Abstract
Autosomal recessive cornea plana is a very rare hereditary ocular disease, characterized by a flattened corneal curvature, marked hyperopia due to low refractive power and frequently consequent accommodative esotropia. Other features include various cornea anterior segment abnormalities, without systemic problems. The purpose of the present study was to investigate the clinical and molecular alterations in a Chinese family with cornea plana. Full ophthalmic examinations of the patients were performed, including slit-lamp examination, fundus examination and ocular ultrasound. Whole-exome sequencing data were screened for pathological variants in the proband, which were confirmed by Sanger sequencing. One novel missense mutation, c.242A>G (p.N81S) and another novel 7 base-pair deletion mutation, c.772-779del (p.G258Cfs*30), were detected in the keratocan (KERA) gene; two affected siblings inherited these variations in a compound heterozygous state, which were derived from the clinically unaffected heterozygous father (c.772_779del) and mother (c.242A>G), respectively. Neither mutation was observed in unrelated healthy controls (n=200). Multiple computer software predictions supported the pathogenicity of the two variants. Furthermore, protein modeling prediction was performed to better understand the molecular basis of cornea plana, particularly the importance of the leucine-rich repeat domain. This study presents the 14th pathogenic KERA mutations identified worldwide and the first in East Asia so far, to the best of our knowledge. These findings guided prenatal diagnosis for the family in question and expand on the variant spectrum of KERA, therefore facilitating genetic counseling.
Keywords: keratocan, leucine-rich repeat domain, cornea plana, protein modeling, novel mutation
Introduction
Cornea plana is a rare disorder characterized by small flat corneas, marked hypermetropia, reduced visual activity, iris abnormalities, a hazy corneal limbus and corneal clouding, with or without associated accommodative esotropia. It is inherited as either an autosomal dominant (CNA1; OMIM121400) or autosomal recessive trait (CNA2; OMIM217300); The defective gene in CNA1 has not been identified. CNA2 are associated with the more severe phenotype and additional ocular manifestations, including the presence of a degree of iris malformation and adhesions between the cornea and iris (1–8). Corneal ectasia, corneal hydrops and idiopathic corneal decompensation are very rare findings (2,9–11).
In humans, bi-allelic mutations in keratocan (KERA) cause CNA2. The gene, which has been localized to chromosome 12q22, contains two introns and three exons; a total of 352 amino acids are translated from the majority of exon 2 and a small portion of exon 3 to form a precursor KERA protein, and exon 1 remains untranslated (12). The gene encodes KERA, an evolutionarily-conserved small leucine-rich proteoglycan (SLRP). The SLRPs are a family of highly conserved extracellular matrix proteoglycans with core proteins composed of leucine-rich repeats (LRRs). LRRs are composed of a series of parallel β-strands, which stack into an arched β-sheet array. The leucine-rich region LXXLXLXXNXL is highly conserved: N represents cysteine or asparagine; L represents a hydrophobic amino acid or leucine; and X represents any amino acid. The interior of the LRR domain is composed of hydrophobic residues, with the exception of the conserved asparagines (Asn, N) which create an Asn-ladder stabilizing the structure, as reported in previous studies (13–15). In addition, the KERA protein is highly expressed in the cornea and is considered to be a key regulator of collagen fibrin production, which regulates the diameter and spacing of fibrils and determines corneal structure and transparency (16). Mouse studies indicate that this gene is exclusively expressed in the cornea and serves an important role during eye development (17). Narrow anterior chamber angles, thin corneal stroma and large-diameter corneal fibers are identified in KERA-knockout mice and no other systemic abnormality (18). To date, 13 different disease-associated KERA mutations have been documented in various populations, all of which manifest as various bilateral anterior segment lesions, including seven missense mutations, three nonsense mutations, one frame shift mutation, one splice-site mutation and one 3 base-pair deletion mutation. Notably, mutations in KERA have not been reported in an Asian population to date (1–4,6–10,19–24).
The present study reports two novel KERA mutations in a Chinese family. The family history was consistent with an autosomal recessive mode of inheritance. Clinical detection and genetic analysis was conducted on five family members.
Materials and methods
Subject recruitment and clinical examination
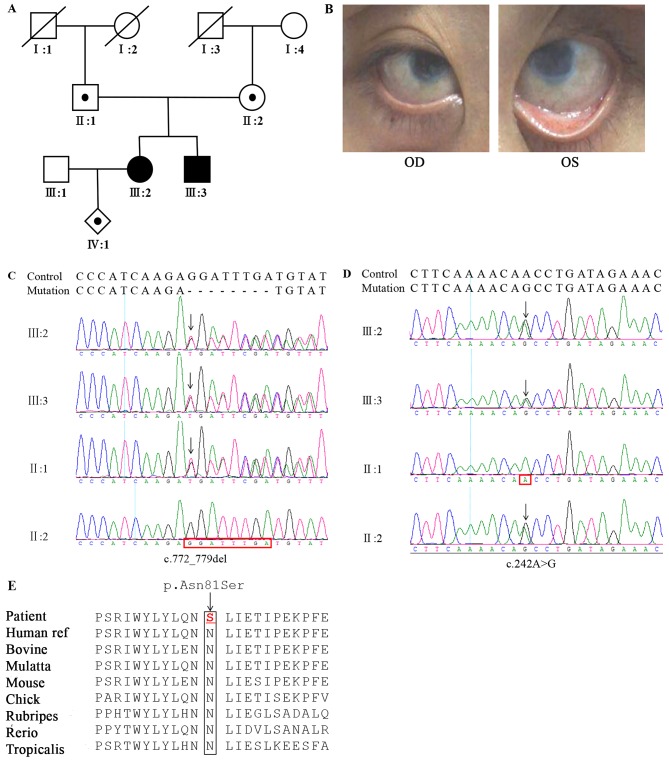
A family with CNA2 was recruited and identified at the Hunan Jiahui Genetics Hospital (Changsha, China) in October 2017. A pedigree of the family is illustrated in Fig. 1A. The proband, III:2, developed visual disorders shortly following birth, but hadn't been diagnosed or treated. She visited the doctor at the age of 26 years due to a considerable decrease in binocular visual acuity and accompanied by photophobia. Clinical and eye examinations demonstrated temporal iridocorneal synechiae, corneal clouding (Fig. 1B) and an unclear fundus. In addition, she was pregnant at the same time. A similar phenotype was observed in the younger brother of the proband, III:3.
Figure 1.
Pedigree structure and sequence chromatograms of detected novel mutations. (A) The pedigree illustrating autosomal recessive inheritance. (B) The left eye of patient III:2 with accommodative esotropia and binocular hazy corneal limbus. In order to better expose the field of vision, the patient assisted in opening the eyelids by hand. (C) The novel mutation c.772_779delGGATTTGA present in III:2, III:3 and II:1. (D) The novel mutation c.242A>G present in III:2, III:3 and II:2. Mutation position and normal sequence are indicated by black arrow and red box, respectively. (E) Multiple-sequence alignment of the Keratocan gene across different species. OD, oculus dextrus; OS, oculus sinister.
Best-corrected visual acuity (BCVA) was established using the Acta Ophthalmologica 2017 LogMAR charts. Visual acuity was examined using the ETDRS chart. Intraocular pressure was measured by Goldmann applanation tonometry. Anterior chamber depth and axial length were measured with ocular A ultrasound. Motility, pupillary reaction, ocular fundus, slit lamp examinations and keratometry (Zeiss keratometer; Zeiss AG) were performed and documented. The growth and development of III:2 and III:3 were in the normal range. Physical examinations were performed to exclude systemic diseases.
Ethical approval
The present study was performed according to the guidelines approved by the Ethics Committee of Hunan Jiahui Genetics Hospital and adhered to the tenets set out in the Helsinki Declaration. Informed consent was obtained from all subjects.
Genomic DNA extraction and variant screening
Venous blood samples were collected from the study subjects (III:2 and III:3), their family members (II:1, II:2 and III:1) and 200 randomly selected normal individuals without any history of ophthalmic disease. In addition, amniotic fluid was collected from III:2 to reflect the condition of the fetus (IV:1). Genomic DNA from all subjects was extracted via the phenol/chloroform method. Whole-exome sequencing data were screened for pathological variants in the patients (III:2 and III:3), and sanger sequencing of the candidate variants in KERA was performed on all subjects.
The extracted DNA was fragmented with DNase and purified via the magnetic bead method, followed by ligation of the adaptor sequence and polymerase chain reaction (PCR) amplification. PCR was conducted using a Phusion® High-Fidelity PCR Master Mix (New England BioLabs, Inc.) and the follows primers: 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ and 5′-CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT-3′. PCR was performed under the following conditions: 98°C for 30 sec; then 12 cycles of 98°C for 10 sec, 65°C for 30 sec and 72°C for 30 sec; and finally 72°C for 5 min. Library preparation was performed as previously described (25). The whole-exome was captured by Nimblegen SeqCap EZ Exome Probes v3.0 (Roche Diagnostics, Basel, Switzerland), extended, purified and finally sequenced on an Illumina HiSeq sequencer (Illumina, Inc.). The coverage of the target sequence was not less than 97% and the average depth of the target area was 130.95.
The original data were obtained following sequencing of all exons. The Burrows Wheeler Aligner algorithm was used to compare the reference sequence (UCSChg19), GATK version 3.7.0 (https://software.broadinstitute.org/gatk/) and VarScan version 2 software (http://dkoboldt.github.io/varscan/) were used to identify mutations, e.g. single nucleotide polymorphisms (SNPs) and InDels, including detection, annotation and statistical analysis. Annovar (http://www.openbioinformatics.org/annovar/) was utilized to catalog the detected variants. The list of mutations was tabulated, with the variants being described according to the HGVS guidelines. Combining this analysis with the public variation database ExAc, mutations with frequencies <0.1% in the population were identified. Mutations were scored for disease effect using a range of predictive tools [e.g. SIFT (http://sift.bii.a-star.edu.sg/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2), Provean (http://provean.jcvi.org/) and MutationTaster (http://www.mutationtaster.org/)]. Based on the results of the data analysis, the mutations were identified with reference to the 1000 Genomes Project (http://browser.1000genomes.org), the Exome Variant Server (http://evs.gs.washington.edu/EVS/), dbSNP database (https://www.ncbi.nlm.nih.gov/projects/SNP/index_old.html) and the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php). HomoloGene (https://www.ncbi.nlm.nih.gov/homologene) and T-Coffee multiple sequence alignment (http://www.tcoffee.org) were utilized to determine the conservation of the mutated amino acid residues between different species. A three-dimensional structure of keratocan was modeled using SWISS-MODEL (https://swissmodel.expasy.org). The analysis was performed in combination with assessment of the clinical manifestations and the mode of inheritance. Finally, the nature of the mutation sites were interpreted according to the guideline by the American College of Medical Genetics and Genomics/the Association for Molecular Pathology (ACMG) (26).
The two novel candidate variations were validated using direct bidirectional sequencing methods. Based on the relevant gene standard sequences listed at the NCBI, primers were designed using Primer Premier 5 software (Premier Biosoft International). Primers forward (5′-3′): CACCCAGTTTTCCTACTGCTTTATA, primers reverse (5′-3′): GCACTGATTCGGGGAACCTT. PCR amplification was performed on the target DNA fragment (700 bp product size, 20 µl reaction system, annealing temperature 62°C) and the product was verified by 1.0% agarose gel electrophoresis. Direct sequencing was performed on an ABI 3130 DNA Sequencer (Applied Biosystems; Thermo Fisher Scientific, Inc.). The sequenced results were aligned with the reference sequence using SeqMan at the NCBI (no. NM_007035).
Results
Clinical findings
III:2 exhibited a BCVA of 0.2 oculus dextrus (OD; right eye) and 0.3 oculus sinister (OS; left eye) with hyperopic refractive values. Additionally, she exhibited esotropia (right eye), microcornea, corneal opacity, short axial length, shallow anterior chamber, iridocorneal adhesion, pupillary irregularities (1×2 mm irregular right pupil shifting to the nasal side; needle-like changes on the left pupil) and slow light reflection. Extraocular motility, confrontation fields and intraocular pressure were normal, and the lens was transparent, although the fundus was difficult to see clearly (Table I). The younger brother of the proband (III:3) presented with similar clinical symptoms, however the parents had not identified any eye problems.
Table I.
Keratocan gene mutations and clinical characteristics of patient with cornea plana.
| Central corneal haze | BCVA | Refraction (DS, DC) | CCT (µm) | ACD (mm) | IOP (mmHg) | Axial length (mm) | K1/K2 (D) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient/origin | Gender/age (y) | Family history | Nucleotide change | RE | LE | RE | LE | RE | LE | RE | LE | RE | LE | RE | LE | RE | LE | RE | LE |
| III:2/China | F/26 | Y | c.242A>G | Y | Y | 0.2 | 0.3 | +12.00/-2.0DC | +14.00 | 473 | 502 | 1.36 | 1.04 | 8.2 | 16.8 | 20.8 | 20.3 | 33.59/35.86 | 27.68/30.36 |
| c.772-779 | |||||||||||||||||||
| delGGATTTGA | |||||||||||||||||||
BCVA, best corrected visual acuity; RE, right eye; LE, left eye; DS, dioptre sphere; DC, dioptre cylinder; CCT, central corneal thickness; ACD, anterior chamber depth; IOP, intraocular pressure; D, dioptre; K1, flat keratometry; K2, steep keratometry; y, years; Y, yes.
Mutation screening and bioinformatics analysis
The family of interest, originating from China, displayed an autosomal recessive pattern of inheritance. A compound heterozygous mutation c.242A>G (p.N81S) and c.772_779delGGATTTGA(P.G258Cfs*30) in exon 2 of the KERA gene was identified in III:2 and III:3, respectively derived from the heterozygous father (c.772_779delGGATTTGA) and mother (c.242A>G; Fig. 1C and D), although neither of the two mutations were present in the normal controls. These variants have not been previously reported in the 1000 Genomes Project, the Human Gene Mutation Database, dbSNP or the Exome Variant Server. The first variation, an N81S amino acid substitution in the KERA protein, was predicted to be likely pathogenic by the in-silico prediction programs, Mutation Taster, Provean, n PolyPhen (27) and SIFT (28) (Table II). The second mutation was a 7 base-pair deletion (G258Cfs*30), leading to the loss of LRR8-11 and producing a strongly truncated protein, which may trigger nonsense-mediated mRNA decay (−66 amino acid/>10% missing); the prediction algorithms supported its pathogenic nature. Multiple sequence alignment indicated that the residues at position 81 of KERA are highly conserved across species (Fig. 1E) and the lost LRR8-11 regions due to 258 residual changes are also highly conserved. According to ACMG, p.N81S was judged likely to be damaging (PM1-3 and PP1-4) and P.G258Cfs*30 was also judged as damaging (PVS1, PM2, PP1, PP3 and PP4) (28).
Table II.
Predicted effect of 2 novel mutations on function of KERA protein.
| Prediction | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation at nucleotide level | Change at protein level | Exon | Predicted protein domain | Conservation | PolyPhen2a | Mutation taster | Proveana | SIFTa | Frameshift and PTC | ACMG/AMP variants classificationb |
| c.242A>G | p.N81S | 2 | LRRs | Highly conserved | Probably damaging | Disease causing | Deleterious | Damaging | No | Likely Pathogenic |
| c.772_779delGGATTTGA | p.G258Cfs*30 | 2 | LRRs | Highly conserved | NA | Disease causing | NA | NA | Yes | damaging |
NA, not available; PTC, premature termination codon
The PolyPhen2, SIFT and Provean algorithm is only applicable to single amino acid substitutions
Classification of each novel variant was following the variant interpretation guideline by the American College of Medical Genetics and Genomics/the Association for Molecular Pathology (26).
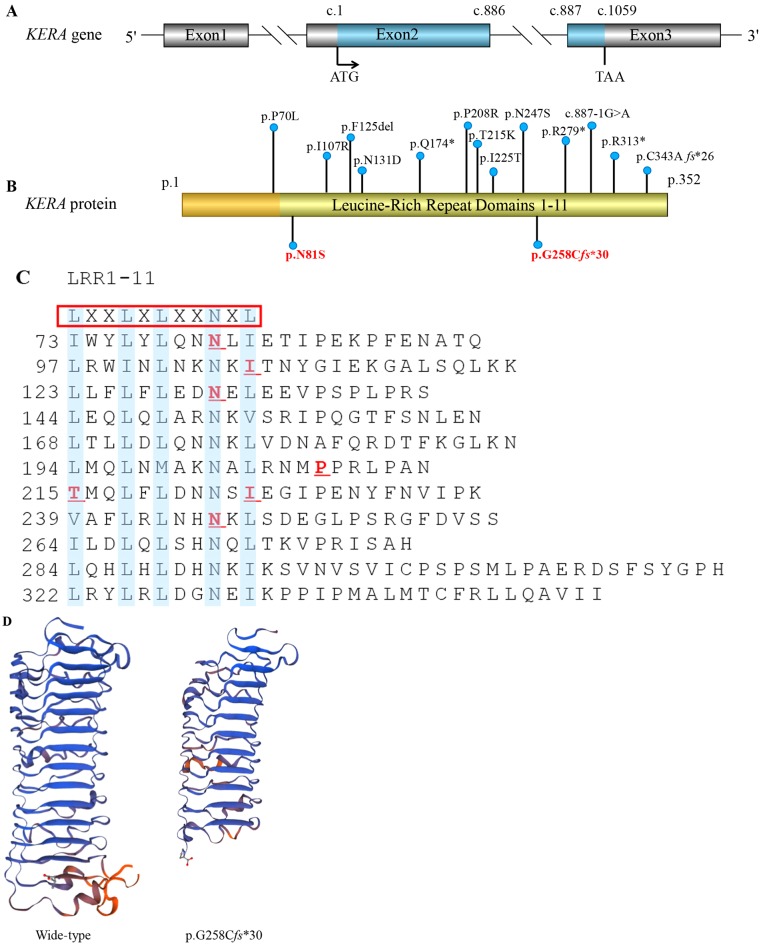
Primary sequence analysis indicated that the N81S substitution occurs at an LRR motif, the first of 11 such motifs present in the sequence (Fig. 2C) (29). G258Cfs*30 indicates a 7 base-pair deletion leading to a frameshift and premature termination codon (PTC), followed by the loss of LRR8-11. The predicted three-dimensional structure of the KERA protein using the SWISS-MODEL online software suggested that the current mutant protein (P.G258Cfs*30) is significantly different from the wild type and these changes may lead to abnormal protein function (Fig. 2D) (30). The architecture of other known LRR proteins reveals that the highly conserved LXXLXLXXNXL repetitive motifs form a series of parallel β-strands and these β-strands form a horseshoe-like concave shape. The asparagines (Asn, N) develop a stable Asn-ladder structure. The present study identified that the p.(Asn81Ser) mutation affects the conserved Asn81 residue of LRR1. This substitution to an aspartic acid residue will alter a hydrogen bond donor to make it either an acceptor or neutral, possibly resulting in the instability of the Asn-ladder structure or abnormal interactions between the mutant KERA and other proteins, including collagen. The second mutation, G258Cfs*30, that is reported in the present study results in the loss of LRR8-11. This deletion may form an abnormal truncated protein, thereby causing disease.
Figure 2.
KERA mutations associated with cornea plana. (A) The structure of the KERA gene. It contains three exons and two introns; the blue marker indicates the translation area, which forms a precursor protein of 352 amino acids. ATG: Initiation codon; TAA: Stop codon. (B) Kera protein schematic, including locations of the variations that have previously been reported. In the present study, amino acid position 81 was changed from asparagine (Asn, N) to serine (Ser, S). The p.G258Cfs*30 mutation leading to premature termination codon is indicated in bold red. Apart from p.P70L, all mutations associated with the disease are located in the LRR domains. (C) The blue highlighting indicates the consensus motifs LXXLXLXXNXL, which form 11 LRR domains. The red underlining indicates the six missense mutations previously identified. (D) Mutant Kera (p.G258Cfs*30) was predicted by Swiss-Model online software to result in the loss of the partial LRR domain and the C-terminal domain, compared with the wild type protein. LRR, leucin-rich repeat; Kera, keratocan.
Discussion
With all the previously reported KERA mutations CNA2 displays a relatively homogeneous phenotype and genetic heterogeneity. Although certain previous studies have documented an atypical phenotype, including superior pellucid corneal marginal degeneration, hypotrichosis and corneal edema (2,9,11,21), they retain a classically identifiable phenotype of CNA2. Any alteration of a highly conserved encoded protein is likely to result in CNA2 with specific characteristics.
The present study reports on the phenotype and identification of suspected pathogenic variations in a Chinese family with CNA2. The proband and her brother, with two novel KERA mutations in a compound heterozygous form, exhibited the typical corneal phenotype, including marked hypermetropia, flat corneas with opacification and iridocorneal synechiae. In addition, the proband exhibited a short axis length. This finding supports the hypothesis proposed in previous studies that the KERA mutation may lead to microphthalmia, although this has not been proven (6,22). At present, there is no known effect on endothelial cell function and no specific studies have been conducted internationally. The unaffected parents in the present case were mutation carriers and did not exhibit corneal abnormalities, suggesting that in the presence of mutant gene products there remains some normal KERA proteins, which allows the cornea to develop normally.
Nearly all the conservative LRRs in the KERA gene are encoded in the larger second exon. Original descriptions of the gene mentioned 11 LRRs. A similar LRR motif exists in the structure of a porcine RNase inhibitor, the crystal structure of which has been elucidated (31). The structure of other proteins containing similar LRR motifs also mimic a β-sheet array and the KERA structural model presented in the present study supports this. These LRRs are flanked by cysteine clusters, a sulfur-containing amino acid that allows for disulfide bonds and the maintenance of the three-dimensional conformation that serves a key role in keratocan functioning.
The most reported mutations in KERA affect the LRR structure/conformation. The present study uncovered a novel missense mutation in KERA exon 2; an Asn81Ser replacement occurs within first LRR motif and disrupts it, potentially altering the stacking or spacing of the β-strand. Another novel deletion mutation, p.G258Cfs*30, in exon 2 leads to a frame shift and PTC, followed by the loss of LRR8-11. The protein is prematurely truncated due to the destruction of 7th LRR and the deficiency of the 8 to 11th LRR motifs, which contain 287 amino acids instead of 353. Conventional LRR motif alignment is believed to be important in the spacing of collagen fibrils on which corneal transparency is dependent. Aberrant KERA in the corneal stroma may not bind to collagen fibrils, therefore damaging the regulatory effect of KERA during the development of the normal corneal structure.
Reviewing the literature, it was identified that CNA2 is a rare disorder with a worldwide distribution and a high prevalence in particular populations, e.g. those of Finnish (19), Saudi (3) and Czech descent (23). Almost no mutations have been identified in the Asian population, except for a Chinese American individual who was mentioned in a previous study, although this case was not documented in detail (8). In historically isolated populations, including Saudi and Finnish populations, the occurrence of CNA2 is not only due to a single common founder effect; certain cultural preferences, including intermarriage or endogamy, may also increase the incidence of recessively inherited diseases (8).
In conclusion, the present study identified two novel KERA variants in a Chinese family with cornea plana. These findings, together with the previously reported mutations, demonstrate the occurrence of CNA2 in different populations and expand the variant spectrum of the KERA gene; therefore, the results of the present study may be valuable for the diagnosis of and genetic counseling for cornea plana.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CNA2
autosomal recessive cornea plana
- L
hydrophobic amino acid or leucine
- X
any amino acid
- N
cysteine or asparagine
- SLRP
small leucine-rich proteoglycan
- LRRs
leucine-rich repeats
- BCVA
best-corrected visual acuity
- ACD
anterior chamber depth
- PTC
premature termination codon
Funding
The present study was supported by grants from the National Key R&D Program of China (grant no. 2017YFC1001802) and the National Natural Science Foundation of China (grant nos. 81501268, 31601035 and 81571450).
Availability of data and materials
All data generated and analyzed during the present study are included in this published article.
Authors' contributions
CZH, XGL, HT collected clinical data from the CNA2 family. CZH, XGL, LQW and PSYL performed molecular genetics experiments on the family and the controls. CZH, XGL, HU TAN and CP conducted the bioinformatics analysis. CZH, CP and LQW designed and supervised the study. CZH drafted the manuscript. WGL contributed in data analysis. CP and PSYL, WGL and LQW revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Informed consent for this investigation was obtained from all participating CNA2 patients and parents, and the principles outlined in the Declaration of Helsinki were followed. The study was conducted in agreement with the Ethical Committee of the Center for Medical Genetics, School of Life Sciences, Central South University (Changsha, China).
Patient consent for publication
The publication of images from the CNA2 patient has the support and informed consent of the patient.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pellegata NS, Dieguez-Lucena JL, Joensuu T, Lau S, Montgomery KT, Krahe R, Kivelä T, Kucherlapati R, Forsius H, de la Chapelle A. Mutations in KERA, encoding keratocan, cause cornea plana. Nat Genet. 2000;25:91–95. doi: 10.1038/75664. [DOI] [PubMed] [Google Scholar]
- 2.Khan AO, Aldahmesh M, Meyer B. Corneal ectasia and hydrops in a patient with autosomal recessive cornea plana. Ophthalmic Genet. 2006;27:99–101. doi: 10.1080/13816810600862469. [DOI] [PubMed] [Google Scholar]
- 3.Khan AO, Aldahmesh M, Meyer B. Recessive cornea plana in the Kingdom of Saudi Arabia. Ophthalmology. 2006;113:1773–1778. doi: 10.1016/j.ophtha.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 4.Khan AO, Aldahmesh MA, Al-Gehedan S, Meyer BF, Alkuraya FS. Corneal decompensation in recessive cornea plana. Ophthalmic Genet. 2009;30:142–145. doi: 10.1080/13816810902937084. [DOI] [PubMed] [Google Scholar]
- 5.Tahvanainen E, Forsius H, Kolehmainen J, Damsten M, Fellman J, de la Chapelle A. The genetics of cornea plana congenita. J Med Genet. 1996;33:116–119. doi: 10.1136/jmg.33.2.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehmann OJ, El-ashry MF, Ebenezer ND, Ocaka L, Francis PJ, Wilkie SE, Patel RJ, Ficker L, Jordan T, Khaw PT, Bhattacharya SS. A novel keratocan mutation causing autosomal recessive cornea plana. Invest Ophthalmol Vis Sci. 2001;42:3118–3122. [PubMed] [Google Scholar]
- 7.Khan AO, Al-Saif A, Kambouris M. A novel KERA mutation associated with autosomal recessive cornea plana. Ophthalmic Genet. 2004;25:147–152. doi: 10.1080/13816810490514397. [DOI] [PubMed] [Google Scholar]
- 8.Ebenezer ND, Patel CB, Hariprasad SM, Chen LL, Patel RJ, Hardcastle AJ, Allen RC. Clinical and molecular characterization of a family with autosomal recessive cornea plana. Arch Ophthalmol. 2005;123:1248–1253. doi: 10.1001/archopht.123.9.1248. [DOI] [PubMed] [Google Scholar]
- 9.Khan AO, Aldahmesh M, Al-Saif A, Meyer B. Pellucid marginal degeneration coexistent with cornea plana in one member of a family exhibiting a novel KERA mutation. Br J Ophthalmol. 2005;89:1538–1540. doi: 10.1136/bjo.2005.073510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liskova P, Hysi PG, Williams D, Ainsworth JR, Shah S, de la Chapelle A, Tuft SJ, Bhattacharya SS. Study of p.N247S KERA mutation in a British family with cornea plana. Mol Vis. 2007;13:1339–1347. [PubMed] [Google Scholar]
- 11.AlBakri A, Khan AO. Regarding corneal decompensation in recessive cornea plana. Ophthalmic Genet. 2016;37:350–351. doi: 10.3109/13816810.2015.1059462. [DOI] [PubMed] [Google Scholar]
- 12.Tasheva ES, Funderburgh JL, Funderburgh ML, Corpuz LM, Conrad GW. Structure and sequence of the gene encoding human keratocan. DNA. 1999;10:67–74. doi: 10.3109/10425179909033939. [DOI] [PubMed] [Google Scholar]
- 13.Kobe B, Deisenhofer J. The leucine-rich repeat: A versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 14.Kobe B, Deisenhofer J. Proteins with leucine-rich repeats. Curr Opin Struct Biol. 1995;5:409–416. doi: 10.1016/0959-440X(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 15.Bella J, Hindle KL, McEwan PA, Lovell SC. The leucine-rich repeat structure. Cell Mol Life Sci. 2008;65:2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao WW, Liu CY. Roles of lumican and keratocan on corneal transparency. Glycoconj J. 2002;19:275–285. doi: 10.1023/A:1025396316169. [DOI] [PubMed] [Google Scholar]
- 17.Liu CY, Shiraishi A, Kao CW, Converse RL, Funderburgh JL, Corpuz LM, Conrad GW, Kao WW. The cloning of mouse keratocan cDNA and genomic DNA and the characterization of its expression during eye development. J Biol Chem. 1998;273:22584–22588. doi: 10.1074/jbc.273.35.22584. [DOI] [PubMed] [Google Scholar]
- 18.Liu CY, Birk DE, Hassell JR, Kane B, Kao WW. Keratocan-deficient mice display alterations in corneal structure. J Biol Chem. 2003;278:21672–21677. doi: 10.1074/jbc.M301169200. [DOI] [PubMed] [Google Scholar]
- 19.Forsius H, Damsten M, Eriksson AW, Fellman J, Lindh S, Tahvanainen E. Autosomal recessive cornea plana. A clinical and genetic study of 78 cases in Finland. Acta Ophthalmol Scand. 1998;76:196–203. doi: 10.1034/j.1600-0420.1998.760215.x. [DOI] [PubMed] [Google Scholar]
- 20.Dudakova L, Palos M, Hardcastle AJ, Liskova P. Corneal endothelial findings in a Czech patient with compound heterozygous mutations in KERA. Ophthalmic Genet. 2014;35:252–254. doi: 10.3109/13816810.2013.811272. [DOI] [PubMed] [Google Scholar]
- 21.Roos L, Bertelsen B, Harris P, Bygum A, Jensen H, Grønskov K, Tümer Z. Case report: A novel KERA mutation associated with cornea plana and its predicted effect on protein function. BMC Med Genet. 2015;16:40. doi: 10.1186/s12881-015-0179-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumari D, Tiwari A, Choudhury M, Kumar A, Rao A, Dixit M. A novel KERA mutation in a case of autosomal recessive cornea plana with primary angle-closure glaucoma. J Glaucoma. 2016;25:e106–e109. doi: 10.1097/IJG.0000000000000258. [DOI] [PubMed] [Google Scholar]
- 23.Dudakova L, Vercruyssen JHJ, Balikova I, Postolache L, Leroy BP, Skalicka P, Liskova P. Analysis of KERA in four families with cornea plana identifies two novel mutations. Acta Ophthalmol. 2018;96:e87–e91. doi: 10.1111/aos.13484. [DOI] [PubMed] [Google Scholar]
- 24.Khan AO. Corneal ectasia in a boy with homozygous KERA mutation. Ophthalmic Genet. 2018;39:141–143. doi: 10.1080/13816810.2017.1350724. [DOI] [PubMed] [Google Scholar]
- 25.Chen R, Im H, Snyder M. Whole-exome enrichment with the Roche nimbleGen SeqCap EZ exome library SR platform. Cold Spring Harb Protoc. 2015;2015:634–641. doi: 10.1101/pdb.prot084855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Offord V, Coffey TJ, Werling D. LRRfinder: A web application for the identification of leucine-rich repeats and an integrative Toll-like receptor database. Dev Comp Immunol. 2010;34:1035–1041. doi: 10.1016/j.dci.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 31.Kobe B, Deisenhofer J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature. 1993;366:751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated and analyzed during the present study are included in this published article.