Abstract
Gastric cancer (GC) is a leading cause of cancer-associated mortality worldwide. Previous studies demonstrated that long noncoding RNAs (lncRNAs) may be dysregulated in GC and may serve important roles in cancer progression. The present study aimed to investigate the role of the novel lncRNA stomach cancer-associated transcript 16 (STCAT16; Assembly Gene ID G038291) in the development and progression of GC. The present data suggested that the expression level of STCAT16 was decreased in GC tissues. The expression level of STCAT16 was identified to be associated with lymph node and tumour node metastasis stages. Furthermore, the expression level of STCAT16 was identified to be significantly associated with poor survival and prognosis. Knockdown of STCAT16 promoted proliferation, colony formation, migration and invasion of BGC-823 cells. In contrast, these features were suppressed in AGS cells following overexpression of STCAT16. In vivo, tumour growth was significantly decreased following STCAT16 overexpression. Collectively, the present data suggested that the lncRNA STCAT16 may act as a tumour suppressor and may inhibit GC tumour cell growth and migration. Additionally, the decreased expression level of STCAT16 was identified to be associated with poor prognosis in patients with GC.
Keywords: gastric carcinoma, long noncoding RNA, stomach cancer-associated transcript 16, cell proliferation, cell movement
Introduction
Gastric cancer (GC) is one of the most common types of cancer worldwide and is one of the leading causes of cancer-associated mortality, particularly in China (1). Recent advances in diagnosis and treatment have improved the long-term survival rate of patients with early-stage GC; however, the prognosis of patients with late-stage GC remains poor, primarily due to cancer invasion and metastasis (2–5). Tumour invasion and metastasis are progressive, multifactorial and multistep processes that involve cell adhesion and migration, local invasion into the adjacent tissue, intravasation, survival in the circulatory system, extravasation and migration into target organs, where tumour cells may further proliferate (6). The invasion of tumour cells into surrounding tissues is an important early event in GC metastasis (7,8). However, the molecular mechanism underlying GC invasion of surrounding tissues remains unclear. Therefore, investigation of this process is required to improve the understanding of tumour metastasis. The present results may facilitate the identification of novel molecular targets that may be used to develop novel therapeutic strategies to inhibit GC metastasis.
Long noncoding RNAs (lncRNAs) are RNA molecules >200 nucleotides in length. These RNAs have limited coding capability; however, they are involved in various biological processes, including epigenetic modifications (9,10), and transcriptional (11) and post-transcriptional regulation (12). Additionally, putative lncRNAs have been identified to encode small peptides (13). Therefore, lncRNAs may serve various roles in multiple diseases. Accumulating evidence has demonstrated that lncRNAs are dysregulated in various types of cancer, serving important roles in signalling pathways controlling tumourigenesis and cancer progression (14). Multiple lncRNAs, including HOX transcript antisense RNA (HOTAIR) (15,16), gastric carcinoma proliferation enhancing transcript 1 (GHET1) (17), hepatocellular carcinoma upregulated long noncoding RNA (HULC) (18), colon cancer associated transcript 1 (CCAT1) (19) and maternally expressed 3 (MEG3) (20,21), were demonstrated to be associated with GC progression, serving as oncogenes or tumour suppressor genes.
A previous study by Iyer et al (22) identified various novel lncRNAs; among these novel lncRNAs, stomach cancer-associated transcript 16 (STCAT16) exhibited a significant downregulation in GC tissues; however, the association between STCAT16 and clinicopathological characteristics of patients with GC remains unknown. In the present study, the expression level of STCAT16 was identified to be downregulated in GC tissues and cell lines. STCAT16 downregulation was significantly associated with poor clinical features and prognosis. Notably, STCAT16 overexpression inhibited proliferative and invasive abilities of GC cells, and multivariate analysis identified that high expression level of STCAT16 was an independent predictor for improved overall survival (OS) rate in patients with GC.
Materials and methods
Human samples
The present study was approved by The Institutional Review Board of The Affiliated Hospital of Nantong University (Nantong, China) and was performed in accordance with the Declaration of Helsinki. Written informed consent was obtained prior to the start of the study. GC tissues and matched normal gastric tissues, located 5 cm from the tumour margin, were collected from 59 patients with GC (age, 32–84 years; female to male ratio, 24:35) from the Department of General Surgery (Affiliated Hospital of Nantong University) between December 2014 and August 2015. Clinical data and follow-up information were obtained from the medical records of the patients. GC stage was assigned according to the tumour, node and metastasis (TNM) classification and staging system recommended by the American Joint Committee on Cancer (23). T and N classification was based on non-invasive examinations and M classification was determined from samples that were surgically removed. OS was defined as the duration between the date of surgery and the date of mortality or the last follow-up. All patients were followed up until December 2017. The patients did not receive radiation therapy or chemotherapy prior to surgery. Patients included in the study did not exhibit long-term use of nonsteroidal anti-inflammatory drugs or corticosteroids prior to the surgery. Cancer classification and pathological types were assessed by two experienced pathologists in a double-blind manner.
Cell culture and transfection
Human GC cell lines (SGC-7901, MKN-45, AGS, MGC-803 and BGC-823) and a human normal gastric mucosa cell line (GES-1) were purchased from the Shanghai Institutes for Biological Sciences of The Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) containing 10% foetal bovine serum (FBS; Zhejiang Tianhang Biotechnology Co., Ltd., Huzhou, China), 100 µg/ml streptomycin and 100 U/ml penicillin at 37°C with 5% CO2. Cells in the logarithmic (log) phase were selected and transfected in 24-well plates at 85% confluence using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The cells were dissociated using a solution containing 0.25% trypsin-EDTA (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Subsequent experiments were performed 72 h post-transfection.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The relative expression level of STCAT16 was determined by RT-qPCR. Total RNA was extracted from 100 mg tissues or cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, RNA was reverse-transcribed into cDNA with a Moloney murine leukaemia virus reverse transcriptase kit (Thermo Fisher Scientific, Inc.). The temperature protocol was as follows: 42°C for 60 min, 70°C for 5 min. According to the manufacturer's instructions, qPCR was executed using SYBR-Green I (Takara Biotechnology, Co., Ltd., Dalian, China) and an Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher Scientific, Inc.). The qPCR reaction mixture contained 10 µl SYBR-Green/ROX qPCR Master Mix (Takara Biotechnology, Co., Ltd.), 1 µl forward primers (STCAT16, 5′-CATCAAGGCTTGTGGGATGT-3′; GAPDH, 5′-CTGGGCTACACTGAGCACC-3′), 1 µl reverse primers (STCAT16, 5′-AAGCCGAAAGGTCAACTGC-3′; GAPDH, 5′-AAGTGGTCGTTGAGGGCAATG-3′), 2 µl cDNA and 6 µl sterile double steamed water. GAPDH was used as a reference gene. The thermocycling conditions were the following: 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 60 sec. Primers of STCAT16 were synthesized by Shanghai Ruian BioTechnologies Co., Ltd. (Shanghai, China). The data were quantified using the 2−ΔΔCq method (24). Each experiment was performed in triplicate.
STCAT16 knockdown and overexpression vectors construction
A total of four plasmids containing sequences for short hairpin RNAs (shRNAs) targeting STCAT16 (STCAT16-shRNA-759, STCAT16-shRNA-1161, STCAT16-shRNA-1356 and STCAT16-shRNA-1693), a plasmid containing the negative control (NC) shRNA and a STCAT16 overexpression plasmid (STCAT16-pcDNA3.1) were constructed by Shanghai GenePharma Co., Ltd (Shanghai, China). Cells were selected in the logarithmic phase and transfected with 4.0 µg/well plasmid DNA in 24-well plates (5×105 cells/well) at 85% confluence using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Empty pcDNA3.1 plasmid was used as NC for the overexpression experiments (mock-NC). The plasmid extraction kit was purchased from Omega Bio-tek, Inc. (Norcross, GA, USA).
Cell proliferation assay
Cell proliferation was assessed using the Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology, Haimen, China) according to the manufacturer's protocol. Cells growing in the exponential phase were plated at a density of 5×104 cells/ml on 96-well plates and four plasmids (STCAT16-shRNA, sh-NC and STCAT16-pcDNA3.1, mock-NC) were transfected with 4.0 µg/well per plasmid DNA using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 0, 24, 48, 72 and 96 h, 10 µl CCK-8 solution was added to each well followed by a 2-h incubation at 37°C. The absorbance was recorded at 450 nm with a microplate reader (Varioskan® Flash; Thermo Fisher Scientific, Inc.). All experiments were performed in triplicate.
Colony formation assay
Colony formation assays were performed as previously described (25). A total of 4.0 µg/well per plasmid DNA of four plasmids (STCAT16-shRNA, sh-NC and STCAT16-pcDNA3.1, mock-NC) were transfected using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Cells were seeded on 6-well plates at a concentration of 100 cells/well and cultured at 37°C for 2–3 weeks. Surviving colonies were counted following staining with Gentian Violet (Amresco, LLC, Solon, OH, USA) for 5 min at room temperature. Colonies with >50 cells were counted under an upright light microscope (magnification, ×100; BX51 Microscope; Olympus Corporation, Tokyo, Japan). All experiments were performed in triplicate.
Wound healing assay
Cells were plated on 6-well plates at a density of 5×105 cells/well and four plasmids (STCAT16-shRNA, sh-NC and STCAT16-pcDNA3.1, mock-NC) were transfected with 4.0 µg/well per plasmid DNA using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The cell monolayers were scratched in a straight line using a pipette tip at 24 h after transfection. The debris was removed with three washes in PBS, and phase-contrast images were acquired using an inverted fluorescence microscope (IX71; Olympus Corporation, Tokyo, Japan; magnification, ×100). Cells were cultured for an additional 24 h, and imaged at 6, 12 and 24 h in that incubation period. Subsequently, the number of migrating cells was counted.
Cell invasion assay
STCAT16-shRNA, sh-NC and STCAT16- pcDNA3.1, mock-NC plasmids (all 4.0 µg/well plasmid DNA) were transfected into GC cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The cell suspension containing 1×105 cells/ml transfected for 48 h was placed in an upper chamber (Transwell®; Corning Inc., Corning, NY, USA) precoated with Matrigel (BD Biosciences, San Jose, CA, USA). A total of 600 µl RPMI-1640 supplemented with 20% FBS was plated in the lower chamber. Invasive cells were able to pass through the Matrigel layer. Subsequently, cells were incubated for 24 h and stained with crystal violet (Beyotime Institute of Biotechnology) at room temperature for 20 min. The field of view (n=15) of each insert was randomly counted using an upright light microscope (BX51; Olympus Corporation, Tokyo, Japan; magnification, ×100) and the average value was calculated.
In vivo tumourigenesis assay
A total of 10 male BALB/c nude mice (age, 4 weeks; weight, 18–20 g) were purchased from The Laboratory Animal Research Centre of Nantong University (Nantong, China). Mice were maintained at 24°C in a temperature-controlled environment with 40–60% relative humidity, under a 12-h light/dark cycle using a dimmer, with food and water ad libitum. Mice were randomly divided into two groups (n=5 in each group) and injected with STCAT16-pcDNA3.1-transfected AGS cells (STCAT16-pcDNA3.1 group) or with empty pcDNA3.1-transfected AGS cells (mock-NC group). The 2×107 cells were subcutaneously injected into the back of the mice. The mice with developing tumours were observed ≥3 times per week. The tumours were allowed to grow for 4 weeks, and the tumour volume (V) was calculated using the following formula: V=0.5 × length × width2. All animal experiments were approved by The Animal Care and Use Committee of Nantong University.
Immunohistochemistry
Tumour tissue sections were fixed in 10% buffered formalin for 24 h at room temperature, embedded in paraffin and cut into 4 µm sections. The sections were deparaffinized in xylene, rehydrated in graded ethanol solutions and treated with 0.1% hydrogen peroxide in methanol for 20 min. Antigen retrieval was performed by microwaving the sections immersed in 10 mM citric acid buffer for 10 min, followed incubation for 1 h at room temperature. Sections were incubated with 5% bovine serum albumin (cat. no. LLBB-1000-01, SurModics, Inc., Eden Prairie, USA) to block non-specific protein binding for 10 min at 37°C, followed by 1-h incubation at room temperature and overnight at 4°C with monoclonal antibody against Ki-67 (1:50 dilution; cat. no. ab16667; Abcam, Cambridge, UK). Sections were washed with PBS and incubated with a horseradish peroxidase-conjugated polyclonal goat anti-rabbit immunoglobulin G biotin secondary antibody (1:500 dilution; cat. no. E030110-01; EarthOx, LLC, San Francisco, CA, USA) for 1 h at room temperature, followed by 0.1% hydrogen peroxide and 0.6 mM 3,3′-Diaminobenzidine (DAB) in PBS for 8 min at room temperature. All sections were stained using a Hematoxylin and Eosin Staining kit (cat. no. C0105; Beyotime Institute of Biotechnology) as previously described (26), and examined using a light microscope (BX51; Olympus Corporation; magnification, ×400). The nuclear was stained blue and cytoplasm was stained red.
Regarding assessment, Ki-67 staining intensity was evaluated using a four rating-level-scheme, where scores ranging from 0 to 3 indicated negative, weak, medium and strong staining, respectively. For extent of staining, a five rating-level-scheme was employed. Thus, based on the total amount of positive stained areas in the whole carcinoma region, the extent of staining was evaluated with scores ranging from 0 to 4 as follows: 0, 0; 1, 1–25; 2, 26–50; 3, 51–75; and 4, 76–100%. In each specimen, five high-power fields (×400) were randomly selected, together with examination of nuclear staining. The product of intensity scores multiplied by degree scores was used as the final staining score (0–12) for Ki-67 assessment. The staining was evaluated as previously described (27).
Statistical analysis
All experiments were performed in triplicate. All statistical analyses were performed using SPSS software (version 17.0; SPSS, Inc., Chicago, IL, USA). Graphs were generated using GraphPad Prism software (version 5.0; GraphPad Software, Inc., La Jolla, CA, USA). Data are presented as the mean ± standard deviation. Differences between two groups and in more than two groups were evaluated using a paired Student's t-test and one-way analysis of variance, respectively. Student-Newman-Keuls post hoc test was performed following ANOVA. The association between the expression level of STCAT16 and clinical characteristics was analysed using χ2 test or Fisher's exact probability. Parameters associated with OS and time to recurrence (TTR) were identified using univariate and multivariate Cox proportional hazards regression models. Kaplan-Meier plots (log-rank test) were used to examine OS and TTR. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression level of STCAT16 is downregulated in human gastric cancer and decreased expression level of STCAT16 is associated with poor OS
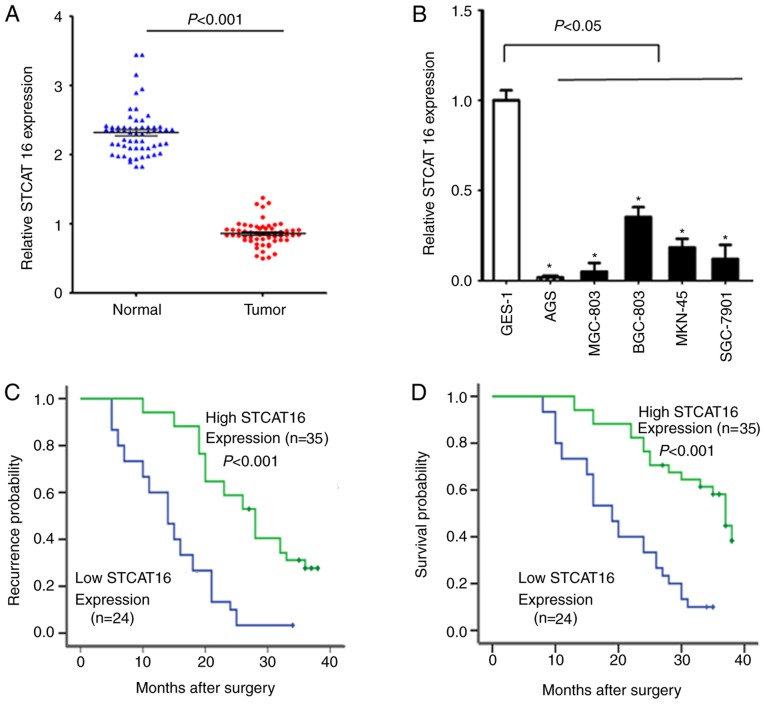
The expression level of STCAT16 in GC tissues was significantly decreased compared with adjacent normal gastric tissues (Fig. 1A). The expression level of STCAT16 varied among GC cell lines; however, all cell lines exhibited a decreased expression level of STCAT16 compared with GES-1 cells (Fig. 1B). Compared with GES-1 cells, AGS and BGC-823 cells exhibited a relative expression level of STCAT16 of 0.015±0.006 and 0.332±0.056, respectively.
Figure 1.
Expression of STCAT16 in GC and its prognostic significance in patients with GC. RT-qPCR was used to detect the expression of STCAT16 in (A) GC tissues and in (B) cell lines. GC cell lines and a human normal gastric mucosa cell line (GES-1) were used for RT-qPCR analysis. (C) Postoperative recurrence and (D) overall survival in patients with increased or decreased expression levels of STCAT16 were assessed by log-rank test. *P<0.05 vs. GES-1 cells. GC, gastric cancer; RT-qPCR, reverse transcription polymerase chain reaction; STCAT16, stomach cancer-associated transcript 16.
The clinical features of the enrolled patients are listed in Table I. If the expression level of STCAT16 in the tumour tissue was decreased >2-folds compared with the matched adjacent normal tissue, the expression level of STCAT16 was considered as low. The expression level of STCAT16 in GC tissues was identified to be associated with TNM stage and lymph node metastasis. The TTR of patients with low STCAT16 expression was significantly decreased compared with patients exhibiting high STCAT16 expression (Fig. 1C). Additionally, OS was significantly decreased in patients exhibiting low expression level of STCAT16 (Fig. 1D).
Table I.
Association between STCAT16 expression and clinical characteristics.
| STCAT16 | ||||
|---|---|---|---|---|
| Clinical characteristics | n | High (n=35) | Low (n=24) | P-value |
| Age (years) | 0.598 | |||
| <60 | 27 | 15 | 12 | |
| ≥60 | 32 | 20 | 12 | |
| Sex | 0.504 | |||
| Male | 35 | 22 | 13 | |
| Female | 24 | 13 | 11 | |
| Tumour size (cm) | 0.679 | |||
| <5 | 40 | 23 | 17 | |
| ≥5 | 19 | 12 | 7 | |
| Tumour, node and metastasis stage | 0.027a | |||
| I/II | 20 | 16 | 4 | |
| III/IV | 39 | 19 | 20 | |
| Differentiation degree | 0.400 | |||
| High or medium | 26 | 17 | 9 | |
| Low | 33 | 18 | 15 | |
| Lymph node metastasis | 0.025a | |||
| Yes | 34 | 16 | 18 | |
| No | 25 | 19 | 6 | |
P<0.05. If the expression level of STCAT16 in the tumour tissue was decreased >2 fold compared with the matched adjacent normal tissue, the expression level of STCAT16 was considered as low. STCAT16, stomach cancer-associated transcript 16.
To further investigate the prognostic potential of STCAT16, univariate and multivariate survival analyses were performed examining clinicopathological characteristics of patients with GC. The univariate analysis was performed using six prognostic factors: Age, sex, differentiation degree, lymph node metastasis, TNM stage and STCAT16 expression level. The multivariate analysis identified that the TNM stage and the expression level of STCAT16 were independent predictors of OS (Table II).
Table II.
Univariate and multivariate analysis of clinicopathological characteristics and overall survival in 59 patients with gastric cancer.
| A, Univariate analysis | |||
|---|---|---|---|
| Clinical characteristics | Hazard ratio | 95% confidence interval | P-value |
| Age (≥60 and <60) | 1.101 | 0.751–2.451 | 0.931 |
| Sex | 0.461 | 0.251–1.072 | 0.442 |
| Differentiation degree (high/middle/low) | 0.908 | 0.742–2.411 | 0.658 |
| Tumour, node and metastasis stage (I/II/III/IV) | 5.455 | 1.890–11.080 | 0.055 |
| Lymph node metastasis (no/yes) | 0.723 | 0.818–3.524 | 0.620 |
| STCAT16 expression level (high/low) | 6.560 | 2.331–17.232 | 0.018 |
| B, Multivariate analysis | |||
| Clinical characteristics | Hazard ratio | 95% confidence interval | P-value |
| Age (≥60 and <60) | 1.224 | 1.331–2.221 | 0.828 |
| Sex | 1.008 | 0.411–1.722 | 0.345 |
| Differentiation degree (high/middle/low) | 1.244 | 0.311–3.616 | 0.120 |
| Tumour, node and metastasis stage (I/II/III/IV) | 6.753 | 1.321–12.381 | 0.039 |
| Lymph node metastasis (no/yes) | 1.232 | 0.223–2.919 | 0.237 |
| STCAT16 expression level (high/low) | 7.393 | 1.928–15.156 | 0.025 |
STCAT16, stomach cancer-associated transcript 16.
STCAT16 interference and overexpression
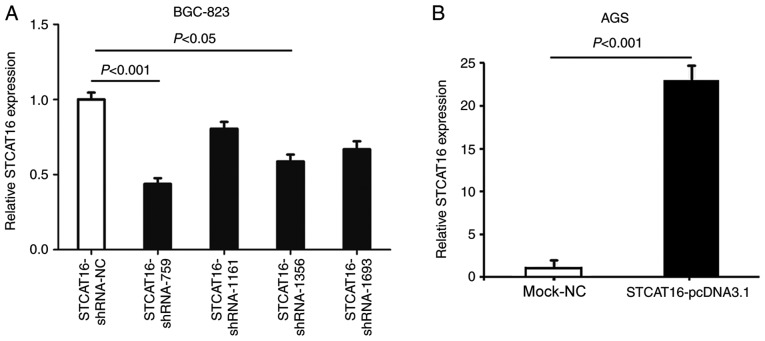
To identify the most effective shRNA, four STCAT16-shRNAs plasmids were transfected into BGC-823 cell line, which exhibited an increased expression level of STCAT16. Among the four shRNAs, STCAT16-shRNA-759 exhibited the most effective knockdown compared with the negative control (STCAT16-sh-NC) group (Fig. 2A). Therefore, STCAT16-shRNA-759 was used in further experiments.
Figure 2.
Construction and selection of vectors used to silence or overexpress STCAT16. (A) STCAT16-shRNA-759 exhibited the most effective knockdown of STCAT16 compared with STCAT16-shRNA-NC in BGC-823 cells. (B) Expression level of STCAT16 was significantly upregulated in the STCAT16-pcDNA3.1 group compared with the negative control (mock-NC) group in AGS cells. STCAT16, stomach cancer-associated transcript 16; sh, short hairpin RNA; NC, negative control.
Furthermore, STCAT16-pcDNA3.1 plasmid was transfected into AGS cells, which exhibited a decreased expression level of STCAT16 compared with other GC cell lines. The expression level of STCAT16 was significantly increased in the STCAT16-pcDNA3.1 group compared with the mock-NC group, which was transfected with an empty vector.
STCAT16 overexpression inhibits gastric cancer cell proliferation and colony formation
To investigate the biological effects of STCAT16 on GC cells, the expression level of STCAT16 was silenced by transfecting sh-STCAT16 into BGC-823 cells and overexpressed by transfecting STCAT16-pcDNA3.1 into AGS cells.
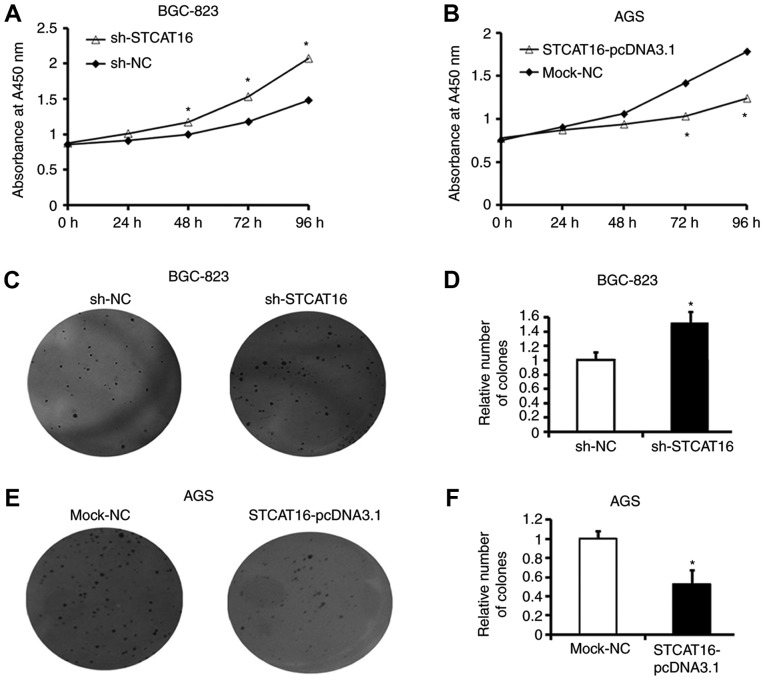
Following transfection with sh-STCAT16, the proliferation of BGC-823 cells was significantly increased at 48, 72 and 96 h compared with the sh-NC group (Fig. 3A). In contrast, the proliferation of AGS cells was significantly decreased at 48, 72 and 96 h following transfection with STCAT16 overexpressing vector Fig. 3B).
Figure 3.
Expression level of STCAT16 affects GC cell proliferation and colony formation in vitro. (A) Proliferative ability of BGC-832 cells following STCAT16 knockdown. (B) Proliferative ability of AGS cells following STCAT16 overexpression. (C and D) Colony formation ability of BGC-832 cells following STCAT16 knockdown. (E and F) Colony formation ability of AGS cells following STCAT16 overexpression. Cells in the sh-STCAT16 group were transfected with STCAT16-shRNA-759. Magnification, ×100. *P<0.05 vs. corresponding control. STCAT16, stomach cancer-associated transcript 16; sh, short hairpin RNA; NC, negative control.
A colony formation assay produced similar results. Following sh-STCAT16 transfection, BGC-823 cells exhibited a significantly increased number of colonies (Fig. 3C and D). Conversely, following STCAT16-pcDNA3.1 transfection, AGS cells exhibited a significantly decreased number of colonies compared with the mock-NC group (Fig. 3E and F).
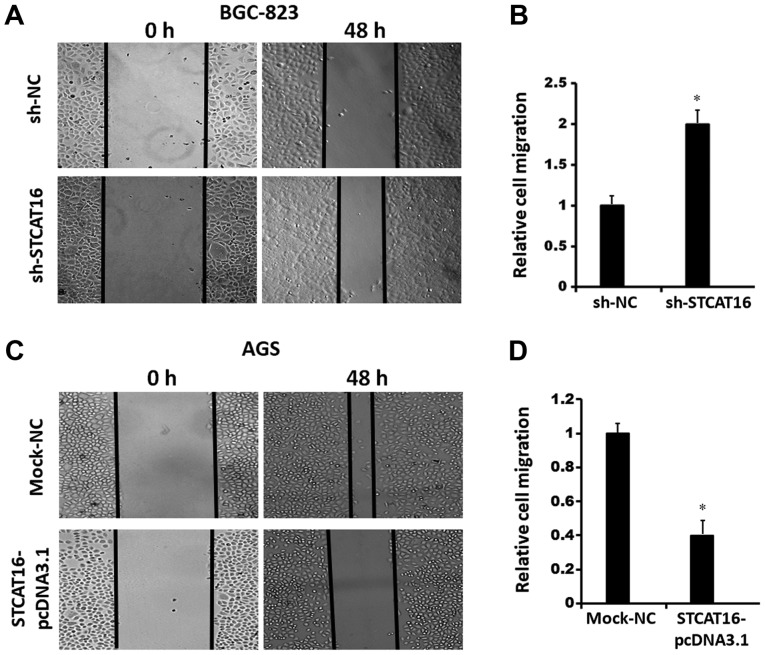
STCAT16 overexpression suppresses gastric cancer cell migration and invasion in vitro
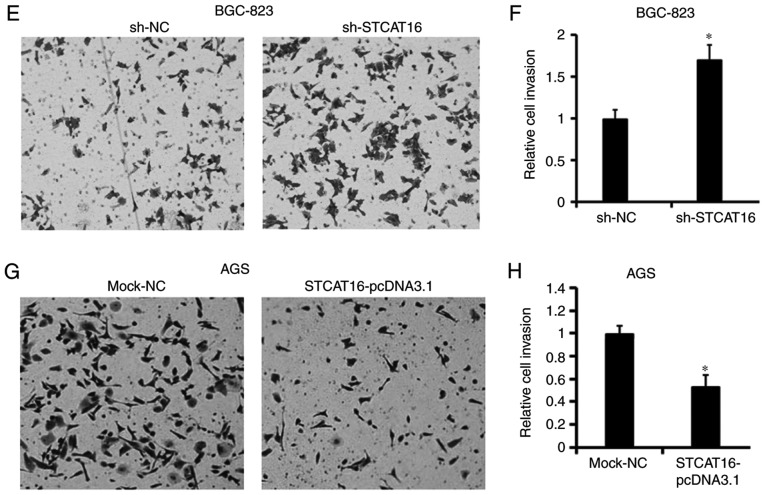
To investigate cell migration and invasion, wound healing assays and a Matrigel assays were performed, respectively (Fig. 4). The results obtained from the wound-healing assay and the invasion assay suggested that decreasing the expression level of STCAT16 promoted BGC-823 cells migration (Fig. 4A and B) and invasion (Fig. 4E and F). Conversely, the migration (Fig. 4C and D) and invasion (Fig. 4G and H) of AGS cells were decreased following STCAT16 overexpression.
Figure 4.
Expression level of STCAT16 affects cell migration and invasion in vitro. (A) Wound healing assay in BGC-832 cells following STCAT16 knockdown and (B) corresponding quantification. (C) Wound healing assay in AGS cells following STCAT16 overexpression and (D) corresponding quantification. Magnification, ×100. (E) Invasion assay in BGC-832 cells following STCAT16 knockdown and (F) corresponding quantification. (G) Invasion assay in AGS cells following STCAT16 overexpression and (H) corresponding quantification. Representative images of invaded cells are presented. Magnification, ×400. Cells in the sh-STCAT16 group were transfected with STCAT16-shRNA-759. *P<0.05 vs. corresponding control. STCAT16, stomach cancer-associated transcript 16; sh, short hairpin RNA; NC, negative control.
Overexpression of STCAT16 inhibits tumour progression in BALB/c nude mice
To further investigate the tumour suppressive role of STCAT16, AGS cells transfected with STCAT16-pcDNA3.1 vector or empty vector were injected into BALB/c nude mice. Tumours were detected in all animals injected with AGS cells.
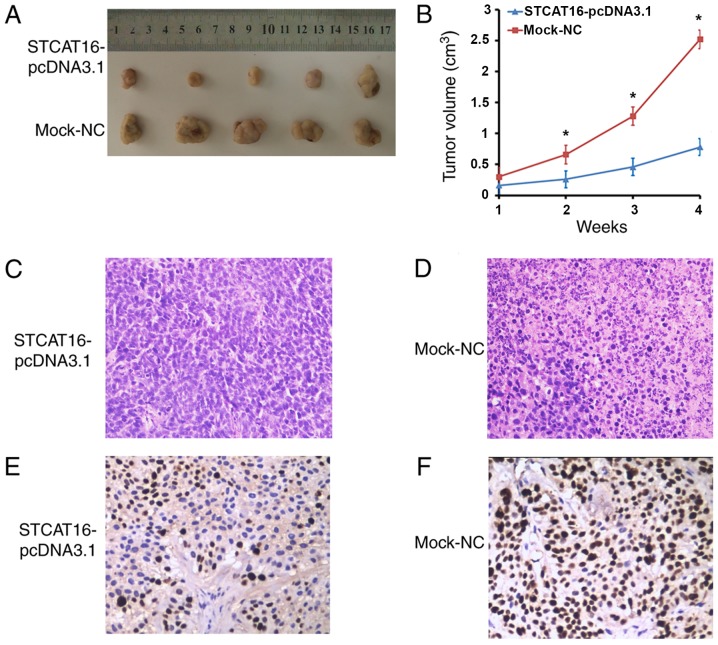
In the mouse xenograft models, the size and the growth of tumours decreased in the STCAT16-pcDNA3.1 group compared with the mock-NC group at 2, 3 and 4 weeks (Fig. 5A and B). Following 4 weeks, the largest tumours exhibited a volume of 3,005.6 and 1,188.1 mm3 in the STCAT16-pcDNA3.1 group and in the mock-NC group, respectively. The xenografts were fixed with formalin, embedded in paraffin, stained with haematoxylin and eosin (H&E), and incubated with MKI67 antibody. H&E staining indicated that the tumor tissue in the STCAT16-pcDNA3.1 transfected group had a smaller area of necrosis, higher differentiation, less pleomorphism and fission, comparing with that in mock-NC group (Fig. 5C and D). Furthermore, tumours derived from cells transfected with STCAT16-pcDNA3.1 exhibited reduced growth compared with the mock group (4.21±0.90 and 9.80±1.23, respectively, P<0.05; Fig. 5E and F). The present result suggested that overexpression of STCAT16 significantly inhibited tumour growth in BALB/c nude mice.
Figure 5.
Effects of STCAT16 overexpression on GC cell growth and proliferation in vivo. (A) Tumour formation in nude mice at 6 weeks following injection with cells transfected with STCAT16-pcDNA3.1 or mock-NC vectors. (B) Growth curve of tumours derived from cells transfected with STCAT16-pcDNA3.1 or mock-NC vectors. Haematoxylin and eosin staining results in tumours derived from cells transfected with (C) STCAT16-pcDNA3.1 or (D) mock-NC vectors. MKI67 staining results in tumours derived from cells transfected with (E) STCAT16-pcDNA3.1 or (F) mock-NC vectors. Magnification, ×400. *P<0.05 vs. corresponding control. GC, gastric cancer; NC, negative control; STCAT16, stomach cancer-associated transcript 16.
Discussion
Accumulating evidence (28,29) has demonstrated that the dysregulation of lncRNAs is involved in the occurrence and development of GC and may promote tumour invasion and metastasis. The overexpression of the lncRNA HOTAIR in GC cells was identified to be associated with TNM stages and lymph node metastasis and to promote colony formation and hepatic metastasis (30). In contrast, knockdown of HOTAIR was identified to reverse epithelial-mesenchymal transition by regulating the expression levels of matrix metalloproteinase (MMP)1 and 3 (31). The upregulation of the lncRNA GHET1 was identified to be associated with gastric tumour growth and invasion and GHET1 overexpression increased the proliferation of GC cells by promoting the interaction between the transcript of MYC proto-oncogene, bHLH transcription factor (MYC) and insulin like growth factor 2 mRNA binding protein 1 (17). HULC expression was identified in hepatocellular cancer; however, it was also demonstrated to be upregulated in GC (32). Additionally, HULC promoted lymph node metastasis and distant metastasis and was identified to be associated with advanced GC (18). A previous study demonstrated that the lncRNA CCAT1 promoted GC progression by upregulating the expression level of MYC (19). The downregulation of the lncRNA growth arrest specific 5 inhibited the proliferation and induced the apoptosis of GC cells by regulating the activities of cyclin dependent kinase inhibitor 1A and E2F transcription factor 1 (33). The lncRNA GATA6 antisense RNA 1 (head to head) was identified to be downregulated in GC, and its downregulation may lead to tumour-associated phenotypes and metastasis by regulating the expression level of MMP9 (34). The downregulation of the lncRNA MEG3 was demonstrated in GC tissues in vitro and in vivo (20,35).
In the present study, the expression level of the novel lncRNA STCAT16 was significantly downregulated in GC and was identified to be associated with poor clinical features. STCAT16 was additionally downregulated in GC cell lines. The downregulation of STCAT16 promoted cellular proliferation, migration and invasion, and colony formation in vitro. Notably, these effects were reversed following overexpression of STCAT16. In vivo, the tumour sizes in the mice injected with cells overexpressing STCAT16 were decreased compared with tumours derived from cells transfected with an empty vector. Furthermore, overexpressing STCAT16 significantly inhibited tumour cell proliferation in BALB/c nude mice. Collectively, the present results suggested that STCAT16 may serve as a tumour suppressor in GC.
Following the identification of dysregulated lncRNAs in GC, it is required to investigate the mechanisms underlying the function of these noncoding RNAs in tumour development and progression. In previous studies, lncRNAs were identified to serve multiple roles by altering gene expression at the transcriptional, by interacting with mRNA molecules, and post-transcriptional levels, by binding to promoter regions (36–38). Additionally, lncRNAs may be involved in post-translational regulation by directly binding to certain proteins involved in tumour progression (36–38). STCAT16 may act as an oncosuppressor via the discussed mechanisms. Further studies are required to investigate the molecular mechanism underlying STCAT16 function in GC.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Youth Foundation of Affiliated Hospital of Nantong University (grant no. TDFY0332), the Hospital Talent Training Foundation (grant no. 2015-68), the Youth Foundation of Nantong Municipal Commission of Health and Family Planning (grant no. WQ2016079) and the China Postdoctoral Science Foundation (grant no. 2018M630592).
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Authors' contributions
JFZ, WJ, ZBM and ZWW conceived and designed the study. JFZ, WJ, QFZ and XLK performed the experiments. JFZ and WJ wrote the manuscript. ZBM and ZWW reviewed and edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Ethical approval was obtained from the independent ethics committees at Affiliated Hospital of Nantong University. All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 5.Nie Y, Wu K, Yu J, Liang Q, Cai X, Shang Y, Zhou J, Pan K, Sun L, Fang J, et al. A global burden of gastric cancer: The major impact of China. Expert Rev Gastroenterol Hepatol. 2017;11:651–661. doi: 10.1080/17474124.2017.1312342. [DOI] [PubMed] [Google Scholar]
- 6.Li Q, Chen C, Kapadia A, Zhou Q, Harper MK, Schaack J, LaBarbera DV. 3D models of epithelial-mesenchymal transition in breast cancer metastasis: High-throughput screening assay development, validation, and pilot screen. J Biomol Screen. 2011;16:141–154. doi: 10.1177/1087057110392995. [DOI] [PubMed] [Google Scholar]
- 7.Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Klein CA. Cancer. The metastasis cascade. Science. 2008;321:1785–1787. doi: 10.1126/science.1164853. [DOI] [PubMed] [Google Scholar]
- 9.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 10.Wang C, Wang L, Ding Y, Lu X, Zhang G, Yang J, Zheng H, Wang H, Jiang Y, Xu L. LncRNA structural characteristics in epigenetic regulation. Int J Mol Sci. 2017;18(pii):E2659. doi: 10.3390/ijms18122659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425:3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang JZ, Chen M, Chen, Gao XC, Zhu S, Huang H, Hu M, Zhu H, Yan GR. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell. 2017;68:171–184.e6. doi: 10.1016/j.molcel.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 14.Huarte M, Rinn JL. Large non-coding RNAs: Missing links in cancer. Hum Mol Genet. 2010;19:R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan W, Liu L, Wei J, Ge Y, Zhang J, Chen H, Zhou L, Yuan Q, Zhou C, Yang M. A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol Carcinog. 2016;55:90–96. doi: 10.1002/mc.22261. [DOI] [PubMed] [Google Scholar]
- 16.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y, Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802–813. doi: 10.1111/febs.12625. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: A clinical and in vitro investigation. Oncol Rep. 2014;31:358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 19.Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–445. doi: 10.1007/s00432-012-1324-x. [DOI] [PubMed] [Google Scholar]
- 20.Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F, Yu J, Ma R. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79. doi: 10.1186/s13046-015-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu G, Meng L, Yuan D, Li K, Zhang Y, Dang C, Zhu K. MEG3/miR-21 axis affects cell mobility by suppressing epithelial-mesenchymal transition in gastric cancer. Oncol Rep. 2018;40:39–48. doi: 10.3892/or.2018.6424. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.In H, Solsky I, Palis B, Langdon-Embry M, Ajani J, Sano T. Validation of the 8th edition of the AJCC TNM staging system for gastric cancer using the national cancer database. Ann Surg Oncol. 2017;24:3683–3691. doi: 10.1245/s10434-017-6078-x. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Zhang JF, Qu LS, Qian XF, Xia BL, Mao ZB, Chen WC. Nuclear transcription factor CDX2 inhibits gastric cancer-cell growth and reverses epithelial-to-mesenchymal transition in vitro and in vivo. Mol Med Rep. 2015;12:5231–5238. doi: 10.3892/mmr.2015.4114. [DOI] [PubMed] [Google Scholar]
- 26.Guo P, Wang J, Gao W, Liu X, Wu S, Wan B, Xu L, Li Y. Salvianolic acid B reverses multidrug resistance in nude mice bearing human colon cancer stem cells. Mol Med Rep. 2018;18:1323–1334. doi: 10.3892/mmr.2018.9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Ding W, Kuai X, Ji Y, Zhu Z, Mao Z, Wang Z. Dermcidin as a novel binding protein of lncRNA STCAT3 and its effect on prognosis in gastric cancer. Oncol Rep. 2018;40:2854–2863. doi: 10.3892/or.2018.6673. [DOI] [PubMed] [Google Scholar]
- 28.Abbastabar M, Sarfi M, Golestani A, Khalili E. lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI J. 2018;17:900–913. doi: 10.17179/excli2018-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong P, Qiao F, Wu H, Cui H, Li Y, Zheng Y, Zhou M, Fan H. LncRNA UCA1 promotes tumor metastasis by inducing miR-203/ZEB2 axis in gastric cancer. Cell Death Dis. 2018;9:1158. doi: 10.1038/s41419-018-1170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao Y, Chen H, Jiang X, Chen S, Li P, Ye M, Li Q, Sun W, Guo J. Low expression of lncRNA-HMlincRNA717 in human gastric cancer and its clinical significances. Tumour Biol. 2014;35:9591–9595. doi: 10.1007/s13277-014-2243-z. [DOI] [PubMed] [Google Scholar]
- 31.Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L, Yu PF, Cheng XD. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci. 2013;9:587–597. doi: 10.7150/ijbs.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klec C, Gutschner T, Panzitt K, Pichler M. Involvement of long non-coding RNA HULC (highly up-regulated in liver cancer) in pathogenesis and implications for therapeutic intervention. Expert Opin Ther Targets. 2019;23:177–186. doi: 10.1080/14728222.2019.1570499. [DOI] [PubMed] [Google Scholar]
- 33.Liu L, Yan B, Yang Z, Zhang X, Gu Q, Yue X. ncRuPAR inhibits gastric cancer progression by down-regulating protease-activated receptor-1. Tumour Biol. 2014;35:7821–7829. doi: 10.1007/s13277-014-2042-6. [DOI] [PubMed] [Google Scholar]
- 34.Park SM, Park SJ, Kim HJ, Kwon OH, Kang TW, Sohn HA, Kim SK, Moo Noh S, Song KS, Jang SJ, et al. A known expressed sequence tag, BM742401, is a potent lincRNA inhibiting cancer metastasis. Exp Mol Med. 2013;45:e31. doi: 10.1038/emm.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dan J, Wang J, Wang Y, Zhu M, Yang X, Peng Z, Jiang H, Chen L. LncRNA-MEG3 inhibits proliferation and metastasis by regulating miRNA-21 in gastric cancer. Biomed Pharmacother. 2018;99:931–938. doi: 10.1016/j.biopha.2018.01.164. [DOI] [PubMed] [Google Scholar]
- 36.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J, Hann SS. Functions and Roles of long-non-coding RNAs in human nasopharyngeal carcinoma. Cell Physiol Biochem. 2018;45:1191–1204. doi: 10.1159/000487451. [DOI] [PubMed] [Google Scholar]
- 38.Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.