Summary
Emerging evidence suggests that Parkinson's disease (PD), besides being an age-associated disorder, might also have a neurodevelopment component. Disruption of mitochondrial homeostasis has been highlighted as a crucial cofactor in its etiology. Here, we show that PD patient-specific human neuroepithelial stem cells (NESCs), carrying the LRRK2-G2019S mutation, recapitulate key mitochondrial defects previously described only in differentiated dopaminergic neurons. By combining high-content imaging approaches, 3D image analysis, and functional mitochondrial readouts we show that LRRK2-G2019S mutation causes aberrations in mitochondrial morphology and functionality compared with isogenic controls. LRRK2-G2019S NESCs display an increased number of mitochondria compared with isogenic control lines. However, these mitochondria are more fragmented and exhibit decreased membrane potential. Functional alterations in LRRK2-G2019S cultures are also accompanied by a reduced mitophagic clearance via lysosomes. These findings support the hypothesis that preceding mitochondrial developmental defects contribute to the manifestation of the PD pathology later in life.
Keywords: Parkinson's disease, LRRK2, neurodevelopment, stem cells, mitochondria, autophagy, mitophagy
Highlights
-
•
Mitochondrial gene expression is altered in NESCs carrying the LRRK2-G2019 mutation
-
•
LRRK2-G2019S mutation induces alterations in mitochondrial morphology in NESCs
-
•
Mitophagy is affected in PD-specific NESCs carrying the LRRK2-G2019S mutation
-
•
Mitochondrial phenotypes in NESC are rescued by genetic correction of LRRK2-G2019S
Walter, Bolognin and colleagues show the detection of mitochondrial phenotypes in NESCs derived from Parkinson's disease (PD) patients carrying the LRRK2-G2019S mutation. This supports the use of stem cells as a relevant model to study PD-associated mitochondrial defects associated to PD.
Introduction
Parkinson's disease (PD) is a neurodegenerative disorder affecting 1% of the population more than 60 years old (Tysnes and Storstein, 2017). The key histopathological hallmark of the disease is the degeneration of dopaminergic neurons in the substantia nigra of the midbrain. The cause of the disease is still unknown. However, a complex interplay between genetic and environmental factors might contribute to its pathogenesis (for review see Burbulla and Kruger, 2011). The dysregulation of mitochondrial function has been highlighted as a crucial player, because complex I activity was shown to be reduced in PD patients (Schapira et al., 1990). More recently, various PD-specific cellular models revealed impaired mitochondrial morphology and transport, increased release of reactive oxygen species (ROS), and reduced mitochondrial motility compared with controls (Hsieh et al., 2016, Smith et al., 2016, Trimmer et al., 2000). Dopaminergic neurons in the substantia nigra have higher energy demands compared with other types of neurons because of the production of the neurotransmitter dopamine, and the particularly long, highly branched, and unmyelinated axons (Pissadaki and Bolam, 2013). Therefore, they are particularly susceptible to mitochondrial dysfunctions compared with other types of neurons (for review see Haddad and Nakamura, 2015). These mitochondrial dysfunctions might be caused by genetic alterations and/or environmental impacts. Several genetic variants are associated with the risk of developing PD (Klein and Westenberger, 2012). Interestingly, mutations in the gene encoding leucine-rich repeat serine/threonine-protein kinase 2 (LRRK2) have been associated with both familial and sporadic PD (Funayama et al., 2002, Lesage et al., 2007, Ozelius et al., 2006). The autosomal-dominant genomic mutation (c.6055 G>A), which results in a LRRK2-p.G2019S (G2019S) substitution, is the most prevalent genetic risk factor for PD (Funayama et al., 2002, Paisan-Ruiz et al., 2004). LRRK2 cellular functions have not yet been fully defined. However, LRRK2 is thought to contribute to autophagy and mitochondrial regulation (Roosen and Cookson, 2016), microtubule dynamics (Krumova et al., 2015), microRNA activity regulation (Gonzalez-Cano et al., 2018), and vesicular trafficking (Cookson, 2016). It is located in the cytoplasm and associated with membranes (Vitte et al., 2010, West et al., 2005). Recently, LRRK2-G2019S has proven to be associated with autophagosomal-endosomal-lysosomal pathways related to mitochondrial health and biogenesis (Roosen and Cookson, 2016, Wallings et al., 2015).
Neural progenitor stem cells have been highlighted as a meaningful model for mitochondria-related disease phenotyping (Lorenz et al., 2017). Here, we use PD patient-specific induced pluripotent stem cell (iPSC)-derived neuroepithelial stem cells (NESCs) to assess mitochondrial features. NESCs are an easily accessible and highly homogeneous model representing early brain development. We observed that mitochondria in NESCs carrying the LRRK2-G2019S mutation were more fragmented and more prone to release ROS compared with isogenic controls. Also, the functionality of mitochondria was impaired in patient-specific stem cells as shown by the decreased membrane potential and respiratory capacity.
Results
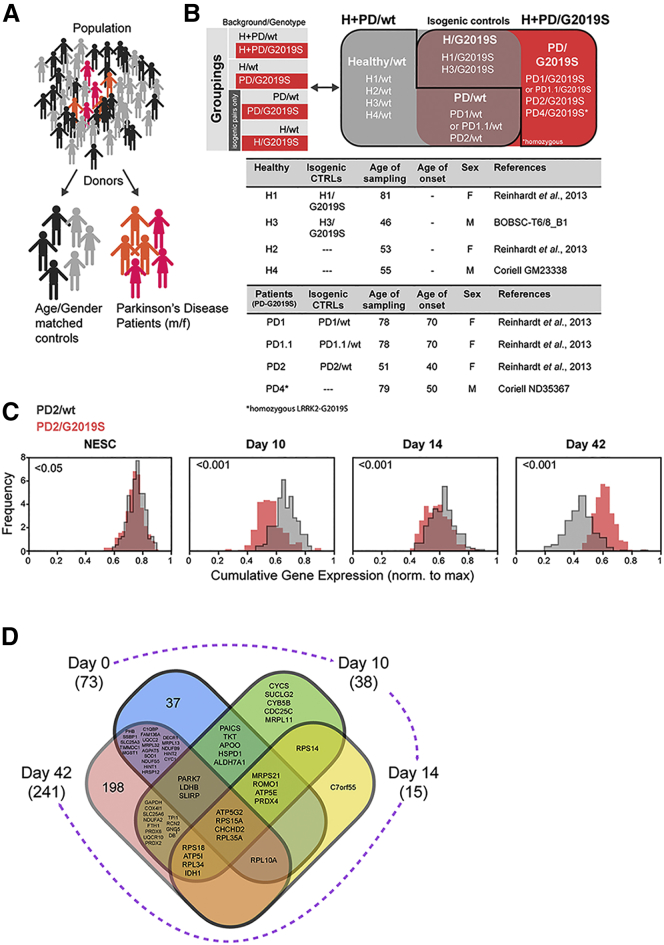
In our study, we used 13 human iPSC-derived NESC lines obtained from three patients carrying the LRRK2-G2019S mutation and four age- and gender-matched healthy individuals. Four isogenic NESC lines (two in which the mutation was introduced and two in which the mutation was corrected) were also generated (Figures 1A and 1B). For patient 1 two different clones were used (PD.1 and PD1.1). The use of isogenic controls enabled us to distinguish between LRRK2-G2019S-dependent and -independent phenotypes. The derivation and characterization of NESCs from iPSCs have been described previously (Reinhardt et al., 2013).
Figure 1.
LRRK2-G2019S Alters Mitochondrial Gene Expression Profile
(A and B) Cartoon and tables illustrating the cell lines used and the grouping applied.
(C) Histograms showing cumulative gene expression distributions for mitochondrial-specific genes in NESCs and during dopaminergic differentiation (10, 14, and 42 days). For each cell, a score called cumulative gene expression was defined as the sum of the gene expression for all the genes associated to mitochondria (details are provided in the Experimental Procedures). These are the genes present in the targeted list of genes specific for mitochondria (Table S1), generated based on the available literature (Calvo et al., 2016). Cells having a higher cumulative gene expression are cells for which the expression of mitochondrial-specific genes is high. For each condition (PD2/WT, PD2/G2019S N = 1, n = 3) and for each day, 250 cells were considered, for a total of 2,000 cells. The statistical significance of the difference in population average between the two genotypes is indicated in each panel and was obtained by means of a z test including Bonferroni correction for multiple hypothesis testing. The frequency reported on the vertical axes represents the number of cells in each bin of the histogram, normalized in a way that each distribution has a surface equal to 1. N indicates the number of experimental repetitions, n indicates the number of technical replicates per cell line.
(D) Venn diagram representing the differentially expressed genes (DEGs) specific for mitochondria. Each ensemble contains the DEGs at a given time point. The total number of mitochondrial DEGs for the corresponding time point is indicated in brackets. The numbers indicate how many DEGs fall into the area of the Venn diagram. Where graphically possible, we report the names of the genes, instead of their number. The names of the genes common to all four time points are reported in bold in the center of the diagram. By definition, the sum of the numbers of genes appearing into each of the four ensembles is equal to the total number of DEGs for that day, reported in brackets.
Throughout the article, unless otherwise stated, we indicate whether the donor was healthy (H) or affected by the pathology (PD). LRRK2 genotype is annotated according to the absence (WT) or presence of the LRRK2-G2019S mutation (G2019S). Cells from a healthy individual are indicated as H/WT, and after the introduction of LRRK2-G2019S as H/G2019S (isogenic control). Cells from a PD patient with the LRRK2-G2019S mutation are annotated as PD/G2019S, and after genetic correction of LRRK2-G2019S as PD/WT. The use of these groupings allowed us to distinguish between effects due to LRRK2-G2019S mutation and effects that depend on the individual's genetic background. We first compared all NESC lines only according to their LRRK2 genotype (H + PD/G2019S versus H + PD/WT). Then, we compared cells from LRRK2-G2019S patients with cells from age- and gender-matched controls (PD/G2019S versus H/WT); third, we compared samples from PD patients with their isogenic controls (PD/G2019S versus PD/WT). Finally, we investigated the impact of introducing the LRRK2-G2019S mutation in a healthy genetic background (H/WT versus H/G2019S).
LRRK2-G2019S Induces Mitochondrial Gene Expression Alterations
Mitochondrial dysfunctions have been previously associated to LRRK2-G2019S in iPSC-derived neurons (Burbulla and Kruger, 2011). To determine whether LRRK2-G2019S mutation affected mitochondrial-related genes also at the NESC level or only after dopaminergic differentiation, we performed droplet-based (Drop-seq) single-cell RNA sequencing (RNA-seq). Here, we focused on the cells from an early-onset patient (PD2/G2019S) and the corresponding isogenic control cells (PD2/WT). We first generated a targeted list specific for mitochondrial genes (Table S1) based on an inventory of human genes encoding mitochondrial-localized proteins (www.broadinstitute.org/pubs/MitoCarta) (Calvo et al., 2016). We profiled 2,000 quality-controlled cells and, after in silico pre-processing of the data, we calculated cumulative gene expression scores for the mitochondrial-based defined gene list (details are provided in the Experimental Procedures section). The analysis of the cumulative gene expression distribution (Figure 1C) showed significant gene expression differences between the genotypes at the different neuronal differentiation time points assessed (10, 14, and 42 days). Interestingly, a significant difference in mitochondria-related genes was already observed in the NESCs carrying the LRRK2-G2019S compared with the LRRK2-WT, before induction of differentiation. Hence, we decided to focus our analysis on NESCs to better characterize the mitochondrial defects appearing already in this cell type. To gain more insights into the dynamics of the mitochondrial gene expression levels, for each day we computed the differentially expressed genes (DEGs) between LRRK2-WT and LRRK2-G2019S. We observed that, among the total genes (approximately 17,000) in common between all the time points in our dataset, the numbers of DEGs at days 0, 10, 14, and 42 were, respectively, 619, 531, 318, and 1,637 (Table S2). This corresponds to approximately 4%, 3%, 2%, and 10% of the total genes. Since our focus is mitochondria, we considered the DEGs that were present in Table S1. Among these mitochondria-related genes, the number, of those differentially expressed at days 0, 10, 14, and 42 were, respectively 73, 38, 15, and 241. These are equivalent to respectively 6%, 3%, 1%, and 21% of the total number of mitochondrial genes in our list. The change of this percentage across the different days reflects the trend observed in the overall percentage of DEGs across the whole genome, thus overall it is not only a feature of the mitochondria-related genes. On the other hand, the most remarkable difference is in the percentage of DEGs at day 42, which is 10% across the whole genome, but 21% (i.e., more than twice) across the list of mitochondrial genes. This indicates that the expression of mitochondria-related genes is dramatically different between LRRK2-WT and LRRK2-G2019S at day 42. We further investigated whether the genes that are differentially expressed between LRRK2-WT and LRRK2-G2019S are different or similar at different time points. We then considered the list of DEGs among the mitochondria-specific genes at each day, and intersect every possible combination of lists, and count the number of DEGs in the intersection (Figure 1D). The majority of the mitochondria-related genes are differentially expressed only at one time point. However, four genes are differentially expressed at every time point: ATP5G2, RPS15A, CHCHD2, and RPL35A. An additional 12 genes are differentially expressed at 3 different time points. Notably, PARK7 (or DJ1) is differentially expressed between LRRK2-WT and LRRK2-G2019S at days 0, 10, and 42. Interestingly, of the 16 genes that are DEGs at 3 or 4 time points, there are 3 that encode components of ATP synthases (ATP5G2, ATP5I, and ATP5E). Perhaps less surprisingly, among these 16 DEGs at 3 or 4 days, there are 5 genes that correspond to ribosomal proteins, namely RPS15A, RPS18, RPL10A, RPL34, and RPL35A, and a sixth one, RPS14, is a DEG at day 10 and 14. We also notice that, among the DEGs that are common between days 10 and 42, we find GAPDH, which codes for an enzyme that catalyzes the sixth step of glycolysis, and has been found to be implicated in several neurodegenerative diseases including PD.
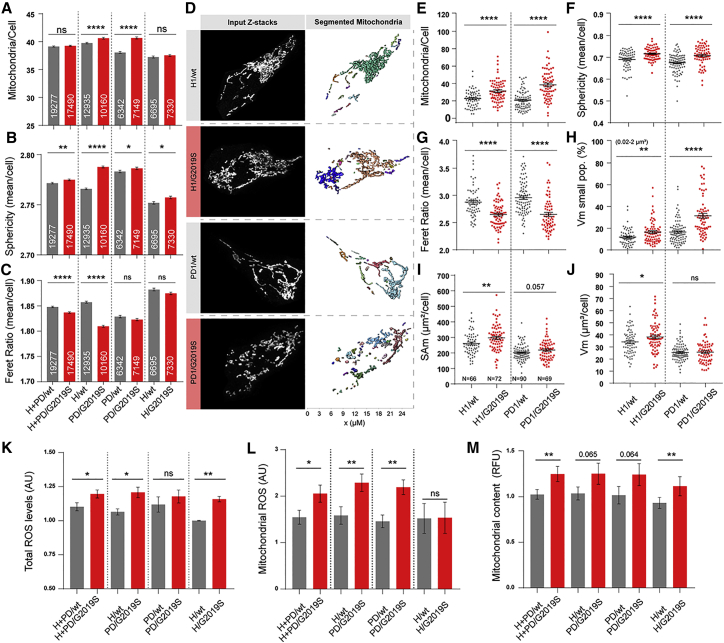
LRRK2-G2019S Induces Mitochondrial Fragmentation in NESCs
Under normal physiological conditions, cells maintain a well-balanced mitochondrial fission/fusion ratio, and any divergence from this stable homeostasis indicates problems in mitochondria functionality (Youle and van der Bliek, 2012). These alterations are usually indicated by fragmented or elongated mitochondrial morphology (Wu et al., 2011). We analyzed mitochondrial morphometrics via immunocytochemistry using an antibody against the translocase of outer mitochondrial membrane 20 protein (TOM20) (Narendra et al., 2008) and established an automated high-content screening (HCS) 2D imaging single-cell analysis approach to unbiasedly quantify mitochondrial features. We analyzed the mitochondrial staining with respect to the number of mitochondria per cell (Figures 2A and S1A). NESCs carrying LRRK2-G2019S from PD patients exhibited significantly more mitochondria per cell than their isogenic controls and H individuals. Insertion of the mutation in the H background did not exert the same effect. Next, we quantified sphericity and elongation of individual mitochondria (Nikolaisen et al., 2014) and compared the mean values of these parameters per cell (Figures 2B and S1B). In agreement with the previous finding, the analysis revealed significantly higher mitochondrial sphericity in LRRK2-G2019S cultures compared with controls. We next used the Feret ratio (Igathinathane et al., 2008), which takes into account the shortest and longest extensions of mitochondria as a measure of their elongation (Figures 2C and S1C). Feret ratio analysis showed significantly smaller ratios of Feret maxima and minima in cells from patients compared with H, indicating that these cells contained less elongated mitochondria than cells from healthy individuals. Correction of the mutation in PD patients and insertion of the mutation in H did not significantly affect this parameter.
Figure 2.
LRRK2-G2019S Induces Mitochondrial Fragmentation in NESCs
(A–C) Automated single-cell analysis of mitochondrial morphology in pooled groups. The number of cells is indicated in each bar (N = 3, n = 3). The following parameters were assessed: (A) number of mitochondria per cell; (B) mean mitochondrial sphericity; (C) Feret ratio.
(D–J) Unbiased/blinded manual 3D single-cell analysis of the mitochondrial features of selected cell lines (H1/WT, H1/G2019S, PD1/WT, and PD1/G2019S). Each dot plotted represents data from a single analyzed cell (n cells per group indicated in I; N = 3, n = 3). Statistical significance was determined using Student's t test. (D) Representative z stack maximum intensity projections of single cells and corresponding post-processing showing segmented mitochondria in different colors. The x-axis indicates the size of the mitochondria in μm. The following parameters were assessed: (E) number of mitochondria per cell; (F) mean mitochondrial sphericity; (G) mean Feret ratio of mitochondria; (H) surface area of mitochondria (SAm) (μm2); (I) mitochondrial volume (Vm) per cell (μm3); (J) percentage of small-volume mitochondria (0.02–2 μm2).
(K) Total ROS levels normalized to H1, N = 5–6, n = 3, Mann-Whitney test was performed within isogenic groups.
(L) Mitochondrial ROS levels normalized to H1, N = 4, n = 1 Mann-Whitney test was performed within isogenic groups.
(M) Mitochondrial content measurements normalized to H1, N = 4, n = 2, Mann-Whitney test was performed within isogenic groups. For all panels, the data are presented as the mean (only 1 median) ± SEM. N indicates the number of experimental repetitions, n indicates the number of technical replicates per cell line.
To verify these findings and explore additional morphological details, we next conducted a detailed 3D analysis of mitochondrial morphology in NESCs. We selected two isogenic pairs derived from a healthy line (H1) and a LRRK2-G2019S carrier PD patient (PD1.1) (Figure 2D). The 3D analysis revealed an increased number of mitochondria (Figure 2E) in LRRK2-G2019S NESCs. When calculated per cell, and under mitochondrial subpopulation analyses, mitochondria from LRRK2-G2019S NESCs displayed greater sphericity (Figures 2F, S1D, and S1E), a reduced Feret ratio (Figures 2G, S1F, and S1G), and an increased volume fraction (percentage) of small mitochondria (Figure 2H) compared with isogenic controls. The total mitochondrial surface area (Figure 2I) and volume (Figures 2J, S1H, and S1I) increased in LRRK2-G2019S NESCs compared with the LRRK2-WT NESCs, suggesting that LRRK2-G2019S NESCs had an increased mitochondrial biomass. The increase did not reach statistical significance in PD patient cells when compared with their isogenic controls. Based on the results of the two different mitochondrial morphology analyses, we concluded that increased levels of mitochondrial fragmentation were present in LRRK2-G2019S NESCs. This suggests that the LRRK2-G2019S mutation interferes with mitochondrial dynamics, suggesting reduced mitochondrial quality.
LRRK2-G2019S Causes Increased Total and Mitochondrial ROS Levels
As LRRK2-G2019S induced aberrant mitochondrial morphologies, we evaluated the possible negative effects in terms of oxidative stress. Mitochondria passively release the main fraction of total cellular ROS (Turrens, 2003), and increased ROS release is a direct indicator of mitochondrial dysfunction (Lambeth et al., 2007). Therefore, we performed a general analysis of total ROS levels using a luminescence-based system. We detected significantly increased ROS levels in LRRK2-G2019S NESCs compared with controls (Figures 2K and S1J). Correction of the LRRK2-G2019S mutation in the patient lines was insufficient to significantly rescue the total ROS levels. On the other hand, the insertion of the LRRK2-G2019S mutation in the healthy background induced increased ROS levels compared with the isogenic controls. We then assessed mitochondria-specific superoxide levels using a MitoSOX probe using flow cytometry (Figures 2L, S1K, and S1L). The resulting data revealed significantly elevated mitochondrial ROS (mROS) levels in cells expressing LRRK2-G2019S. Correction of the LRRK2-G2019S mutation rescued mROS levels, but healthy control cells with the inserted mutation showed no elevation in mROS (Figure 2L). We next measured the mitochondrial content of NESCs using flow cytometry with the MitoTracker probe and observed an increased amount in NESCs carrying the LRRK2-G2019S mutation (Figures 2M, S1M, and S1N). This increase did not reach statistical significance in the comparisons H versus P and P/WT versus P/G2019S.
Overall, these results indicate that LRRK2-G2019S-expressing NESCs exhibited a higher degree of mitochondrial fragmentation and morphological alterations than mitochondria in LRRK2-WT NESCs. When comparing H/WT with H/G2019S, it was apparent that the amplitude of the phenotypes was smaller than when comparing H/WT with P/G2019S. This suggests that LRRK2-G2019S mutation plays a deleterious effect on mitochondria, which can be exacerbated by a permissive genetic background.
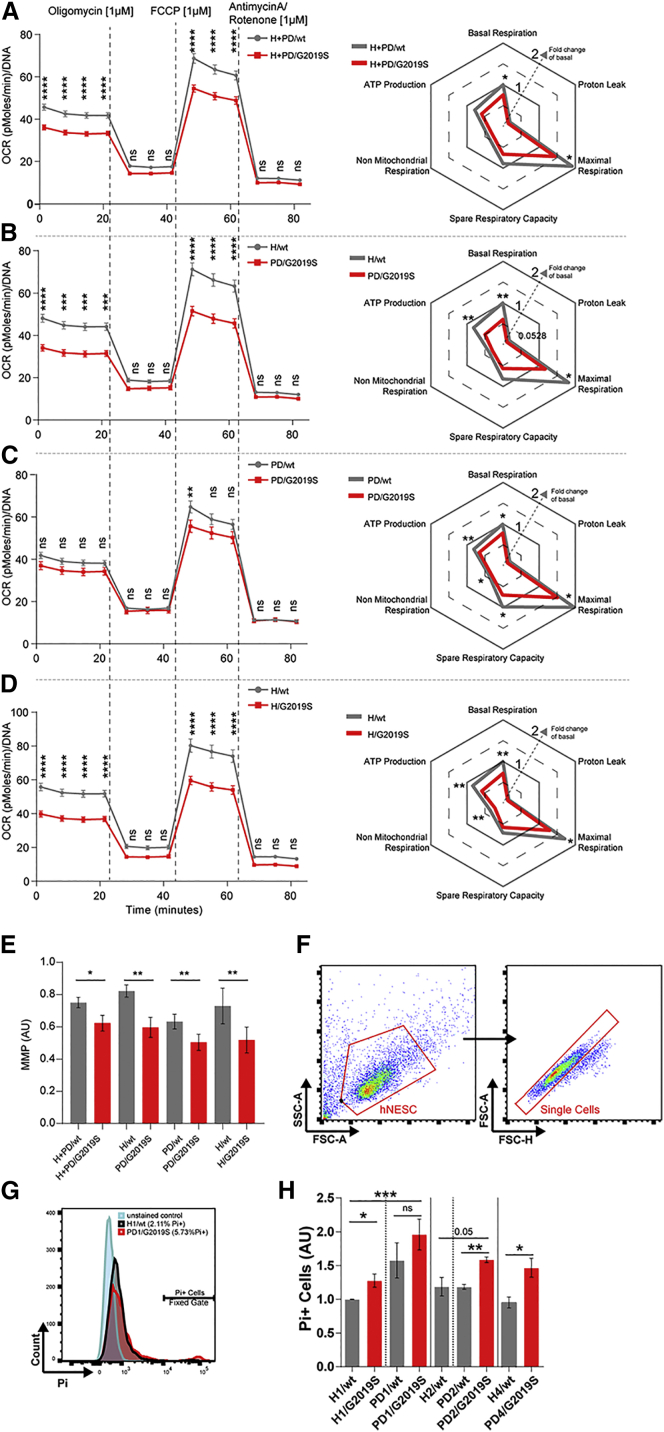
LRRK2-G2019S Alters Mitochondrial Functionality
To address whether LRRK2-G2019S also functionally altered mitochondria, we analyzed mitochondrial respiration and mitochondrial membrane potential (MMP). The oxygen consumption rate (OCR) was measured with the Seahorse Extracellular Flux Analyzer (Figures 3A–3D and S2A–S2E). LRRK2-G2019S NESCs showed a significantly decreased basal respiration compared with LRRK2-WT (Figures 3A and S2A). We injected oligomycin to determine the proportion of ATP-linked OCR, followed by FCCP, an uncoupling compound, used to induce maximal respiration. LRRK2-G2019S NESCs showed a significantly reduced maximal respiration capacity compared with LRRK2-WT. The same differences were observed when comparing H/WT and PD/G2019S (Figures 3B and S2B), PD/WT and PD/G2019S patients (Figures 3C and S2C), and H/WT and H/G2019S lines (Figures 3D and S2D). Proton leakage was unchanged between the tested conditions. We next analyzed MMP, another parameter indicating mitochondrial quality (Brand and Nicholls, 2011). Using TMRM dye, we detected significantly lower MMP in LRRK2-G2019S-expressing NESCs compared with LRRK2-WT (Figures 3E and S2F).
Figure 3.
LRRK2-G2019S Induces Loss of Respiratory Capacity and Cell Death
(A–D) Mitochondrial total respiratory capacity measurements obtained using Seahorse, N = 3, n = 8 of the following cell groupings (A) H + PD/WT and H + PD/G2019S, (B) H/WT and PD/G2019S, (C) PD/WT and PD/G2019S, (D) H/WT and H/G2019S. Maximal respiration, proton leakage, basal respiration, ATP production, and non-mitochondrial respiration are represented with OCR graphs shown on the left and with spider plots on the right. Data in the spider plot were calculated based on the grouped data points of the oxygen consumption rates. Significant differences between the groups were tested using a two-way ANOVA, as indicated in the graphs.
(E) Mitochondrial membrane potential (MMP) quantification. The data were normalized to MitoTracker 488 live staining and H1 data. Statistical significance was tested using Student's t-test.
(F–H), (F) Flow cytometry scatter plots showing the gating strategy for the Pi quantification of the NESC population. (G) Representative histograms showing the Pi+ cell fraction in the H1/WT and PD1/G2019S NESCs. (H) Bar graphs showing the resulting Pi+ cell quantification in all the applied groupings, N = 3–4, n = 1. In all panels, the data are presented as the mean ± SEM. N indicates the number of experimental repetitions, n indicates the number of technical replicates per cell line.
We also assessed the viability of the NESCs using propidium iodide (Pi) staining via flow cytometry. The Pi+ quantification revealed increased cell death in LRRK2-G2019S NESC cultures compared with LRRK2-WT (Figures 3F–H). In a previous study, increased cell death was observed in NSC LRRK2-G2019S cultures after the addition of a proteasome inhibitor (Liu et al., 2012). It was previously reported that neural stem cells are competent to differentiate into neurons, astrocytes, and oligodendrocytes (Conti et al., 2005), but they do not efficiently differentiate in midbrain dopaminergic neurons (Reinhardt et al., 2013). This different behavior stresses the relevance of NESCs for the study of PD. Comparing PD1/WT with PD1/G2019S, the increase did not reach statistical significance.
Together, these findings suggest that mitochondria are functionally altered in LRRK2-G2019S NESCs and this is associated with increased cell death.
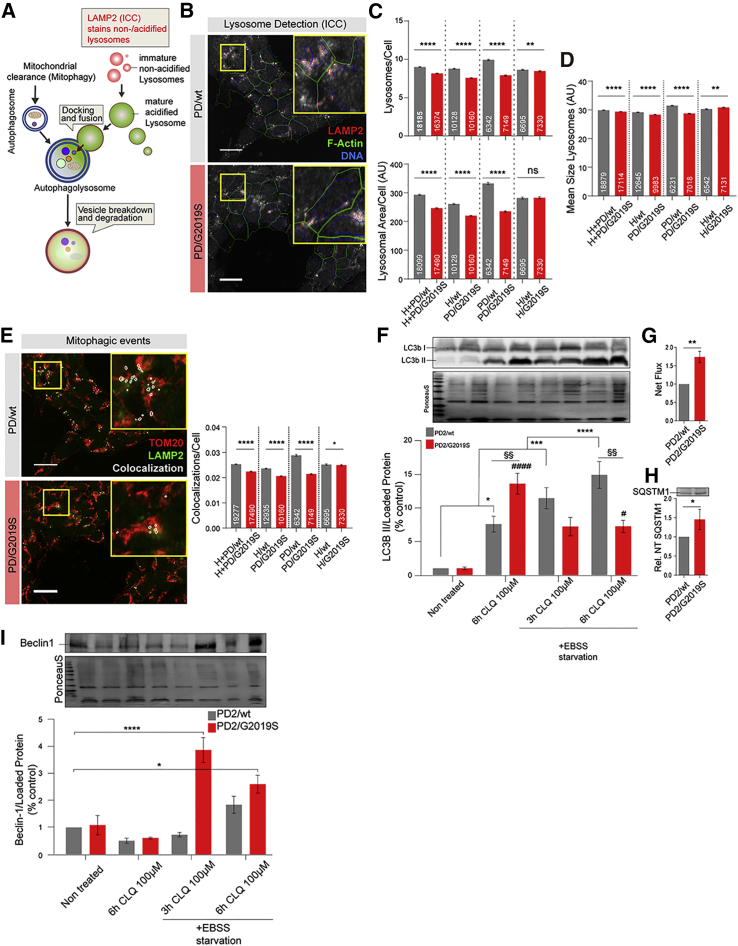
LRRK2-G2019S Triggers Aberrant Mitochondrial Quality Control
Mitochondrial functionality and turnover are ultimately linked. The effective clearance of dysfunctional mitochondria is strongly dependent on the autophagosomal-lysosomal pathway (ALP). The mitochondrial phenotypes we observed in LRRK2-G2019S-expressing NESCs are potentially the result of ineffective mitochondrial clearance, which might be the result of insufficient ALP function (Youle and van der Bliek, 2012). Macromitophagy is ALP mediated (Figure 4A). Thus, we speculated that ALP in LRRK2-G2019S NESCs could be affected, potentially resulting in insufficient mitochondrial clearance. Accordingly, we performed HCS-based lysosome quantification to detect differences in the final and rate-limiting step of ALP.
Figure 4.
LRRK2-G2019S Impairs Mitochondrial Quality Control
(A) Illustration depicting the mitochondrial clearance pathway via lysosomes (adapted from Fullgrabe et al., 2014), highlighting staining methods and chloroquine perturbation.
(B) Representative photomicrographs of LAMP2 ICC-based lysosome quantification; HCS-based feature detection is indicated.
(C–E) HCS-based quantification of lysosomal morphological features per cell, grouped comparisons (N = 3, n = 3); the number of single cells analyzed is indicated in each bar. Statistical significance was analyzed using Mann-Whitney test. The following parameters were assessed: (C) number of lysosomes and lysosomal area per cell; (D) mean lysosome size per cell; (E) mitophagy level estimation using quantification of lysosomal (LAMP2) and mitochondrial (TOM20) staining co-localization events. Representative TOM20 and LAMP2 photomicrographs are shown with co-localization in white (the co-localization events may be in another z plane). The number of cells analyzed is indicated in the bars.
(F–I) Western blot-based autophagy activity and stress activation analysis in NESCs (N = 3, n = 3). Statistical significance was determined using two-way ANOVA and Tukey multiple comparisons test (∗comparison with untreated PD2.GC cells; #comparison with untreated PD2.G2019S cells; and §comparison with PD2.GC cells—same condition). (F) Quantification of LC3B I/II protein levels under basal conditions and during blocking of autophagosomal-lysosomal fusion (CLQ, chloroquine) in combination with activation of autophagy using Earle's balanced salt solution-based starvation. (G) Calculation of net flux autophagic activity. (H) Relative basal SQSTM1 protein levels. (I) Upstream autophagy activation signal analysis using Beclin-1. Scale bars, 20 μm. For all panels, the data are presented as the mean ± SEM. N indicates the number of experimental repetitions, n indicates the number of technical replicates per cell line.
After unbiased identification of all lysosomal-associated membrane protein 2-positive (LAMP2+) areas in single cells (Figure 4B), we quantified the LAMP2+ puncta representing the total pool of lysosomes, including potential non-acidified pre-lysosomes. We quantified lysosome number and total lysosomal area (Figures 4C, S3A, and S3B). Based on these features, we calculated the mean size of all lysosomes (Figures 4D and S2C). The approach revealed a reduced number of lysosomes, reduced total lysosomal area, and reduced mean lysosomal size in LRRK2-G2019S-expressing NESCs compared with LRRK2-WT. In most comparisons, these lysosomal phenotypes were directly associated with LRRK2-G2019S mutation. However, the results obtained with H/G2019S isogenic controls indicated that LRRK2-G2019S itself is not sufficient to affect the total lysosomal area per NESC feature. We next addressed the potential mitophagy events by quantifying the co-localization of LAMP2 puncta with the mitochondrial TOM20 area in single NESCs (Figures 4E and S2D). We detected a significantly reduced number of co-localization in LRRK2-G2019S-expressing NESCs compared with controls, indicating reduced macro-mitophagic clearance via lysosomes.
To investigate the potential dysregulation of the ALP upstream of the lysosome, we performed a protein-based analysis of autophagosomal capacity using cells of one patient and its isogenic control (PD2/WT and PD2/G2019S). Using western blotting analysis (Figures 4F–4I), we measured the protein levels of the autophagosomal membrane-bound form of LC3B (LC3B-II) (Figure 4F), sequestesome 1 (SQSTM1, also known as p62) (Figures 4H and S3E), and Beclin-1 (Figure 4I), widely used markers for monitoring the autophagic process, in the absence or in the presence of chloroquine, an inhibitor of ALP degradation.
We observed significantly elevated basal autophagic flux activity in LRRK2-G2019S-expressing NESCs compared with LRRK2-WT (Figure 4G). Consistently, the autophagy substrate SQSTM1 levels were elevated in these cells (Figures 4H and S3G). Next, we tested the cellular autophagy response to Earle's balanced salt solution-induced starvation. Despite the observed elevated basal autophagic level in the patient cells, these levels were not sustained during starvation (Figure 4F). Although starvation resulted in continuously elevated LC3BII levels in controls, the presence of CLQ, the LC3BII levels in LRRK2-G2019S-expressing NESCs were markedly decreased compared with controls. Strikingly, parallel quantification of Beclin-1 (Figure 4I), which acts upstream of autophagy signaling and activation (Kang et al., 2011), indicated a strong activation signal, as reflected by elevated Beclin-1 levels (3 and 6 h) and in the presence of CLQ 100 μM. However, the Beclin-1 signal did not result in autophagosome formation because of the lack of LC3BII abundance under the same conditions (Figure 4I). These results show that LRRK2-G2019S negatively affects autophagy upstream of the observed lysosomal limitations.
Collectively, the ALP analysis of NESCs revealed decreased ALP functionality in LRRK2-G2019S-expressing NESCs compared with isogenic controls.
Discussion
In this study, we observed that mitochondria in NESC cells carrying the LRRK2-G2019S mutation were more fragmented and were more prone to release ROS compared with isogenic controls. Beside morphology, also the functionality of mitochondria was impaired in the presence of LRRK2-G2019S, as shown by the decreased membrane potential and respiratory capacity.
Evidence from human neurons has previously shown that LRRK2 influences mitochondrial function (Cooper et al., 2012, Sanders et al., 2014) and dynamics (Wang et al., 2012). An increase of fragmented mitochondria has been observed in iPSC-derived dopaminergic neurons along with impaired clearance of unhealthy mitochondria, and alteration in mitochondria respiration and bioenergetics, as reviewed in Sison et al. (2018). However, no information was previously available regarding the status of the mitochondria at the stem cell level in the context of LRRK2. Here, we showed that already before neuronal differentiation at the level of NESCs, mitochondrial phenotypes are detectable, suggesting that this is a relevant model to study PD mitochondrial alterations. Moreover, mitochondria were recently shown to be of crucial importance in neural stem cells (Lorenz et al., 2017, Sanders et al., 2014), highlighting their key role in cell fate decisions during development (Beckervordersandforth et al., 2017, Khacho et al., 2016). Mitochondria are critically involved in numerous cellular processes that rely on energy, such as cell growth, maintenance, proliferation, activity, and neurogenesis (Beckervordersandforth et al., 2017). Based on these findings, it is tempting to speculate that the described mitochondrial alterations in NESCs are important contributors to the PD-specific dynamics of dopaminergic differentiation.
Neurogenic niches are characterized by particularly elevated metabolic activity, which associates with high ROS levels (Walton et al., 2012). Beside their detrimental effect on cell viability, ROS play a role in the regulation of neural cell fate (Khacho et al., 2016, Orford and Scadden, 2008, Sarsour et al., 2009). Consequently, the observed elevated ROS levels might contribute to the increased cell death observed in LRRK2-G2019S NESCs. LRRK2 mutation can alter the interaction with the antioxidant protein peroxiredoxin 3, reducing the ability to scavenge ROS (Angeles et al., 2011). It has been shown that mitochondria-targeted antioxidants effectively sequester ROS and protect against mitochondrial damage (Oyewole and Birch-Machin, 2015). NESCs are stable and highly proliferative cultures, which permit quick evaluation of functional and morphological mitochondrial features. The present model and the here-described assays can leverage pharmacological screenings that aim to ameliorate bioenergetics defects and reduce mitochondrial ROS in the context of PD.
Control of mitochondrial homeostasis is of major importance in cellular homeostasis (Dias et al., 2013). Insufficient clearance of dysfunctional mitochondria is associated with accumulation of damaged mitochondria and high ROS levels, ultimately resulting in reduced cell viability. Clearance of malfunctioning mitochondria largely occurs through lysosomal-mediated mitophagy. Lysosomal dysfunctionality has been associated to PD in several studies (Chang et al., 2017, Roosen and Cookson, 2016). The status of mitochondrial biogenesis can be directly linked to the functionality of mitochondrial quality control mechanisms (Tronstad et al., 2014). Lysosomal phenotypes in PD context have mostly been described in terminally differentiated neurons (Migdalska-Richards et al., 2017). Here, we also found LRRK2-G2019S-associated alterations in the ALP in NESCs, indicating that ALP dysregulation can already occur at a developmental stage.
In summary, the data presented in this study highlight disease-specific phenotypes of patient-derived NESCs carrying the LRRK2-G2019S mutation. The detection of these phenotypes in a developmentally early neural stem cell model supports the hypothesis that PD might have a developmental component.
Experimental Procedures
The work with human iPSCs was approved by the local ethical committee (Ethic Review Panel of the University of Luxembourg, PDiPS project and CNER No 201305/04).
Single-Cell RNA-Seq Using Drop-Seq
Microfluidics Fabrication
Microfluidics devices were fabricated using a previously published design (Macosko et al., 2015). Soft lithography was performed using previously published protocols using SU-8 2050 photoresist (MicroChem) on a 4″ silicon substrate (Macosko et al., 2015, Mazutis et al., 2013). Drop-seq chips were fabricated using silicon-based polymerization chemistry. In brief, a polydimethylsiloxane (PDMS) base and a crosslinker (Dow Corning), were combined at a 10:1 ratio, mixed, and degassed, before pouring the mix onto the Drop-seq master template. PDMS was cured on the master template, at 80°C for 2 h. After the incubation and cooling, the PDMS stamps were cut and the inlet/outlet ports were punched with 1.25-mm biopsy punchers (World Precision Instruments). The PDMS monolith was plasma-bonded to a clean microscopic glass slide using Harrick plasma cleaner. The flow channels of the Drop-seq chip were then subject to hydrophobicity treatment.
Single-Cell Suspension and RNA-Seq
The key steps of the protocol used here align with the original Drop-seq work (Macosko et al., 2015) with minor changes as described below. After the enzymatic digestion of the cells using Accutase at 37°C (10–20 min) single cells were pelleted by centrifugation at 300 rcf for 5 min. Cell clumps and debris were excluded from the suspension using a 40-μm nylon cell strainer (BD). The single-cell suspension was diluted with 1% BSA in 1× PBS to achieve a cell density of ∼150 cells/mL. Diluted cells were placed on ice until loaded onto the pre-fabricated Drop-seq chip. Specially synthesized barcoded beads (ChemGenes) were co-encapsulated with cells inside the droplets, having optimized lysis reagents adapted from Macosko et al. (2015). This procedure allows capturing the cellular mRNA by barcoded oligo (dT) handles on the surface of the specialized beads. The microfluidic chip generates monodispersed droplets of ∼1 nL volume. The bead concentration used for optimal single bead encapsulation within the droplet was 200 beads/mL, and beads were prepared in Drop-seq lysis buffer (Macosko et al., 2015).
One ml of the cell and the bead suspension was loaded into 3-mL syringes (BD). A micro-stirrer was used (V&P Scientific) to keep the beads in stable suspension. The QX200 carrier oil (Bio-Rad) was used as a continuous phase in the droplet generation. Oil was loaded into a 20-mL syringe (BD). For droplet generation, 3.6 and 13 mL/h were used for the dispersed and continuous phases, respectively, using KD Scientific Legato syringe pumps. This generated droplets with a diameter of ∼115 μm (∼1 nL volume). The droplet suspension was collected in a 50-mL Falcon tube. Then, the droplet consistency and stability were documented by bright-field microscopy to check on multiple bead occupancy.
The subsequent droplet breakage, reverse transcription, and the exonuclease treatment were carried out in accordance with the original Drop-seq work (Macosko et al., 2015). The buffer for the reverse transcription contained: 1× Maxima RT buffer, 4% Ficoll PM 400 (Sigma), 1 μM dNTPs (Thermo Scientific), 1 U/mL RNase Inhibitor (Lucigen), 2.5 μM Template Switch Oligo, and 10 U/mL Maxima H Minus RT (Thermo Scientific).
Following Exo I treatment, the bead counts were estimated using an INCYTO C-Chip disposable hemacytometer. Then, aliquots of 10,000 beads were prepared in 0.2-mL Eppendorf PCR tubes. PCR mix was dispensed in a volume of 50 mL using 1× Hifi HotStart ReadyMix (Kapa Biosystems) and 0.8 mM Template-Switch-PCR primer. The thermocycling program for the PCR amplification was adapted from the previous work, except for the final PCR cycles: 95°C for 3 min; four cycles of: 98°C for 20 s, 65°C for 45 s, 72°C for 3 min; 10 cycles of: 98°C for 20 s, 67°C for 20 s, 72°C for 3 min; followed by a final extension step of 72°C for 5 min. After PCR amplification, libraries were purified with 0.6× Agencourt AMPure XP beads (Beckman Coulter), in accordance with the manufacturer's protocol. Finally, the purified libraries were eluted in 20 μL of molecular-grade water. Before the sequencing library preparation, the quality and the concentration of the libraries was assessed using Bioanalyzer High Sensitivity Chip (Agilent Technologies).
3D Blinded Unbiased Mitochondrial Morphology Analysis
3D image processing and analysis based on LSM confocal z stacks were performed using Image-Pro Plus software (version 7.0) with SharpStack Total deconvolution and 3D Constructor modules (Media Cybernetics, Washington, USA). Analysis of mitochondrial shape properties was conducted by adapting previously described protocols (Nikolaisen et al., 2014). The mitochondrial channel of the confocal z stack was extracted, background-corrected (fixed level), and spatially calibrated and 3D blind deconvolution (10 iterations) was performed. Flat 2D projections (maximum intensity composite) of the mitochondrial z stacks were employed for manual segmentation of single cells, but we also compared with the nuclear and cytoplasmic channels (F-actin) as guidance in this step. By drawing the cellular outline, a binary black and white image was created, and this was subsequently used as a mask for making a z stack with only one (or two) cell(s). The processed single-cell z stacks were loaded into the 3D Constructor module using no sub-sampling, and an isosurface (surface level = 900) was created without further filtering or simplification. Mitochondrial shape-parameters were then obtained and analyzed. Objects larger than 0.02 μm3 were quantified as mitochondrial objects.
The quantitative image analysis was done with blinded samples. Pooled single-cell data obtained from three replicated experiments are presented. The inter-experimental variation was evaluated and found insignificant (not shown). Statistical analysis of two groups was performed using t test. Only cells that were possible to segment into single cells or doublets (only a few cases) were included in this analysis. For doublets, the mean value was used for statistical analysis. Cells that were clearly apoptotic (condensed nucleus, fragmented cell body) were excluded. Samples in this analysis (n = number of cells analyzed): PD1.G2019S n = 69, PD1.GC: n = 90, H1.G2019S: n = 72, H1: n = 66.
Quantification and Statistical Analysis
Statistical analysis was performed as follows: unpaired or paired (isogenic controls) Student's t test when the sample size was small and data normally distributed. For HCS imaging data that was not normally distributed we applied Mann-Whitney test. Two-way ANOVA followed by Bonferroni test was applied when indicated. All data are presented as mean ± SEM. Significance levels were set at ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
For plotting, GraphPad Prism was used. For single-cell RNA-seq data, the statistical significance of the differences in cumulative gene expressions (Figure 1C) was assessed using a z test, with Bonferroni correction to account for multiple testing. More details on the different statistical analyses of single-cell RNA-seq data are explained in the section titled “Analysis of Mitochondrial genes expression at the single cell level” of the Supplemental Information.
Author Contributions
Conceptualization, J.W. and J.C.S.; Cell Culture and Assay Design, J.W.; Methodology, J.W., P.M.A.A., S.L.N., S.K.P., L.S., S.M., R.P., F.H., X.Q., J.J, J.A.-F, T.I., A.M., L.G.C., S.B., A.S., D.G., T.S., and K.J.T.; Investigation, J.W., P.M.A.A., S.N., S.K.P., S.B., S.M., R.P., F.H., A.S., and K.J.T.; Writing – Original Draft, S.B., J.W., and J.C.S.; Writing – Review & Editing, all the co-authors; Supervision, L.P.D.A., A.S., K.J.T., and J.C.S.; J.W. and S.B. contributed equally; P.M.A.A., S.L.N., and S.K.B. contributed equally.
Acknowledgments
The authors would like to thank Thea van Wuellen, Marie Fossepre, Antoine Treff, and Annegraet Daujeumont for technical assistance. We thank Prof. Dr. Hans R. Schöler (Max-Planck-Gesellschaft), Dr. Jared Sterneckert (CRTD), Prof. Dr. Thomas Gasser (Hertie Institute in Tuebingen), and the Coriell Institute for providing the cell lines. The LAMP-2 monoclonal antibody was developed by J.T. August and J.E.K. Hildreth and was obtained from the Developmental Studies Hybridoma Bank created by the NICHD of the NIH and maintained at the University of Iowa, Department of Biology, Iowa City, IA 52242, USA. This project was supported by the LCSB pluripotent stem cell core facility. The JCS lab is supported by the Fonds National de la Recherche (FNR) (CORE, C13/BM/5791363 and Proof-of-Concept program PoC15/11180855 & PoC16/11559169). This is an EU Joint Programme - Neurodegenerative Disease Research (JPND) project (INTER/JPND/14/02; INTER/JPND/15/11092422). Further support comes from the SysMedPD project, which has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement no. 668738. J.W., X.Q., L.G.-C., J.J., and A.S.M. were supported by fellowships from the FNR (AFR, Aides à la Formation-Recherche). S.M. is supported by the FNR through the PRIDE DTU CriTiCS, reference 10907093. We also thank the private donors who support our work at the LCSB.
Published: April 11, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.03.004.
Accession Numbers
The accession number for the FASQT files reported in this paper is number GEO: GSE128040.
Supplemental Information
References
- Angeles D.C., Gan B.H., Onstead L., Zhao Y., Lim K.L., Dachsel J., Melrose H., Farrer M., Wszolek Z.K., Dickson D.W. Mutations in LRRK2 increase phosphorylation of peroxiredoxin 3 exacerbating oxidative stress-induced neuronal death. Hum. Mutat. 2011;32:1390–1397. doi: 10.1002/humu.21582. [DOI] [PubMed] [Google Scholar]
- Beckervordersandforth R., Ebert B., Schaffner I., Moss J., Fiebig C., Shin J., Moore D.L., Ghosh L., Trinchero M.F., Stockburger C. Role of mitochondrial metabolism in the control of early lineage progression and aging phenotypes in adult hippocampal neurogenesis. Neuron. 2017;93:1518. doi: 10.1016/j.neuron.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M.D., Nicholls D.G. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulla L.F., Kruger R. Converging environmental and genetic pathways in the pathogenesis of Parkinson's disease. J. Neurol. Sci. 2011;306:1–8. doi: 10.1016/j.jns.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Calvo S.E., Clauser K.R., Mootha V.K. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D., Nalls M.A., Hallgrimsdottir I.B., Hunkapiller J., van der Brug M., Cai F., International Parkinson's Disease Genomics C., Me Research T., Kerchner G.A., Ayalon G. A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat. Genet. 2017;49:1511–1516. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L., Pollard S.M., Gorba T., Reitano E., Toselli M., Biella G., Sun Y., Sanzone S., Ying Q.L., Cattaneo E. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson M.R. Cellular functions of LRRK2 implicate vesicular trafficking pathways in Parkinson's disease. Biochem. Soc. Trans. 2016;44:1603–1610. doi: 10.1042/BST20160228. [DOI] [PubMed] [Google Scholar]
- Cooper O., Seo H., Andrabi S., Guardia-Laguarta C., Graziotto J., Sundberg M., McLean J.R., Carrillo-Reid L., Xie Z., Osborn T. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson's disease. Sci. Transl. Med. 2012;4:141ra190. doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias V., Junn E., Mouradian M.M. The role of oxidative stress in Parkinson's disease. J. Parkinsons Dis. 2013;3:461–491. doi: 10.3233/JPD-130230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullgrabe J., Klionsky D.J., Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- Funayama M., Hasegawa K., Kowa H., Saito M., Tsuji S., Obata F. A new locus for Parkinson's disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann. Neurol. 2002;51:296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cano L., Menzl I., Tisserand J., Nicklas S., Schwamborn J.C. Parkinson's disease-associated mutant LRRK2-mediated inhibition of miRNA activity is antagonized by TRIM32. Mol. Neurobiol. 2018;55:3490–3498. doi: 10.1007/s12035-017-0570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad D., Nakamura K. Understanding the susceptibility of dopamine neurons to mitochondrial stressors in Parkinson's disease. FEBS Lett. 2015;589:3702–3713. doi: 10.1016/j.febslet.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.H., Shaltouki A., Gonzalez A.E., Bettencourt da Cruz A., Burbulla L.F., St Lawrence E., Schule B., Krainc D., Palmer T.D., Wang X. Functional impairment in miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson's disease. Cell Stem Cell. 2016;19:709–724. doi: 10.1016/j.stem.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igathinathane C., Pordesimo L.O., Columbus E.P., Batchelor W.D., Methuku S.R. Shape identification and particles size distribution from basic shape parameters using ImageJ. Comput. Electron. Agric. 2008;63:168–182. [Google Scholar]
- Kang R., Zeh H.J., Lotze M.T., Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khacho M., Clark A., Svoboda D.S., Azzi J., MacLaurin J.G., Meghaizel C., Sesaki H., Lagace D.C., Germain M., Harper M.E. Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Cell Stem Cell. 2016;19:232–247. doi: 10.1016/j.stem.2016.04.015. [DOI] [PubMed] [Google Scholar]
- Klein C., Westenberger A. Genetics of Parkinson's disease. Cold Spring Harb. Perspect. Med. 2012;2:a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumova P., Reyniers L., Meyer M., Lobbestael E., Stauffer D., Gerrits B., Muller L., Hoving S., Kaupmann K., Voshol J. Chemical genetic approach identifies microtubule affinity-regulating kinase 1 as a leucine-rich repeat kinase 2 substrate. FASEB J. 2015;29:2980–2992. doi: 10.1096/fj.14-262329. [DOI] [PubMed] [Google Scholar]
- Lambeth J.D., Kawahara T., Diebold B. Regulation of Nox and Duox enzymatic activity and expression. Free Radic. Biol. Med. 2007;43:319–331. doi: 10.1016/j.freeradbiomed.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage S., Durr A., Brice A. LRRK2: a link between familial and sporadic Parkinson's disease? Pathol. Biol. (Paris) 2007;55:107–110. doi: 10.1016/j.patbio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Liu G.H., Qu J., Suzuki K., Nivet E., Li M., Montserrat N., Yi F., Xu X., Ruiz S., Zhang W. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012;491:603–607. doi: 10.1038/nature11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C., Lesimple P., Bukowiecki R., Zink A., Inak G., Mlody B., Singh M., Semtner M., Mah N., Aure K. Human iPSC-derived neural progenitors are an effective drug discovery model for neurological mtDNA disorders. Cell Stem Cell. 2017;20:659–674.e9. doi: 10.1016/j.stem.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazutis L., Gilbert J., Ung W.L., Weitz D.A., Griffiths A.D., Heyman J.A. Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc. 2013;8:870–891. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migdalska-Richards A., Wegrzynowicz M., Rusconi R., Deangeli G., Di Monte D.A., Spillantini M.G., Schapira A.H.V. The L444P Gba1 mutation enhances alpha-synuclein induced loss of nigral dopaminergic neurons in mice. Brain. 2017;140:2706–2721. doi: 10.1093/brain/awx221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaisen J., Nilsson L.I., Pettersen I.K., Willems P.H., Lorens J.B., Koopman W.J., Tronstad K.J. Automated quantification and integrative analysis of 2D and 3D mitochondrial shape and network properties. PLoS One. 2014;9:e101365. doi: 10.1371/journal.pone.0101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford K.W., Scadden D.T. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- Oyewole A.O., Birch-Machin M.A. Mitochondria-targeted antioxidants. FASEB J. 2015;29:4766–4771. doi: 10.1096/fj.15-275404. [DOI] [PubMed] [Google Scholar]
- Ozelius L.J., Senthil G., Saunders-Pullman R., Ohmann E., Deligtisch A., Tagliati M., Hunt A.L., Klein C., Henick B., Hailpern S.M. LRRK2 G2019S as a cause of Parkinson's disease in Ashkenazi Jews. N. Engl. J. Med. 2006;354:424–425. doi: 10.1056/NEJMc055509. [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C., Jain S., Evans E.W., Gilks W.P., Simon J., van der Brug M., Lopez de Munain A., Aparicio S., Gil A.M., Khan N. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Pissadaki E.K., Bolam J.P. The energy cost of action potential propagation in dopamine neurons: clues to susceptibility in Parkinson's disease. Front. Comput. Neurosci. 2013;7:13. doi: 10.3389/fncom.2013.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt P., Glatza M., Hemmer K., Tsytsyura Y., Thiel C.S., Hoing S., Moritz S., Parga J.A., Wagner L., Bruder J.M. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS One. 2013;8:e59252. doi: 10.1371/journal.pone.0059252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosen D.A., Cookson M.R. LRRK2 at the interface of autophagosomes, endosomes and lysosomes. Mol. Neurodegener. 2016;11:73. doi: 10.1186/s13024-016-0140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders L.H., Laganiere J., Cooper O., Mak S.K., Vu B.J., Huang Y.A., Paschon D.E., Vangipuram M., Sundararajan R., Urnov F.D. LRRK2 mutations cause mitochondrial DNA damage in iPSC-derived neural cells from Parkinson's disease patients: reversal by gene correction. Neurobiol. Dis. 2014;62:381–386. doi: 10.1016/j.nbd.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarsour E.H., Kumar M.G., Chaudhuri L., Kalen A.L., Goswami P.C. Redox control of the cell cycle in health and disease. Antioxid. Redox Signal. 2009;11:2985–3011. doi: 10.1089/ars.2009.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira A.H., Cooper J.M., Dexter D., Clark J.B., Jenner P., Marsden C.D. Mitochondrial complex I deficiency in Parkinson's disease. J. Neurochem. 1990;54:823–827. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- Sison S.L., Vermilyea S.C., Emborg M.E., Ebert A.D. Using patient-derived induced pluripotent stem cells to identify Parkinson's disease-relevant phenotypes. Curr. Neurol. Neurosci. Rep. 2018;18:84. doi: 10.1007/s11910-018-0893-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.A., Jansson J., Rocha E.M., Osborn T., Hallett P.J., Isacson O. Fibroblast biomarkers of sporadic Parkinson's disease and LRRK2 kinase inhibition. Mol. Neurobiol. 2016;53:5161–5177. doi: 10.1007/s12035-015-9435-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer P.A., Swerdlow R.H., Parks J.K., Keeney P., Bennett J.P., Jr., Miller S.W., Davis R.E., Parker W.D., Jr. Abnormal mitochondrial morphology in sporadic Parkinson's and Alzheimer's disease cybrid cell lines. Exp. Neurol. 2000;162:37–50. doi: 10.1006/exnr.2000.7333. [DOI] [PubMed] [Google Scholar]
- Tronstad K.J., Nooteboom M., Nilsson L.I., Nikolaisen J., Sokolewicz M., Grefte S., Pettersen I.K., Dyrstad S., Hoel F., Willems P.H. Regulation and quantification of cellular mitochondrial morphology and content. Curr. Pharm. Des. 2014;20:5634–5652. doi: 10.2174/1381612820666140305230546. [DOI] [PubMed] [Google Scholar]
- Turrens J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tysnes O.B., Storstein A. Epidemiology of Parkinson's disease. J. Neural Transm. (Vienna) 2017;124:901–905. doi: 10.1007/s00702-017-1686-y. [DOI] [PubMed] [Google Scholar]
- Vitte J., Traver S., Maues De Paula A., Lesage S., Rovelli G., Corti O., Duyckaerts C., Brice A. Leucine-rich repeat kinase 2 is associated with the endoplasmic reticulum in dopaminergic neurons and accumulates in the core of Lewy bodies in Parkinson disease. J. Neuropathol. Exp. Neurol. 2010;69:959–972. doi: 10.1097/NEN.0b013e3181efc01c. [DOI] [PubMed] [Google Scholar]
- Wallings R., Manzoni C., Bandopadhyay R. Cellular processes associated with LRRK2 function and dysfunction. FEBS J. 2015;282:2806–2826. doi: 10.1111/febs.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton N.M., Shin R., Tajinda K., Heusner C.L., Kogan J.H., Miyake S., Chen Q., Tamura K., Matsumoto M. Adult neurogenesis transiently generates oxidative stress. PLoS One. 2012;7:e35264. doi: 10.1371/journal.pone.0035264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yan M.H., Fujioka H., Liu J., Wilson-Delfosse A., Chen S.G., Perry G., Casadesus G., Zhu X. LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum. Mol. Genet. 2012;21:1931–1944. doi: 10.1093/hmg/dds003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.B., Moore D.J., Biskup S., Bugayenko A., Smith W.W., Ross C.A., Dawson V.L., Dawson T.M. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. U S A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Zhou F., Zhang Z., Xing D. Mitochondrial oxidative stress causes mitochondrial fragmentation via differential modulation of mitochondrial fission-fusion proteins. FEBS J. 2011;278:941–954. doi: 10.1111/j.1742-4658.2011.08010.x. [DOI] [PubMed] [Google Scholar]
- Youle R.J., van der Bliek A.M. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.