Abstract
Background & Aims
The lysosome is an acidic organelle that is important for maintaining cellular and metabolic homeostasis in hepatocytes. Lysosomal dysfunction and chronic inflammation coexist, and both contribute to obesity-associated hepatic insulin resistance. However, in the context of obesity, the interplay between inflammatory signals and hepatic lysosomal function remains largely unknown. Inducible nitric oxide synthase (iNOS) is a hallmark for inflammation, and is activated in obesity. The aim of this study is to understand the molecular link between iNOS-mediated lysosomal nitric oxide (NO) production, hepatic lysosomal function, and autophagy in the context of obesity-associated insulin resistance.
Methods
The role of iNOS in hepatic autophagy, as related to insulin and glucose homeostasis were studied in mice with diet-induced obesity (DIO). The effects and mechanisms of iNOS-mediated lysosomal NO production on lysosomal function and hepatic autophagy were studied in primary hepatocytes as well as in a mouse model of DIO.
Results
We demonstrate that obesity promotes iNOS localization to the lysosome and decreases levels of lysosomal arginine, resulting in an accumulation of NO in hepatic lysosomes. This lysosomal NO production is attenuated by treatment with a NO scavenger, while co-overexpression of mTOR and a lysosomal arginine transporter (SLC38A9) enhances lysosomal NO production and suppresses autophagy. In addition, we show that deletion of iNOS ameliorates lysosomal nitrosative stress in the livers of DIO mice, promotes lysosomal biogenesis by activating transcription factor EB (TFEB), and enhances lysosomal function and autophagy. Lastly, deletion of iNOS in mice with DIO improves hepatic insulin sensitivity, which is diminished by suppression of TFEB or autophagy related 7 (Atg7).
Conclusions
Our studies suggest that lysosomal iNOS-mediated NO signaling disrupts hepatic lysosomal function, contributing to obesity-associated defective hepatic autophagy and insulin resistance.
Keywords: Autophagy, Insulin Resistance, Inducible Nitric Oxide Synthase, Lysosome, Nitric Oxide
Abbreviations used in this paper: Atg7, autophagy related 7; CQ, chloroquine; CTSB, cathepsin B; DIO, diet-induced obesity; EBSS, Earle's balanced salt solution; HFD, high-fat diet; iNOS, inducible nitric oxide synthase; iNOSL, inducible nitric oxide synthase pool localized at lysosomes; IR, insulin resistance; KO, knockout; LC3, microtubule-associated protein 1A/1B-light chain 3; LPS, lipopolysaccharide; mTOR, mammalian target of rapamycin; NO, nitric oxide; OA, oleic acid; PTIO, 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide; RD, regular diet; SNAP, S-Nitroso-N-acetylpenicillamine; TFEB, transcription factor EB; TNF, tumor necrosis factor; WT, wild-type
Graphical abstract
See editorial on page 153.
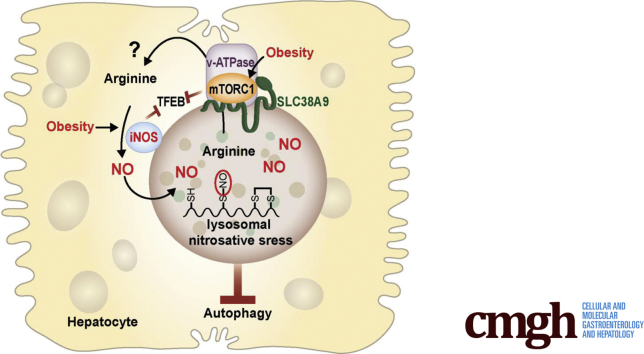
Summary.
Inducible nitric oxide synthase (iNOS) plays a key role in the pathogenesis of liver diseases. We demonstrated that obesity promotes hepatic lysosomal iNOS localization and subsequent overproduction of lysosomal nitric oxide. This lysosomal nitric oxide bioactivates compromises hepatic autophagy, contributing to hepatic insulin resistance in obese mice.
The lysosome is an acidic organelle that orchestrates the disposal of intracellular and extracellular components. Each hepatocyte contains approximately 250 lysosomes, which act as recycling centers and regulate glycogen metabolism, cholesterol trafficking, and viral defense.1 Recent studies show that the lysosome also serves as a platform for the mammalian target of rapamycin 1 to sense nutritional flux,2, 3 demonstrating a crucial lysosomal function for maintaining metabolic homeostasis. Furthermore, the lysosome processes and degrades autophagic cargoes, maintaining organelle and protein quality control for the liver.4, 5 In the liver, dysfunction of lysosomes and autophagy lead to metabolic abnormalities. For example, defective autophagy contributes to obesity-associated hepatic insulin resistance.6, 7 Moreover, deletion of lysosome-associated membrane protein type 2A alters hepatic carbohydrate and lipid metabolism.8 In contrast, overexpression of the transcription factor EB (TFEB), a master regulator of lysosomal biogenesis that is tightly regulated by mammalian target of rapamycin (mTOR), ameliorates steatosis and improves glucose homeostasis in the liver of obese mice.9
It is well known that in the context of obesity, chronic inflammation and lysosome dysfunction coexist in the liver.10, 11 Emerging evidence indicates that lysosomes play important roles in inflammatory responses.12, 13 However, the physiological relevance of the interaction between inflammatory signaling and hepatic lysosome dysfunction in obesity-associated liver pathologies remains largely unknown. Recently, Zhao et al14 found that lysosome-mediated toll-like receptor 4 degradation prevented nonalcoholic fatty liver disease and nonalcoholic steatohepatitis in both mice and monkeys. In addition, overexpression of TFEB in the liver ameliorated ethanol-induced liver injury and inflammation by increasing lysosomal biogenesis.15 Moreover, we recently demonstrated that increased nitrosative stress in hepatic lysosomes of obese mice and patients with high steatosis.11 These findings highlight a key role between lysosomes and inflammatory signaling in the pathophysiology of obesity-mediated hepatic dysfunction.
Activation of inducible nitric oxide synthase (iNOS) is a hallmark of inflammation and plays a key role in the pathogenesis of many liver diseases.16 Importantly, hepatic iNOS is elevated in the livers of patients with chronic liver diseases such as hepatitis C,17 alcoholic cirrhosis, and α1-antitrypsin disorder.18 Moreover, activation of iNOS contributes to hepatic insulin resistance (IR) and diabetes in mice with either diet-induced obesity (DIO) or genetically induced obesity.19, 20 At the cellular level, iNOS produces pathological NO, which triggers downstream effects, such as aberrant S-nitrosylation. These downstream effects can disrupt the function of organelles such as the mitochondria21 and the endoplasmic reticulum.20
Studies, including those from our laboratory, show that aberrant NO bioactivities compromise lysosomal function in neurodegenerative diseases,22 cardiovascular disease,23 nonalcoholic fatty liver disease,11 and kidney disease.24 However, the source of the NO that promotes lysosomal nitrosative stress in the liver in the context of obesity is not known. NO is abundant in the lysosomes of eosinophils and neutrophils, and iNOS can translocate to the phagosomal membrane of macrophages.25 However, it is currently unknown if NO in hepatocytes can localize in lysosomes or if it is generated by a local iNOS. Here, we provide the first evidence that obesity induces hepatic lysosomal iNOS localization and subsequent overproduction of lysosomal NO, which in turn impairs hepatic autophagy and contributes to hepatic insulin resistance in obese mice.
Results
Obesity Promotes Lysosomal Localization of iNOS and Lysosomal NO Production, Impairing Hepatic Autophagy
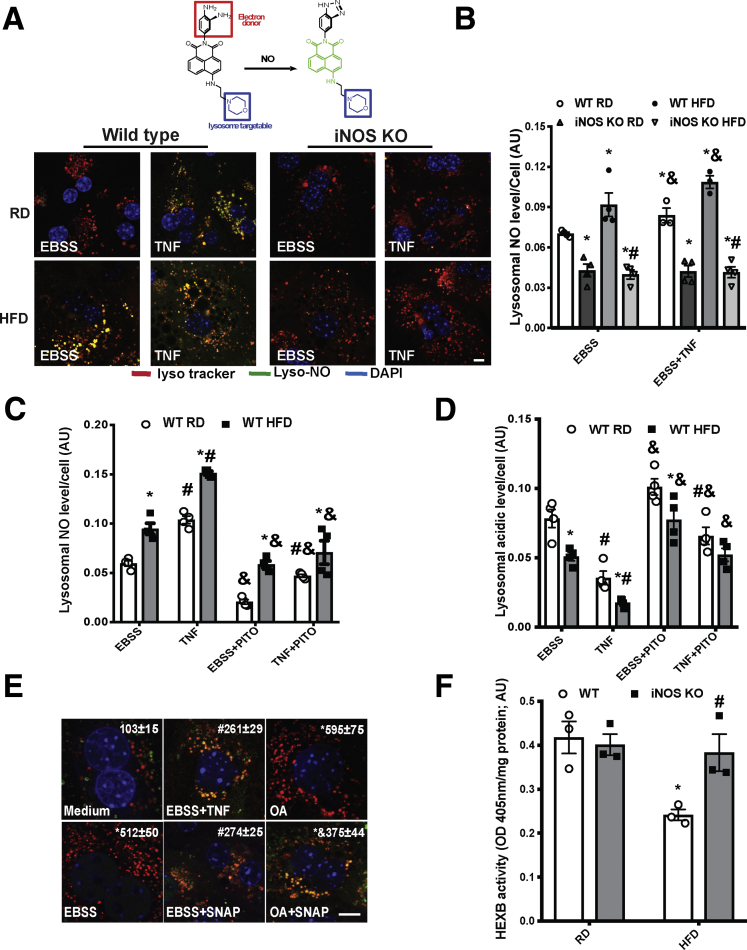
We recently showed that obesity promotes lysosomal protein S-nitrosylation in the livers of obese mice and in patients with high steatosis.11 To address whether obesity elevates NO levels specifically in lysosomes, we measured the level of NO in the lysosomes of primary hepatocytes isolated from wild-type (WT) and iNOS knockout (KO) mice on a regular diet (RD) or high-fat diet (HFD) using a lysosome-specific NO probe.26 Obesity significantly enhanced NO levels in the lysosome (Figure 1A and B). This effect was abolished by deletion of iNOS, suggesting that activation of iNOS contributes to accumulation of NO in lysosomes. To test whether altering lysosomal NO production directly influences lysosomal function and hepatic autophagy, hepatocytes isolated from lean or obese mice were treated ex vivo with the proinflammatory cytokine tumor necrosis factor (TNF),27 in the presence or absence of the chemical NO scavenger, 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide (PTIO). In primary hepatocytes from obese mice, lysosomal NO levels were significantly increased compared with control mice while TNF treatment increased NO levels from both groups as expected (Figure 1C). Cells exposed to PTIO alone or with TNF decreased lysosomal NO production and improved lysosomal function (Figure 1C and D). To determine whether NO is directly involved in the iNOS-mediated blockage of autophagic flux, primary hepatocytes from RFP-GFP-LC3 transgenic mice28 were pretreated with TNF to induce NO production or a chemical NO donor (S-Nitroso-N-acetylpenicillamine [SNAP]), and then exposed to Earle's balanced salt solution (EBSS) or oleic acid (OA) to induce autophagy.6 TNF- and SNAP-inhibited autophagic vesicle formation and autophagic flux (Figure 1E). To further establish the effect of iNOS-mediated signaling on lysosomal function in vivo, we assessed hexosaminidase subunit beta activities in the lysosomal fractions isolated from livers of lean and DIO mice. As shown in Figure 1F, obesity decreased hepatic hexosaminidase subunit beta enzyme activities, which is restored by iNOS deletion.
Figure 1.
iNOS elevates lysosomal NO production in hepatocytes in obesity. (A) Top: Structure of Lyso-NINO and the reaction of Lyso-NINO with NO. Bottom: Representative confocal images (63×) of live primary hepatocytes from WT or iNOS-KO mice stained with 5.0-μM Lyso-NINO (15 minutes). Cells were treated with EBSS (4 hours) and pretreated with TNF (10 ng/mL, 16 hours). Scale bar: 10 μm. (B) Quantification of lysosomal NO production in primary hepatocytes. Data are presented as mean ± SEM. * indicates statistical significance compared with WT RD in EBSS, # indicates statistical significance between HFD groups, and & indicates statistical significance between EBSS- and TNF-treatment in the same cell type, as determined by analysis of variance (ANOVA) followed by a post hoc test (P < .05; n = 3, biological replicates; 12 weeks on HFD). (C) Lysosomal NO levels and (D) lysosomal acidity in live primary hepatocytes from WT mice. Autophagy was induced by EBSS (4 hours) and pretreatment with TNF (10 ng/mL, 16 hours) in the presence or absence of PTIO (100 nM, 30 minutes). * indicates statistical significance between RD and HFD groups in the same treatment, # indicates statistical significance between EBSS and TNF in the same cell type, and & indicates statistical significance between measurements taken before and after PTIO treatment in the same cell type, as determined by ANOVA followed by a post hoc test (P < .05; n = 3, biological replicates; 12 weeks on HFD). (E) Representative confocal images (63×) of primary hepatocytes isolated from RFP-GFP-LC3 transgenic mice. Autophagy was induced by EBSS (4 hours) or OA (200 μM, 16 hours), ± treatment with TNF (10 ng/mL, 16 hours) or SNAP (50 μM, 4 hours). The number of the red LC3 puncta/field are noted at the top of the image. All data are presented as mean ± SEM. * indicates statistical significance compared with medium, # indicates statistical significance between EBSS groups, and & indicates statistical significance between OA groups determined by ANOVA followed by post hoc test (P < .05; 10 fields/group; n = 3, biological replicates). Scale bar: 10 μm. (F) Hexosaminidase subunit beta activity in lysosomal fractions from WT and iNOS KO mice fed an RD and HFD (16 weeks on HFD). An equivalent amount of liver tissues from lean and obese mice were used. Data are presented as mean ± SEM. * indicates statistically significant difference relative to lean condition in the same type of mice, # indicates statistical significance between HFD groups, by determined by ANOVA followed by post hoc test (P < .05; each sample is the combined fraction from livers of 4 mice; n = 12, biological replicates).
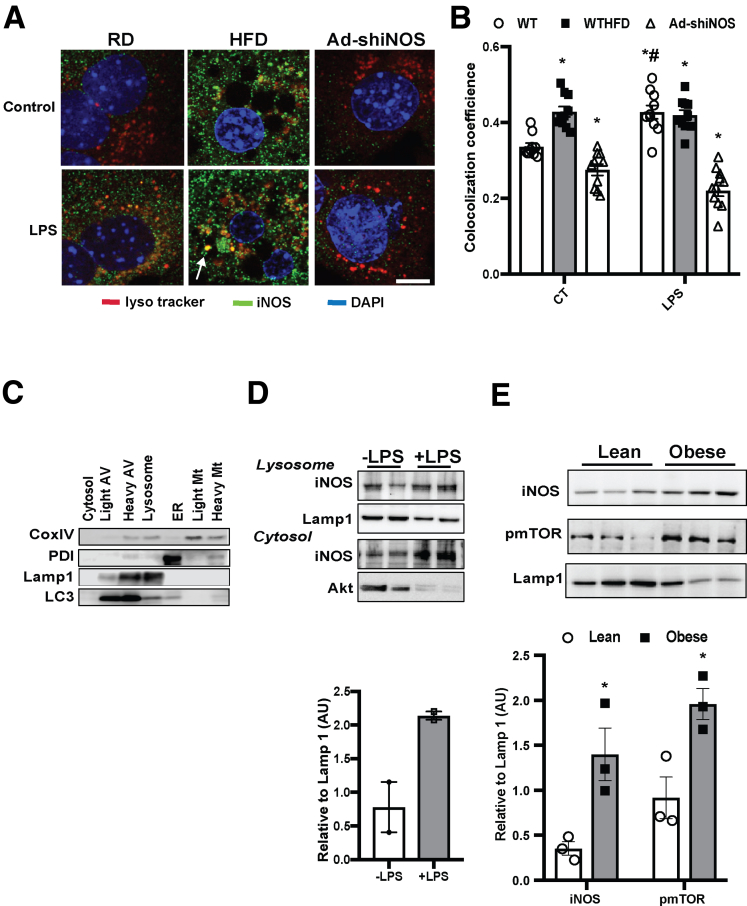
Membrane-associated NOS promotes subcellular NO production.25, 29 To determine whether obesity-associated lysosomal nitrosative stress could be regulated by a local iNOS, we first examined the potential lysosomal localization of iNOS in hepatocytes. In primary hepatocytes from mice fed an RD, there was a detectable intracellular iNOS pool localized at lysosomes (iNOSL) (Figure 2A and B). Treatment of mice with lipopolysaccharide (LPS) (a commonly used agent to induce hepatic NO production in rodents)30 resulted in increased expression of iNOSL which was decreased after iNOS suppression (Figure 2A and B). Moreover, mice with DIO had significantly enhanced levels of both basal and LPS-induced iNOSL in primary hepatocytes (Figure 2A and B). Similar results were observed in lysosomal fractions from livers of lean mice treated with LPS (a well-known nitric oxide inducer; Figure 2C and D), and from livers of ob/ob mice (Figure 2E). Notably, this elevated iNOSL in the livers of obese mice was associated with activation of lysosomal mTOR (Figure 2E). To further define the localization of endogenous iNOSL in the lysosome, we purified lysosomes from livers of lean mice treated with LPS. These lysosomes were then subjected to trypsin digestion to remove the outer lysosomal membrane proteins. Trypsin treatment greatly diminished iNOSL expression (Figure 2F), indicating that iNOSL is mainly localized at the outer membrane of the lysosome.
Figure 2.
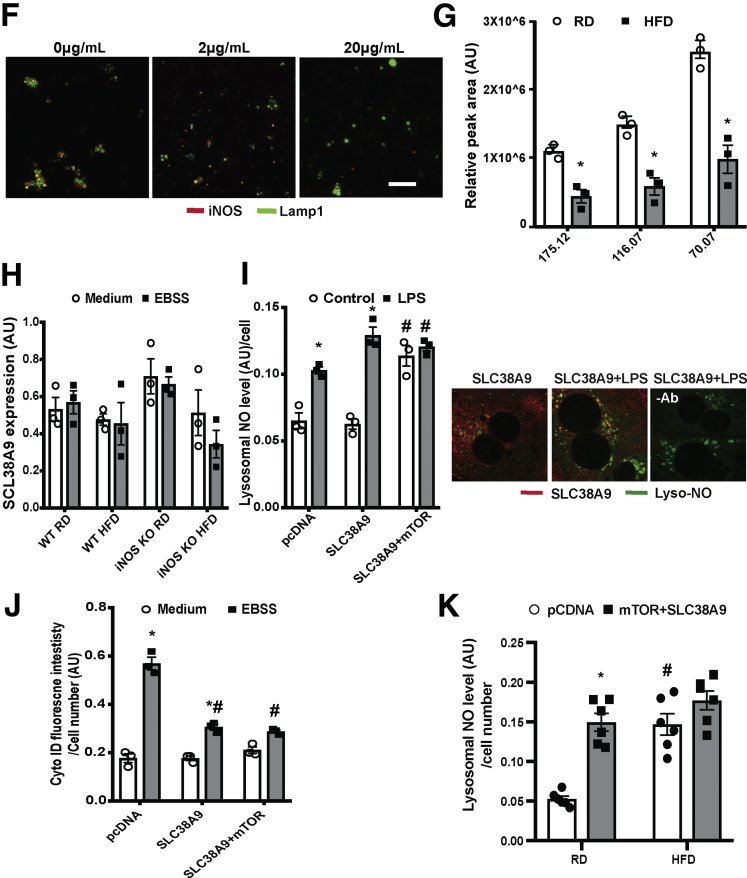
Obesity and inflammation promote lysosomal-localization of iNOS. (A) Representative confocal images (63×) and (B) quantification of lysosomal iNOS in primary hepatocytes from WT mice fed an RD and HFD, and WT fed an HFD and transduced with Ad-shiNOS. Lysotracker was added to cells for 15 minutes; LPS: 1 μg/mL, 6 hours. Scale bar: 10 μm. Data are shown as mean ± SEM as determined by Person’s correlation coefficient. * indicates statistically significant difference relative to -LPS in WT RD, and # indicates statistical significance between +LPS and –LPS groups in same type of mice, as determined by analysis of variance (ANOVA) followed by post hoc test (P < .05; n = 3, biological replicates; 12 weeks on HFD). (C) Western blots of organelle fractions, CoxIV: mitochondrial (Mt) marker; PDI: endoplasmic reticulum (ER) marker; Akt: cytosol marker; Lamp1: lysosomal marker; LC3: autophagy vesicle marker. (D) Lysosomal iNOS expression in livers of WT mice or WT mice treated with LPS (1 mg/kg/mouse; each lane is the combined fraction from livers of 4 mice) and (E) in the livers of lean and obese mice (ob/ob 10 weeks). The densitometry of Western blot analysis of panel E is shown on the bottom of the panel. * indicates statistically significant difference relative to lean, as determined by Student’s t test (P < .05; each lane is the combined fraction from livers of 3 mice; n = 9, biological replicates). (F) Representative confocal images (63×) of lysosomes isolated from WT mice treated with LPS (1 mg/kg/mouse). The lysosomes were treated with trypsin at 0, 2, and 20 μg/mL for 10 minutes at room temperature. Scale bar: 5 μm. (G) Lysosomal arginine profile in the livers of mice fed with an RD and HFD. Data are normalized to liver weight and presented as mean ± SEM. * statistically significant difference relative to RD group, as determined by Student’s t test (P < .05; each sample is the combined fraction from livers of 4 mice; n = 12, biological replicates; 16 weeks on HFD). (H) Levels of messenger RNAs encoding SLC38A9 in the primary hepatocytes from WT mice or iNOS KO mice fed with an RD and HFD, as assessed by quantitative real-time reverse-transcriptase polymerase chain reaction. Autophagy was induced by EBSS (4 hours). Data are presented as mean ± SEM. Statistically significant difference relative to medium is determined by ANOVA followed by a post hoc test (n = 3, biological replicates). (I) Lysosomal NO levels and (J) autophagic vesicle numbers in live primary hepatocytes from WT mice. Cells were transfected with indicated constructs for 48 hours, pretreated with LPS (1μg/mL, 6 hours) and autophagy was induced by EBSS (4 hours). * indicates statistical significance between control and treated cells that were transfected with the same plasmid; # indicates statistical significance between cells of the same treatment group compared with cells transfected with pcDNA, as determined by ANOVA followed by a post hoc test (P < .05; n = 3, biological replicates; 12 weeks on HFD). Representative confocal images (63×) of primary hepatocytes transfected with SLC38A9-flag (48 hours) stained with 5.0 μM Lyso-NINO (15 m, green) and anti-flag (red) are shown on the right panel of panel I. Scale bar: 10 μm. (K) Lysosomal NO levels in live primary hepatocytes from WT mice fed with an RD and HFD. Cells were transfected with the indicated constructs for 48 hours and autophagy was induced by EBSS (4 hours). * indicates statistical significance between pcDNA control and mTOR+SLC38A9 in the same type of cells, and # indicates statistical significance between RD and HFD cells that were transfected with the same plasmid, as determined by ANOVA followed by a post hoc test (P < .05; n = 6, biological replicates; 12 weeks on HFD).
Arginine is required for NOSs to catalyze NO production. Lysosomes are enriched with arginine,31 which can be transported by a lysosomal arginine transporter to provide cellular fuel by activating mTOR.32 To address whether iNOSL could utilize the potential local arginine as an NO substrate, we performed a metabolome analysis to quantify lysosomal arginine in livers of WT mice fed an RD or HFD. Mice with DIO showed markedly reduced levels of arginine in the lysosome compared with lean mice (Figure 2G). These data indicate that local iNOS in the lysosome could potentially utilize lysosomal arginine, to generate local NO. SLC38A9 is a lysosome-specific arginine transporter.32 Although obesity and iNOS deletion did not significantly alter hepatic SLC38A9 expression (Figure 2H), overexpression of SLC38A9 increased lysosomal NO production in primary hepatocytes pretreated with LPS (Figure 2I). Coexpression of mTOR and SLC38A9 augmented this effect by inducing lysosomal NO production in the absence of inflammation (Figure 2I) and suppressed starvation-induced autophagy in primary hepatocytes (Figure 2J). We further determined this potential regulation in the setting of obesity. Although the overexpression of mTOR and SLC38A9 increased lysosomal NO production in primary hepatocytes from obese mice, this did not reach a statistical significance (Figure 2K). We reason that in the liver of obese mice, the hepatic iNOS enzymatic activity has reached a maximal level for utilizing endogenous lysosomal arginine, suggesting a threshold effect. Taken together, these data suggest that obesity and inflammation promote the localization of iNOS to the lysosome, and that this elevates lysosomal NO production, impairing the process of lysosome autophagy.
iNOS Regulates Hepatic Lysosomal Nitrosative Stress and Impairs Lysosomal Function in Obese Mice
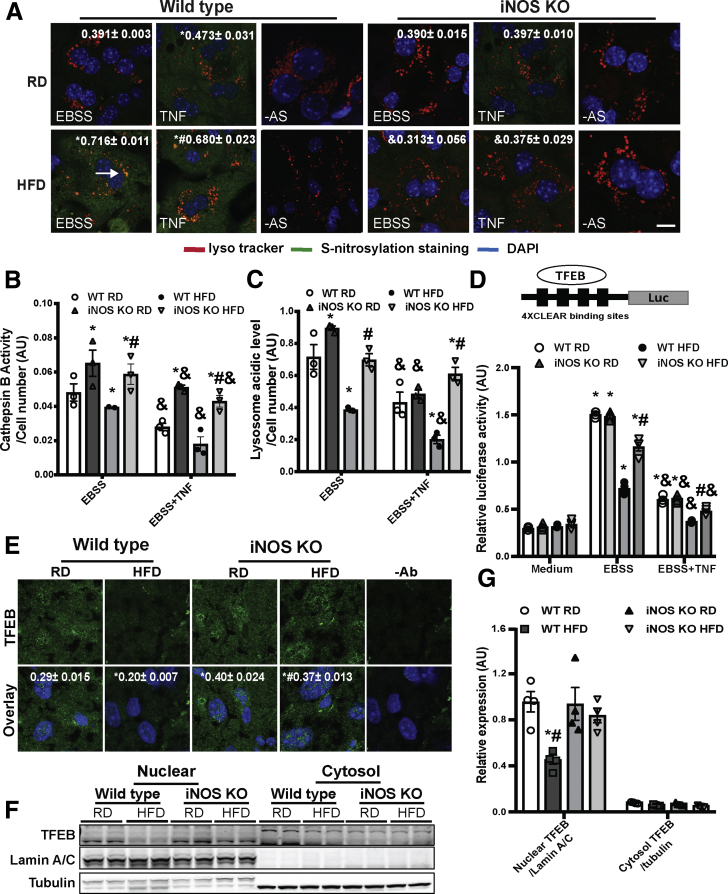
We previously showed that in the liver, obesity elevated S-nitrosylated targets in the lysosome.11 To determine whether the obesity-associated lysosomal nitrosative stress is regulated by iNOS, the S-nitrosylation status of the lysosome was studied in situ in primary hepatocytes from WT and iNOS-deficient mice.33 Deleting iNOS suppressed lysosomal nitrosative stress in the livers of obese mice (Figure 3A). Importantly, in primary hepatocytes from obese mice, deletion of iNOS restored cathepsin B (CTSB) activity and lysosomal pH (Figure 3B and C). We further evaluated the effect of iNOS on lysosome biogenesis by measuring the transcriptional activity of TFEB, an autophagy/lysosomal master regulator,9 in primary hepatocytes from WT or iNOS-KO mice fed an RD or HFD. In obese mice, TFEB activity (Figure 3D) and its nuclear translocation (Figure 3E–G) were suppressed, and this effect was ameliorated by iNOS deletion. Together, these data indicate that activation of iNOS contributes to hepatic lysosomal nitrosative stress in obesity.
Figure 3.
iNOS induces hepatic lysosomal nitrosative stress. (A) Representative confocal images (40×) of S-nitrosylated proteins in primary hepatocytes from WT and iNOS-KO mice (n = 3) using a modified in situ biotin-switch method. Autophagy was induced by EBSS (4 hours) and cells were pretreated with TNF (10 ng/mL, 16 hours). –AS: no ascorbate as a negative control for biotin-switch assay. Scale bar: 10 μm. Quantification of the S-nitrosylated protein and lysotracker are shown on the top of each image. Data are shown as mean ± SEM as determined by Person’s correlation coefficient. * indicates statistically significant difference relative to EBSS in WT RD, and # indicates statistical significance between HFD group determined by analysis of variance (ANOVA) followed by post hoc test (P < .05; 10 fields/group; n = 3, biological replicates; 12 weeks on HFD). (B) CTSB activity and (C) lysosomal acidity in live primary hepatocytes from WT and iNOS-KO mice. * indicates statistical significance compared with WT RD in the same treatment, # indicates statistical significance within HFD groups in the same treatment, and & indicates statistical significance between EBSS- and TNF-treated groups in the same cell type, determined by ANOVA followed by a post hoc test (P < .05; n = 3, biological replicates; 12 weeks on HFD). (D) TFEB activity in primary hepatocytes from WT and iNOS-KO mice. Cells were transfected with the indicated constructs for 48 hours, and treated with EBSS (4 hours) ± TNF (10 ng/mL, 16 hours). The data were normalized to Renilla luciferase. * indicates statistical significance compared with WT RD medium from the same cell type, # indicates statistical significance between HFD groups in the same treatment, and & indicates statistical significance between EBSS and TNF in the same cell type determined by ANOVA followed by a post hoc test (P < .05; n = 3, biological replicates; 12 weeks on HFD). The 4XCLEAR construct is shown on the top of the panel. (E) Representative confocal images (63×) for TFEB staining in livers of WT mice and iNOS KO mice. Green: TFEB; blue: DAPI. –Ab: nonantibody controls. Scale bar: 10 μm. Quantified colocalization of TFEB with DAPI are shown at the top of each image. Data are shown as Pearson’s correlation coefficient as mean ± SEM. * indicates statistically significant difference relative to WT RD, and # indicates statistical significance between HFD group determined by ANOVA followed by a post hoc test (P < .05; 10 fields/group; n = 3, biological replicates; 16 weeks on HFD). (F) Representative Western blot analysis of TFEB nuclear translocation in panel E. Each sample is the combined fraction from livers of 6 mice. (G) Densitometry of Western blot analysis in panel F. * indicates statistical significance between HFD and RD in same type of mouse, # indicates statistical significance between HFD group, determined by ANOVA followed by post hoc test (P < .05; each lane is the combined fraction from livers of 3 mice; n = 12, biological replicates).
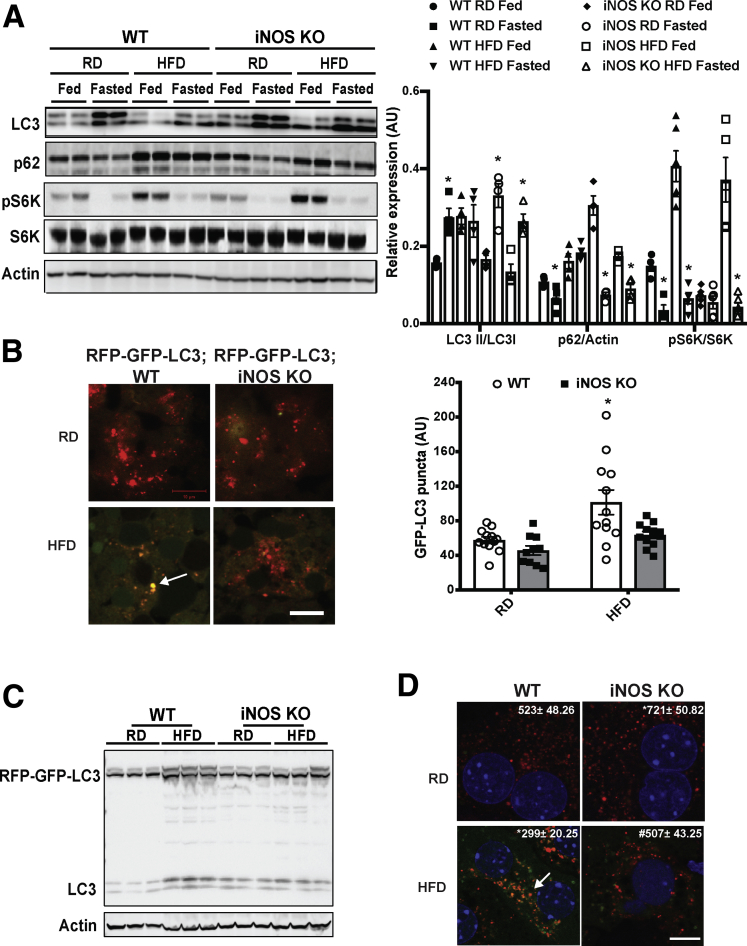
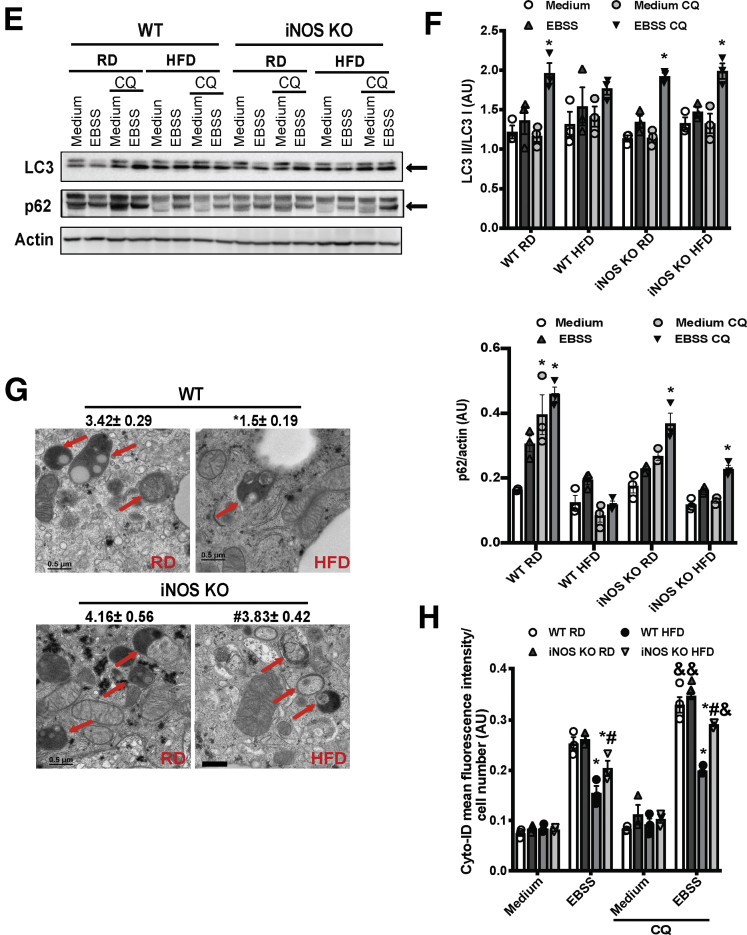
The lysosome is the major site for processing autophagic cargoes.34 To determine whether the iNOS-mediated lysosomal nitrosative stress contributes to autophagic dysfunction, we examined microtubule-associated protein 1A/1B-light chain 3 (LC3) conversion in livers from WT and iNOS-KO mice fed an RD or an HFD. A HFD suppressed a fasting-induced conversion of LC3 and the degradation of p62, but these effects were ameliorated in the absence of iNOS (Figure 4A). This iNOS-mediated autophagic flux was likely independent of mTOR, as phosphorylation of S6K was maintained in the livers of iNOS-KO mice fed an HFD (Figure 4A). We also evaluated if the hepatic autophagic flux was modified by iNOS in vivo in the livers of RFP-GFP-LC3 transgenic and RFP-GFP-LC3 iNOS-KO mice (Figure 4B and C). In mice fed an HFD, the autophagic flux induced by fasting was blocked, whereas this blockage was relieved by iNOS deletion (Figure 4B and C). We next determined whether iNOS alters autophagy in primary hepatocytes transduced with adeno-RFP-GFP-LC3. Deletion of iNOS significantly improved lysosome-mediated GFP degradation in hepatocytes from obese mice (Figure 4D). Moreover, treatment with a lysosome inhibitor (chloroquine [CQ]) blocked autophagic flux in primary hepatocytes from WT mice fed an RD (Figure 4E and F). This effect was blunted when WT mice were fed an HFD, but iNOS deletion ameliorated the obesity-induced blockage of autophagy flux (Figure 4E and F). Given the fact that we observed iNOS deletion activates TFEB, which controls autophagic gene expressions, we postulated that the regulation of autophagy by iNOS might also occur at an early stage of autophagic process. Indeed, electron microscopy analysis showed that the number of autophagic vesicles was decreased in mice fed an HFD compared with mice fed an RD (Figure 4G), an effect that was enhanced in the absence of iNOS (Figure 4G). Similar results were also observed by assessing the formation of starvation-induced autophagic vacuoles using a CytoID assay (Figure 4H).
Figure 4.
iNOS suppresses hepatic autophagy. (A) Representative Western blots of LC3 conversion, p62 and S6K (T421/S424) phosphorylation in livers from WT and iNOS-KO mice under fed or fasting (16 hours) conditions. Densitometry of Western blot analysis in A is shown on the right of the panel. *indicates statistical significance between fasting and fed in the same type of mice fed with same type of diet, determined by Student’s t test (P < .05; n = 4, biological replicates; 16 weeks on HFD). (B) Representative images (40×) of RFP-GFP-LC3 puncta in the livers of RFP-GFP-LC3 and RFP-GFP-LC3;iNOS KO mice fed an RD or HFD (16 weeks on HFD, fasted for 16 hours). Quantified numbers of the GFP-LC3 puncta/field are shown on the right of the panel. Data are shown as mean ± SEM. * indicates statistically significant difference relative to WT RD. determined by analysis of variance (ANOVA) followed by post hoc test (P < .05; 10 fields/group; n = 3, biological replicates). Scale bar: 10μm. (C) LC3 expression in livers from RFP-GFP-LC3 and RFP-GFP-LC3;iNOS-KO mice fed an RD or HFD (16 weeks on HFD, fasted for 16 hours). (D) Representative confocal images (63×) of primary hepatocytes isolated from WT or iNOS-KO mice (n = 3) transduced with Ad-mRFP-GFP-LC3 (MOI=2). Cells were treated with EBSS (4 hours). The number of the red LC3 puncta/field is noted at the top of each image. All data are presented as mean ± SEM. * indicates statistical significance compared with WT, # indicates statistical significance between HFD groups determined by ANOVA followed by post hoc test (P < .05; 10 fields/group; n = 3, biological replicates; 12 weeks on HFD). Scale bar: 10 μm. (E) Representative Western blots of LC3 conversion (arrow indicates LC3-II) and p62 expression in primary hepatocytes from livers of WT and iNOS-KO mice. EBSS (4 hours) was used to induce autophagy, and CQ (20 μM, 4 hours) was used to inhibit lysosomal degradation. Each lane contains a mixture of protein lysates from 3 mice. (F) Densitometry of Western blot analysis in E. * indicates statistical significance between EBSS and EBSS CQ groups in same type of mouse, determined by ANOVA followed by post hoc test (P < .05; n = 9, biological replicates; 12 weeks on HFD). (G) Representative transmission electron microscopy images (5,000×) of livers from fasting (16 hours) WT and iNOS-KO mice. Arrows: autophagic vesicles. Scale bar: 0.5 μm. Numbers at the top of each graph represent the average number of autophagic vesicles/7.5 μm2. All data are presented as mean ± SEM. * indicates statistical significance compared with WT RD; # indicates statistical significance between HFD group, determined by ANOVA followed by post hoc test (P < .05 n = 3, biological replicates; 12 images/mouse; 16 weeks on HFD). (H) Autophagic vacuoles in live primary hepatocytes from mice described in panel E, as detected by the Cyto-ID kit. Data are presented as mean ± SEM. * indicates statistical significance compared with WT RD in EBSS, # indicates statistical significance between HFD groups in the same treatment, and & indicates statistical significance compared with EBSS groups without CQ, determined by ANOVA followed by a post hoc test (P < .05; n = 3, biological replicates; 12 weeks on HFD).
iNOS Contributes to Obesity-Associated Hepatic Insulin Resistance by Regulating Autophagy
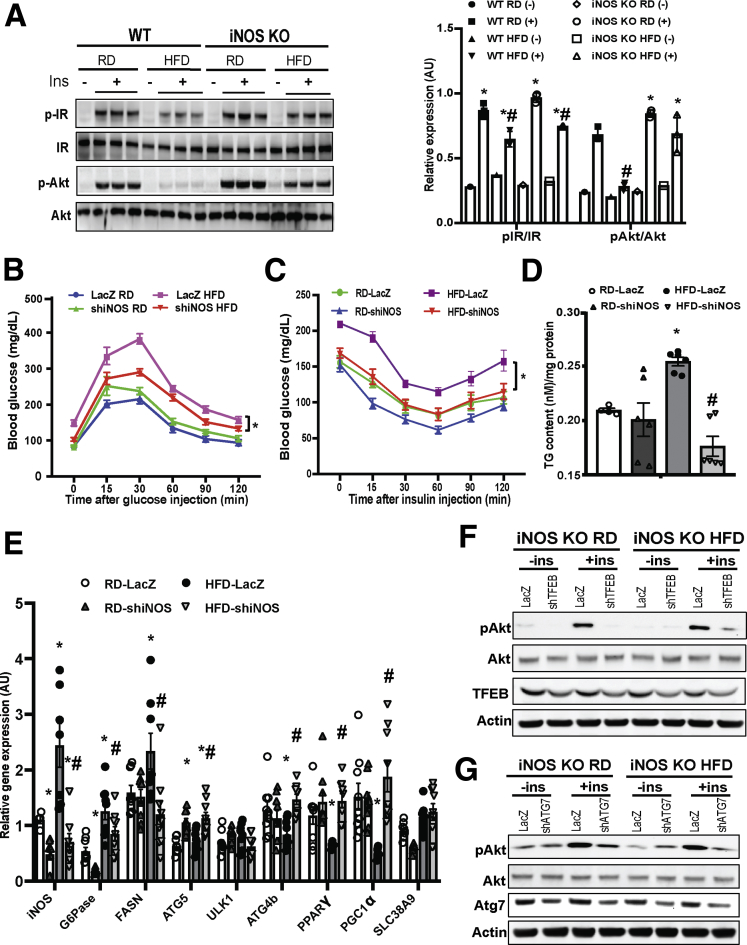
Obesity elevates iNOS expression in the liver,20, 35 skeletal muscle, and adipose tissue.36, 37 To evaluate the physiological relevance of iNOS-mediated lysosomal nitrosative stress in the liver, insulin action was assessed in hepatocytes isolated from WT and iNOS-KO mice. Deletion of iNOS ameliorated obesity-associated hepatic insulin resistance (Figure 5A). We then examined the effect of iNOS on systemic glucose homeostasis and insulin sensitivity using adenovirus-mediated iNOS suppression in the liver. Suppression of hepatic iNOS improved systemic glucose and insulin homeostasis (Figure 5B and C), which was associated with a significant decrease in hepatic steatosis (Figure 5D and E) and gluconeogenesis (Figure 5E), as well as increased expression of TFEB-target genes (Figure 5E). To link the defect in iNOS-mediated lysosomal function and autophagy with hepatic insulin resistance, we knocked down TFEB using adenoviral mediated gene silencing and examined insulin action in iNOS-KO primary hepatocytes. Suppression of TFEB significantly mitigated the improved hepatic insulin sensitivity in iNOS-KO cells (Figure 5F). Similar results were observed in primary hepatocytes of iNOS-KO mice transduced with adenoviral mediated gene silencing of key autophagy regulator, autophagy related 7 (Atg7) (Figure 5G). Together, these data indicate that iNOS desensitizes hepatic insulin signaling, in part, by modulating autophagy.
Figure 5.
iNOS-mediated impairment of hepatic autophagy contributes to obesity-associated insulin resistance. (A) Hepatic insulin action in primary hepatocytes from WT and iNOS-KO mice (10 weeks on HFD). Ins: insulin, 5 nM for 10 minutes; p-IR: IRtyr1150/1151; p-AKT: Aktser473. Densitometry of Western blot analysis of panel A is shown on the right of the panel. * indicates statistical significance between +Ins and –Ins in the same type of mice, and # indicates statistical significance between RD and HFD group in the same type of mice, determined by 2-way analysis of variance (ANOVA) with a post hoc test (P < .05; n = 3, biological replicates). (B) Glucose tolerance and (C) insulin tolerance tests in WT mice transduced with control virus (Ad-lacZ) or Ad-shiNOS and fed an RD or HFD (n = 8–10, biological replicates; 8 weeks on HFD). Data are presented as mean ± SEM. * indicates statistical analysis of area under the curve between HFD groups, determined by 2-way ANOVA with a post hoc test (P < .05). (D) Hepatic triglyceride levels in WT and iNOS-KO mice on an RD and HFD. Data are presented as mean ± SEM. * indicates a statistically significant difference relative to WT RD group; # indicates a statistically significant difference between HFD groups determined by ANOVA followed by post hoc test (P < .05; n = 3–4, biological replicates; 12 weeks on HFD). (E) Levels of messenger RNAs encoding gluconeogenesis, lipogenesis proteins and autophagy regulators in the livers of WT mice and WT mice transduced with Ad-shiNOS, as assessed by quantitative real-time RT-PCR. Data are presented as mean ± SEM. * indicates statistical significance compared with LacZ RD group, # indicates statistical significance between HFD groups determined by ANOVA followed by post hoc test (P < .05; n = 4–6, biological replicates; 12 weeks on HFD). (F, G) Hepatic insulin action in primary hepatocytes isolated from WT and iNOS-KO mice transduced with (F) Ad-shTFEB, (G) Ad-shAtg7, or control virus (Ad-lacZ). Each lane contains a mixture of protein lysates from 3 mice. IN: insulin, 5 nM for 10 minutes.
Discussion
Autophagy malfunction is recognized as a major factor leading to obesity-associated metabolic disorders, but the physiological cause of this aberrant autophagy remains unknown. Our results offer novel insight into the mechanistic basis of hepatic autophagy dysfunction in obesity and related insulin resistance. We found that obesity promotes local lysosomal iNOS-mediated NO production in the hepatocyte, leading to impaired lysosomal function and autophagy in the liver, thus contributing to the development of metabolic defects associated with obesity.
Under physiological conditions, NO promotes liver function and cell survival.38, 39 In contrast, iNOS generates excessive NO under HFD conditions, contributing to insulin resistance in the liver,35 muscle,37 and adipose tissues.37 Accumulating evidence indicates that NO signaling affects multiple stages of autophagy. For example, the initiation of autophagy is inhibited by various triggers in different cell types.23, 40 We recently showed that hepatic autophagic flux is blocked by lysosomal nitrosative stress in obesity.11 However, the source of excess NO required for protein S-nitrosylation in the lysosome is unknown. Data from this study answers this question by demonstrating that obesity decreases arginine in the lysosome, activating iNOSL (Figure 2), which in turn elevates the level of lysosomal NO (Figure 1A and B). This elevated lysosomal NO leads to lysosomal nitrosative stress and impaired autophagy flux.
The regulation of autophagy is clearly complex, involving in many steps and regulators. We found that in addition to modulating lysosomal nitrosative stress and autophagic flux, iNOS regulates the activity of TFEB and several of its target genes involved in autophagy (Figure 5F). Therefore, we do not rule out the possibility that iNOS-mediated signaling affects other steps in the autophagic process, including the activation of autophagy. Moreover, although we found that deletion of iNOS significantly decreased obesity-induced lysosomal NO production (Figure 2) and activated TFEB (Figure 3D–G), iNOS deletion did not affect mTOR activity in the liver of mice fed an HFD (Figure 4A). This result indicates that in the context of obesity, iNOS-mediated NO production is downstream of mTOR signaling, and that the iNOS-mediated suppression of TFEB is independent of mTOR. In fact, TFEB phosphorylation could be regulated by other kinases (eg, adenosine 5′monophosphate-activated protein kinase, Ca2+/calmodulin-dependent protein kinase) as well as by phosphatases.41 Thus, future studies are required to determine the molecular mechanism of iNOS-mediated suppression of TFEB in obesity.
Production of lysosomal NO was previously demonstrated in granulocytes, where it contributes to pathogen killing and inflammation.25, 42 Here, we provide the first evidence that obesity elevates NO in the lysosome (Figure 1A and B), impairing lysosomal function and disrupting autophagy in hepatocytes (Figures 3 and 4). It has been hypothesized that compartmentalization of NO synthase with its target proteins for S-nitrosylation could elevate NO concentrations locally.43 It has been shown that activated iNOS is associated with lysosomal vesicles, increasing membrane association of the molecule in immune cells.25, 44 We now add to this phenomenon by demonstrating in hepatocytes a portion of cellular iNOS is localized in the outer membrane of lysosomes (Figure 2F). Moreover, a recent study showed that lysosomes can process the turnover of iNOS.45 In our study, we observed that after trypsin digestion, residual iNOS remained in the lysosomal matrix in the presence of LPS treatment (Figure 2F). Future studies are needed to determine whether the iNOS that localized in the lysosomal matrix remains enzymatically function.
Typically, iNOS is expressed predominantly in Kupffer cells in the liver,46 but obesity elevates iNOS expression in hepatocytes, contributing to hepatic insulin resistance and inflammation.47 Furthermore, the role of iNOS in hepatic insulin resistance in obesity is complicated. Although deletion of iNOS protects from the obesity-associated hepatic steatosis and insulin resistance early on, these beneficial effects decrease over time (unpublished data), despite a significant reduction in hepatic steatosis.48 Nevertheless, our data showed that liver-specific iNOS suppression has a protective role. Specifically, we showed that iNOS inactivates TFEB, and that suppression of TFEB and Atg7 diminishes the improved hepatic insulin sensitivity by iNOS deletion (Figure 5E and F). In addition to regulating the lysosome-autophagy process, TFEB also serves as a key player in lipid metabolism.9 In our study, although we observed significant effects on expression of several tested TFEB targets due to deletion of iNOS (Figure 5E), and hepatic insulin insensitivity was clearly mediated by the effects of iNOS on TFEB and autophagy (Figure 5E and F), an autophagy-independent metabolic action of the iNOS-TFEB axis cannot be ruled out at this time.
In summary, our study demonstrates that obesity activates iNOSL in the liver, promoting excessive NO accumulation in the lysosome, which drives lysosomal nitrosative stress and blocks hepatic autophagy. Insights into the mechanisms by which inflammatory signaling impairs lysosomal function and drives the pathologies associated with obesity should accelerate the development of novel therapies for metabolic diseases.
Materials and Methods
Cell Culture
Primary hepatocytes were isolated from mice using the collagenase type X (Wako, Japan; 039-17864) perfusion method.20 Briefly, primary hepatocytes were isolated by collagenase type X (Wako Pure Chemical Industries, ltd, Japan; 039-17864) perfusion method. Cells were washed with hepatocyte wash medium (Thermo Fisher Scientific, Waltham, MA; 17704024), purified by Percoll (GE Healthcare, Chicago, IL; 17089101) density gradient separation, and resuspended in William’s E medium (Thermo Fisher Scientific; 12551032) with 5% fetal bovine serum, 10-nM dexamethasone, and 20-nM insulin. Cells were then seeded on collagen-coated plates at a final density of 3.5 × 104 cells/cm2. After 4 hours, attached cells were cultured with fresh medium and transduced with the indicated adenoviruses. Primary hepatocytes were treated for 4 hours with EBSS (Sigma-Aldrich, St. Louis, MO; E3024), containing 200-μM OA (16 hours; Sigma-Aldrich; O1008), 10-ng/mL TNF (16 hours; PeproTech), 1-μg/mL LPS (6 hours; Sigma-Aldrich; L2630), 50-μM SNAP (4 hours; Sigma-Aldrich, N3398), 100-nM PTIO (30 minutes; Sigma-Aldrich; P5084), and 20-μM CQ (Sigma-Aldrich, C6628). Insulin signaling in primary hepatocytes was stimulated with a 10-minutes exposure to 5-nM insulin (Sigma-Aldrich; I5500).
Mouse Models
Animal care and experimental procedures were performed with approval from the University of Iowa's Institutional Animal Care and Use Committee. C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME; 000664), iNOS-KO mice (The Jackson Laboratory; 002609), RFP-GFP-LC3 mice (provided by Dr Joseph A. Hill at the University of Texas, Southwestern Medical Center), and littermate WT lean controls were kept on a 12-hour light cycle, and were fed an RD (2920X Teklad global diet, Madison, WI). Mice used for the DIO model were placed on a 60% kcal HFD (Research Diets, New Brunswick, NJ; D12492) immediately after weaning at 3 weeks of age. Adenovirus-shiNOS was delivered into mice via retro-orbital injection at a titer of 1 × 1011 ifu/mouse.20 For LPS treatment in mice, 1 dose of LPS (1 mg/kg/mouse/day) was administered daily for 3 days.
Intracellular NO Measurements
For lysosomal NO imaging, live primary hepatocytes were stained with 5.0 μM of a lysosome-specific fluorescent probe, Lyso-NINO, for 5 minutes at 37°C.26 The fluorescence intensity was measured by a microplate reader (Ex = 440 nm, Em = 530 nm) at 37°C, and images were taken with a Zeiss 700 confocal microscope (Oberkochen, Germany).
Immunoblotting
Primary antibodies for immunoblotting were anti-LC3 (Novus, Centennial, CO; NB600), anti-p62 (Abnova, Walnut, CA; H00008878-M03 for primary hepatocytes; and MBL, Woburn, MA; PM066 for liver tissue), anti-pAKT(S473) (Cell Signaling Technology, Danvers, MA; 9271), anti-pIR (Millipore Sigma, Burlington, MA; 407707), anti-AKT (Santa Cruz Biotechnology, Dallas, TX; sc-8312), anti-IR (Santa Cruz Biotechnology; sc-711), anti-Atg7 (Abgent, San Diego, CA; AP1813a), anti-iNOS (Santa Cruz Biotechnology; sc-8310), anti-TFEB (Bethyl, Montgomery County, TX; A303-673A), anti-Lamp1 (Cell Signaling Technology, Danvers, MA; 3243), anti-pmTOR (S2448) (Cell Signaling Technology; 2971), anti-pS6K (T421/S424) (Cell Signaling Technology; 9204), anti-S6K (Cell Signaling Technology; 9202), anti-Lamin A/C (Cell Signaling Technology; 4777), anti-flag (Sigma-Aldrich; F9291), anti-actin (Abcam, Cambridge, UK, ab8227), or anti-β-tubulin (Santa Cruz Biotechnology; sc-9104) used at a 1:1000 dilution, and secondary antibodies were conjugated with horseradish peroxidase goat–anti-mouse–IgG (Santa Cruz Biotechnology; sc-2005), horseradish peroxidase mouse-anti-rabbit-IgG (Santa Cruz Biotechnology; sc-2357), or horseradish peroxidase goat-anti-guinea pig-IgG (Santa Cruz Biotechnology; sc-2438). The images were detected by ChemiDoc Touch Imaging System (Bio-Rad, Hercules, CA). Densitometry analyses of Western blot images were performed by using the Image Lab (Bio-Rad).
Immunohistology, Immunocytochemistry, and Electron Microscopy
Frozen liver sections were fixed with 4% paraformaldehyde and stained with an anti-TFEB antibody (Bethyl; A303-673A) and an Alexa-488–conjugated secondary antibody (Invitrogen, Waltham, MA; A11008). For iNOS staining in the lysosomal fractions, purified lysosomes were treated with trypsin for 10 minutes at room temperature and stained with an anti-iNOS antibody (BD Biosciences, San Jose, CA, USA; 610329) and an anti-Lamp1 antibody (Cell Signaling Technology, 3243). Images were taken using a Zeiss 700 confocal microscope, and images were quantified using ImarisColoc (Bitplane, Concord, MA). For electron microscopy analysis, liver sections were fixed with 2.5% formaldehyde/glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4; Emsdiasum, Hatfield, PA) and 1% OsO4 (Electron Microscopy Science, Hatfield, PA; 15949), followed by dehydration and staining with uranyl acetate/lead citrate. Images were taken using a JEOL JEM 1230 electron microscope (Peabody, MA).
Biotin-Switch Assay
S-nitrosylated proteins were detected in situ as described previously.11 The images were observed using a Zeiss 700 confocal microscope and quantified using an ImarisColoc (Bitplane).
Quantitative Real-Time Reverse-Transcriptase Polymerase Chain Reaction
Total RNA was isolated using the Trizol reagent (Invitrogen; 15596026) and reverse transcribed into cDNA using an iScript complementary DNA synthesis kit (Bio-Rad, 1708891). Quantitative real-time reverse-transcriptase polymerase chain reaction analysis was performed using SYBR Green (Bio-Rad; 1725121). The primers used were Atg4b: forward 5′- TGGGTGTTATTGGAGGGAAG-3′, reverse 5′- CAGAAAAACCCCACAGCAAT-3′; Atg5: forward 5′-AGATGGACAGCTGCACACAC-3′, reverse 5′-GCTGGGGGACAATGCTAATA-3′; Atg7: forward 5′-TGCCTATGATGATCTGTGTC-3′, reverse 5′-CACCAACTGTTATCTTTGTCC-3′; FASN: forward 5′-AGAGATCCCGAGACGCTTCTA-3′, reverse 5′-GCCTGGTAGGCATTCTGTAGT-3′; G6Pase: forward 5′-CGACTCGCTATCTCCAAGTGA-3′, reverse-5′-ACAAAGTTGCTCTGAAAACAAATCA-3′; iNOS: forward 5′-GTTCTCAGCCCAACAATACAAGA-3′, reverse 5′-GTGGACGGGTCGATGTCAC-3; PEPCK: forward 5′-CTGCATAACGGTCTGGACTTC-3′, reverse 5′-CAGCAACTGCCCGTACTCC-3′; PPARa: forward 5′- AGAGCCCCATCTGTCCTCTC-3′ reverse 5′- ACTGGTAGTCTGCAAAACCAAA-3′; PPARr: forward 5′- TATGGAGTGACATAGAGTGTGCT-3′ reverse 5′- CCACTTCAATCCACCCAGAAAG-3′; SLC38A9: forward 5′-CTCTCTGAGGTGGAACATGAAGT-3′, reverse 5′-GCATGGTCACTAACCCTCTGAAT-3′; ULK1: forward 5′-AACATCGTGGCGCTGTATGA-3′, reverse 5′-TGCGCATAGTGTGCAGGTAG-3′; and 18S: forward 5′-AGTCCCTGCCCTTTGTACACA-3′, reverse 5′- CGATCCGAGGGCCTCACTA-3′.
Adenovirus Transduction and Luciferase Assay
Adenovirus-mRFP-GFP-LC3, adenovirus-shTFEB, and adenovirus-shAtg7 were transduced into primary hepatocytes at a titer of 10 MOI11 for 48 hours. mTOR (73384) and SLC38A9 (71869) constructs32 were purchased from Addgene, Watertown, MA and transfected into primary hepatocytes by polyethylenimine (Polysciences INC, Warrington, PA; 23966). Primary hepatocytes were transfected with the 4X-CLEAR luciferase reporter49 using polyethylenimine. Firefly luciferase and Renilla luciferase activities were measured by Dual-Glo Luciferase Assay (Promega, Madison, WI, USA; E2920) at 48 hours post-transfection.
CytoID Analysis
Primary hepatocytes were stained with CytoID following the manufacturer's protocol (ENZO Lifesciences, Farmingdale, NY; ENZ-51031).11 Briefly, Cyto-ID Green Detection Reagent was added to primary hepatocytes and incubated at 37°C for 30 minutes. The level of autophagy was expressed as fluorescent intensity, and normalized by nuclear intensity. The nuclei were stained by DAPI (Thermo Fisher Scientific; D1306).
CTSB Activity Assays and Measurement of Lysosomal pH
Lysosomal function was measured in live primary hepatocytes. The CTSB assay was performed using a CTSB Detection Kit (Magic Red Cathepsin-B assay kit, ImmunoChemistry Technologies; Bloomington, MN, U.S.A. 938) and lysosomal pH was measured using LysoSensr Green DND-189 (Thermo Fisher Scientific; L7535).11
Subcellular Fractionation
Lysosomal fractions were prepared as previously described.50 Equivalent amounts of liver tissue from lean and obese mice were extracted and separated by density gradient centrifugation on a multistep Nycodenz (Sigma-Aldrich; D2158) gradient. Briefly, the endoplasmic reticulum was prepared as a pellet by centrifugation of the supernatant at 100,000 g. Autophagic vacuoles were collected from the 15%–20% and 20%–24% interfaces, lysosomes from the 24%–26% interface and mitochondria from 26% to 50% interface. Fractions were further washed in 0.25-M sucrose (Sigma-Aldrich; S0389) and collected by centrifugation. The nuclear fractions were prepared as previously described.20
Lysosomal Arginine Profile
Lysosomal fractions from liver samples were lyophilized and lysosomal metabolites were extracted using a methanol/acetonitrile/water (4.5:4.5:1). Cleared extracts were dried and reconstituted in acetonitrile/water (1:1). A total of 10 μL of sample was subjected to hydrophilic interaction chromatography with a Phenomenex Luna column at a flow rate of 300 μL/min as previously described.51 Tandem mass spectrometry was performed using a Q Exactive Hybrid Quadrpole-Orbitrap mass spectrometer (Thermo Fisher Scientific) in positive mode at the FOEDRC Metabolic Phenotyping Core (University of Iowa, Iowa City, IA). Targeted identification of arginine was accomplished using parallel reaction monitoring of parent ion (175.1189 m/z) and daughter ions (116.0708 m/z and 70.0659 m/z) following fragmentation using a collision energy of 30.
Glucose Tolerance Tests, Insulin Tolerance Tests, Hepatic Triglyceride Content, and Insulin Infusions
Glucose tolerance tests were performed by intraperitoneal glucose injection (1 g/kg, 50% dextrose; Hospra Inc, Nashville, TN; 0409-6648-02). Insulin tolerance tests were performed by intraperitoneal insulin injection (0.75 IU/kg Humulin; Lilly USA; 002-8215-01), and insulin infusion studies were performed in mice with portal vein insulin infusion (0.75 IU/kg Humulin; Lilly USA, LLC, 002-8215-01).20
Statistical Analysis
Results are expressed as mean ± the SEM; n represents the number of individual mice (biological replicates), or individual experiments as indicated in the figure legends. Statistical analysis was performed by Student's t test, or one-way analysis of variance followed by Tukey’s post hoc test, or 2-way analysis of variance followed by Bonferroni’s post hoc test, using GraphPad Prism (GraphPad Software Prism 8, San Diego, CA) as indicated in the figure legends.
Acknowledgments
The authors thank Drs Vitor A. Lira, Long-sheng Song, and Sara Sebag at the University of Iowa for scientific discussions and insights. They are grateful to Dr Albert La Spada (University of California, San Diego) for providing the transcription factor EB–luciferase construct, Dr Ana Maria Cuervo (Albert Einstein College of Medicine) for providing technical supports on lysosomal biology, and Dr Joseph Hill (University of Texas, Southwestern Medical Center) for providing RFP-GFP-LC3 transgenic mice. The authors also thank Dr David Sabatini (Whitehead Institute) for donating the mammalian target of rapamycin 1 and SLC38A9 constructs to Addgene.
Footnotes
Author contributions L.Y., Q.Q., and Z.Z. designed the study. Q.Q., Z.Z., M.L., K.S., D.C., A.J.R., H.J.C. and L.Y. performed the experiments. E.B.T., J.A.A., W.X.D., H.J.C. and Y.X. provided critical reagents and scientific suggestions on the manuscript. L.Y. conceived and supervised the study and wrote the manuscript.
Conflicts of interest The authors declares no conflict.
Funding Zeyuan Zhang is supported by an American Heart Association Predoctoral Award (19PRE34380258); Adam J. Rauckhorst is supported by an American Diabetes Association Postdoctoral Award (1-18-PDF-060); Eric B. Taylor is supported by R01 DK104998; Wen-Xing Ding is supported by R01 AA020518, R01 DK102142, U01 AA024733, a KUMC Frontier-Lied Pilot Grant, and P20GM103549 & P30GM118247; Ling Yang is supported by an American Heart Association Scientist Development Grant (15SDG25510016), an American Diabetes Association Innovative Basic Science Award (1-18-IBS-149), and R01 DK108835-01A1.
References
- 1.Kuntz E., Kuntz H.D. Springer; Berlin: 2002. Hepatology, Principles, and Practice: History, Morphology, Biochemistry, Diagnostics, Clinic, Therapy. [Google Scholar]
- 2.Korolchuk V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M., O'Kane C.J., Deretic V., Rubinsztein D.C. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13:453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D.M. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Codogno P., Meijer A.J. Autophagy in the liver. J Hepatol. 2013;59:389–391. doi: 10.1016/j.jhep.2013.02.031. [DOI] [PubMed] [Google Scholar]
- 5.Doria A., Gatto M., Punzi L. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- 6.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., Czaja M.J. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang L., Li P., Fu S., Calay E.S., Hotamisligil G.S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider J.L., Suh Y., Cuervo A.M. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014;20:417–432. doi: 10.1016/j.cmet.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Settembre C., De Cegli R., Mansueto G., Saha P.K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T.J., Wollenberg A.C., Di Bernardo D., Chan L., Irazoqui J.E., Ballabio A. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuo Y., Yamashina S., Sonoue H., Arakawa A., Nakadera E., Aoyama T., Uchiyama A., Kon K., Ikejima K., Watanabe S. Abnormality of autophagic function and cathepsin expression in the liver from patients with non-alcoholic fatty liver disease. Hepatol Res. 2014;44:1026–1036. doi: 10.1111/hepr.12282. [DOI] [PubMed] [Google Scholar]
- 11.Qian Q., Zhang Z., Orwig A., Chen S., Ding W.X., Xu Y., Kunz R.C., Lind N.R.L., StamLer J.S., Yang L. S-nitrosoglutathione reductase dysfunction contributes to obesity-associated hepatic insulin resistance via regulating autophagy. Diabetes. 2018;67:193–207. doi: 10.2337/db17-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Persellin R.H. Role of lysosomes in the pathogenesis of rheumatoid arthritis. Med Clin North Am. 1968;52:635–641. [PubMed] [Google Scholar]
- 13.Weissmann G. The role of lysosomes in inflammation and disease. Annu Rev Med. 1967;18:97–112. doi: 10.1146/annurev.me.18.020167.000525. [DOI] [PubMed] [Google Scholar]
- 14.Zhao G.N., Zhang P., Gong J., Zhang X.J., Wang P.X., Yin M., Jiang Z., Shen L.J., Ji Y.X., Tong J., Wang Y., Wei Q.F., Wang Y., Zhu X.Y., Zhang X., Fang J., Xie Q., She Z.G., Wang Z., Huang Z., Li H. Tmbim1 is a multivesicular body regulator that protects against non-alcoholic fatty liver disease in mice and monkeys by targeting the lysosomal degradation of Tlr4. Nat Med. 2017;23:742–752. doi: 10.1038/nm.4334. [DOI] [PubMed] [Google Scholar]
- 15.Chao X., Wang S., Zhao K., Li Y., Williams J.A., Li T., Chavan H., Krishnamurthy P., He X.C., Li L., Ballabio A., Ni H.M., Ding W.X. Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology. 2018;155:865–879.e12. doi: 10.1053/j.gastro.2018.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwakiri Y., Kim M.Y. Nitric oxide in liver diseases. Trends Pharmacol Sci. 2015;36:524–536. doi: 10.1016/j.tips.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandemir O., Polat A., Kaya A. Inducible nitric oxide synthase expression in chronic viral hepatitis and its relation with histological severity of disease. J Viral Hepat. 2002;9:419–423. doi: 10.1046/j.1365-2893.2002.00382.x. [DOI] [PubMed] [Google Scholar]
- 18.McNaughton L., Puttagunta L., Martinez-Cuesta M.A., Kneteman N., Mayers I., Moqbel R., Hamid Q., Radomski M.W. Distribution of nitric oxide synthase in normal and cirrhotic human liver. Proc Natl Acad Sci U S A. 2002;99:17161–17166. doi: 10.1073/pnas.0134112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinozaki S., Choi C.S., Shimizu N., Yamada M., Kim M., Zhang T., Shiota G., Dong H.H., Kim Y.B., Kaneki M. Liver-specific inducible nitric-oxide synthase expression is sufficient to cause hepatic insulin resistance and mild hyperglycemia in mice. The J Biol Chem. 2012;286:34959–34975. doi: 10.1074/jbc.M110.187666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L., Calay E.S., Fan J., Arduini A., Kunz R.C., Gygi S.P., Yalcin A., Fu S., Hotamisligil G.S. METABOLISM. S-Nitrosylation links obesity-associated inflammation to endoplasmic reticulum dysfunction. Science. 2015;349:500–506. doi: 10.1126/science.aaa0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho D.H., Nakamura T., Fang J., Cieplak P., Godzik A., Gu Z., Lipton S.A. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butler D., Bahr B.A. Oxidative stress and lysosomes: CNS-related consequences and implications for lysosomal enhancement strategies and induction of autophagy. Antioxid Redox Signal. 2006;8:185–196. doi: 10.1089/ars.2006.8.185. [DOI] [PubMed] [Google Scholar]
- 23.Jaishy B., Zhang Q., Chung H.S., Riehle C., Soto J., Jenkins S., Abel P., Cowart L.A., Van Eyk J.E., Abel E.D. Lipid-induced NOX2 activation inhibits autophagic flux by impairing lysosomal enzyme activity. J Lipid Res. 2015;56:546–561. doi: 10.1194/jlr.M055152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tojo A., Guzman N.J., Garg L.C., Tisher C.C., Madsen K.M. Nitric oxide inhibits bafilomycin-sensitive H(+)-ATPase activity in rat cortical collecting duct. Am J Physiol. 1994;267:F509–F515. doi: 10.1152/ajprenal.1994.267.4.F509. [DOI] [PubMed] [Google Scholar]
- 25.Roszer T. Springer; Amsterdam: 2012. The Biology of Subcellular Nitric Oxide. [Google Scholar]
- 26.Yu H., Xiao Y., Jin L. A lysosome-targetable and two-photon fluorescent probe for monitoring endogenous and exogenous nitric oxide in living cells. J Am Chem Soc. 2012;134:17486–17489. doi: 10.1021/ja308967u. [DOI] [PubMed] [Google Scholar]
- 27.Sass G., Koerber K., Bang R., Guehring H., Tiegs G. Inducible nitric oxide synthase is critical for immune-mediated liver injury in mice. The Journal of clinical investigation. 2001;107:439–447. doi: 10.1172/JCI10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L., Wang Z.V., Hill J.A., Lin F. New autophagy reporter mice reveal dynamics of proximal tubular autophagy. J Am Soc Nephrol. 2014;25:305–315. doi: 10.1681/ASN.2013040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villanueva C., Giulivi C. Subcellular and cellular locations of nitric oxide synthase isoforms as determinants of health and disease. Free Radic Biol Med. 2010;49:307–316. doi: 10.1016/j.freeradbiomed.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskin D.L., Rodriguez del Valle M., Heck D.E., Hwang S.M., Ohnishi S.T., Durham S.K., Goller N.L., Laskin J.D. Hepatic nitric oxide production following acute endotoxemia in rats is mediated by increased inducible nitric oxide synthase gene expression. Hepatology. 1995;22:223–234. [PubMed] [Google Scholar]
- 31.Harms E., Gochman N., Schneider J.A. Lysosomal pool of free-amino acids. Biochem Biophys Res Commun. 1981;99:830–836. doi: 10.1016/0006-291x(81)91239-0. [DOI] [PubMed] [Google Scholar]
- 32.Wang S., Tsun Z.Y., Wolfson R.L., Shen K., Wyant G.A., Plovanich M.E., Yuan E.D., Jones T.D., Chantranupong L., Comb W., Wang T., Bar-Peled L., Zoncu R., Straub C., Kim C., Park J., Sabatini B.L., Sabatini D.M. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thibeault S., Rautureau Y., Oubaha M., Faubert D., Wilkes B.C., Delisle C., Gratton J.P. S-nitrosylation of beta-catenin by eNOS-derived NO promotes VEGF-induced endothelial cell permeability. Mol Cell. 2010;39:468–476. doi: 10.1016/j.molcel.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 34.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 35.Shinozaki S., Choi C.S., Shimizu N., Yamada M., Kim M., Zhang T., Dong H.H., Kim Y.B., Kaneki M. Liver-specific inducible nitric-oxide synthase expression is sufficient to cause hepatic insulin resistance and mild hyperglycemia in mice. J Biol Chem. 2011;286:34959–34975. doi: 10.1074/jbc.M110.187666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elizalde M., Ryden M., van Harmelen V., Eneroth P., Gyllenhammar H., Holm C., Ramel S., Olund A., Arner P., Andersson K. Expression of nitric oxide synthases in subcutaneous adipose tissue of nonobese and obese humans. J Lipid Res. 2000;41:1244–1251. [PubMed] [Google Scholar]
- 37.Perreault M., Marette A. Targeted disruption of inducible nitric oxide synthase protects against obesity-linked insulin resistance in muscle. Nat Med. 2001;7:1138–1143. doi: 10.1038/nm1001-1138. [DOI] [PubMed] [Google Scholar]
- 38.Anggard E. Nitric oxide: mediator, murderer, and medicine. Lancet. 1994;343:1199–1206. doi: 10.1016/s0140-6736(94)92405-8. [DOI] [PubMed] [Google Scholar]
- 39.Iwakiri Y., Grisham M., Shah V. Vascular biology and pathobiology of the liver: Report of a single-topic symposium. Hepatology. 2008;47:1754–1763. doi: 10.1002/hep.22203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Criollo A., Senovilla L., Authier H., Maiuri M.C., Morselli E., Vitale I., Kepp O., Tasdemir E., Galluzzi L., Shen S., Tailler M., Delahaye N., Tesniere A., De Stefano D., Younes A.B., Harper F., Pierron G., Lavandero S., Zitvogel L., Israel A., Baud V., Kroemer G. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–631. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puertollano R., Ferguson S.M., Brugarolas J., Ballabio A. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 2018;37:e98804. doi: 10.15252/embj.201798804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takemoto K., Ogino K., Wang D.H., Takigawa T., Kurosawa C.M., Kamyabashi Y., Hibino Y., Hitomi Y., Ichimura H. Biochemical characterization of reactive nitrogen species by eosinophil peroxidase in tyrosine nitration. Acta Med Okayama. 2007;61:17–30. doi: 10.18926/AMO/32911. [DOI] [PubMed] [Google Scholar]
- 43.Jia J., Arif A., Terenzi F., Willard B., Plow E.F., Hazen S.L., Fox P.L. Target-selective protein S-nitrosylation by sequence motif recognition. Cell. 2014;159:623–634. doi: 10.1016/j.cell.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winston B.W., Krein P.M., Mowat C., Huang Y. Cytokine-induced macrophage differentiation: a tale of 2 genes. Clin Invest Med. 1999;22:236–255. [PubMed] [Google Scholar]
- 45.Jia L., Wang Y., Wang Y., Ma Y., Shen J., Fu Z., Wu Y., Su S., Zhang Y., Cai Z., Wang J., Xiang M. Heme oxygenase-1 in macrophages drives septic cardiac dysfunction via suppressing lysosomal degradation of inducible nitric oxide synthase. Circ Res. 2018;122:1532–1544. doi: 10.1161/CIRCRESAHA.118.312910. [DOI] [PubMed] [Google Scholar]
- 46.Leifeld L., Fielenbach M., Dumoulin F.L., Speidel N., Sauerbruch T., Spengler U. Inducible nitric oxide synthase (iNOS) and endothelial nitric oxide synthase (eNOS) expression in fulminant hepatic failure. J Hepatol. 2002;37:613–619. doi: 10.1016/s0168-8278(02)00271-4. [DOI] [PubMed] [Google Scholar]
- 47.Fujimoto M., Shimizu N., Kunii K., Martyn J.A., Ueki K., Kaneki M. A role for iNOS in fasting hyperglycemia and impaired insulin signaling in the liver of obese diabetic mice. Diabetes. 2005;54:1340–1348. doi: 10.2337/diabetes.54.5.1340. [DOI] [PubMed] [Google Scholar]
- 48.Nozaki Y., Fujita K., Wada K., Yoneda M., Kessoku T., Shinohara Y., Imajo K., Ogawa Y., Nakamuta M., Saito S., Masaki N., Nagashima Y., Terauchi Y., Nakajima A. Deficiency of iNOS-derived NO accelerates lipid accumulation-independent liver fibrosis in non-alcoholic steatohepatitis mouse model. BMC Gastroenterol. 2015;15:42. doi: 10.1186/s12876-015-0269-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cortes C.J., Miranda H.C., Frankowski H., Batlevi Y., Young J.E., Le A., Ivanov N., Sopher B.L., Carromeu C., Muotri A.R., Garden G.A., La Spada A.R. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nat Neurosci. 2014;17:1180–1189. doi: 10.1038/nn.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel B., Cuervo A.M. Methods to study chaperone-mediated autophagy. Methods. 2015;75:133–140. doi: 10.1016/j.ymeth.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathew A.V., Zeng L., Byun J., Pennathur S. Metabolomic profiling of arginine metabolome links altered methylation to chronic kidney disease accelerated atherosclerosis. J Proteomics Bioinform. 2015;(suppl 14):001. doi: 10.4172/jpb.S14-001. [DOI] [PMC free article] [PubMed] [Google Scholar]