Abstract
NORAD (non-coding RNA activated by DNA damage) is a long non-coding RNA (lncRNA) that is upregulated and promotes cell progression in various human types of cancer; however, its function in non-small cell lung cancer (NSCLC) remains unclear. The present study investigated the regulatory function and underlying mechanisms of NORAD in NSCLC. NORAD and miR-136-5p expression were assessed by reverse transcription-quantitative polymerase chain reaction, and proliferation and glycolysis-associated markers were also assessed. Direct miR-136-5p regulation by NORAD was detected using luciferase reporter assay and RNA immunoprecipitation. NORAD was highly expressed in NSCLC tissues and cell lines. NORAD overexpression increased NSCLC proliferation and glycolysis. Further investigation revealed that NORAD serves as a competing endogenous RNA for miR-136-5p. Gain- and loss-of-function experiments confirmed that miR-136-5p reversed the promoting effects of NORAD in NSCLC. Results of the present study indicate that NORAD serves as a growth-promoting lncRNA in NSCLC by suppressing the function of miR-136-5p. NORAD and miR-136-5p interaction may provide a potential target for NSCLC treatment.
Keywords: NORAD, miR-136-5p, competing endogenous RNA, non-small cell lung cancer
Introduction
Lung cancer is the most commonly diagnosed type of cancer worldwide (1), with a 5-year survival rate of ~20% (2). With increased air pollution exposure and smoking, the incidence of non-small cell lung cancer (NSCLC) and its respective mortality rate have risen, threatening the health of individuals worldwide. Therefore, obtaining a clear understanding of the underlying mechanisms of NSCLC is critical for improving the efficacy of clinical treatment. Long non-coding RNAs (lncRNAs) are transcripts that are less than 200 nucleotides in length (3). However, lncRNAs are involved in various functions in cancer cells, including cell growth and metastasis (4), which are implicated in cancer progression.
The functions of a novel cytoplasmic lncRNA, NORAD (non-coding RNA activated by DNA damage), also known as LINC00657 or LOC647979, have been investigated (5). NORAD serves as an oncogene and is associated with overall survival in breast cancer (6) and pancreatic cancer (7). However, its underlying mechanisms have not been revealed. Existing evidence suggests that lncRNAs could interact with microRNAs as competing endogenous RNAs (ceRNAs) or ‘RNA sponges’, recruiting these molecules and reducing their regulatory effect on target mRNAs (8,9). In patients with pancreatic cancer, NORAD is believed to serve as a sponge for miR-125a-3p to regulate Ras homolog family member A (7). NORAD was reported to be associated with epithelial-mesenchymal transition, metastasis and poor prognosis in patients with colorectal cancer, by interacting with miR-202-5p (10). In the present study, NORAD was found to function as a ceRNA to inhibit miR-136-5p. Upregulation of NORAD expression in NSCLC and tissues was associated with increased lung cancer cell viability and anaerobic glycolysis. This study provides novel insight on the possible mechanism of lncRNA NORAD in regulating NSCLC.
Materials and methods
Statement of ethics
Informed consents were obtained from all of the participating patients, and the study was approved by the Clinical Research Ethics Committee of Suqian People's Hospital of Nanjing Drum Tower Hospital Group (Nanjing, China).
Cell culture
The NSCLC cell lines A549, H1975, H1650, LK-2, H1299, H460 and epithelial cell line HBE were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). A549 cells were cultured in F-12K medium supplemented with 10% fetal bovine serum (FBS) (all purchased from Gibco/Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in 5% CO2. H1975, H1650, LK-2, H1299, H460 and HBE cell lines were cultured in RPMI-1640 medium, supplemented with 10% FBS (all purchased from Gibco/Thermo Fisher Scientific, Inc.) at 37°C in 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using Trizol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and synthesized into cDNA using a reverse transcription kit (Invitrogen; Thermo Fisher Scientific, Inc.). RT-qPCR was performed using the 7500 Fast Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc) using SYBR-Green PCR kit (Toyobo Life Science, Osaka, Japan), according to the manufacturer's protocols. PCR amplification conditions were: 95°C for 5 sec, 60°C for 30 sec, 72°C for 30 sec for 40 cycles. The results were normalized to the internal reference gene GAPDH. The primer sequences used for RT-qPCR assays were as follows: NORAD forward, 5′-TGATAGGATACATCTTGGACATGGA-3′ and reverse, 5′-AACCTAATGAACAAGTCCTGACATACA-3′; GAPDH forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′. For the detection of miRNA expression, reverse transcription was performed and microRNAs were detected with stem-loop primers purchased from RiboBio (Guangzhou, China): miR-136-5p, F: ACTCCATTTGTTTTGATGATGGA. U6 snoRNA was used as the endogenous control: U6, F: CTCGCTTCGGCAGCACA and R: ACGCTTCACGAATTTGCGT. Relative fold changes were calculated using the 2−ΔΔCq method (11). All PCR assays were repeated three times.
Plasmid construction
NORAD cDNA fragments containing either the predicted potential microRNA binding sites, wild-type (wt) or scrambled microRNA binding site sequences, mutation (mut) were amplified by PCR. The plasmid was constructed by cloning NORAD cDNA into the pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.). Mimics and inhibitors of miR-136-5p were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
CCK-8 assay
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to detect A549 and H460 cell proliferation. The cells (1×104 cells/well) were seeded into 96-well plates at 37°C in 5% CO2, and transfected with the indicated plasmid. A total of 10 µl CCK-8 solution was subsequently added and incubated was carried out for another 4 h at 37°C. CCK-8 reagent was added at 0, 24, 48 and 72 h, according to the manufacturer's protocol. Absorbance rate was measured at a wavelength of 450 nm using a microplate reader.
Lactate dehydrogenase (LDH) activity, lactate production, glucose utilization assay and intracellular ATP level
A total of 1×106 transfected cells were used for LDH activity and lactate production assay using the Lactate Dehydrogenase Activity Assay kit and Lactate Assay kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), according to the manufacturer's protocols. For glucose utilization assay, transfected cells were incubated for 24 h and replaced with phenol-red free RPMI with 1% FBS or phenol-red free RPMI with 1% FBS and cultured for 3 days. Glucose concentrations in media were measured using a colorimetric glucose assay kit (BioVision, Inc., Milpitas, CA, USA) and normalized to the cell number. Intracellular ATP was measured using a firefly luciferase-based ATP assay kit (Beyotime Institute of Biotechnology, Haimen, China). The relative ATP level is expressed as ATP value/protein value.
Dual-luciferase reporter assay
Recombinant plasmids or an empty plasmid encoding the firefly luciferase reporter were constructed and transfected into cells as well as the Renilla luciferase reporter pRL-CMV plasmid (Promega Corporation, Madison, WI, USA) as an internal control. miR-136-5p mimic or a negative control were subsequently transfected into each group. Luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega Corporation), according to the manufacturer's protocols.
Western blot analysis
Lysis buffer RIPA was used to harvest cell protein. Equal amounts of protein (20 µg) were added onto a 10% SDS-PAGE gel and transferred onto polyvinylidene difluoride membranes. The membranes were incubated with primary antibodies overnight at 4°C following blocking with skim milk, and washed with TBST buffer 3 times. They were subsequently incubated with secondary antibody for 2 h at room temperature. Enhanced chemiluminescence reagent (Thermo Fisher Scientific, Inc.) was added to the membranes and images were captured using a Tanon 6100 chemiluminescent imaging system (Tanon Science and Technology Co., Ltd., Shanghai, China). The primary antibodies were E2F1 (dilution 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA; cat. no. 3742) and β-actin (dilution 1:2,000; Cell Signaling Technology, Inc., cat. no. 4970). The secondary antibody was horseradish peroxidase-conjugated secondary antibody (dilution 1:2,000, GTVision III Detection kit, GeneTech, Shanghai, China; cat. no. GK500710).
RNA immunoprecipitation (RIP)
A549 and H460 cell lysates containing NORAD and miRNAs were incubated with RIP buffer using EZ-Magna RIP RNA-binding protein immunoprecipitation kit, (EMD Millipore, Billerica, MA, USA), according to the manufacturer's protocols, containing magnetic beads conjugated to human anti-argonaute2 (Ago2) antibody (EMD Millipore). Normal mouse immunoglobulin G (IgG; EMD Millipore) was used as a negative control. NORAD and miRNAs presented in the precipitates were assessed by RT-qPCR.
Statistical analysis
All the data are presented as means ± standard deviation. The Student t-test, χ2 test, log-rank test and Fisher's exact test was performed for comparisons between each group. The correlation between NORAD, miR-136-5p and E2F1 expression was analyzed using Pearson Correlation. For comparison between two groups, we used two-tailed Student's t-test. Multiple group comparisons were calculated with one-way ANOVA analysis. Least-significant difference (LSD) was used as a post hoc test. Kaplan-Meier analysis was used to calculate overall survival. P<0.05 was used to indicate statistically significant results. The results are presented as the mean ± standard error of the mean.
Results
High NORAD expression in NSCLC
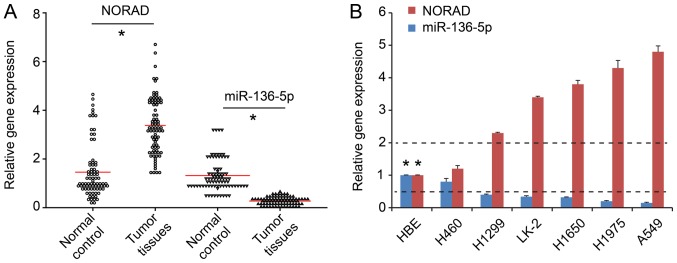
RT-qPCR demonstrated higher NORAD expression in NSCLC tumor tissues compared with adjacent tissues (n=80; P<0.05; Fig. 1A). Additionally, the expression of NORAD in the NSCLC lines was significantly higher compared with that in normal epithelial HBE cells (P<0.05; Fig. 1B). To further elucidate the underlying mechanism of NORAD, A549 and H460 cell lines were selected for subsequent experiments, as they exhibited the highest and lowest NORAD expression levels, respectively (Fig. 1B).
Figure 1.
High NORAD expression and low miR-136-5p expression in NSCLC. (A) NORAD expression was increased while miR-136-5p expression was decreased in the NSCLC tissues compared with these levels in the non-tumor adjacent tissues. (B) The expression of NORAD in NSCLC cell lines was higher compared with that in normal epithelial HBE cells, while miR-136-5p was lower. *P<0.05 vs. HBE cell line. NSCLC, non-small cell lung cancer; NORAD, non-coding RNA activated by DNA damage.
Effect of NORAD expression on NSCLC cell proliferation and glycolysis
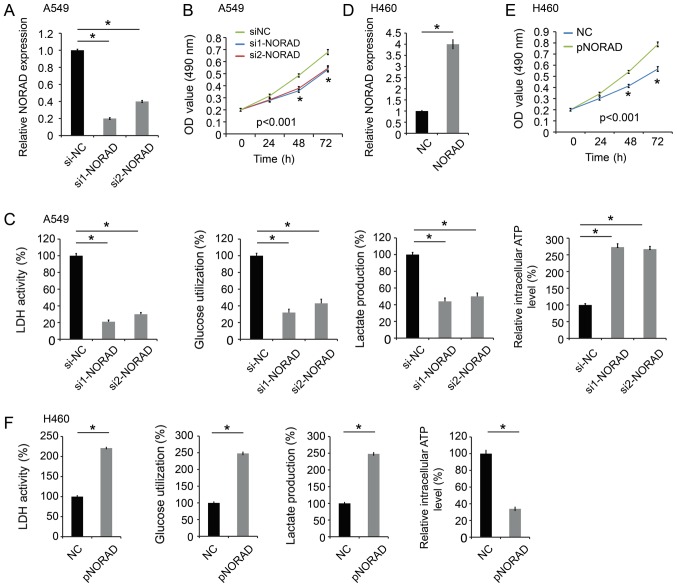
qPCR analysis indicated that the expression level of NORAD was decreased in A549 cells transfected with the NORAD small interfering RNAs (si1-NORAD and si2-NORAD) (Fig. 2A), and transfection with these siRNAs inhibited A549 cell proliferation (Fig. 2B). Since aerobic glycolysis is the primary aspect of altered cancer metabolism, a significant decrease was indicated in LDH activity, glucose utilization, lactate production, and an increase in intracellular ATP level in A549 cells following NORAD expression silencing (Fig. 2C).
Figure 2.
NORAD promotes cell proliferation and glycolysis. (A) The level of NORAD was decreased in A549 cells following transfection with NORAD siRNAs (si1-NORAD and si2-NORAD). (B) NORAD-knockdown inhibited A549 cell proliferation. (C) NORAD-knockdown significantly decreased LDH activity, glucose utilization, lactate production, and increased intracellular ATP level in A549 cells. (D) The expression level of NORAD was increased in H460 cells transfected with pcDNA3-NORAD (pNORAD). (E) NORAD overexpression promoted cell proliferation in H460 cells. (F) NORAD overexpression increased LDH activity, glucose utilization, lactate production, and decreased the intracellular ATP level in H460 cells. *P<0.05 vs. control group. NORAD, non-coding RNA activated by DNA damage; NC, negative control; siRNA, small interfering RNA; LDH, lactate dehydrogenase.
In addition, the function of ectopic expression of NORAD was examined in NSCLC. The H460 cell line was transfected with pcDNA3-NORAD (pNORAD) (Fig. 2D). The proliferation of H460 cells with NORAD overexpression was significantly increased compared with the control groups (Fig. 2E). Furthermore, NORAD overexpression increased LDH activity, glucose utilization, lactate production, and decreased intracellular ATP level in H460 cells (Fig. 2F). Therefore, these results indicated that NORAD could promote NSCLC proliferation and glycolysis.
NORAD functions as a competing endogenous RNA and sponges miR-136-5p in NSCLC
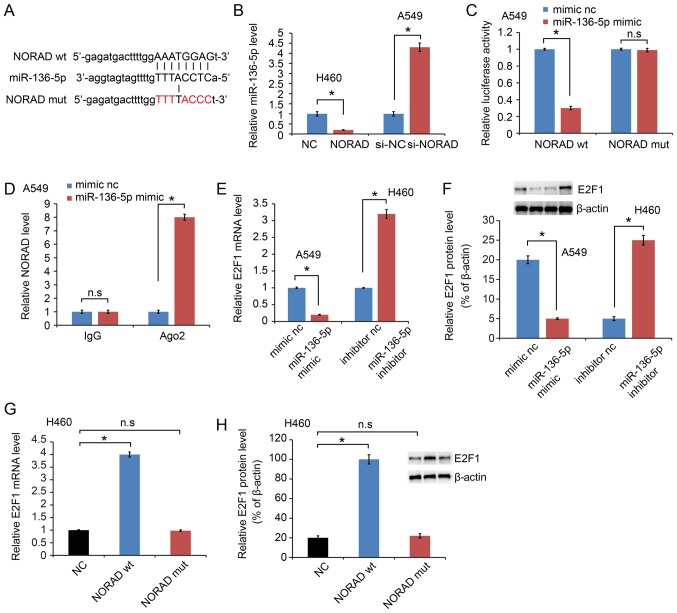
LncRNAs have been reported to sponge miRNAs and functionally liberate other RNA transcripts (9). A Starbase 2.0 software (http://starbase.sysu.edu.cn/) was used in the present study to predict the potential miRNA binding sites in NORAD and a sequence complementary to miRNA-136-5p was identified (Fig. 3A). In addition, miR-136-5p was upregulated in the si-NORAD-treated cells compared with that noted in the negative control-treated cells, while miR-136-5p was downregulated in phosphorylated-NORAD cells (Fig. 3B). A dual-luciferase reporter assay was performed in A549 cells to test the binding site of miR-136-5p in NORAD. The activity of luciferase reporters containing the theoretical binding site of NORAD in miR-136-5p was significantly decreased in the NORAD wt constructs, while no effect was observed in the NORAD mut constructs (Fig. 3C). NORAD expression was further examined on magnetic beads conjugated to the anti-Ago2 antibody and it was indicated that NORAD was highly enriched in cells transfected with the miR-136-5p mimic (Fig. 3D). This result suggested that NORAD and miR-136-5p can combine with Ago2 and form an increased RNA-induced silencing complex (RISC) and that miR-136-5p can bind to NORAD in NSCLC cells.
Figure 3.
NORAD serves as a competing endogenous RNA of miR-136-5p. (A) Bioinformatics analysis indicated that miR-136-5p directly targets the 3′-UTR of NORAD wt. NORAD mut means mutation of binding sites in the 3’-UTR of NORAD. (B) NORAD negatively regulated miR-136-5p expression in NSCLC cell lines. (C) miR-136-5p negatively regulated the luciferase activity of NORAD wt, rather than NORAD mut. (D) Detection of NORAD expression on magnetic beads conjugated to anti-Ago2 antibody. NORAD was highly enriched in cells transfected with the miR-136-5p mimic. The (E) mRNA expression and (F) protein expression level of E2F1 was detected following transfection with miR-136-5p mimics or inhibitor. The (G) mRNA expression and (H) protein expression level of E2F1 was examined in NORAD wt or NORAD mut cells. *P<0.05 vs. control group. NSCLC, non-small cell lung cancer; NORAD, non-coding RNA activated by DNA damage; wt, wild-type; mut, mutated; E2F1, E2F transcription factor 1. n.s. not significant.
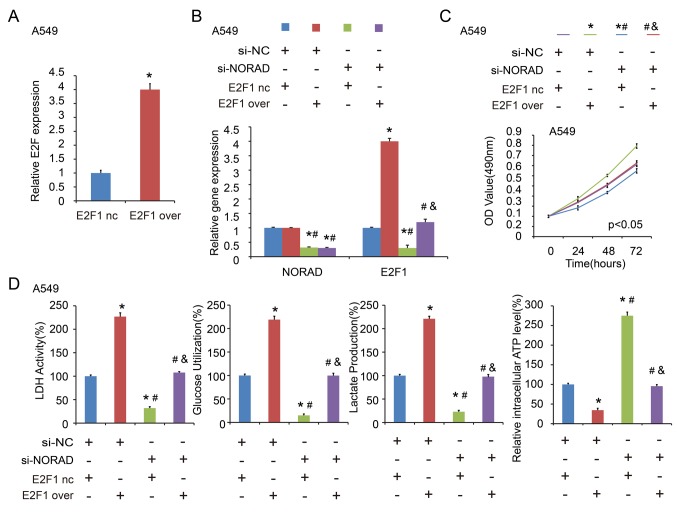
E2F transcription factor 1 (E2F1) is a potential target of miR-136-5p (12). The results of the present study also confirmed decreased E2F1 expression following transfection with miR-136-5p mimics, while upregulated expression was observed with the miR-136-5p inhibitor at the mRNA (Fig. 3E) and protein level (Fig. 3F). It was further indicated that E2F1 was increased by NORAD wt, while NORAD mut failed to increase E2F1 at the mRNA (Fig. 3G) and protein expression level (Fig. 3H).
NORAD is a target of miR-136-5p
We firstly validated miR-136-5p expression after transfection with the miR-136-5p mimic or inhibitor (Fig. 4A and B). Then NORAD was detected. Transfection with miR-136-5p mimics inhibited NORAD expression, while transfection with inhibitor exhibited an opposite effect (Fig. 4A and B).
Figure 4.
miR-136-5p regulates NORAD expression level. (A) Detection of miR-136-5p expression following transfection with miR-136-5p mimics. Transfection with miR-136-5p mimics inhibited NORAD expression. (B) Detection of miR-136-5p expression following transfection with the miR-136-5p inhibitor. Transfection with miR-136-5p inhibitor increased NORAD expression. *P<0.05 vs. control group. NORAD, non-coding RNA activated by DNA damage; nc, negative control.
miR-136-5p reverses the promoting effects of NORAD in NSCLC
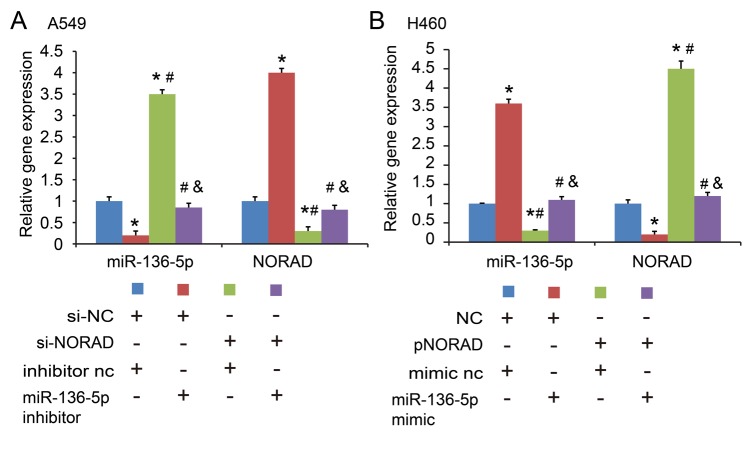
To understand the importance of miR-136-5p binding in NORAD promoting NSCLC progression, the present study co-transfected A549 cells with NORAD nc/si1 or miR-136-5p nc/inhibitor, and H460 cells with miR-136-5p mimic. The gene expression levels are indicated in Fig. 5A and B. It was indicated that miR-136-5p inhibitor reversed the inhibitory effect of NORAD-knockdown on cell proliferation (Fig. 6A) and glycolysis (Fig. 6B) in the A549 cell line. In addition, transfection with miR-136-5p mimic abrogated the enhanced effect of ectopic expression of NORAD on cell proliferation (Fig. 6C) and glycolysis (Fig. 6D) in H460 cells. E2F1 expression was confirmed by RT-qPCR after transfection of the E2F1 overexpression plasmid (Fig. 7A). Similarly, in the co-transfected A549 cells with NORAD nc/si1 or E2F1 nc/over (negative control/overexpression), it was indicated that E2F1 overexpression abolished the si-NORAD-mediated cell proliferation and glycolysis (Fig. 7B-D). These findings indicated that NORAD promoted NSCLC progression in part via competitively binding with miR-136-5p.
Figure 5.
Detection of NORAD and miR-136-5p expression levels. (A) Detection of NORAD and miR-136-5p expression levels by co-transfecting A549 cells with NORAD nc/si1 and miR-136-5p nc/inhibitor. *P<0.05 vs. si-NC+inhibitor nc group; #P<0.05 vs. si-NC+miR-136-5p inhibitor group; &P<0.05 vs. si-NORAD+ inhibitor nc group. (B) Detection of NORAD and miR-136-5p expression levels by co-transfecting H460 cells with NORAD nc/over and miR-136-5p nc/mimic. *P<0.05 vs. NC+mimic nc group; #P<0.05 vs. NC+miR-136-5p mimic group; &P<0.05 vs. pNORAD+mimic nc group. NORAD, non-coding RNA activated by DNA damage.
Figure 6.
miR-136-5p reversed the promoting effects of NORAD in NSCLC cells. (A) Cell proliferation and (B) glycolysis were examined following co-transfection with NORAD nc/si1 and miR-136-5p nc/inhibitor in A549 cells. *P<0.05 vs. si-NC+inhibitor nc group; #P<0.05 vs. si-NC+miR-136-5p inhibitor group; &P<0.05 vs. si-NORAD+ inhibitor nc group. (C) Cell proliferation and (D) glycolysis were detected following co-transfection with NORAD nc/over and miR-136-5p nc/mimics in H460 cells. *P<0.05 vs. NC+mimic nc group; #P<0.05 vs. NC+miR-136-5p mimic group; &P<0.05 vs. pNORAD+mimic nc group. NORAD, non-coding RNA activated by DNA damage; NSCLC, non-small-cell lung cancer.
Figure 7.
E2F1 overexpression abolishes the si-NORAD-mediated cell proliferation and glycolysis. (A) Detection of E2F1 expression by RT-qPCR. (B) Detection of NORAD and E2F1 expression levels by co-transfecting A549 cells with NORAD nc/si1 and E2F1 nc/over. (C) Cell proliferation and (D) glycolysis were detected following co-transfection with NORAD nc/si1 and E2F1 nc/over in A549 cells. *P<0.05 vs. si-NC+E2F1 nc group; #P<0.05 vs. si-NC+E2F1 over group; &P<0.05 vs. si-NORAD+ E2F1 nc group. NORAD, non-coding RNA activated by DNA damage; E2F1, E2F transcription factor 1.
Association of NORAD, miR-136-5p and E2F1 in NSCLC
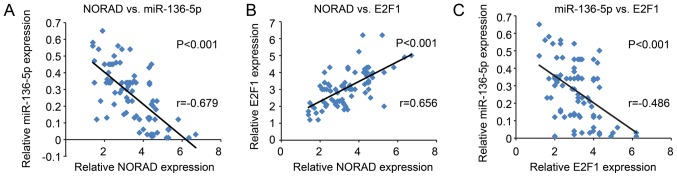
After analyzing their expression and association, NORAD was found to be inversely correlated with miR-136-5p, while positively correlated with E2F1 expression (Fig. 8A and B). In addition, miR-136-5p was inversely correlated with E2F1, all of which were indicated to be statistically significant (Fig. 8C).
Figure 8.
Correlation between NORAD, miR-136-5p and E2F1 in NSCLC tissues. Correlations between (A) NORAD and miR-136-5p, (B) NORAD and E2F1 and (C) miR-136-5p and E2F1. NORAD, non-coding RNA activated by DNA damage; E2F1, E2F transcription factor 1; NSCLC, non-small cell lung cancer.
Discussion
Accumulating evidence has revealed that lncRNA deregulation promotes the growth of human malignancies (13), resulting in tumor progression and uncontrolled tumor growth, providing insights into novel strategies for cancer treatment. In lung cancers, numerous lncRNAs have been investigated, in order to understand the complex underlying mechanisms, including Hox transcript antisense RNA (HOTAIR) (14), taurine upregulated 1 (15), tubulin α 4b (TUBA4B) (16) and metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) (17). However, the underlying mechanisms of lncRNAs in lung cancer remain unclear. The present study identified an lncRNA, named NORAD, which was increased in lung cancer tissues, as well as in NSCLC cell lines, demonstrating that NORAD serves oncogenic functions in NSCLC.
Cancer metabolism, characterized by aerobic glycolysis or Warburg effect, is characterized by the unusual use of glucose in the production of lactate even under sufficient oxygen conditions (18). However, the functions of lncRNAs in cancer metabolism remain unclear. Previous research has revealed that lncRNA crystalline β-gamma domain containing 3 (CRYBG3) interacts with lactate dehydrogenase A to affect glycolysis (18). In ovarian cancer, lncRNA small nucleolar RNA host gene 3 (SNHG3) regulates mitochondrial proteomes, affecting cancer metabolism (19). In the present study, a significant decrease in LDH activity, glucose utilization, lactate production, and an increase in intracellular ATP level, was observed in lung cancer cells when the NORAD expression level was modified. NORAD affects glycolysis, leading to a decrease in ATP. However, we found that ATP was elevated in our data. First, we speculate that NORAD may affect the Krebs cycle, which in turn increases ATP. Moreover, a large amount of lactic acid is produced during the glycolysis of malignant tumors, and is transported into the liver through blood to provide sufficient raw materials for gluconeogenesis, resulting in increased conversion of glucose conversion and utilization of glucose barrier by peripheral tissues. These procedures consume a large amount of ATP. Therefore, when lactic acid is reduced, gluconeogenesis is reduced and ATP is increased. The specific mechanism needs to be further elaborated. These results indicate that NORAD could regulate metabolism in NSCLC mainly by affecting glycolysis.
In the past decade, investigation of miRNAs has dominated the field of non-coding RNA regulation (20,21). miRNAs have been reported to impact, in various manners, genetic alteration and signaling pathways (22). It is well known that miRNAs exert their gene silencing functions through a ribonucleoprotein complex called the RNA induced silencing complex (RISC) (23). Potential microRNA targets can be isolated from this complex after Ago2 co-immunoprecipitation as Ago2 is a vital component of the RISC complex necessary for siRNA or miRNA-mediated gene silencing (24). In the present study, the data showed that high miR-136-5p and NORAD both bind to the Ago2 complex. We proposed that miR-136-5p inhibited the level of NORAD expression in a way similar to miRNA-mediated silencing of protein-coding genes. Therefore, a high level of miR-136-5p leads to RISC cleavage, resulting in decreased NORAD expression. Conversely, low miR-136-5p leads to high NORAD expression. Our results are consistent with a previous report of functional interactions among microRNAs and long non-coding RNAs (25–27).
The activity of the dual-luciferase reporter assay containing the theoretical binding site of NORAD in miR-136-5p was significantly decreased in NORAD wt constructs, while there was no effect in NORAD mut constructs. It was also demonstrated that miR-136-5p mimics arrested lung cancer cell proliferation and glycolysis, and NORAD reversed this function. These findings demonstrated that NORAD interacts with miR-136-5p in NSCLC proliferation. Previous research has verified that E2F1 is a target of NORAD (28). lncRNAs act as ceRNA to bind specific miRNAs and regulate their function. NORAD regulates E2F1 expression directly. We hypothesized that NORAD may inhibit miR-136-5p expression in such a manner. This suggests that NORAD may enhance E2F1 by repressing miR-136-5p. These results suggest a NORAD/miR-136-5p/E2F1 axis that regulates cell proliferation and glycolysis in NSCLC. A number of limitations in our experiments must be mentioned, including the insufficient variety of functional experiments performed (i.e. migration and invasion assays). Thus, further investigation is warranted.
In conclusion, the present study demonstrated that the lncRNA NORAD promotes NSCLC cell proliferation and glycolysis by competitively binding miR-136-5p. NORAD may be valuable as a targeted therapy. We identified crosstalk between miR-136-5p and NORAD, shedding novel light on the potential treatment of NSCLC.
Acknowledgements
Not applicable.
Funding
This study was funded by the Natural Science Foundation of Nanjing Drum Tower Hospital Group Suqian People's Hospital (grant no. S201518).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WG and LFW performed the in vitro studies. TW made substantial contributions to the design of the present study, and interpreted the data. BS and YW wrote the manuscript and interpreted the data. WSM and XYW collected tumor tissues, acquired data and revised the article critically for important intellectual content. LJ and LJF participated in designing the study. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Informed consents were obtained from all of the participating patients, and the study was approved by the Clinical Research Ethics Committee of Suqian People's Hospital of Nanjing Drum Tower Hospital Group (Nanjing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Chang JY, Senan S, Paul MA, Mehran RJ, Louie AV, Balter P, Groen HJ, McRae SE, Widder J, Feng L, et al. Stereotactic ablative radiotherapy versus lobectomy for operable stage I non-small-cell lung cancer: A pooled analysis of two randomised trials. Lancet Oncol. 2015;16:630–637. doi: 10.1016/S1470-2045(15)70168-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khandelwal A, Bacolla A, Vasquez KM, Jain A. Long non-coding RNA: A new paradigm for lung cancer. Mol Carcinog. 2015;54:1235–1251. doi: 10.1002/mc.22362. [DOI] [PubMed] [Google Scholar]
- 4.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 5.Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Li J, Koirala P, Ding X, Chen B, Wang Y, Wang Z, Wang C, Zhang X, Mo YY. Long non-coding RNAs as prognostic markers in human breast cancer. Oncotarget. 2016;7:20584–20596. doi: 10.18632/oncotarget.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Wang X, Wen C, Huo Z, Wang W, Zhan Q, Cheng D, Chen H, Deng X, Peng C, Shen B. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol Cancer. 2017;16:169. doi: 10.1186/s12943-017-0738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Li XY, Hu P, Ding YS. LncRNA NORAD contributes to colorectal cancer progression by inhibition of miR-202-5p. Oncol Res. 2018 doi: 10.3727/096504018X15190844870055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Chen W, Yang Y, Chen B, Lu P, Zhan L, Yu Q, Cao K, Li Q. MiR-136 targets E2F1 to reverse cisplatin chemosensitivity in glioma cells. J Neurooncol. 2014;120:43–53. doi: 10.1007/s11060-014-1535-x. [DOI] [PubMed] [Google Scholar]
- 13.Lalevée S, Feil R. Long noncoding RNAs in human disease: Emerging mechanisms and therapeutic strategies. Epigenomics. 2015;7:877–879. doi: 10.2217/epi.15.55. [DOI] [PubMed] [Google Scholar]
- 14.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, Han L, Xia R, Wang KM, Yang JS, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Hu L, Wang J, Zhang F, Chen J, Xu G, Wang Y, Pan Q. Low expression LncRNA TUBA4B is a poor predictor of prognosis and regulates cell proliferation in non-small cell lung cancer. Pathol Oncol Res. 2017;23:265–270. doi: 10.1007/s12253-016-0089-y. [DOI] [PubMed] [Google Scholar]
- 17.Han T, Jiao F, Hu H, Yuan C, Wang L, Jin ZL, Song WF, Wang LW. EZH2 promotes cell migration and invasion but not alters cell proliferation by suppressing E-cadherin, partly through association with MALAT-1 in pancreatic cancer. Oncotarget. 2016;7:11194–11207. doi: 10.18632/oncotarget.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaitheesvaran B, Xu J, Yee J, Q-Y L, Go VL, Xiao GG, Lee WN. The Warburg effect: A balance of flux analysis. Metabolomics. 2015;11:787–796. doi: 10.1007/s11306-014-0760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li N, Zhan X, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. 2018;150:343–354. doi: 10.1016/j.ygyno.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Mirzaei H, Masoudifar A, Sahebkar A, Zare N, Sadri Nahand J, Rashidi B, Mehrabian E, Mohammadi M, Mirzaei HR, Jaafari MR. MicroRNA: A novel target of curcumin in cancer therapy. J Cell Physiol. 2018;233:3004–3015. doi: 10.1002/jcp.26055. [DOI] [PubMed] [Google Scholar]
- 21.Kumar R, Xi Y. MicroRNA, epigenetic machinery and lung cancer. Thorac Cancer. 2011;2:35–44. doi: 10.1111/j.1759-7714.2011.00043.x. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, Kang C. miRNA interventions serve as ‘magic bullets’ in the reversal of glioblastoma hallmarks. Oncotarget. 2015;6:38628–38642. doi: 10.18632/oncotarget.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang H, Huang G, Zhao N, Zhang T, Jiang M, He Y, Zhou X, Jiang X. Long non-coding RNA TPT1-AS1 promotes cell growth and metastasis in cervical cancer via acting AS a sponge for miR-324-5p. J Exp Clin Cancer Res. 2018;37:169. doi: 10.1186/s13046-018-0846-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Zhuo M, Yuan C, Han T, Cui J, Jiao F, Wang L. A novel feedback loop between high MALAT-1 and low miR-200c-3p promotes cell migration and invasion in pancreatic ductal adenocarcinoma and is predictive of poor prognosis. BMC Cancer. 2018;18:1032. doi: 10.1186/s12885-018-4954-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu HJ, Jin PY, Tang Y, Fan SH, Zhang ZF, Wang F, Wu DM, Lu J, Zheng YL. microRNA-136 inhibits proliferation and promotes apoptosis and radiosensitivity of cervical carcinoma through the NF-κB pathway by targeting E2F1. Life Sci. 2018;199:167–178. doi: 10.1016/j.lfs.2018.02.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.