:272–278. doi: 10.1016/j.bbrc.2017.02.094. [DOI] [PubMed] [Google Scholar]
- 41.Kong Q, Qiu M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem Biophys Res Commun. 2018;495:1594–1600. doi: 10.1016/j.bbrc.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, Luo P, Guo W, Shi Y, Xu D, Zheng H, Jia L. lncRNA SNHG20 knockdown suppresses the osteosarcoma tumorigenesis through the mitochondrial apoptosis pathway by miR-139/RUNX2 axis. Biochem Biophys Res Commun. 2018;503:1927–1933. doi: 10.1016/j.bbrc.2018.07.137. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, Wu F, Mo YY. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan X, Wang S, Liu M, Lu Z, Zhan Y, Wang W, Xu AM. Histological and pathological assessment of miR-204 and SOX4 levels in gastric cancer patients. Biomed Res Int. 2017;2017:6894675. doi: 10.1155/2017/6894675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Todorova K, Metodiev MV, Metodieva G, Mincheff M, Fernández N, Hayrabedyan S. Micro-RNA-204 participates in TMPRSS2/ERG regulation and androgen receptor reprogramming in prostate cancer. Horm Cancer. 2017;8:28–48. doi: 10.1007/s12672-016-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abmutalib NS, Othman SN, Mohamad Yusof A, Abdullah Suhaimi SN, Muhammad R, Jamal R. Integrated microRNA, gene expression and transcription factors signature in papillary thyroid cancer with lymph node metastasis. Peer J. 2016;4:e2119. doi: 10.7717/peerj.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geng Y, Deng L, Su D, Xiao J, Ge D, Bao Y, Jing H. Identification of crucial microRNAs and genes in hypoxia-induced human lung adenocarcinoma cells. Onco Targets Ther. 2016;9:4605–4616. doi: 10.2147/OTT.S103430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data analyzed during this study are included in this published article.