Abstract
The poor outcome of patients with esophageal squamous cell carcinoma (ESCC) highlights the importance of the identification of novel effective prognostic biomarkers. Long non-coding RNAs (lncRNAs) serve regulatory roles in various types of cancer. The aim of the present study was to investigate the lncRNA expression profile in ESCC and to identify lncRNAs associated with the prognosis of ESCC by performing comprehensive bioinformatics analyses. The RNA-sequencing (Seq) expression dataset GSE53625 generated from ESCC samples was used as a training dataset. Additional RNA-Seq datasets relative to ESCC samples were downloaded from The Cancer Genome Atlas and used as a validation dataset. Data were screened using the limma package, and differentially expressed lncRNAs between early- and late-stage ESCC were identified. A random forest algorithm was used to select the optimal lncRNA biomarkers, which were then analyzed using the support vector machine (SVM) algorithm with R software. The identified lncRNA biomarkers were examined in the validation dataset by bidirectional hierarchical clustering and using an SVM classifier. Subsequently, univariate and multivariate Cox regression analyses were performed to analyze the potential ability lncRNAs to predict the survival rate of patients with ESCC. By examining the training group, 259 deregulated lncRNAs between early- and advanced-stage ESCC were identified. Further bioinformatics analyses identified a nine-lncRNA signature, including AC098973, AL133493, RP11-51M24, RP11-317N8, RP11-834C11, RP11-69C17, LINC00471, LINC01193 and RP1-124C. This nine-lncRNA signature was used to predict the tumor stage and patient survival rate with high reliability and accuracy in the training and validation datasets. Furthermore, these nine lncRNA biomarkers were primarily involved in regulating the cell cycle and DNA replication, and these processes were previously identified to be associated with the progression of ESCC. The identified nine-lncRNA signature was identified to be associated with the tumor stage, and could be used as predictor of the survival rate of patients with ESCC.
Keywords: esophageal squamous cell carcinoma, long non-coding RNA signature, survival, tumor stage
Introduction
Esophageal cancer is one of the most common lethal malignancies worldwide (1). Esophageal cancer is the sixth most common cause of cancer-associated mortality, causing >400,000 mortalities per year (2). Esophageal cancer presents as two principal types: Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma. Although the incidence rate of Barrett's adenocarcinoma is increasing in Western countries, the incidence of ESCC is increasing at a fast rate in East Asian populations (3). Due to the lack of specific symptoms and effective early diagnostic methods, the 5-year survival rate of patients with ESCC remains low, ranging between 10 and 25% (4). Current biomarkers, such as serum squamous cell carcinoma antigen, carbohydrate antigen 19-9, carcinoembryonic antigen and cytokeratin-19 fragments, are commonly used in the diagnosis of patients with ESCC. However, these tumor markers are not used in the early detection of ESCC, due to insufficient diagnostic sensitivity and specificity (5,6). Therefore, understanding the molecular mechanisms underlying ESCC tumorigenesis would facilitate the identification and the development of novel biomarkers with high sensitivity and specificity that may be able to improve the early detection and prognosis of ESCC.
The development and progression of cancer involve various types of genomic alterations, including DNA mutations, epigenetic modifications, changes in gene expression and the complex interplay of these processes (7). Epigenetic modifications, such as DNA methylation, histone deacetylation, chromatin remodeling and non-coding RNA (ncRNA) regulation, are critical for the development and metastasis of various types of cancer, including ESCC (8,9). ncRNAs have attracted increasing interest over the past decade. Long ncRNAs (lncRNAs) are a large class of ncRNAs of >200 nucleotides in length, and lncRNA genes are interspersed within the genome (10–12). lncRNAs have been shown to serve critical roles in cancer initiation and progression, mediating oncogenic or tumor suppressing effects at the transcriptional and post-transcriptional levels (13,14). Aberrantly expressed lncRNAs have been reported to serve as potential biomarkers for cancer diagnosis and prognosis (15). For example, the increased expression of HOX transcript antisense RNA in metastatic breast cancer (16), CDKN2B antisense RNA 1-induced epigenetic silencing of cyclin dependent kinase inhibitor 2B in leukemia (17) and the increased expression of metastasis associated lung adenocarcinoma transcript 1 in metastatic non-small cell lung cancer are lncRNA-mediated processes associated with the development or progression of cancer (18,19). Dysregulated lncRNAs are frequently observed in ESCC (20), but the functions of most lncRNAs in ESCC remain unclear.
Systems biology approaches can facilitate the understanding of the pathogenesis of ESCC and the identification of potential novel biomarkers. Many transcriptome analyses and datasets of ESCC samples have been generated, and several lncRNAs have been identified as ESCC-associated lncRNAs (21–25). Nevertheless, compared with coding genes and microRNAs, the specific lncRNAs involved in the onset and development of ESCC remain unknown.
The main aim of the present study was to identify effective biomarkers or therapeutic targets associated with ESCC. An ESCC gene expression profile dataset was downloaded from the Gene Expression Omnibus (26) (GEO; accession no. GSE53625) and was used as a training dataset. Additionally, expression profiles were downloaded from The Cancer Genome Atlas (27) (TCGA) and were used as a validation dataset. Tumor samples were then divided into early- and advanced-stage ESCC, according to the Tumor-Node-Metastasis (TNM) staging system (28), and differentially expressed lncRNAs (DElncRs) between early- and advanced-stage tumor samples were identified. A random forest algorithm was used to select optimal lncRNA biomarkers, which were then analyzed via the support vector machine (SVM) algorithm in R. The identified lncRNA biomarkers were also examined in the validation dataset by performing bidirectional hierarchical clustering and classification using an SVM classifier. Univariate and multivariate Cox regression analyses were then performed to determine the ability of the identified lncRNAs to predict the patient survival rates.
Materials and methods
RNA expression data
RNA expression profiles and patient information from the GSE53625 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM1296956) were downloaded from the GEO (http://www.ncbi.nlm.nih.gov/geo/) database, which is based on the Affymetrix (Thermo Fisher Scientific, Inc.) GPL18109 platform. In the original study, tumor tissues were collected from 179 patients with ESCC (29). In the present study, the GSE53625 dataset was used as a training dataset. RNA-Seq expression profiling data generated using the Illumina HiSeq 2000 (Illumina, Inc.) and ESCC patient information (http://www.cbioportal.org/study?id=hnsc_tcga#clinical) were downloaded from TCGA (https://gdc-portal.nci.nih.gov/). The TCGA dataset was used as a validation dataset. The GSE53625 dataset contained normalized gene expression data. The TCGA RNA-Seq expression data were quantified using an Expectation-Maximization algorithm (30) of normalized read counts (log2 transformed). The demographic and clinical data of patients in both the training and validation datasets are presented in Table I.
Table I.
Demographic and clinical information of patients in the training dataset GSE53625 and in the validation dataset downloaded from TCGA.
| Patient characteristics | GSE53625 dataset, n=179 | TCGA dataset, n=86 |
|---|---|---|
| Age, years (mean ± SD) | 59.34±9.03 | 58.29±10.63 |
| Gender | ||
| Male | 146 | 75 |
| Female | 33 | 11 |
| Alcohol use | ||
| Yes | 106 | 62 |
| No | 73 | 23 |
| Unavailable | 0 | 1 |
| Tobacco use | ||
| Yes | 114 | 57 |
| No | 65 | 27 |
| Unavailable | 0 | 2 |
| Pathological grade N | ||
| N0 | 83 | 49 |
| N1 | 62 | 26 |
| N2 | 22 | 6 |
| N3 | 12 | 2 |
| Unavailable | 0 | 3 |
| Pathological grade T | ||
| T1 | 12 | 7 |
| T2 | 27 | 25 |
| T3 | 110 | 48 |
| T4 | 30 | 4 |
| Unavailable | 0 | 2 |
| Tumor stage | ||
| I | 10 | 7 |
| II | 77 | 47 |
| III | 92 | 27 |
| IV | 0 | 3 |
| Unavailable | 0 | 2 |
| Adjuvant therapy | ||
| Yes | 104 | 0 |
| No | 45 | 0 |
| Unavailable | 30 | 86 |
| Survival status | ||
| Deceased | 106 | 30 |
| Alive | 73 | 54 |
| Unavailable | 0 | 2 |
| Overall survival, months (mean ± SD) | 36.25±22.86 | 13.67±11.82 |
TCGA, The Cancer Genome Atlas.
DElncR identification
According to the pathological disease stage of patients in the training dataset, tumor tissues were divided into early-stage (stage I and II) and advanced-stage (stage III and IV) ESCC. The limma software (version 3.34.9) package of R/Bioconductor (version 3.6) (31) was used to analyze DElncRs between early- and advanced-stage ESCC. False discovery rate <5% was used as the cutoff value, based on a permutation test, as previously described (32). The fold-change (FC) values of gene expression between tumor tissues and normal esophageal tissues were calculated, and |log2FC|>0.263 was used as the cutoff value.
DElncR clustering analysis
Bidirectional hierarchical clustering based on the expression profile of the DElncRs identified in the GSE53625 dataset was performed by calculating the centered Pearson correlation coefficient (33). A heatmap was then constructed using the R package pheatmap (version 1.0.12) (34). To determine whether the Pearson correlation coefficient was appropriate for hierarchical clustering, the chisq.test (χ2) function in R and the Kaplan-Meier method in the R survival package (version 2.43–3; http://cran.r-project.org/web/packages/survival/index.html) were used for further evaluation. Specifically, the associations between the clusters classified by hierarchical clustering and the stages of ESCC were analyzed using the chisq.test function in R. Subsequently, the Kaplan-Meier method in the R survival package was used to estimate the associations between clusters and patient survival rates based on the patient information in the different clusters.
Identification of the optimal combination of lncRNAs using a random forest algorithm
Optimal lncRNA biomarkers for ESCC were selected using a random forest algorithm, which was calculated using bootstrap methods (35). The random forest prediction model was generated using the R package randomForest (version 4.6–14) (36). In the bootstrap method, out-of-bag (OBB) error rates were used to evaluate the selection performance of the random forest algorithm, and lower OBB error rates indicated higher prediction accuracy.
Construction of a SVM classifier
The optimal combination of lncRNAs was further analyzed using the SVM function in the e1071 package of R, and a SVM classifier was constructed (37). The SVM classifier was used to separate the data points from the two classification groups using a decision surface. To evaluate the robustness of the SVM model, a 10-fold cross-validation was performed (38). The SVM classifier was then used to distinguish between patients with early- and advanced-stage-like ESCC. Moreover, Kaplan-Meier curves were plotted for patients with early- and advanced-stage ESCC and compared using the log-rank test.
Validation of optimal lncRNA biomarkers
The identified optimal lncRNA biomarkers were hierarchically clustered by calculating Pearson's correlation coefficient. The SVM classifier, generated using the training dataset, was used to distinguish between patients with early- and advanced-stage ESCC in the validation dataset.
Univariate and multivariate Cox regression analysis
Univariate and multivariate Cox regression analyses were performed to identify independent prognostic factors associated with patient survival rates. The tumor stages were classified using the SVM classifier. Then, demographic and clinical information, including age, gender, alcohol use, tobacco use, TNM grade and adjuvant therapy, were analyzed using univariate and multivariate analyses. In addition, the patients were separated based on age, gender, alcohol use, tobacco use, TNM grade and adjuvant therapy and the relationship between the tumor stage, as classified by the SVM classifier, and patient prognosis was analyzed.
Functional enrichment analysis of genes associated with the identified lncRNA biomarkers
The correlation between optimal lncRNA biomarkers and the expression of all genes in the training dataset was evaluated by calculating Pearson correlation coefficients using the cor.test function in R (39). Each lncRNA was associated with ≥1 gene. The genes were arranged in descending order based on the absolute value of the Pearson coefficient, and an lncRNA-mRNA network was generated using the top 1% of lncRNA-mRNA gene pairs.
Next, the mRNA-mRNA interactions of the top 1% of genes were identified using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (40). Using a STRING score >0.8, the genetic interactions predicted by the protein-protein interaction network were further analyzed. Finally, Kyoto Encyclopedia of Genes and Genomes (KEGG) (41) pathway enrichment analysis was performed for all mRNAs that correlated with the expression of the identified lncRNAs using the Database for Annotation, Visualization and Integrated Discovery (version 6.8) bioinformatics resources (42), and P<0.05 was considered to indicate a statistically significant difference.
Results
DElncR screening
A flow chart of the present study is shown in Fig. 1. According to the pathological disease stage of patients in the training dataset, the tumor samples collected from 179 patients were divided into early- and advanced-stage ESCC, and the two groups consisted of 87 and 92 patients, respectively. Using the limma R package, a total of 259 DElncRs were identified, including 175 downregulated and 84 upregulated lncRNAs.
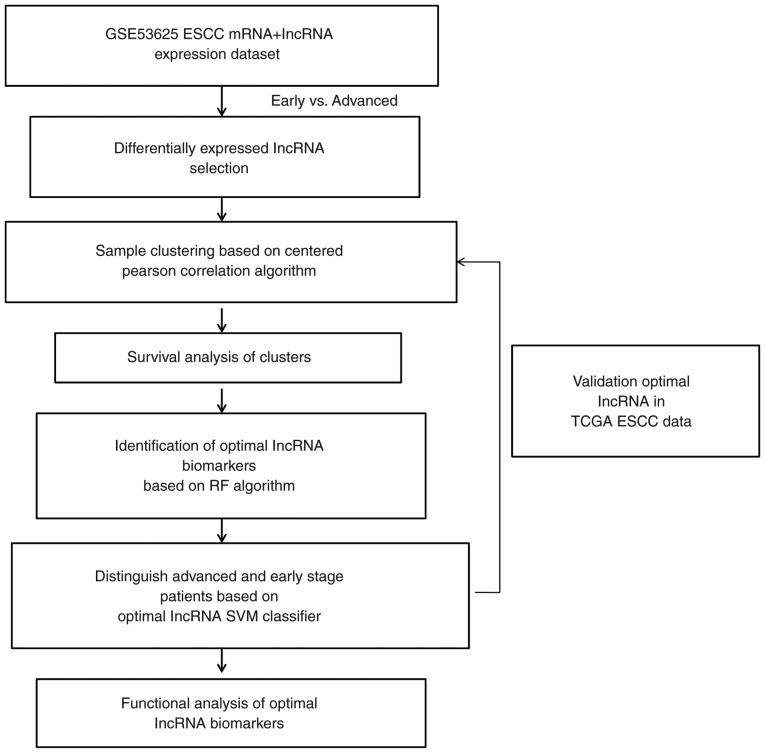
Figure 1.
Flow chart of the present study. TCGA, The Cancer Genome Atlas; lncRNA, long non-coding RNA; ESCC, esophageal squamous cell carcinoma; RF, random forest; SVM, support vector machine.
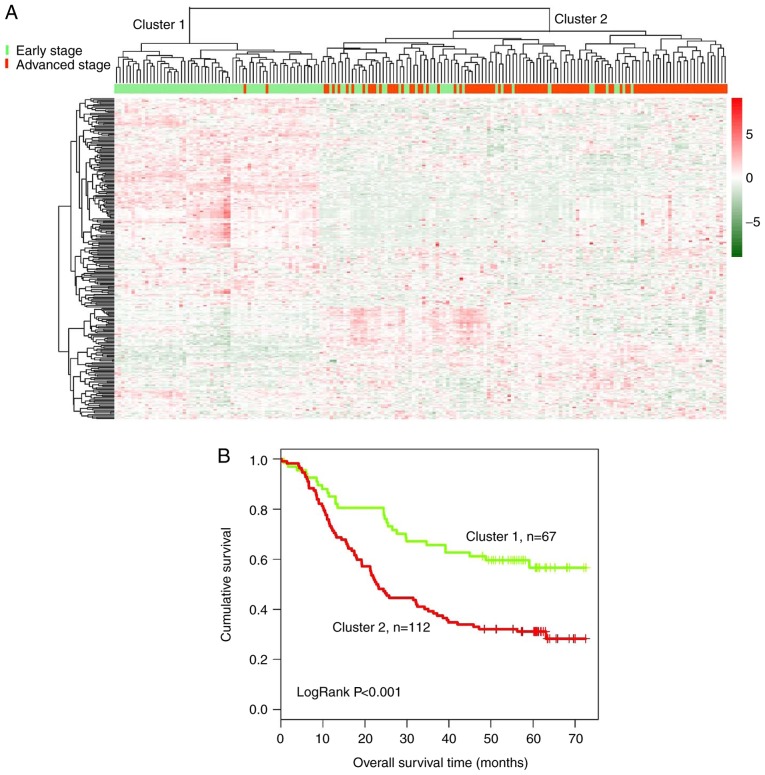
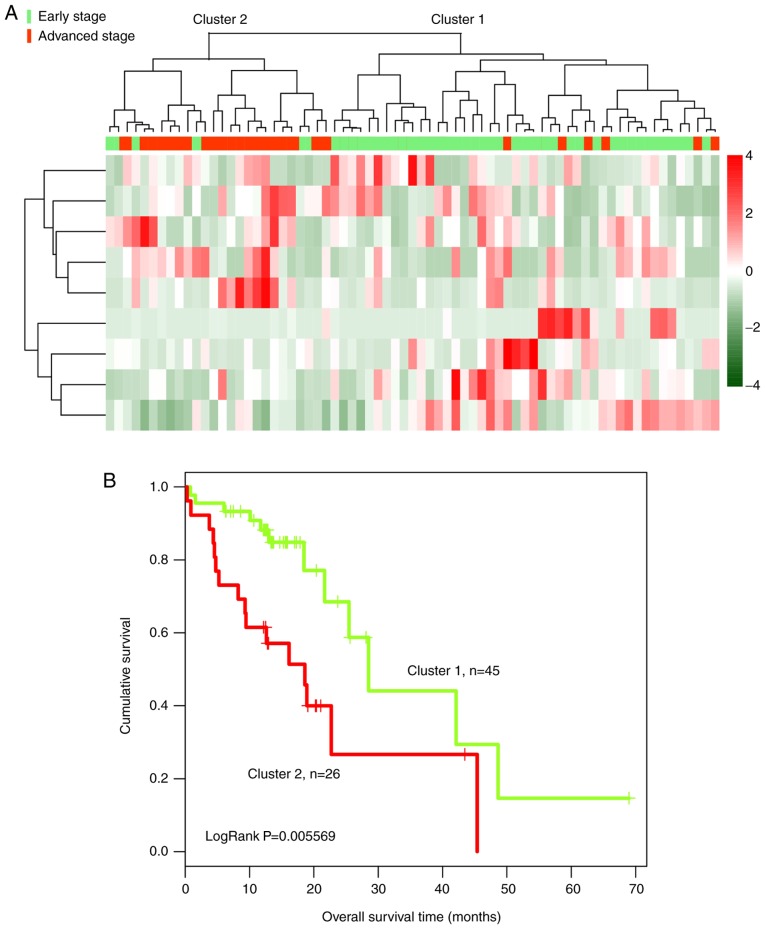
Subsequently, bidirectional hierarchical clustering was performed based on the expression profiles of 259 DElncRs in 179 tumor tissue samples, by calculating the centered Pearson correlation. Samples were separated into two clusters (Fig. 2A). In cluster 1, 65 of 67 tumor samples presented at an early stage. In cluster 2, most of the tumor samples (90/112) presented at an advanced stage, whereas 22 samples presented at an early stage. The accuracy of tumor stage identification was 86.59% (155/179; χ2=97.39; P=2.20×10−16).
Figure 2.
Clustering and survival analysis of 179 esophageal squamous cell carcinoma samples based on the expression of 259 differentially expressed lncRNAs in the training dataset. (A) Bidirectional hierarchical clustering of 259 lncRNAs in 179 tumor tissue samples. Green indicates early-stage tumors and red indicates advanced-stage tumors. (B) Kaplan-Meier survival curves of patients from cluster 1 and cluster 2. The green curve represents patients from cluster 1 and the red curve represents patients from cluster 2. lncRNA, long non-coding RNA.
Kaplan-Meier analysis suggested that the patients in cluster 1 exhibited a significantly longer survival time compared with the patients in cluster 2 (43.50±21.51 and 31.92±22.64 months, respectively; P=2.639×10−4; Fig. 2B).
Identification of lncRNA biomarkers for early-stage ESCC
A random forest algorithm was used to identify the most important lncRNAs. The optimal lncRNA combination was obtained using the smallest OBB error rate (OBB error=0.183; Fig. 3). A total of nine optimal lncRNA biomarkers of ESCC were identified, including AC098973, AL133493, RP11-51M24, RP11-317N8, RP11-834C11, RP11-69C17, LINC00471, LINC01193 and RP1-124C. The expression levels of AC098973, AL133493, RP11-51M24 and RP11-317N8 were significantly increased in early-stage tumors compared with advanced-stage tumors, whereas the expression levels of the remaining five lncRNAs were significantly decreased in early-stage tumors (Table II).
Figure 3.
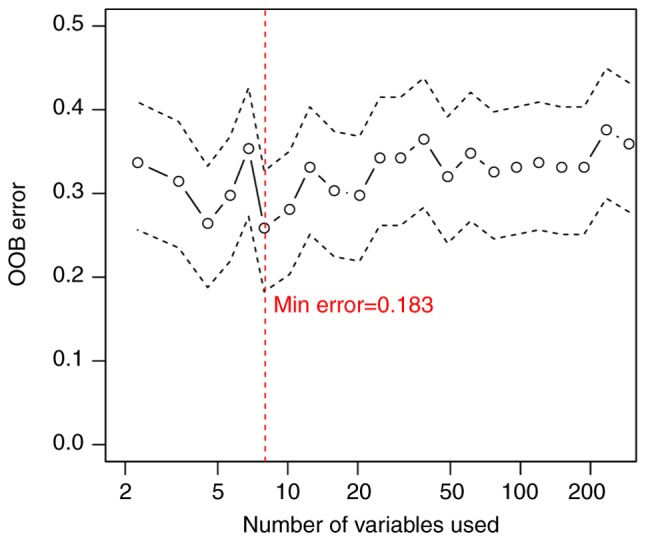
Random forest prediction model. The x-axis represents the number of long non-coding RNAs. The y-axis represents the OOB error rate. OOB, out-of-bag.
Table II.
lncRNA biomarkers associated with the progression of esophageal squamous cell carcinoma.
| Ensembl ID | Gene name | Genomic coordinates | P-value | False discovery rate | Log2 fold change |
|---|---|---|---|---|---|
| ENSG00000225548 | AC098973 | Chr3: 27,802,762-27,891,301(+) | 0.0002 | 0.0078 | −0.5822 |
| ENSG00000233922 | AL133493 | Chr21: 45,593,654-45,603,056(+) | 0.0008 | 0.0340 | −0.5420 |
| ENSG00000249875 | RP11-51M24 | Chr4: 174,354,854-174,376,445(−) | <0.0001 | 0.0012 | −0.3375 |
| ENSG00000257272 | RP11-317N8 | Chr14: 35,873,857-35,875,303(+) | 0.0001 | 0.0043 | −0.3017 |
| ENSG00000249388 | RP11-834C11 | Chr12: 54,082,118-54,102,693(+) | 0.0002 | 0.0079 | 0.2818 |
| ENSG00000227912 | RP11-69C17 | Chr10: 2,166,332-2,169,460(+) | 0.0003 | 0.0139 | 0.2981 |
| ENSG00000181798 | LINC00471 | Chr2: 231,508,426-231,514,339(+) | <0.0001 | 0.0014 | 0.4400 |
| ENSG00000258710 | LINC01193 | Chr15: 20,940,438-20,993,303(+) | 0.0002 | 0.0083 | 0.5258 |
| ENSG00000232316 | RP1-124C6 | Chr6: 113,428,540-113,433,421(−) | <0.0001 | 0.0002 | 0.5867 |
lncRNA, long non-coding RNA.
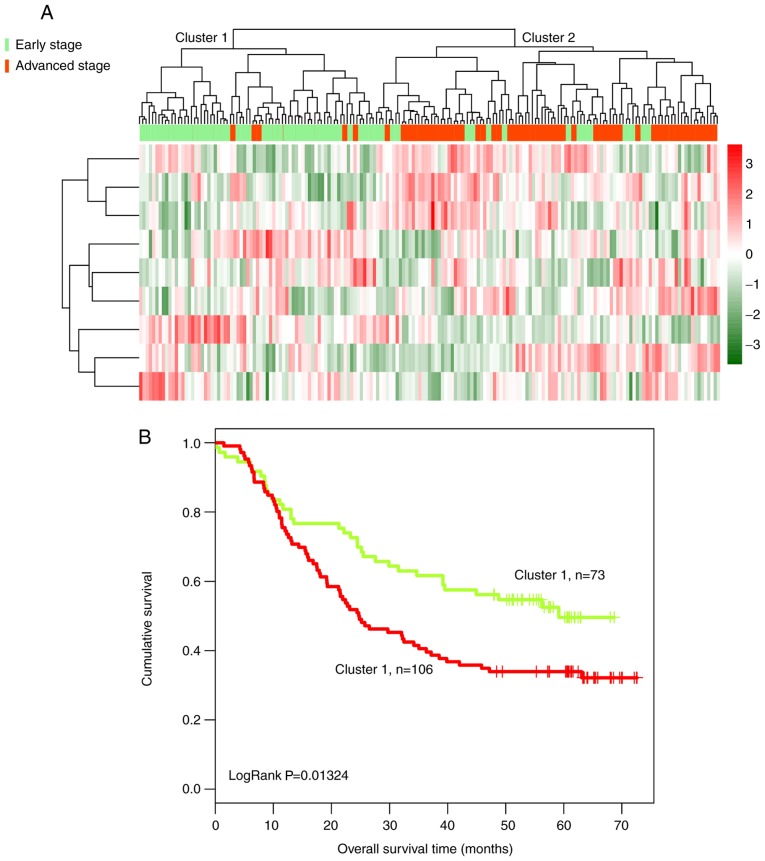
All tumor samples were hierarchically clustered based on the expression level of the nine identified lncRNAs, by calculating Pearson correlation coefficients. Tumor samples were divided into two clusters (Fig. 4A). In cluster 1, 68 of 73 tumor samples presented at an early stage and only five tumor samples presented at an advanced stage. By contrast, in cluster 2, 87 of 106 tumor samples presented at an advanced stage, and 19 samples presented at an early stage. The clusters exhibited an overall accuracy of 86.59% (155/179). The patients in cluster 1 had a significantly longer overall survival time compared with the patients in cluster 2 (40.25±21.61 and 33.50±23.39 months, respectively; Fig. 4B). The present results suggested that these nine lncRNAs could be used to predict the survival outcome of patients with ESCC.
Figure 4.
Clustering and survival analysis of 179 ESCC tumors based on the expression of nine lncRNA biomarkers in the training dataset. (A) Bidirectional hierarchical clustering of nine lncRNA biomarkers of ESCC in 179 tumor samples in the training dataset. Green indicates early-stage tumors and red indicates advanced-stage tumors. (B) Kaplan-Meier survival curves of patients from cluster 1 and cluster 2. The green curve represents patients from cluster 1 and the red curve represents patients from cluster 2. ESCC, esophageal squamous cell carcinoma; lncRNA, long non-coding RNA.
lncRNA classification by SVM classifier
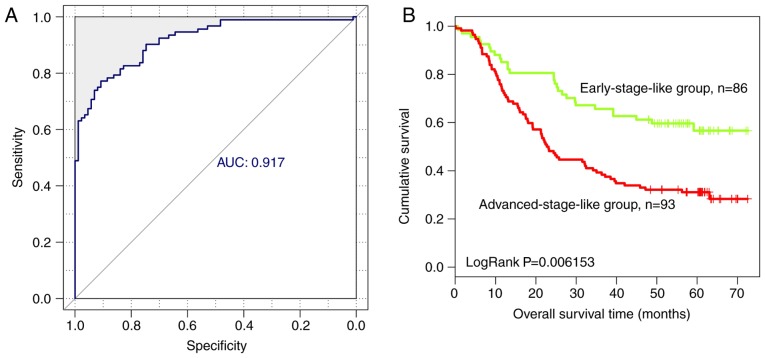
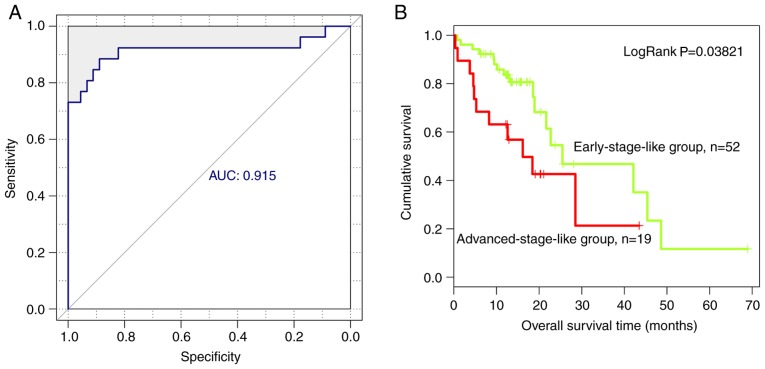
Based on the expression level of the nine optimal lncRNAs identified in the present study, an SVM classifier was established to identify tumors at different stages. The resulting SVM classifier could distinguish the progression stage of ESCC in 160 of 179 samples, exhibiting an overall accuracy of 89.39%, a sensitivity of 90.22%, a specificity of 88.51%, a positive predictive value (PPV) of 89.25%, a negative predictive value (NPV) of 89.53% and a receiver operating characteristic area under the curve (AUC) of 0.917 (Fig. 5A).
Figure 5.
Analysis of nine lncRNA biomarkers using an SVM classifier in the training dataset. (A) Receiver operating characteristic curve of the SVM classifier based on nine lncRNA biomarkers of esophageal squamous cell carcinoma. (B) Kaplan-Meier survival curves of patients with early-stage-like and advanced-stage-like tumors. The green curve represents patients with early-stage-like tumors and the red curve represents patients with advanced-stage-like tumors. AUC, area under the curve; SVM, support vector machine; lncRNA, long non-coding RNA.
In addition, overall survival in patients with early-stage-like and advanced-stage-like tumors, as defined by the SVM classifier, was calculated using the Kaplan-Meier method and compared using the log-rank test. Patients with early-stage-like tumors exhibited a significantly longer overall survival time compared with patients with advanced-stage-like tumors (40.15±22.46 and 32.81±22.78 months, respectively; Fig. 5B). The present results suggested that the tumor progression stage identified by the SVM classifier on the basis of the expression levels of the nine identified lncRNAs was associated with patient survival rate.
Validation of optimal lncRNA biomarkers in ESCC
Additional RNA-Seq expression profiling datasets associated with ESCC were downloaded from TCGA. Specifically, transcriptomic data from 86 tumor samples and survival data from 71 patients with ESCC were downloaded. Therefore, 71 ESCC samples were used to validate the ability of the nine identified lncRNAs to predict the progression of ESCC.
By calculating Pearson correlation coefficients, 71 tumor samples, including 45 at an early stage and 26 at an advanced stage, were hierarchically clustered based on the expression levels of the nine identified lncRNAs in the TCGA dataset (Fig. 6A). In the validation dataset, tumor samples were divided into two clusters based on the expression levels of the nine lncRNAs. Cluster 1 consisted of 45 samples, including 39 at an early stage and six at an advanced stage. Cluster 2 consisted of 26 samples, including six at an early stage and 20 at an advanced stage. The overall accuracy of the identified clusters was 83.10% (59/71). The patients in cluster 1 exhibited a significantly longer overall survival time than patients in cluster 2 (16.70±11.96 and 14.32±11.01 months, respectively; Fig. 6B).
Figure 6.
Clustering and survival analysis of 71 ESCC samples based on the expression of nine lncRNA biomarkers in the validation dataset. (A) Bidirectional hierarchical clustering of nine lncRNA biomarkers of ESCC in 71 tumor tissue samples in the validation dataset. Green represents early-stage tumors and red represents advanced-stage tumors. (B) Kaplan-Meier survival curves of patients from cluster 1 and cluster 2. The green curve represents patients from cluster 1 and the red curve represents patients from cluster 2. lncRNA, long non-coding RNA; ESCC, esophageal squamous cell carcinoma.
In addition, the SVM classifier based on the expression levels of the nine identified lncRNAs was used to discriminate different tumor progression stages in the validation dataset. The SVM classifier could accurately distinguish the progression stage of ESCC in 64/71 samples, exhibiting an overall accuracy of 90.14%. The sensitivity, specificity, PPV and NPV of the SVM classifier were 73.08, 100, 100 and 86.54%, respectively, with an AUC of 0.915 (Fig. 7A). Furthermore, after performing Kaplan-Meier survival analysis followed by log-rank test, patients with early-stage-like ESCC, as classified by the SVM classifier, exhibited a significantly longer overall survival time compared with patients with advanced-stage-like ESCC (16.51±11.97 and 13.96±10.59 months, respectively; P=0.038; Fig. 7B).
Figure 7.
Analysis of nine lncRNA biomarkers using an SVM classifier in the validation dataset. (A) Receiver operating characteristic curve of the SVM classifier based on nine lncRNA biomarkers associated with ESCC. (B) Kaplan-Meier survival curves of patients with early- and advanced-stage-like ESCC. The green curve represents patients with early-stage-like tumors and the red curve represents patients with advanced-stage-like tumors. lncRNA, long non-coding RNA; ESCC, esophageal squamous cell carcinoma; AUC, area under the curve; SVM, support vector machine.
Based on the results from the bidirectional hierarchical clustering and the SVM classification, the optimal combination of lncRNAs was able to reliably predict the survival time of patients with ESCC.
Identification of independent prognostic factors associated with patient survival rates
Univariate and multivariate Cox regression analyses were performed to identify independent prognostic factors associated with patient survival rates (Table III). The SVM classification, tobacco use and adjuvant therapy were significantly correlated with overall patient survival time.
Table III.
Univariate and multivariate Cox regression analysis for SVM prediction model and clinical features.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variables | HR | P-value | HR | P-value |
| SVM prediction (Early/advanced stage like) | 1.723 | 0.006a | 1.637 | 0.005a |
| Age (≤60 or >60) | 1.681 | 0.007a | 1.416 | 0.140 |
| Gender | 0.782 | 0.305 | 1.366 | 0.390 |
| Alcohol use | 0.864 | 0.455 | 0.967 | 0.916 |
| Tobacco use | 0.750 | 0.014 | 0.456 | 0.010a |
| Pathological grade (N0+N1) or (N2+N3) | 1.645 | 0.028a | 1.079 | 0.794 |
| Pathological grade (T1+T2) or (T3+T4) | 1.091 | 0.711 | 0.834 | 0.528 |
| Adjuvant therapy | 2.264 | 0.003a | 2.492 | 0.002a |
P<0.05. SVM, support vector machine; HR, hazard ratio.
Additionally, the hazard ratios of the SVM classifier and stratified clinical factors were calculated, including age, gender, alcohol use, tobacco use, tumor grade and adjuvant therapy (Table IV). Patients with early-stage-like tumors exhibited a longer survival time than patients with advanced-stage-like tumors. According to the present results, the tumor progression stage predicted by the constructed SVM classifier was significantly correlated with the patient survival rate in the following subgroups: Male patients, patients <60 years old, alcohol consumers, smokers, patients with tumors at TNM stages of N0+N1 or T3+T4 and patients who did not receive adjuvant therapy.
Table IV.
Univariate regression analysis for each clinicopathological characteristic in the training dataset.
| Univariate analysis | |||
|---|---|---|---|
| Variables | HR | 95% CI | P-value |
| Age | |||
| ≤60, n=99 | 1.966 | 1.113–3.473 | 0.0176a |
| >60, n=80 | 1.433 | 0.829–2.477 | 0.1954 |
| Gender | |||
| Male, n=146 | 1.705 | 1.099–2.645 | 0.0160a |
| Female, n=33 | 1.707 | 0.691–4.219 | 0.2413 |
| Alcohol use | |||
| Yes, n=106 | 1.767 | 1.038–3.007 | 0.0335a |
| No, n=73 | 1.771 | 0.9799–3.199 | 0.0552 |
| Tobacco use | |||
| Yes, n=114 | 1.714 | 1.025–2.865 | 0.0375a |
| No, n=65 | 1.805 | 0.973–3.35 | 0.0576 |
| Pathological grade | |||
| N0+N1, n=34 | 1.640 | 1.058–2.543 | 0.0256a |
| N2+N3, n=145 | 1.497 | 0.447–5.015 | 0.5099 |
| Pathological grade | |||
| T1+T2, n=39 | 0.896 | 0.348–2.309 | 0.8198 |
| T3+T4, n=140 | 1.998 | 1.24–3.219 | 0.0038a |
| Adjuvant therapy | |||
| Yes, n=104 | 1.197 | 0.742–1.932 | 0.4609 |
| No, n=45 | 4.890 | 1.687–14.22 | 0.0013a |
P<0.05. HR, Hazard ratio.
Functional analysis of genes associated with the nine optimal lncRNAs
A total of 1,656 genes were identified to be significantly correlated with the nine identified lncRNAs. Notably, 728 genes were positively correlated, and 928 genes were negatively correlated. A co-expression network of lncRNAs and genes was then established. After screening for gene-gene interaction pairs in the STRING database, lncRNA-mRNA co-expression networks were constructed (data not shown).
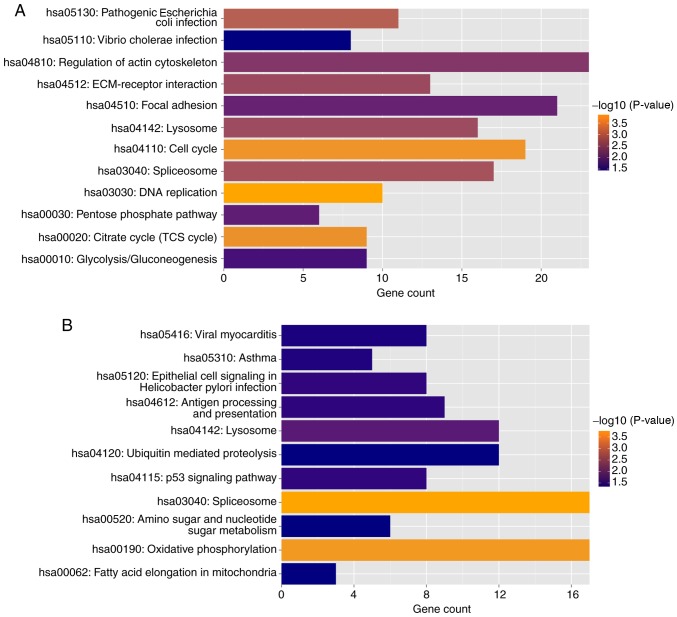
The genes significantly correlated with the nine identified lncRNA were involved in several KEGG pathways, such as ‘cell cycle’ and ‘DNA replication’, indicating that these lncRNAs may be involved in the progression of ESCC by regulating these cellular processes (Fig. 8). Specifically, the present analysis identified the enriched KEGG pathways that were negatively and positively associated with lncRNAs (Fig. 8A and B, respectively).
Figure 8.
KEGG pathway enrichment analysis. (A) Enriched KEGG pathways of protein-coding genes negatively correlated with lncRNAs. (B) Enriched KEGG pathways of protein-coding genes positively correlated with lncRNAs. X axis represents the number of genes involved in the indicated functional pathways. KEGG, Kyoto Encyclopedia of Genes and Genomes; lncRNA, long non-coding RNA.
Discussion
ESCC is a neoplastic diseases with one of the highest mortality rates worldwide, which exhibits a particularly high incidence in certain regions of China (43). However, the etiology of ESCC remains poorly understood. The present study analyzed public databases in order to identify novel effective biomarkers or therapeutic targets involved in the pathogenesis of ESCC.
In the present study, 259 DElncRs between early- and advanced-stage ESCC were identified. These 259 lncRNAs were used to predict the tumor stage in the training dataset with high accuracy. Using a random forest algorithm, a total of nine lncRNA biomarkers associated with ESCC were identified, including AC098973, AL133493, RP11-51M24, RP11-317N8, RP11-834C11, RP11-69C17, LINC00471, LINC01193 and RP1-124C. In addition, the present results suggested that the combination of these nine lncRNAs was able to predict tumor stage and patient survival rate in the training dataset. These nine lncRNA biomarkers associated with ESCC were subsequently validated. In the validation dataset, the nine lncRNAs were used to predict the tumor stage and patient survival rate with high reliability and accuracy. Furthermore, these nine lncRNA biomarkers were identified to be involved in regulating ‘cell cycle’ and ‘DNA replication’, which were previously identified to be associated with the progression of ESCC (44,45). Collectively, the present study identified nine candidate lncRNAs associated with the progression and prognosis of ESCC. Additionally, data enrichment analysis identified the possible molecular mechanism underlying their function.
The association between the dysregulation of certain lncRNAs and the prognosis of patients with cancer has been reported for several malignancies, such as hepatocellular carcinoma (46), breast cancer (16) and colorectal cancer (47). In addition, many previous transcriptome analyses of ESCC samples have been performed (48–50). Several groups have reported the aberrant expression of various lncRNAs in ESCC and multiple ESCC-associated lncRNAs have been identified, some of which may be used as biomarkers for the diagnosis or prognosis of ESCC (21–25). Li et al (29) compared the expression levels of lncRNAs in ESCC tissues with paired adjacent normal tissues and identified a three-lncRNA signature, consisting of ENST00000435885.1, XLOC_013014 and ENST00000547963.1, which was identified to be associated with the prognosis of patients with ESCC (GEO accession no. GSE53625). By analyzing the datasets generated by Li et al (29), a nine-lncRNA signature was identified in the present study. The nine identified lncRNAs were able to predict the tumor stage and survival time of patients with ESCC. In addition, the nine-lncRNA signature identified in the training dataset showed reliable prognostic ability in the validation dataset downloaded from ATCG. Therefore, the identified lncRNA signature may be used to determine the prognosis of patients with ESCC.
To the best of our knowledge, the lncRNAs identified in the present study, including AC098973, AL133493, RP11-51M24, RP11-317N8, RP11-834C11, RP11-69C17, LINC00471, LINC01193 and RP1-124C have not been functionally annotated. However, in the present study, the possible functions of these lncRNAs were predicted using mRNA expression data from the same group of patients. The genes correlated with the signature lncRNAs were identified to be involved in several KEGG pathways, such as ‘cell cycle’ and ‘DNA replication’, suggesting that these lncRNAs may be involved in the progression of ESCC by regulating these cellular processes.
Notably, the present study presents certain limitations. Although the nine-lncRNA signature identified in the present study was generated and tested in a large cohort of patients with ESCC, datasets from other institutions and other countries are required to verify its clinical application. The training and validation datasets used in the present study exhibited differences in the survival rates, possibly due to the different tumor stages. In particular, the training dataset contained no ESCC at stage IV. Therefore, the validity of the nine lncRNAs identified in the present study should be confirmed in additional prospective studies. Further studies are needed to validate the prognostic ability of these nine lncRNAs in an independent cohort of patients with ESCC. In the present study, a nine-lncRNA signature associated with tumor stage was identified. Notably, these nine lncRNAs were able to predict the survival time of patients with ESCC. However, the prognostic ability of the nine-lncRNA signature identified in the present study should be validated in further prospective studies in order to use it in clinical settings.
Acknowledgements
Not applicable.
Funding
The present work was supported by The Jiangsu Natural Science Foundation (grant no. BK20161598), The Jiangsu Province Health and Family Planning Commission (project no. H2017035), The Science and Technology of Nanjing Science Committee (project no. 201605006) and The Jiangsu Provincial Science and Technology Department (project no. BE2017759).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
JY and XW performed data analyses and wrote the manuscript. KH, MZ, XZ, YZ and SC contributed significantly to data analyses and manuscript revision. QZ and XX conceived and designed the study. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.van Hagen P, Hulshof M, van Lanschot J, Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, Richel DJ, Nieuwenhuijzen GA, Hospers GA, Bonenkamp JJ, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;366:2074–2084. doi: 10.1056/NEJMoa1112088. [DOI] [PubMed] [Google Scholar]
- 3.Matsushima K, Isomoto H, Yamaguchi N, Inoue N, Machida H, Nakayama T, Hayashi T, Kunizaki M, Hidaka S, Nagayasu T, et al. MiRNA-205 modulates cellular invasion and migration via regulating zinc finger E-box binding homeobox 2 expression in esophageal squamous cell carcinoma cells. J Transl Med. 2011;9:30. doi: 10.1186/1479-5876-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y, Muto M. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. 2015;149:1700–1715. doi: 10.1053/j.gastro.2015.08.054. [DOI] [PubMed] [Google Scholar]
- 5.Hirajima S, Komatsu S, Ichikawa D, Takeshita H, Konishi H, Shiozaki A, Morimura R, Tsujiura M, Nagata H, Kawaguchi T, et al. Clinical impact of circulating miR-18a in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:1822–1829. doi: 10.1038/bjc.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosugi S, Nishimaki T, Kanda T, Nakagawa S, Ohashi M, Hatakeyama K. Clinical significance of serum carcinoembryonic antigen, carbohydrate antigen 19-9, and squamous cell carcinoma antigen levels in esophageal cancer patients. World J Surg. 2004;28:680–685. doi: 10.1007/s00268-004-6865-y. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 8.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126:2775–2782. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J, Wei M, Xu C, Wu C, Zhang Z, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49:2949–2959. doi: 10.1016/j.ejca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 19.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 20.Li CQ, Huang GW, Wu ZY, Xu YJ, Li XC, Xue YJ, Zhu Y, Zhao JM, Li M, Zhang J, et al. Integrative analyses of transcriptome sequencing identify novel functional lncRNAs in esophageal squamous cell carcinoma. Oncogenesis. 2017;6:e297. doi: 10.1038/oncsis.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao W, Wu W, Shi F, Chen X, Wu L, Yang K, Tian F, Zhu M, Chen G, Wang W, et al. Integrated analysis of long noncoding RNA and coding RNA expression in esophageal squamous cell carcinoma. Int J Genomics. 2013;2013:480534. doi: 10.1155/2013/480534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan Z, Mao W, Bao Y, Zhang M, Su X, Xu X. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med. 2016;5:2442–2447. doi: 10.1002/cam4.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao J, Huang JX, Lin M, Wu ZD, Yu H, Wang PC, Ye J, Chen P, Wu J, Zhao GJ. Microarray expression profile analysis of aberrant long non-coding RNAs in esophageal squamous cell carcinoma. Int J Oncol. 2016;48:2543–2557. doi: 10.3892/ijo.2016.3457. [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Wei C, Li P, Wang L, Li W, Chen K, Zhang J, Zhang W, Jiang G. Integrative analysis of mRNA and lncRNA profiles identified pathogenetic lncRNAs in esophageal squamous cell carcinoma. Gene. 2018;661:169–175. doi: 10.1016/j.gene.2017.12.053. [DOI] [PubMed] [Google Scholar]
- 25.Mathé EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: Associations with survival. Clin Cancer Res. 2009;15:6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomczak K, Czerwinska P, Wiznerowicz M. The cancer genome atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, Guo W, Shi S, He J. Evaluation of the 7(th) edition of the UICC-AJCC tumor, node, metastasis classification for esophageal cancer in a Chinese cohort. J Thorac Dis. 2016;8:1672–1680. doi: 10.21037/jtd.2016.06.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, Wang S, Zhou F, Shi S, Feng X, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63:1700–1710. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon TK. The expectation maximization algorithm. IEEE Signal Process Mag. 1996;13:47–60. doi: 10.1109/79.543975. [DOI] [Google Scholar]
- 31.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiao S, Zhang S. On correcting the overestimation of the permutation-based false discovery rate estimator. Bioinformatics. 2008;24:1655–1661. doi: 10.1093/bioinformatics/btn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng Z, Zhu G, Qi J, Ma H, Nian H, Wang Y. RNA-seq analyses of multiple meristems of soybean: Novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol. 2014;14:169. doi: 10.1186/1471-2229-14-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zapf A, Brunner E, Konietschke F. A wild bootstrap approach for the selection of biomarkers in early diagnostic trials. BMC Med Res Methodol. 2015;15:43. doi: 10.1186/s12874-015-0025-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cutler A, Stevens JR. Random forests for microarrays. Methods Enzymol. 2006;411:422–432. doi: 10.1016/S0076-6879(06)11023-X. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Liu X. Screening of feature genes in distinguishing different types of breast cancer using support vector machine. OncoTargets Ther. 2015;8:2311–2317. doi: 10.2147/OTT.S85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fushiki T. Estimation of prediction error by using K-fold cross-validation. Statistics Computing. 2011;21:137–146. doi: 10.1007/s11222-009-9153-8. [DOI] [Google Scholar]
- 39.Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J Stat Softw. 2012;46(pii):i11. [PMC free article] [PubMed] [Google Scholar]
- 40.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43 (Database Issue) 2015:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 43.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 44.Dai L, Li JL, Liang XQ, Li L, Feng Y, Liu HZ, Wei WE, Ning SF, Zhang LT. Flowers of Camellia nitidissima cause growth inhibition, cell-cycle dysregulation and apoptosis in a human esophageal squamous cell carcinoma cell line. Mol Med Rep. 2016;14:1117–1122. doi: 10.3892/mmr.2016.5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dadkhah E, Naseh H, Farshchian M, Memar B, Sankian M, Bagheri R, Forghanifard MM, Montazer M, Kazemi Noughabi M, Hashemi M, Abbaszadegan MR. A cancer-array approach elucidates the immune escape mechanism and defects in the DNA repair system in esophageal squamous cell carcinoma. Arch Iran Med. 2013;16:463–470. [PubMed] [Google Scholar]
- 46.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 47.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. Long non-coding RNA HOTAIR regulates Polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 48.Ito T, Shimada Y, Kan T, David S, Cheng Y, Mori Y, Agarwal R, Paun B, Jin Z, Olaru A, et al. Pituitary tumor-transforming 1 increases cell motility and promotes lymph node metastasis in esophageal squamous cell carcinoma. Cancer Res. 2008;68:3214–3224. doi: 10.1158/0008-5472.CAN-07-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma S, Bao JYJ, Kwan PS, Chan YP, Tong CM, Fu L, Zhang N, Tong AHY, Qin YR, Tsao SW, et al. Identification of PTK6, via RNA sequencing analysis, as a suppressor of esophageal squamous cell carcinoma. Gastroenterology. 2012;143:675–686.e12. doi: 10.1053/j.gastro.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 50.Sawada G, Niida A, Uchi R, Hirata H, Shimamura T, Suzuki Y, Shiraishi Y, Chiba K, Imoto S, Takahashi Y, et al. Genomic landscape of esophageal squamous cell carcinoma in a Japanese population. Gastroenterology. 2016;150:1171–1182. doi: 10.1053/j.gastro.2016.01.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.