Abstract
Long noncoding RNAs (lncRNAs) are a novel class of gene regulators involved in tumor biogenesis. Glioblastoma is the most common and malignant type of brain tumor. The function and prognostic significance of lncRNAs in glioblastoma remain unclear. In the present study, updated gene annotations were adopted to investigate lncRNA expression profiles in publicly available glioma microarray datasets from the Gene Expression Omnibus and the Repository for Molecular Brain Neoplasia Data. In a training set of 108 samples of glioblastoma, using univariate Cox regression analysis with a permutation P<0.005, four lncRNAs, including insulin-like growth factor binding protein 7-antisense 1 (IGFBP7-AS1), were significantly associated with patient overall survival. These four lncRNAs were integrated as an expression-based molecular signature to divide patients in the training set into high-risk and low-risk subgroups, with distinct survival rates (hazard ratio, 2.72; 95% CI, 1.71–4.31; P<0.001). The prognostic value of the lncRNA signature was confirmed in two additional datasets comprising a total of 147 samples from patients with glioblastoma. The prognostic value of this signature was independent of age and Karnofsky performance status. This signature was also able to predict different outcomes in cases of glioblastoma associated with an isocitrate dehydrogenase 1 mutation. Further bioinformatics analyses revealed that ‘epithelial-mesenchymal transition’ and ‘p53 pathway’ gene sets were enriched in glioblastoma samples with higher IGFBP7-AS1 expression. Furthermore, in vitro experiments demonstrated that knockdown of IGFBP7-AS1 inhibited the viability, migration and invasion of U87 and U251 glioma cells. In conclusion, the present study identified a lncRNA signature able to predict glioblastoma outcomes, and provided novel information regarding the role of IGFBP7-AS1 in glioma development.
Keywords: glioblastoma, prognosis, microarray, long noncoding RNA, insulin-like growth factor binding protein 7-antisense 1
Introduction
Glioma is the most common type of primary brain tumor, with an annual incidence of ~6.6/100,000 individuals in the USA (1). Approximately half of all glioma cases are classified as glioblastoma, which is the most malignant type of brain tumor (1). Despite current treatment standards involving maximal safe resection, followed by radiotherapy with concomitant and adjuvant temozolomide (TMZ) (2), the median survival of patients with glioblastoma is only 14–17 months (2,3). As a recognized disease with high heterogeneity, survival variability has been observed in cases of glioblastoma with similar clinical and histological features (4). Traditional clinicopathological factors, including histological grade, age and Karnofsky performance status (KPS), do not appear to be sufficient for precise outcome prediction (3,4). However, with the characterization of specific genetic alterations using advanced sequencing and microarray technologies, several genes and multi-gene expression-based molecular signatures have been discovered. These have since been used for the identification of novel glioblastoma subtypes, and may also have better prognostic significance (3–5).
Proteins are primary functional effectors in cells that represent the vast majority of prognostic biomarkers currently used for glioma (3). Over 68% of the human genome is transcribed to long noncoding RNAs (lncRNAs), a class of transcripts >200 nucleotides in length that lack protein-coding ability. These have emerged as important regulators of tissue physiology and disease processes, including tumorigenesis (6). lncRNAs can regulate the expression levels of oncogenes or tumor suppressors through various mechanisms, including chromatin modification, transcriptional control and post-transcriptional processing, and affect various aspects of cellular homeostasis (7). The expression pattern of lncRNAs is highly tissue- and cell type-specific, a characteristic that may be applied for accurate molecular cancer subclassification and outcome prediction (7). A successfully established example is the lncRNA prostate cancer antigen 3, which is currently used for early diagnosis and treatment response surveillance in prostate cancer (8).
Reports have revealed that lncRNAs are aberrantly expressed in glioma tissue relative to normal brain tissue (9,10). Some lncRNAs have been implicated in glioma proliferation, apoptosis, motility and stemness maintenance (11,12). However, the detailed mechanisms and clinical application of most lncRNAs remain largely unknown. By virtue of the constant updating of gene annotation databases, such as GENCODE and RefSeq, more gene probes from previously published microarray studies are likely to be repurposed as lncRNAs, providing novel information for investigating the expression profiles and prognostic value of lncRNAs in cancer.
In the present study, the updated gene annotations were adopted to conduct lncRNA profiling on publicly available microarray datasets of glioma, and a four-lncRNA signature was identified to be able to predict the outcome of glioblastoma. Insulin-like growth factor binding protein 7-antisense 1 (IGFBP7-AS1), one of the survival-associated lncRNAs, was further investigated by bioinformatics analysis and functional experiments.
Materials and methods
Glioma datasets preparation
Gene expression profiles generated by the Affymetrix HG-U133 Plus 2.0 microarray platform (Affymetrix; Thermo Fisher Scientific, Inc.) and corresponding clinical data were downloaded from the publicly available Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo) and the Repository for Molecular Brain Neoplasia Data (Rembrandt) project (13). The Rembrandt database is available at the Georgetown Database of Cancer web portal (https://gdoc.georgetown.edu). Rembrandt database (13) includes 227 grade IV glioblastomas, 84 grade III gliomas, 99 grade II gliomas and 28 normal brain (NB) samples, according to the world health organization grading system (14). GEO dataset GSE4290 (15) includes 81 glioblastomas, 31 grade III gliomas, 45 grade II gliomas and 23 NB. All these samples were used for lncRNA expression analysis. To avoid interference from different treatment regimens on the analysis of survival-associated lncRNAs, only glioblastoma samples from patients who received post-operative radiation with or without chemotherapy were analyzed, including 108 samples from the GEO dataset GSE16011 (4), 69 from the GEO dataset GSE7696 (16) and 78 from Rembrandt database. The clinical characteristics of the 255 patients with glioblastoma used for survival analysis were described in Table SI.
Microarray data processing and analysis
The raw Affymetrix microarray expression data were processed and normalized using the Robust Multichip Average algorithm (17). The custom Chip Definition File was used to reorganize probes to Entrez Gene identifiers (version 21; National Center for Biotechnology Information, National Institutes of Health) and exclude inaccurate or wrongly annotated probes (18). To avoid systematic error across different experiments, each dataset was standardized independently by the Z-score method to transform the expression of each gene into having a mean of 0 and a standard deviation of 1 (19). Similar to previous methods (9), genes on microarray platforms were identified as lncRNAs according to their RefSeq transcript ID (National Center for Biotechnology Information, National Institutes of Health) and Ensembl gene ID (20) from the updated NetAffx annotation files (HG-U133_Plus_2 Annotations, Release 36; Affymetrix; Thermo Fisher Scientific, Inc.) and then filtered by removing pseudogenes, ribosomal RNAs, transfer RNAs, microRNAs (miRNAs) and other short noncoding RNAs. Finally, a gene list containing 1,895 lncRNAs was created for further analysis. Among these lncRNAs, 1,495 transcripts were annotated on RefSeq and Ensembl databases and 400 transcripts were annotated selectively on RefSeq (Table SII).
Hierarchical clustering analysis was performed to divide patients based on multi-lncRNA expression levels using centered correlation metrics and the average linkage method. Gene set enrichment analysis (GSEA) was implemented according to lncRNA median expression, with the gene sets from the Molecular Signatures Database (software.broadinstitute.org/gsea) (21).
Cell culture and transient transfection
The human glioma cell lines U87 (cat. no. HTB-14, glioblastoma from unknown origin) and U251 were purchased from the Chinese Academy of Sciences. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 50 µg/ml streptomycin, and were incubated at 37°C in an atmosphere containing 5% CO2.
The small interfering RNA (siRNA) targeting IGFBP7-AS1 and nonspecific negative control (NC) sequence were synthesized by Shanghai GenePharma Co., Ltd. Cell transient transfection was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) 24 h after seeding cells, at 70% confluency. Transfection complexes were prepared according to the manufacturer's protocol and added to the cells to a final oligonucleotide concentration of 100 nM. The transfection medium was replaced 6 h post-transfection. Cells were incubated for 2 days prior to subsequent experimentation. The siRNA sequences are presented in Table I.
Table I.
Small interfering RNA sequences of long noncoding RNA IGFBP7-AS1 and nonspecific NC.
| Gene symbol | Positive-sense strand (5′-3′) | Antisense strand (5′-3′) |
|---|---|---|
| IGFBP7-AS1 | CUAAGUUUCUGGAAGAUAAAG | UUAUCUUCCAGAAACUUAGAA |
| NC | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
IGFBP7-AS1, insulin-like growth factor binding protein 7-antisense 1; NC, negative control.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted using RNAiso Plus® (Takara Bio, Inc.) according to the manufacturer's protocol. Total RNA (1 µg) was used to synthesize cDNA using the PrimeScript® RT reagent kit with gDNA Eraser (both from Takara Bio, Inc.) according to the manufacturer's protocol. Then, 1 µl cDNA was used to determine the quantification cycle (Cq) value of each sample using the SYBR® Premix Ex Taq II kit (Takara Bio, Inc.) in a LightCycler 480 system (Roche Applied Science). GAPDH served as the reference gene. Relative gene expression was calculated using the 2−ΔΔCq method (22). The PCR primer sequences are in Table II.
Table II.
Primer sequences of IGFBP7-AS1 and the reference gene GAPDH.
| Gene symbol | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| IGFBP7-AS1 | GGAAAGCTCTTCCTGACCCA | CCTGCTAATCTCAGGCAGCA |
| GAPDH | CCCATCACCATCTTCCAGGAG | GTTGTCATGGATGACCTTGGC |
IGFBP7-AS1, insulin-like growth factor binding protein 7-antisense 1.
Cell viability assay
Cells were seeded into 96-well plates at a density of 1,000-1,500 cells/well, for 1–5 days. The cell viability in each well was assessed at 1, 2, 3, 4 and 5 days using the Cell Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) according to the manufacturer's protocol. Absorbance was measured at 450 nm to calculate the number of viable cells.
Wound healing and Transwell assays
A scratch was made on 80% confluent monolayer cell cultures, and culture medium was replaced with serum-free DMEM. The healing process was monitored for 24 h, and the percentage of wound closure was calculated using ImageJ software (version 1.51; National Institutes of Health).
In total, 5×104 cells in 200 µl serum-free DMEM medium were seeded onto the upper chamber of Transwell apparatus (Costar; Corning, Inc.) with a Matrigel-coated membrane. A total of 600 µl DMEM containing 10% FBS was added to the lower chamber as a chemoattractant. After a 12-h incubation at 37°C in a 5% CO2. atmosphere, the cells that invaded the lower chamber through the membrane were fixed with 4% methanol for 30 min at room temperature, stained with 0.1% crystal violet for 15 min at room temperature and counted under a light microscope (magnification, ×100).
Statistical analysis
SPSS (version 20.0; IBM Corp.) and GraphPad Prism (version 7.0; GraphPad Software Inc.) were used for statistical analysis. ANOVA was performed for multiple comparisons followed by Dunnett's post-hoc test. Data are presented as the mean ± SD, where appropriate. Student's t-test was used to compare two groups. χ2 test was used to compare qualitative variables. Survival-related lncRNAs were screened via univariate Cox regression analysis using Biometric Research Branch-Array Tools version 4.5.0 (National Center Institute, National Institutes of Health) and a permutation test method with 10,000 permutations (23). The selected lncRNAs were analyzed using a multivariate Cox regression analysis in the training set and a risk score formula was established including each identified lncRNA, and weighted using the estimated regression coefficients in the multivariate model (24,25). Survival differences were evaluated by the Kaplan-Meier method, and curves were compared using a log-rank test. Time-dependent receiver operating characteristic (ROC) curves were created with the R-package ‘survivalROC’ and Nearest Neighbor Estimation method (26). P<0.05 was considered to indicate a statistically significant difference.
Results
Four lncRNAs, tumor protein P73-antisense 1 (TP73-AS1), IGFBP7-AS1, PAX interacting protein 1-antisense 2 (PAXIP1-AS2) and long intergenic non-protein coding RNA 672 (LINC00672), are associated with glioblastoma outcomes
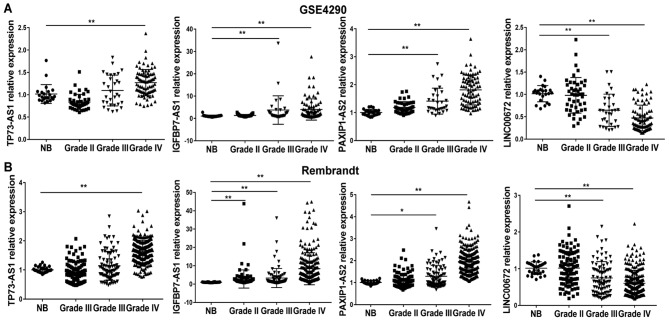
Among the 159 glioblastoma patients in the GSE16011 dataset, 108 patients received post-operative radiation without chemotherapy. These 108 patients were used as a training set to screen survival-associated lncRNAs. A panel of four lncRNAs strongly associated with overall survival was identified via univariate Cox regression analysis, when the permutation was P<0.005, as listed in Table III. In addition, the upregulation of lncRNAs with a positive regression coefficient (TP73-AS1, IGFBP7-AS1 and PAXIP1-AS2), and the downregulation of lncRNAs with a negative coefficient (LINC00672) were observed in glioblastoma tissues compared with in normal brain tissues in the GSE4290 and Rembrandt datasets (Fig. 1).
Table III.
Long noncoding RNAs associated with overall survival of 108 cases of glioblastoma in the training set.
| Gene symbol | Permutation P-value | Hazard ratio | Coefficient | Ensembl ID | Entrez Gene ID | RefSeq ID |
|---|---|---|---|---|---|---|
| TP73-AS1 | 0.001 | 3.229 | 1.172 | ENSG00000227372 | 57212 | NR_033708/ |
| NR_033709/ | ||||||
| NR_033710/ | ||||||
| NR_033711/ | ||||||
| NR_033712 | ||||||
| IGFBP7-AS1 | <0.001 | 1.447 | 0.369 | ENSG00000245067 | 255130 | NR_034081 |
| PAXIP1-AS2 | <0.001 | 2.537 | 0.931 | ENSG00000214106 | 100132707 | NR_024476/ |
| NR_024477 | ||||||
| LINC00672 | 0.003 | 0.605 | −0.502 | ENSG00000263874 | 100505576 | NR_038847 |
IGFBP7-AS1, insulin-like growth factor binding protein 7-antisense 1; LINC00672, long intergenic non-protein coding RNA 672; PAXIP1-AS2, PAX interacting protein 1-antisense 2; TP73-AS1, tumor protein P73-antisense 1.
Figure 1.
Expression levels of four lncRNAs in glioma samples from two different datasets. The expression of TP73-AS1, IGFBP7-AS1, PAXIP1-AS2 and LINC00672 lncRNAs across glioma of different malignant grades relative to NB in the (A) GSE4290 and (B) Rembrandt datasets is presented. Each bar represents the mean ± SD. *P<0.05 and **P<0.01. IGFBP7-AS1, insulin-like growth factor binding protein 7-antisense 1; LINC00672, long intergenic non-protein coding RNA 672; lncRNA, long noncoding RNA; NB, normal brain; PAXIP1-AS2, PAX interacting protein 1-antisense 2; TP73-AS1, tumor protein P73-antisense 1.
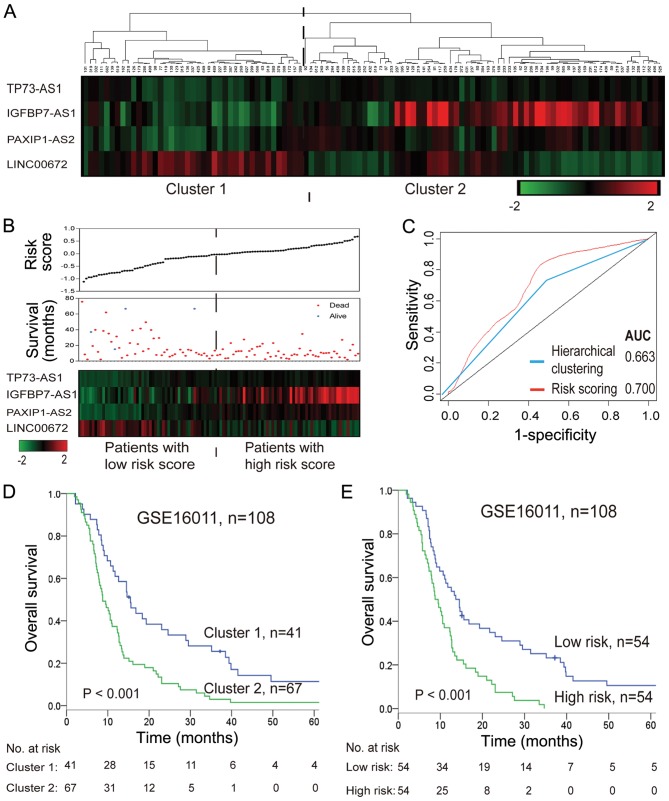
As shown in Fig. 2A and D, hierarchical clustering with these four survival-associated lncRNAs subdivided patients in the training set into two clusters with distinct survival rates (median 8.76 months vs. 15.48 months). The four identified lncRNAs were integrated into a molecular signature using a risk score formula to weigh their prognostic effects. The risk score was calculated as follows: Risk score = (0.397× expression value of TP73-AS1) + (0.191× expression value of IGFBP7-AS1) + (0.468× expression value of PAXIP1-AS2) + (−0.129× expression value of LINC00672). The risk score of each patient was calculated, and then patients were divided into a high-risk or low-risk group according to the median risk score (−0.0254) of the training set. Patients in the high-risk group exhibited significantly shorter survival times than those in the low-risk group (median 9.48 months vs. 13.68 months; log-rank P<0.001; Fig. 2B and E). The correlation of the risk score with overall survival was also significant when assessed in the univariate model (Table IV). A larger area under the curve in time-dependent ROC analysis was achieved using the risk scoring method, illustrating that this constituted a better algorithm for predicting outcomes, with good sensitivity and specificity compared with hierarchical clustering (Fig. 2C).
Figure 2.
Analysis of the four lncRNAs associated with overall survival of the 108 patients with glioblastoma in the training set. (A) Hierarchical clustering analysis for patients based on expressions of lncRNAs. (B) Distribution of risk score, survival status and lncRNA expression. (C) Time-independent receiver operating characteristic curves for risk scoring and hierarchical clustering method to predict overall survival. (D) Kaplan-Meier survival curves for patients divided by hierarchical clustering. (E) Kaplan-Meier survival curves for patients divided by risk score. AUC, area under the curve; LINC00672, long intergenic non-protein coding RNA 672; IGFBP7-AS1, insulin-like growth factor binding protein 7-antisense 1; lncRNA, long noncoding RNA; PAXIP1-AS2, PAX interacting protein 1-antisense 2; TP73-AS1, tumor protein P73-antisense 1.
Table IV.
Univariate and multivariate Cox regression analyses for overall survival of patients with glioblastoma in public datasets.
| A, GSE16011 (training set, n=108) | ||||||
|---|---|---|---|---|---|---|
| Univariate model | Multivariate model | |||||
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| lncRNA-risk score | 2.716 | 1.712–4.308 | <0.001a | 1.906 | 1.124–3.230 | 0.017a |
| Sex | 0.856 | 0.565–1.298 | 0.465 | 0.841 | 0.552–1.282 | 0.421 |
| Age | 1.033 | 1.016–1.051 | <0.001a | 1.019 | 0.999–1.039 | 0.057 |
| KPS | 0.980 | 0.967–0.994 | 0.004a | 0.984 | 0.970–0.998 | 0.029a |
| B, Entire independent dataset (n=147) | ||||||
| Univariate model | Multivariate model | |||||
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| lncRNA-risk score | 2.560 | 1.586–4.133 | <0.001a | 2.450 | 1.539–3.901 | <0.001a |
| Sex | 0.893 | 0.610–1.307 | 0.560 | 1.030 | 1.011–1.050 | 0.658 |
| Age | 1.029 | 1.011–1.048 | 0.002a | 0.917 | 0.625–1.346 | 0.002a |
| C, GSE7696 dataset (n=69) | ||||||
| Univariate model | Multivariate model | |||||
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| lncRNA-risk score | 3.709 | 1.463–9.403 | 0.006a | 2.600 | 0.899–7.520 | 0.042a |
| Sex | 0.887 | 0.491–1.602 | 0.690 | 0.540 | 0.457–1.507 | 0.540 |
| Age | 1.041 | 1.008–1.075 | 0.015a | 1.026 | 0.989–1.063 | 0.167 |
| D, Rembrandt dataset (n=78) | ||||||
| Univariate model | Multivariate model | |||||
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| lncRNA-risk score | 2.197 | 1.216–3.969 | 0.009a | 2.451 | 1.335–4.501 | 0.004a |
| Sex | 0.950 | 0.575–1.571 | 0.842 | 1.189 | 0.706–2.002 | 0.516 |
| Age | 1.031 | 1.007–1.055 | 0.011a | 1.028 | 1.003–1.053 | 0.027a |
| KPS | 0.994 | 0.988–1.000 | 0.060 | 0.993 | 0.987–1.000 | 0.043a |
P<0.05. KPS, Karnofsky performance status; CI, confidence interval; HR, hazard ratio.
Further validation of the four-lncRNA signature in two independent datasets
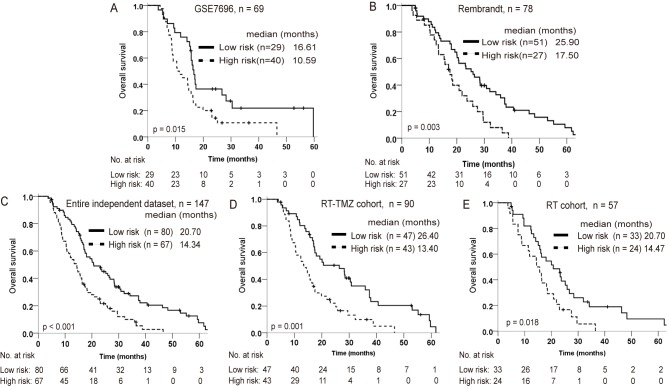
From the GSE7696 and Rembrandt datasets, 69 and 78 patients with glioblastoma, who had received postoperative radiation, respectively, were considered as independent datasets for validating the prognostic value of the lncRNA signature. Of these, a total of 57 patients accepted radiotherapy only (RT cohort), and 90 patients accepted radiotherapy combined with TMZ chemotherapy (RT-TMZ cohort). Using the same cutoff point for the lncRNA-risk score as for the training set, patients in the GSE7696 and Rembrandt datasets could be also divided into distinct survival subgroups (Fig. 3A and B). Risk score-based subclassification of the entire independent cohort (both the GSE7697 and Rembrandt patients) had similar results, whether the patients accepted TMZ treatment or not. Furthermore, for patients with a high-risk score, radiation combined with TMZ did not appear to significantly prolong the survival time relative to radiotherapy alone (Fig. 3C-E).
Figure 3.
Prognostic value of the four-lncRNA signature in independent datasets of 148 glioblastoma cases. Kaplan-Meier survival analysis for patients in (A) GSE7696, (B) Rembrandt and (C) the entire independent dataset. The prognostic value of these lncRNAs was then evaluated in a subset of patients that received (D) RT-TMZ combined treatment and (E) RT alone. lncRNA, long noncoding RNA; RT, radiotherapy; TMZ, temozolomide.
Prognostic value of the four-lncRNA signature is independent of conventional clinicopathological factors
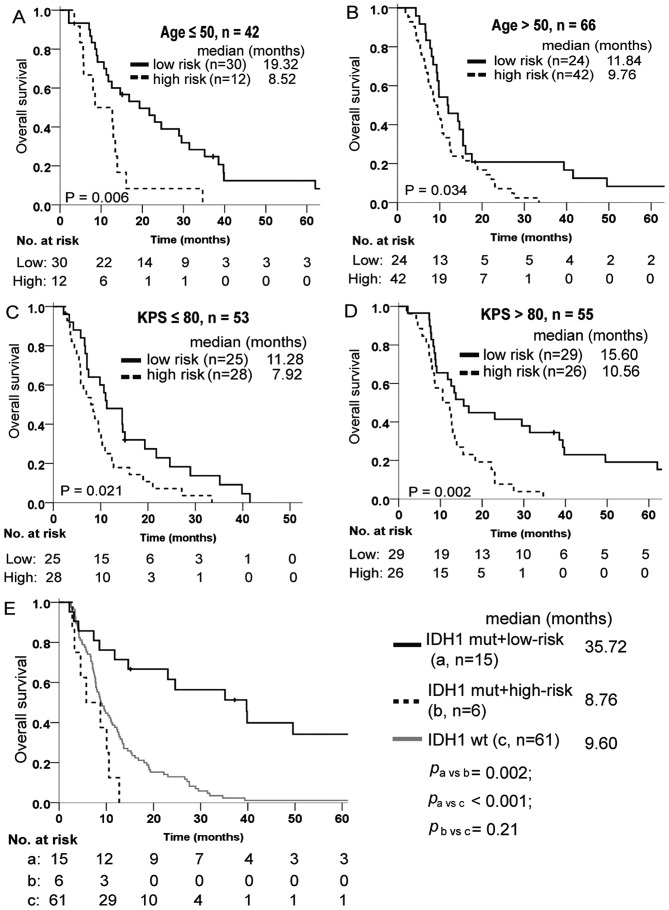
Multivariate Cox regression analysis was adopted to estimate whether the prognostic value of the lncRNA signature was independent of the clinical characteristics of patients with glioblastoma. As shown in Table IV, the lncRNA-risk score remained significantly associated with overall survival when adjusted by age and KPS, when available, in every cohort. Data stratification analysis was also performed to assess the prognostic value of this signature with respect to the same clinical factors. For this, patients in the training set were first stratified into a younger group (age ≤50) and an older group (age >50), then into a poor performance group (KPS ≤80) and a good performance group (KPS >80). The results indicated that the lncRNA signature remained a useful prognostic indicator within each age and KPS stratum (Fig. 4A-D).
Figure 4.
Stratification analysis for the four-lncRNA signature in the training set of glioblastoma cases. (A) Kaplan-Meier survival analysis for younger patients (age ≤50) and (B) older patients (age >50), as well as (C) poor performance patients (KPS ≤80) and (D) good performance patients (KPS >80). (E) Kaplan-Meier survival analysis for patients with IDH1 mutation subdivided into (a) low-risk and (b) high-risk compared with (c) patients with wt IDH1. lncRNA, long noncoding RNA; IDH1, isocitrate dehydrogenase 1; wt, wild type.
The isocitrate dehydrogenase 1 (IDH1) mutation is one of the most valuable biomarkers for glioma diagnosis and prognosis. In the GSE16011 dataset, the lncRNA signature could predict different outcomes of glioblastoma carrying the IDH1 mutation. Patients exhibiting mutations in IDH1 gene and high-risk signature exhibited a poor survival, similarly to those with wild-type IDH1 (Fig. 4E).
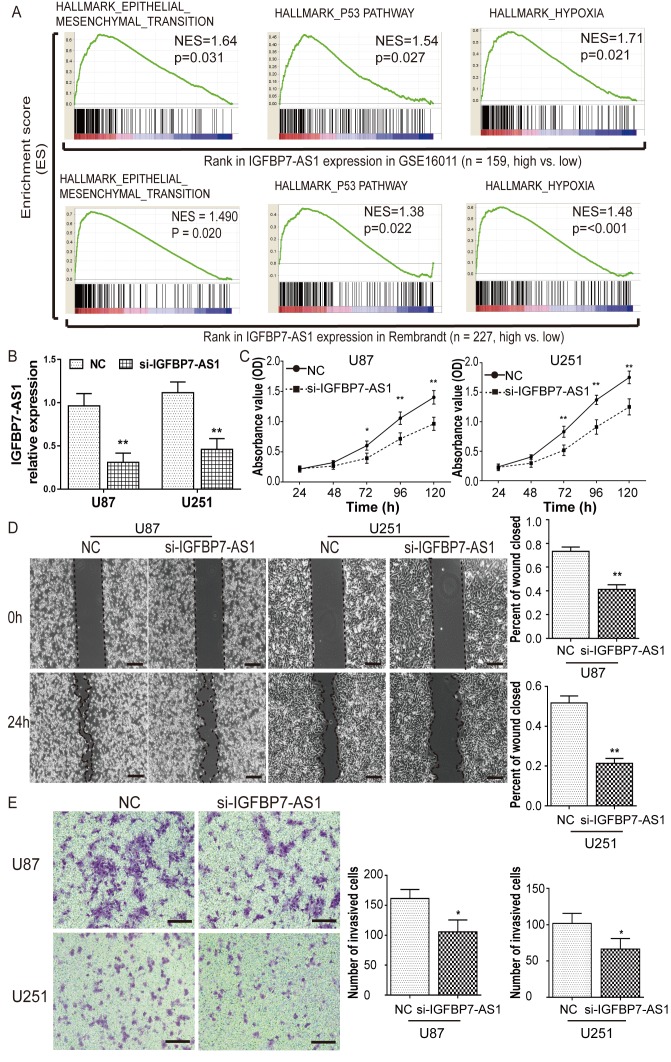
Knockdown of IGFBP7-AS1 inhibits the viability and invasion of U87 and U251 glioma cells
As the oncogenic function of TP73-AS1 in glioma has been reported in recent studies (27–29), GSEA was performed to identify the biological processes and pathways of IGFBP7-AS1, PAXIP1-AS2 and LINC00672 lncRNAs involved in glioma pathogenesis by dividing glioblastoma cases from the GSE16011 and Rembrandt datasets into high- and low-expression groups according to the median lncRNA expression. The results demonstrated that the ‘epithelial-mesenchymal transition’, ‘p53 pathway’ and ‘hypoxia’ gene sets were significantly enriched in glioblastoma samples with high IGFBP7-AS1 expression relative to those with low expression (Fig. 5A). IGFBP7-AS1 expression was also the most upregulated of the four prognostic lncRNAs in glioblastoma compared with NB (Fig. 1). Therefore, the role of IGFBP7-AS1 with regards to glioma viability and invasion was further assessed following the transfection of U87 and U251 glioma cell lines with a siRNA targeting IGFBP7-AS1 (Fig. 5B). IGFBP7-AS1 siRNA markedly reduced U87 and U251 cell viability compared with in NC-transfected cells, as determined using a CCK-8 assay (Fig. 5C). After knockdown of IGFBP7-AS1, a wound-healing assay showed that the wound closure percentage was visibly reduced, and a Transwell assay revealed that the number of cells that invaded through the Matrigel matrix was significantly decreased (Fig. 5D and E). These results further demonstrated that IGFBP7-AS1 may serve a role in the regulation of glioma cell migration and invasion.
Figure 5.
Knockdown of IGFBP7-AS1 reduces the viability and invasion of U87 and U251 glioma cells. (A) Gene set enrichment analysis of IGFBP7-AS1 in glioblastoma samples from GSE16011 and Rembrandt datasets. (B) Interference efficiency of IGFBP7-AS1 siRNA in glioma cells analyzed via reverse transcription-quantitative PCR. (C) Cell viability of glioma cell lines following transfection with siRNA or NC measured by Cell Counting kit-8 assay. (D) Migration of glioma cells following transfection with siRNA or NC measured by wound-healing assay. Scale bar, 200 µm. (E) Invasion of glioma cells following transfection with siRNA or NC detected by Transwell assay. Scale bar, 100 µm. Each bar represents the mean ± SD of three independent experiments. *P<0.05 and **P<0.01. IGFBP7-AS1, insulin-like growth factor binding protein 7-antisense 1; NC, negative control; NES, Normalized enrichment score; OD, optical density; siRNA, small interfering RNA.
Discussion
lncRNAs are a new class of noncoding gene regulators involved in cancer pathogenesis and prognosis (6,7). Although analyses of global expression patterns and functional characterization of lncRNAs have been performed, the molecular mechanisms and clinical applications of lncRNAs in the context of glioblastoma remain unclear (9–12). Previous studies demonstrated that lncRNA expression profiles can be obtained by mining existing microarray data, as thousands of probes on commonly used arrays are likely to be repurposed as lncRNAs (9,30). Moreover, the number of probes available to map lncRNAs is increasing rapidly due to the updating of lncRNA databases, such as GENCODE, in order to provide a more comprehensive documentation of sequence information, functional annotation and expression profiles (31). By combining the microarray mining method and updated gene annotations, the present study identified 1,895 lncRNAs on the Affymetrix platform. To the best of our knowledge, more than half of these genes were not previously annotated as lncRNAs; therefore, the present study provided novel information for investigating lncRNA profiles in cancer, and a four-lncRNA signature was identified with potential for predicting outcomes in glioblastoma.
The current standard treatment for glioblastoma is maximal safe resection followed by radiotherapy and TMZ chemotherapy. Postoperative radio-chemotherapy confers better prognosis than surgery alone, or than radiation without chemotherapy (2). To avoid interference from different treatment regimens, only the patients who had received postoperative radiotherapy were chosen for survival analysis in the training set. The results revealed that four lncRNAs were significantly associated with overall survival in 108 cases of glioblastoma. Upregulation of TP73-AS1, IGFBP7-AS1 and PAXIP1-AS2, with a positive regression coefficient, and downregulation of LINC00672, with a negative coefficient, was observed in glioblastoma samples relative to normal brain tissue. The expression trends were also associated with the malignancy grades of glioma, indicating their potential use in glioma diagnosis and prognosis.
Based on the expression levels of the four lncRNAs, patients with glioblastoma in the training set could be divided into two groups with distinct survival by both hierarchical clustering and a risk scoring method. Risk scoring is a widely used and effective method to establish a multi-gene expression-based molecular signature to predict outcomes of various types of cancer (24,25). By performing ROC analysis, risk scoring was determined as a better estimator of overall survival, with better sensitivity and specificity compared with hierarchical clustering. Overall, patients in the training set with high-risk lncRNA signatures exhibited poorer survival. The reproducibility of this signature was validated in additional independent datasets, including patients who received radiotherapy alone, and those treated with radiation combined with TMZ. The results indicated that the prognostic value of the lncRNA signature in cases of glioblastoma was not restricted by different treatment options. In addition, the prognostic value of this signature was independent of well-known clinical prognostic factors, including age and KPS, further attesting to its potential usefulness in clinical practice. It is worth noting that, compared with radiotherapy alone, postoperative radiation combined with TMZ may have prolonged the survival of patients with glioblastoma, who exhibited a low-risk lncRNA signature. For high-risk patients, TMZ failed to improve prognosis. This observation implies that the roles of these lncRNAs in glioma progression may be associated with treatment resistance.
IDH1 and IDH2 mutations were previously identified as key biomarkers for molecular pathology and outcome prediction in glioblastoma. The R132H mutation in the IDH1 gene is the most frequent type of IDH mutation (14,32). Mutations in IDH mainly occur in lower grade glioma and recurrent glioblastoma, which have improved survival rates compared with primary glioblastoma. In primary glioblastoma, patients exhibiting IDH mutations also possess better survival rates than patients without mutation (32). IDH mutation can induce global DNA hypermethylation and deregulate genes, including lncRNAs, inhibiting cellular differentiation, which is involve in tumor initiation and progression (32,33). These genetic findings promoted the emergence of therapeutic approaches aimed to target the mutant form of IDH in glioma and other types of cancer (32). In the GSE16011 dataset, the expression levels of the four prognostic lncRNAs were significantly different between glioblastoma cases with and without the IDH1 mutation. A correlation between these lncRNA levels and the IDH mutation has also been reported in a RNA-sequencing report (10). Furthermore, in the present study, IDH1 mutant glioblastoma with a high-risk lncRNA signature exhibited poor survival, which was comparable with IDH wild type glioblastoma. This may have clinical implications for identifying at-risk patients, among those with IDH mutant glioblastoma, who may therefore require more intensive treatment, such as IDH-targeted therapy in clinical trials (32). These results also indicated the possibility that these lncRNAs are involved in IDH mutation-associated glioma tumorigenesis. Further studies are needed to elucidate the relationship between the IDH mutation and lncRNA deregulation.
The functions of lncRNAs are closely associated with their expression level, as they do not encode proteins (30). The expression of TP73-AS1 is increased in a variety of malignancies, including hepatocellular carcinoma, breast cancer and glioblastoma, and the upregulation of TP73-AS1 also predicts the poor prognosis of cancer (28,34,35). TP73-AS1 may combine with miRNAs to regulate glioma growth, apoptosis and invasion (27–29). To the best of our knowledge, the remaining three lncRNAs were identified as survival predictors in glioma for the first time in the present study. However, in endometrial cancer, LINC00672 has been reported to be downregulated and involved in p53-mediated gene suppression and malignant progression (36). With respect to IGFBP7-AS1 and PAXIP1-AS2, no functional studies on cancer have been reported so far, to the best of our knowledge. IGFBP7-AS1 and PAXIP1-AS2 are the antisense transcripts of IGFBP7 and PAXIP1 genes, respectively. IGFBP7 has been reported to inhibit insulin-like growth factor signaling, and induce senescence and apoptosis in hepatocellular carcinoma (37), whereas PAXIP1 has been reported to participate in DNA damage repair and chemosensitivity in lung cancer (38). It is worth noting that antisense lncRNAs could affect cellular homeostasis by interacting with their positive sense gene, and regulating their expression and function. For instance, the natural antisense transcript of zinc finger E-box binding homeobox 2 (ZEB2) interacts with ZEB2 mRNA to modulate alternative splicing, enhance protein translation and promote tumor invasion (39). However, their exact functions in glioma are still unclear. GSEA was thus performed, and it revealed that ‘p53 pathway’ and ‘epithelial-mesenchymal transition’ were enriched in glioblastoma cases with higher IGFBP7-AS1 expression. p53 is a vital gene, involved in regulation of the cell cycle and tumor growth (40). Epithelial-mesenchymal transition (EMT) is a complex cellular process contributing to the switch of epithelial cells into motile mesenchymal cells, which promotes the invasion and metastasis of epithelial tumors (41). Notably, reports have described an EMT-like phenomenon in glioma, and its association with tumor invasiveness and poorer prognosis (41,42). The present results revealed that IGFBP7-AS1 may affect glioma cell survival by regulating tumor growth and migration. Moreover, IGFBP7-AS1 expression was the most upregulated of the four prognostic lncRNAs in the glioblastoma cases analyzed, which indicated that IGFBP7-AS1 may be comparatively more important and more deserving of research focus. Further functional experiments demonstrated that knockdown of IGFBP7-AS1 reduced the viability, migration and invasion of U87 and U251 glioma cells, providing a mechanistic explanation for the prognostic ability of IGFBP7-AS1. IGFBP7-AS1 may therefore constitute a novel target for future molecular therapy development.
In conclusion, a combination of microarray mining and gene re-annotation may be considered an effective method to investigate the expression profiles of lncRNAs in cancer. A novel four-lncRNA signature was identified as a composite biomarker for glioblastoma outcome prediction. The role of IGFBP7-AS1 in influencing glioma cell viability, migration and invasion was demonstrated with further in vitro experiments. The present study provided a feasible method and novel information for understanding the functions of lncRNAs in glioma pathogenesis and prognosis.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by The National Natural Science Foundation of China (grant no. 81372692) and National Key Clinical Specialist Construction Program of China.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
DL and LY designed this study and wrote the manuscript. DL analyzed and interpreted the datasets. JL and HL performed the cell experiments. SQ analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the united states in 2009–2013. Neuro Oncol. 2016;18(Suppl 5):v1–v75. doi: 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas-implications for classification and therapy. Nat Rev Clin Oncol. 2017;14:434–452. doi: 10.1038/nrclinonc.2016.204. [DOI] [PubMed] [Google Scholar]
- 4.Gravendeel LA, Kouwenhoven MC, Gevaert O, de Rooi JJ, Stubbs AP, Duijm JE, Daemen A, Bleeker FE, Bralten LB, Kloosterhof NK, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 5.Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 8.Hessels D, Schalken JA. The use of PCA3 in the diagnosis of prostate cancer. Nat Rev Urol. 2009;6:255–261. doi: 10.1038/nrurol.2009.40. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, Lui WM, Wong ST, Leung GK. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Reon BJ, Anaya J, Zhang Y, Mandell J, Purow B, Abounader R, Dutta A. Expression of lncRNAs in low-grade gliomas and glioblastoma multiforme: An in silico analysis. PLoS Med. 2016;13:e1002192. doi: 10.1371/journal.pmed.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XQ, Leung GK. Long non-coding RNAs in glioma: Functional roles and clinical perspectives. Neurochem Int. 2014;77:78–85. doi: 10.1016/j.neuint.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Kiang KM, Zhang GP, Leung GK. Long non-coding RNAs dysregulation and function in glioblastoma stem cells. Noncoding RNA. 2015;1:69–86. doi: 10.3390/ncrna1010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: Helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7:157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 15.Sun LX, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, et al. Stem cell-related ‘self-renewal’ signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 17.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 18.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–17. doi: 10.1177/117693510700300022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, Levy R. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 25.Zhang XQ, Sun S, Lam KF, Kiang KM, Pu JK, Ho AS, Lui WM, Fung CF, Wong TS, Leung GK. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol Dis. 2013;58:123–131. doi: 10.1016/j.nbd.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 26.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341X.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 27.Xiao S, Wang R, Wu X, Liu W, Ma S. The long noncoding RNA TP73-AS1 interacted with miR-124 to modulate glioma growth by targeting inhibitor of apoptosis-stimulating protein of p53. DNA Cell Biol. 2018;37:117–125. doi: 10.1089/dna.2017.3941. [DOI] [PubMed] [Google Scholar]
- 28.Zhang R, Jin H, Lou F. The long non-coding RNA TP73-AS1 interacted with miR-142 to modulate brain glioma growth through HMGB1/RAGE pathway. J Cell Biochem. 2018;119:3007–3016. doi: 10.1002/jcb.26021. [DOI] [PubMed] [Google Scholar]
- 29.Wang JB, Liu FH, Chen JH, Ge HT, Mu LY, Bao HB, Lin ZG. Identifying survival-associated modules from the dysregulated triplet network in glioblastoma multiforme. J Cancer Res Clin Oncol. 2017;143:661–671. doi: 10.1007/s00432-016-2332-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagarde J, Uszczynska-Ratajczak B, Carbonell S, Pérez-Lluch S, Abad A, Davis C, Gingeras TR, Frankish A, Harrow J, Guigo R, Johnson R. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat Genet. 2017;49:1731–1740. doi: 10.1038/ng.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol. 2016;27:599–608. doi: 10.1093/annonc/mdw013. [DOI] [PubMed] [Google Scholar]
- 33.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Huang Y, Huang Y, Fu Y, Tang D, Kang R, Zhou R, Fan XG. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 2017;36:51. doi: 10.1186/s13046-017-0519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao J, Xu F, Zhang D, Yi W, Chen X, Chen G, Zhou E. TP73-AS1 promotes breast cancer cell proliferation through miR-200a-mediated TFAM inhibition. J Cell Biochem. 2018;119:680–690. doi: 10.1002/jcb.26231. [DOI] [PubMed] [Google Scholar]
- 36.Li W, Li H, Zhang L, Hu M, Li F, Deng J, An M, Wu S, Ma R, Lu J, Zhou Y. Long non-coding RNA LINC00672 contributes to p53 protein-mediated gene suppression and promotes endometrial cancer chemosensitivity. J Biol Chem. 2017;292:5801–5813. doi: 10.1074/jbc.M116.758508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akiel M, Guo C, Li X, Rajasekaran D, Mendoza RG, Robertson CL, Jariwala N, Yuan F, Subler MA, Windle J, et al. IGFBP7 deletion promotes hepatocellular carcinoma. Cancer Res. 2017;77:4014–4025. doi: 10.1158/0008-5472.CAN-16-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jhuraney A, Woods NT, Wright G, Rix L, Kinose F, Kroeger JL, Remily-Wood E, Cress WD, Koomen JM, Brantley SG, et al. PAXIP1 potentiates the combination of WEE1 inhibitor AZD1775 and platinum agents in lung cancer. Mol Cancer Ther. 2016;15:1669–1681. doi: 10.1158/1535-7163.MCT-15-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beltran M, Puig I, Peña C, García JM, Alvarez AB, Peña R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki K, Matsubara H. Recent advances in p53 research and cancer treatment. J Biomed Biotechnol. 2011;2011:978312. doi: 10.1155/2011/978312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahlert UD, Nikkhah G, Maciaczyk J. Epithelial-to-mesenchymal(-like) transition as a relevant molecular event in malignant gliomas. Cancer Lett. 2013;331:131–138. doi: 10.1016/j.canlet.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 42.Pećina-Šlaus N, Kafka A, Varošanec AM, Marković L, Krsnik Ž, Njirić N, Mrak G. Expression patterns of Wnt signaling component, secreted frizzled-related protein 3 in astrocytoma and glioblastoma. Mol Med Rep. 2016;13:4245–4251. doi: 10.3892/mmr.2016.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.