Abstract
Histone deacetylase 6 (HDAC6) is an attractive target for cancer therapeutic intervention. Selective HDAC6 inhibitors is important to minimise the side effects of pan inhibition. Thus, new class of hydroxamic acid-based derivatives were designed on structural basis to perform preferential activity against HDAC6 targeting solid tumours. Interestingly, 1-benzylbenzimidazole-2-thio-N-hydroxybutanamide 10a showed impressive preference with submicromolar potency against HDAC6 (IC50 = 510 nM). 10a showed cytotoxic activity with interesting profile against CCHE-45 at (IC50 = 112.76 µM) when compared to standard inhibitor Tubacin (IC50 = 20 µM). Western blot analysis of acetylated-α-tubulin verified the HDAC6 inhibiting activity of 10a. Moreover, the insignificant difference in acetylated-α-tubulin induced by 10a and Tubacin implied the on-target cytotoxic activity of 10a. Docking of 10a in the binding site of HDAC6 attributed the activity of 10a to π-π stacking with the amino acids of the hydrophobic channel of HDAC6 and capture of zinc metal in bidentate fashion. The therapeutic usefulness besides the on-target activity may define 10a as an interesting safe-lead inhibitor for future development.
Keywords: Preferential HDAC6 inhibitor, acetylated-α-tubulin, on-target activity, benzimidazole, acute promyeloblastic leukemia, choroid plexus carcinoma, cytotoxicity
Graphical Abstract

1. Introduction
Mechanisms and signalling pathways that lead to transformation of normal cell into cancer cell occupy a remarkable space in the experimental oncology researches. This type of biological studies succeeded to identify some important genes1,2, proteins3, transcriptional and epigenetic factors4–6 that contribute to the hallmarks of cancer7. Epigenetic modifications are associated with changes in gene transcription, and alteration in chromatin structure8. The main epigenetic modifications include histone methylation and acetylation8 the reversible addition and removal of acetyl group is governed by the controlled expression and activity of histone acetyltransferases (HATs) as well as histone deacetylases (HDACs)9,10. Regularly, chromatin is switched between two states: the loose state and condensed state. The loose form of the chromatin (euchromatin) in which the histone protein is acetylated in lysine residues by HATs, exposes genes for transcription11. On contrary, HDACs are associated with chromatin condensed form12, this heterochromatin structure intervenes with gene expression.
There are four classes and two families of human HDACs involving 18 mammalian isoforms with different physiological functions and distinct cellular compartments; some are either solely present in the nucleus or the cytosol, while other isoforms are shuttling between the nucleus and the cytosol13–15. Eleven isoforms of deacetylases (HDAC 1–11) which constitute the classical family, share similar structure, and require Zn2+ for their activity onset. While the sirtuin family contains 7 isoforms (SIRT 1–7), which are structurally different from the classical family, and they are NAD-dependent16,17. HDACs are deregulated in different cancer types,8 however, the intricate physiological role of HDAC and its involvement in normal cell proliferation18, mitosis, 19 development, cardiac morphogenesis20, signal transduction pathways21,22, and apoptosis19,23–25 made pan inhibition of HDACs leads to exhibition of toxicity26–30. Researchers worked extensively on providing structural determinants for the generation of selective or even preferential inhibitors against one isoform12,31–33 or group of isoforms13,34,35. Selective HDAC inhibition reduces the toxicity that results from complete shutdown of normal physiological functions of HDACs due to pan inhibition. It was reported that the zinc-dependent HDAC isoforms require an inhibitor with the following characterisations: hydrophobic cap, metal deter and a linker to link between the metal deter and the cap36. Only HDAC6 and HDAC8 that can accommodate large hydrophobic cap due to the presence of a second tube-like pocket with a different shape and close to the active hydrophobic pocket37,38 when compared to the other isoforms, but the difference between HDAC6 and HDAC8 is the protein surface flexibility that was identified as a factor of selectivity36. The other isoforms are closely similar and the elaboration of selective inhibitors against them is yet, very limited and not feasible to achieve36. Thus, many selective inhibitors that were discovered where almost against HDAC6 and/or HDAC8 due to the distinct protein structural differences between these two specific isoforms and other isoforms.
HDAC6 is found to be overexpressed in several cancer cell types39–44, it is also implicated in the onset or the progression of many neurodegenerative diseases45–47 and autoimmune disorders48–50. HDAC6 protein is the only isoform with two active deacetylase domains that are identical and function independently51; the linker between the two domains is the dynein motor binding (DMB) domain, and cytoplasmic retention signal (SE14) motif that enables the enzyme to reside in the cytoplasm to perform its functions regularly (Figure 1)52. Zinc finger ubiquitin binding domain (BUZ) is located at the C-terminal and is absent in the other HDAC isoforms14. Due to that unique structure, and cytoplasmic localisation, HDAC6 is able to deacetylate non-histone proteins; such as α-tubulin, heat shock protein 90 (Hsp90), and cortactin53. The post-translational modification of these non-histone proteins contribute to cancer cell proliferation, migration, protein homoeostasis, regulation of expression of critical immune modulators, the stability and activity of transcriptional factors such as hypoxia inducible factors (HIFs), the activity of estrogenic and androgenic receptors, and platelet aggregation in coagulation process14,36,44,53,54.
Figure 1.
Schematic representation and functional domains of human HDAC6. HDAC6 is the only HDAC with two tandem deacetylase domains (DD1 and DD2) including catalytic activity. A nuclear export signal (NES) prevents the accumulation of the protein in the nucleus and the Ser-Glu-containing tetrapeptide (SE14) region ensures stable anchorage of the enzyme in the cytoplasm. The nuclear localisation signal (NLS) translocates HDAC6 into nucleus. The linker (dynein motor binding, DMB) between both CATs can bind to dynein and the high affinity ubiquitin-binding zinc finger domain (BUZ). aa, amino acid.
Upon surveying the literature on the chemical structures of the reported selective HDAC6 inhibitors, we have found that all are with large rigid or non-rigid cap; a common structural feature as depicted in the representative examples; Tubacin55, Rocillinostat39 and Tubastatin A56 (Figure 2). The linker properties regarding the length and saturation/unsaturation varied from structure to another, and ZBG group was conserved with all the reported structures57. Tubacin that is the first reported as potent selective HDAC6 inhibitor (HDAC6 IC50 = 4 nM)55,56; however, it has never been able to be tested in vivo due to its complex structure and high lipophilicity44. Rocillinostat; although it is the considered the first selective inhibitor of HDAC6, which is orally available and has moved to phase I/II of clinical trials for treating multiple myeloma as well as lymphoma58–60, its potency in solid tumours is very low as a single agent61 and has not been extensively investigated. Tubastatin A was reported as selective HDAC6 inhibitor with neuroprotective activity56 and no study proved its potential antiproliferative activity against any type of cancer, but it increased the sensitivity of non-small cell lung cancer (NSCLC) to cisplatin62. Furthermore, Tubastatin A is of rigid tertahydro-γ-carboline hydrophobic cap that is not accessible to introduce different variables to develop but ring extension31. This gap encouraged scientists to initiate serious attempts to identify new HDAC6 selective inhibitors to combat specific types of cancer. Accordingly, we aimed at generating a lead compound that initially performs preferential HDAC6 inhibition, with new chemical entity, and is feasible to be synthesised and developed. Enzymatic assay against all human HDAC isoforms are going to be conducted to determine the preferential inhibition activity of the synthesised compounds, which will be coupled with the cytotoxic activity against specific type of children brain cancer; primary choroid plexus carcinoma (CCHE-45) and cell-based assays for detection of acetylated α-tubulin.
Figure 2.
Representative examples of most popular selective HDAC6 inhibitors with the corresponding potencies.
Thus, it is important to emphasise the critical approaches toward suggesting a structural design for elaboration of new lead with preferential HDAC6 inhibiting activity: i) large hydrophobic cap for possible accommodation in the HDAC6 hydrophobic channel which reduces the chance of perfect fitting with the other HDAC isoforms, ii) different linker lengths were used because no definite report generally defines the recommended length of linker, iii) zinc binding group (ZBG) is included as a conservative fragment63,64that is essential to capture the zinc metal of the enzyme to stop its catalysis and enhance the stable residence of inhibitor into the catalytic domain.
Consequently, it was suggested to work on binuclear aromatic heterocycle with flexible chemical entity to accommodate different variables for further modifications that might be useful for future development of the generated lead inhibitor to improve both the potency and selectivity. 2-Mercaptobenzimidazole was chosen as the scaffold of the target inhibitor based on the accessibility of two structural active sites for introduction of different variables which are N1-benzimidazole and 2-mercaptobenzimidazole. Both sites, are acidic with different pKa values, would play a role in optimisation of the reaction conditions to introduce the different variables at the target site and avoid multiple alkylation or even getting undesirable regioisomers (Figure 3).
Figure 3.
Rational design of preferential HDAC6 lead inhibitor.
2. Materials and methods
2.1. Chemistry
Melting points were determined on digital Gallen-Kamp MFB-595 instrument using open capillary tubes and are uncorrected. IR spectra were recorded as potassium bromide discs on Schimadzu FT-IR 440 spectrometer. 1H NMR spectra were recorded on Bruker spectrophotometer at 400 MHz in DMSO-d6; values (δ) are given in parts per million (ppm) downfield from tetramethylsilane (TMS) as internal reference. 13 C NMR spectra were recorded using the same spectrophotometer that used for recording 1H NMR at 101 MHz in DMSO-d6. Mass spectra were recorded on ACQUITY UPC2 System mass spectrometer for ES detection and Schimadzu Triple Quadrupole GC-MS mass spectrometer for EI detection. The elemental analyses were performed at the Microanalytical Center, Cairo University, Cairo, Egypt. Reactions were followed up by thin layer chromatography (TLC) using Merck Silica gel/TLC cards with fluorescent indicator UV254 using Hexane:Ethyl acetate (EtOAc) 4:1 and DCM:MeOH 9.5:0.5 as the eluting systems and the spots were visualised using Spectroline E series dual wavelength UV lamp at λ = 254 nm.
2.1.1. Synthesis of ethyl (1H-benzo[d]imidazol-2-yl)thioacetate (3)
A mixture of 2-mercaptobenzimidazole 1 (0.75 g, 5 mmol) and ethyl chloroacetate 2 (0.75 ml, 7.5 mmol) was added to a stirred solution of absolute ethanol at 60° C, then anhydrous K2CO3 (1.035 g, 7.5 mmol) was added. The reaction mixture was left stirring overnight. The inorganic salts were filtered off, the solvent was evaporated under reduced pressure and the white organic residue was washed with water several times to furnish white fibrous crystals that was pure enough to be submitted to the next step of the reactions.
Yield: 34%, m.p.: 60 °C, white microcrystals; 1H NMR (DMSO-d6, 400 MHz): δ 12.60 (s, 1H, NH), 7.48 (d, J = 5.1 Hz, 1H, 8-H), 7.38 (s, 1H, 5-H), 7.11 (dd, J = 5.7, 2.2 Hz, 2H, 6,7-H), 4.20 (s, 2H, S-CH2), 4.12 (q, J = 7.1 Hz, 2H, COOCH2CH3), 1.17 (t, J = 7.1 Hz, 3H, COOCH2CH3).
2.1.2. General synthesis of ethyl 2-((1-(benzyl/cycloalkyl-1H-benzo[d]imidazol-2-yl)thio)acetates (5a-d)
Anhydrous K2CO3 (0.21 g, 1.5 mmol) was added to a stirred mixture of ethyl (benzimidadol-2-yl)thioacetate 3 (0.236 g, 1 mmol) and benzyl chloride 4a (0.12 ml, 1 mmol) or 4-bromobenzyl bromide 4b (0.25 g, 1 mmol) or cyclohexylmethyl bromide 4c (0.14 ml, 1 mmol) or cyclohexylethyl bromide 4d (0.16 ml, 1 mmol) in DMF and the temperature was raised to 100° C. The reaction mixture was judged complete after 1 h upon checking with TLC using Hexane:EtOAc 4:1 as eluting system. The reaction mixture was poured onto water and the organic product was extracted by ethyl acetate, the organic extract was washed with water several times then dried over anhydrous sodium sulphate. The solvent was evaporated under reduced pressure to give the product as dark oil residue. All trials to recrystallise the product from Methanol or ethanol furnished only Ethyl 2-((1–(4-bromobenzyl)-1H-benzo[d]imidazol-2-yl)thio)acetate (5b) from methanol as golden yellow microcrystals but all the other derivatives 5a, 5c, and 5d were obtained as oil products in yields 45%, 54% and 65% respectively with enough purity to be submitted to the next step of reactions.
2.1.3. Ethyl 2-((1–(4-bromobenzyl)-1H-benzo[d]imidazol-2-yl)thio)acetate (5b)
Yield: 40%, m.p.: 107 °C, golden yellow needles; IR (KBr) ʋmax/cm−1:1741 (CO). 1H NMR (DMSO-d6, 400 MHz): δ 7.56–7.52 (m, 3H, Ar-H), 7.50–7.46 (m, 1H, Ar-H), 7.20–7.14 (m, 4H, Ar-H), 5.41 (s, 2H, 4-BrC6H4CH2), 4.26 (s, 2H, S-CH2), 4.11 (q, J = 7.1 Hz, 2H, COOCH2CH3), 1.16 (t, J = 7.1 Hz, 3H, COOCH2CH3). 13 C NMR (DMSO-d6, 101 MHz): δ 168.35 (CO), 150.63 (S-C), 142.86, 136.22, 135.73, 131.68, 129.29, 122.01, 121.88, 120.94, 117.85, and 109.85 (Ar-C), 61.27 (COOCH2CH3), 46.11(4-BrC6H4CH2), 33.83(S-CH2), 14.01 (COOCH2CH3). MS LRMS (EI): (calc) 404.02 (found) 404.08 (M)+. Anal. Calcd for C18H17BrN2O2S: C, 53.34; H, 4.23; N, 6.91. Found: C, 53.56; H, 4.11; N, 7.19.
2.1.4. General synthesis of 2-((1-benzyl/cycloalkyl-1H-benzo[d]imidazol-2-yl)thio)-N-hydroxyacetamides (6a-d)
Hydroxylamine hydrochloride (10 equivalent) was neutralised in 1 M NaOMe solution in methanol (14 equivalent) via stirring for 10 min and filtered off to discard sodium chloride. Hydroxylamine solution was added to a stirred solution of the respective ester (5a-d) (1 equivalent) in methanol (5 ml) at room temperature. The reaction mixture was left stirring until judged complete upon observing disappearance of the spot of ester starting material on TLC after 10 min using DCM:MeOH 9.5:0.5 as eluting system. The solvent was evaporated under reduced pressure and the crude residue was quenched with distilled water (15–20 ml). All the organic dirt of the aqueous reaction mixture was scavenged while vigorous shaking with EtOAc and the aqueous extract of the product was separated free from the organic dirt remained from the reaction. The aqueous extract of the product was acidified with drop wise addition of 10% HCl until observing precipitation at pH = 5.0. The resulting suspension was left in the fridge overnight for the product to complete its precipitation. The precipitate was filtered off, washed several times with distilled water and dried under vacuum to give crystalline product.
2.1.5. 2-((1-benzyl-1H-benzo[d]imidazol-2-yl)thio)-N-hydroxyacetamide (6a)
Yield: 22.0%, m.p.: 97 °C, golden yellow microcrystals; IR (KBr) ʋmax/cm−1: 3427 (broad band NH/OH), 1717 (CO). 1H NMR (DMSO-d6, 400 MHz): δ 7.57 – 7.51 (m, 1H, Ar-H)), 7.51–7.44 (m, 1H, Ar-H)), 7.36–7.22 (m, 5H, Ar-H), 7.19–7.12 (m, 2H, Ar-H), 5.41 (s, 2H, C6H5CH2), 4.20 (s, 2H, S-CH2), 3.60 (broad s, 2H). 13 C NMR (DMSO-d6, 101 MHz):δ 169.68 (CO), 151.16 (S-C), 142.91, 136.32, 128.82, 127.85, 127.20, 121.92, 121.82, 117.77, 109.92 (Ar-C), 46.82 (C6H5CH2), 34.37 (S-CH2). MS LCMS (ES): (calc) 313.09 (found) 314.1561 (MH)+. Anal. Calcd for C16H15N3O2S: C, 61.32; H, 4.82; N, 13.41. Found: C, 61.49; H, 5.02; N, 13.25.
2.1.6. 2-((1–(4-bromobenzyl)-1H-benzo[d]imidazol-2-yl)thio)-N-hydroxyacetamide (6b)
Yield: 19.0%, m.p.: 167 °C, golden yellow microcrystals; IR (KBr) ʋmax/cm−1: 3405 (broad band NH/OH), 1713 (CO). 1H NMR (DMSO-d6, 400 MHz): δ 7.52 (d, J = 8.1 Hz, 3H, Ar-H), 7.45 (d, J = 4.8 Hz, 1H, Ar-H), 7.16 (dd, J = 11.6, 6.1 Hz, 4H, Ar-H), 5.39 (s, 2H, 4-BrC6H4CH2), 4.46 (broad s, 2H), 4.14 (s, 2H, S-CH2). 13 C NMR (DMSO-d6, 101 MHz): δ 169.84 (CO), 151.73 (S-C), 142.97, 136.09, 135.81, 131.65, 129.31, 121.79, 121.72, 120.86, 117.70, and 109.68 (Ar-C), 46.08 (4-BrC6H4CH2), 36.04 (S-CH2). MS LCMS (ES): (calc) 391.00 (found) 392.0124(MH)+. Anal. Calcd for C16H14BrN3O2S: C, 48.99; H, 3.60; N, 10.71. Found: C,48.71; H, 3.81; N, 10.94.
2.1.7. 2-((1-(Cyclohexylmethyl)-1H-benzo[d]imidazol-2-yl)thio)-N-hydroxyacetamide (6c)
Yield: 10.0%, m.p.: 160 °C, golden yellow needle crystals; IR (KBr) ʋmax/cm−1: 3429 (broad band NH/OH), 1712 (CO). 1H NMR (DMSO-d6, 400 MHz): δ 7.50 (dd, J = 8.4, 6.8 Hz, 2H, 6,7-H), 7.21 – 7.09 (m, 2H, 6,7-H), 4.18 (s, 2H, S-CH2), 3.98 (d, J = 7.3 Hz, 2H, C6H11CH2), 3.45 (broad s, 2H), 1.82–1.86 (m, 1H, C6H11-H), 1.66–1.51 (m, 5H, C6H11-H), 1.13–1.04 (m, 5H, C6H11-H). 13 C NMR (DMSO-d6, 101 MHz): δ 169.68 (CO), 151.06 (S-CH2), 142.64, 136.72, 121.64, 121.50, 117.60, and 110.02 (Ar-C), 49.60 (C6H11CH2), 37.81(S-CH2), 34.29 (C6H11-C-1), 30.23(C6H11-C-2,6), 25.80 (C6H11-C-3,5), 25.22 (C6H11-C-4). MS LCMS (ES): (calc) 319.14 (found) 320.1853 (MH)+. Anal. Calcd for C16H21N3O2S: C, 60.16; H, 6.63; N, 13.16. Found: C, 60.37; H, 6.94; N, 13.38.
2.1.8. 2-((1–(2-cyclohexylethyl)-1H-benzo[d]imidazol-2-yl)thio)-N-hydroxyacetamide (6d)
Yield: 9.0%, m.p.: 157 °C, light brown fibrous crystals; IR (KBr) ʋmax/cm−1: 3430 (broad band NH/OH), 1712 (CO). 1H NMR (DMSO-d6, 400 MHz): δ 7.50 (dd, J = 18.6, 6.7 Hz, 2H, 5,8-H), 7.18 (d, J = 6.3 Hz, 2H, 6,7-H), 4.18 (m, 4H, S-CH2, C6H11CH2CH2), 3.41 (broad s, 2H), 1.78–1.61 (m, 7H, C6H11-H), 1.30–1.18 (m, 4H, C6H11-H), 0.96 (q, J = 10.9 Hz, 2H, C6H11-H). 13 C NMR (DMSO-d6, 101 MHz): δ 169.65 (CO), 150.56 (S-C), 142.83, 136.05, 121.68, 121.53, 117.68, and 109.52 (Ar-C), 41.57 (C6H11CH2CH2), 36.32 (S-CH2), 34.55 (C6H11CH2CH2), 34.10 (C6H11-C-1), 32.62 (C6H11-C-2,6), 26.01 (C6H11-C-3,5), 25.65 (C6H11-C-4). MS LCMS (ES): (calc) 333.15 (found) 334.2125 (MH)+. Anal. Calcd for C17H23N3O2S: C, 61.23; H, 6.95; N, 12.60. Found: C, 61.47; H, 7.20; N, 12.91.
2.1.9. Methyl 4-((1H-benzo[d]imidazol-2-yl)thio)butanoate (8)
Potassium hydroxide (0.112 g, 2 mmol) was dissolved in DMF (5 ml) upon stirring at 90° C, the resulting solution left to cool to room temperature, then benzimidazole-2-thiol (1) (0.15 g, 1 mmol) and methyl 4-chlorobutyrate (7) (0.145 ml, 1.20 mmol) were added to the solution and left stirring overnight at room temperature. The reaction was found very complete upon checking with TLC using Hexane:EtOAc3:1. The resulting reaction mixture was poured onto ice-cold water to give milky solution of oil product. The product was extracted with EtOAc and the organic extract was washed several times with water and dried over anhydrous sodium sulphate. The solvent was evaporated under reduced pressure to give yellowish oil product that upon standing at room temperature for 10 h furnished off-white needle crystals of the product.
Yield: 76.0%, m.p.: 122 °C, off-white needle crystals; IR (KBr) ʋmax/cm−1: 3435 (NH), 1752 (CO). 1H NMR (DMSO-d6, 400 MHz): δ 12.54 (s, 1H, NH), 7.43 (s, 2H, 5,8-H), 7.11 (dd, J = 5.9, 3.2 Hz, 2H, 6,7-H), 3.58 (s, 3H, OCH3), 3.29 (t, J = 7.1 Hz, 2H, S-CH2), 2.47 (t, J = 7.5 Hz, 2H, 2-CH2), 2.01–1.97 (p, J = 7.2, 2H, 3-CH2).13C NMR (DMSO-d6, 101 MHz): δ 172.81 (CO), 149.83 (S-C), 121.35 (Ar-C), 51.37 (COOCH3), 32.00 (S-CH2), 30.43 (2-CH2), 24.79 (3-CH2). MS LRMS (EI): (calc) 250.08 (found) 250.12 (M)+. Anal. Calcd for C12H14N2O2S: C, 57.58; H, 5.64; N, 11.19. Found: C, 57.82; H, 5.31; N, 11.45.
2.1.10. General synthesis of methyl 4-((1-benzyl/cycloalkyl-1H-benzo[d]imidazol-2-yl)thio)butanoates (9a-d)
Anhydrous K2CO3 (0.21 g, 1.5 mmol) was added to a stirred mixture of Methyl 4-((1H-benzo[d]imidazol-2-yl)thio)butanoate 8 (0.25 g, 1 mmol) and benzyl chloride 4a (0.12 ml, 1 mmol) or 4-bromobenzyl bromide 4b (0.25 g, 1 mmol) or cyclohexylmethyl bromide 4c (0.14 ml, 1 mmol) or cyclohexylethyl bromide 4d (0.16 ml, 1 mmol) in DMF and the temperature was raised to100 oC. The reaction mixture was judged complete after 2 h upon checking with TLC using Hexane:EtOAc 4:1 as eluting system. The reaction mixture was poured onto water and the organic product was extracted by ethyl acetate, the organic extract was washed with water several times and dried over anhydrous sodium sulfate. The solvent was evaporated under reduced pressure to give the product as yellowish oil residue. The intermediate ester derivatives (9a-d) were obtained in yields 85%, 69%, 90%, and 97% respectively with enough purity to be submitted to the next step of reactions without further purification.
2.1.11. General synthesis of 4-((1-benzyl/cycloalkyl-1H-benzo[d]imidazol-2-yl)thio)-N-hydroxybutanamides (10a-d)
Hydroxylamine hydrochloride (10 equivalent) was neutralised in 1 M NaOMe solution in methanol (14 equivalent) via stirring for 10 min and filtered off to discard sodium chloride. Hydroxylamine solution was added to a stirred solution of the respective ester (5a-d) (1 equivalent) in methanol (5 ml) at room temperature. The reaction mixture was left stirring until judged complete upon observing disappearance of the spot of ester starting material on TLC after 2 h using DCM:MeOH 9.5:0.5 as eluting system. The solvent was evaporated under reduced pressure and the crude residue was quenched with distilled water (15–20 ml). All the organic dirt of the aqueous reaction mixture was scavenged while vigorous shaking with EtOAc and the aqueous extract of the product was separated free from the organic dirt that remained from the reaction. The aqueous extract of the product was acidified with drop wise addition of 10% HCl until observing precipitation at pH = 5.0. The resulting suspension was left in the fridge overnight for the product to complete its precipitation. The resulting precipitate was filtered off, washed several times with distilled water and dried under vacuum to give powder product. The resulting products were recrystallised from Hexane:EtOAc 1:1 to afford crystalline products.
2.1.12. 4-((1-benzyl-1H-benzo[d]imidazol-2-yl)thio)-N-hydroxybutanamide (10a)
Yield: 25.0%, m.p.: 145 °C, off-white microcrystals; IR (KBr) ʋmax/cm−1: 3426 (broad band NH/OH), 1710 (CO). 1H NMR (DMSO-d6, 400 MHz): δ 7.61–7.53 (m, 1H, Ar-H), 7.50–7.43 (m, 1H, Ar-H), 7.36–7.22 (m, 3H, Ar-H), 7.22–7.09 (m, 4H, Ar-H), 5.39 (s, 2H, C6H5CH2), 3.33–3.37 (m, 2H, S-CH2), 2.37 (t, J = 7.3 Hz, 2H, 2-CH2), 1.95 (m, 2H, 3-CH2). 13C NMR (DMSO-d6, 101 MHz): δ 173.98 (CO), 151.43 (S-C), 143.09, 136.43, 136.43, 136.20, 136.20, 128.80, 128.80, 127.75, 127.75, 127.01, 127.01, 121.82, 121.73, 117.76, and 109.85 (Ar-C), 46.69 (C6H5-CH2), 32.42 (S-CH2), 31.25 (2-CH2), 24.56 (3-CH2). MS LCMS (ES): (calc) 341.12 (found) 342.2353 (MH)+. Anal. Calcd for C18H19N3O2S: C, 63.32; H, 5.61; N, 12.31. Found: C, 63.50; H, 5.46; N, 12.57.
2.1.13. 4-((1–(4-bromobenzyl)-1H-benzo[d]imidazol-2-yl)thio)-N-hydroxybutanamide (10b)
Yield: 30.0%, m.p.: 153 °C, off-white macrocrystals; IR (KBr) ʋmax/cm−1: 3430 (broad band NH/OH), 1710 (CO). 1H NMR (DMSO-d6, 400 MHz): δ 12.19 (s, 1H), 7.62–7.56 (m, 1H, Ar-H), 7.53 (d, J = 8.3 Hz, 2H), 7.49–7.43 (m, 1H), 7.15 (dt, J = 18.5, 7.4 Hz, 4H), 5.38 (s, 2H, 4-BrC6H4-CH2), 3.33–3.37 (m, 2H, S-CH2), 2.37 (t, J = 7.3 Hz, 2H, 2-CH2), 2.00–1.90 (m, 2H, 3-CH2). 13 C NMR (DMSO-d6, 101 MHz): δ 173.90 (CO), 151.33 (S-C), 143.04, 136.08, 135.86, 131.67, 129.16, 121.84, 121.76, 120.82, 117.77, and 109.74 (Ar-C), 45.99 (4-BrC6H4-CH2), 32.36 (S-CH2), 31.22 (2-CH2), 24.49 (3-CH2). MS LCMS (ES): (calc) 419.03 (found) 420.1528 (MH)+. Anal. Calcd for C18H18BrN3O2S: C, 51.44; H, 4.32; N, 10.00. Found: C, 51.69; H, 4.08; N, 9.77.
2.1.14. 4-((1-(Cyclohexylmethyl)-1H-benzo[d]imidazol-2-yl)thio)-N-hydroxybutanamide (10c)
Yield: 35.0%, m.p.: 149 °C, off-white microcrystals; IR (KBr) ʋmax/cm−1: 3426 (broad band NH/OH), 1716 (CO). 1H NMR (DMSO-d6, 400 MHz): δ 12.18 (s, 1H), 7.54 (d, J = 6.6 Hz, 1H, 8-H), 7.45 (d, J = 6.3 Hz, 1H, 5-H), 7.22–7.03 (m, 2H, 6,7-H), 3.93 (d, J = 7.0 Hz, 2H, C6H11-CH2), 3.35 (t, J = 6.8 Hz, 2H, S-CH2), 2.39 (t, J = 7.1 Hz, 2H, 2-CH2), 2.05–1.87 (m, 2H, 3-CH2), 1.81 (m, 1H, C6H11-H), 1.62–1.48 (m, 5H, C6H11-H), 1.10–1.01 (m, 5H, C6H11-H). 13 C NMR (DMSO-d6, 101 MHz): δ 173.91(CO), 151.41(S-C), 142.90, 136.56, 121.47, 121.35, 117.58, 109.85 (Ar-C), 49.49 (C6H11-CH2), 37.75 (S-CH2), 32.45 (2-CH2), 31.26 (C6H11-C-1), 30.23 (C6H11-C-2,6), 25.78 C6H11-C-3,5), 25.20 (C6H11-C-4), 24.58 (3-CH2). MS LCMS (ES): (calc) 347.17 (found) 348.2614 (MH)+. Anal. Calcd for C18H25N3O2S: C, 62.22; H, 7.25; N, 12.09. Found: C, 62.53; H, 6.98; N, 12.31.
2.1.15. 4-((1–(2-cyclohexylethyl)-1H-benzo[d]imidazol-2-yl)thio)-N-hydroxybutanamide(10d)
Yield: 31.0%, m.p.: 132 °C, off-white microcrystals; IR (KBr) ʋmax/cm−1: 3431 (broad band NH/OH), 1715 (CO). 1H NMR (DMSO-d6, 400 MHz): δ 12.21 (s, 1H), 7.53 (dd, J = 6.7, 2.0 Hz, 1H, 8-H), 7.48–7.41 (m, 1H, 5-H), 7.21–7.08 (m, 2H, 6,7-H), 4.12 (t, J = 7.5 Hz, 2H, C6H11-CH2CH2), 3.36–3.33 (m, 2H, S-CH2), 2.38 (t, J = 7.4 Hz, 2H, 2-CH2), 1.95 (p, J = 7.3 Hz, 2H, 3-CH2), 1.75–1.54 (m, 7H, C6H11-CH2CH2,C6H11-H), 1.29–1.10 (m, 4H, C6H11-H), 0.97–0.89 (m, 2H, C6H11-H). 13 C NMR (DMSO-d6, 101 MHz): δ 173.91(CO), 150.84 (S-C), 143.04, 135.89, 121.51, 121.39, 117.63, and 109.41(Ar-C), 41.44 (C6H11-CH2CH2), 38.87 (S-CH2), 36.28 (C6H11-CH2CH2), 34.57(2-CH2), 32.58 (C6H11-CH2CH2), 32.39 (C6H11-C-1), 31.03 (C6H11-C-2,6), 25.97 (C6H11-C-3,5), 25.64 (C6H11-C-4), 24.60 (3-CH2). MS LCMS (ES): (calc) 361.18 (found) 362.2741 (MH)+. Anal. Calcd for C19H27N3O2S: C, 63.13; H, 7.53; N, 11.62. Found: C, 63.41; H, 7.18; N, 11.87.
2.2. Biological activity
2.2.1. Histone deacetylase inhibitor assays
All enzymatic assays were conducted in BPS Bioscience Inc. in 6042 Cornerstone Court West, Ste. B, San Diego, California, USA using the biochemicals, buffers and reagents according to BPS Bioscience catalog numbers as provided in the supplementary materials.
2.2.1.1. % inhibition of HDAC enzyme at 10 µM of the test inhibitor
The test compounds were dissolved in DMSO. A series of dilutions for each test compound was prepared with 10% DMSO in HDAC assay buffer and 5 µL of the final dilution (10 µM) was added to a 50 µL reaction so that the final concentration of DMSO is 1% in all of the reactions. The enzymatic reactions for the HDAC enzymes were conducted in duplicate at 37 °C for 30 min in a 50 µL mixture containing HDAC assay buffer, 5 µg BSA (bovine serum albumin), HDAC substrate (10 µM), HDAC enzyme (varied in ng/reaction according to the HDAC subtype as presented in supplementary materials) and a test compound (10 µM). After enzymatic reactions, 50 µL of 2 × HDAC Developer was added to each well and the plate was incubated at room temperature for an additional 15 min. Fluorescence intensity was measured at an excitation of 360 nm and an emission of 460 nm using a Tecan Infinite M1000 microplate reader.
HDAC activity assays were performed in duplicate. The fluorescent intensity data were analyzed using the computer software, GraphPad Prism. In the absence of the compound, the fluorescent intensity (Ft) in each data set was defined as 100% activity. In the absence of HDAC, the fluorescent intensity (Fb) in each data set was defined as 0% activity. The percent activity in the presence of each compound was calculated according to the following equation: % activity = (F-Fb)/(Ft-Fb), where F = the fluorescent intensity in the presence of the compound.
2.2.1.2. IC50 of the test inhibitor against HDAC6
All of the compounds are dissolved in DMSO. The serial dilution of the compounds was first performed in 100% DMSO with the highest concentration at 1 mM. Each intermediate compound dilution (in 100% DMSO) will then get directly diluted 10x fold into assay buffer for an intermediate dilution of 10% DMSO in HDAC assay buffer and 5 µL of the dilution was added to a 50 µL reaction so that the final concentration of DMSO is 1% in all of reactions. The enzymatic reactions for the HDAC enzymes were conducted in duplicate at 37 °C for 30 min in a 50 µL mixture containing HDAC assay buffer, 5 µg BSA, an HDAC substrate (10 µM), a HDAC enzyme (10 ng/reaction) and a test compound (10 doses range from 0.0003 µM to 10 µM upon 3-fold dilution)/standard inhibitor (10 doses range from 0.00003 µM to 1 µM upon 3-fold dilution). After enzymatic reactions, 50 µL of 2 x HDAC Developer was added to each well for the HDAC enzymes and the plate was incubated at room temperature for an additional 15 min. Fluorescence intensity was measured at an excitation of 360 nm and an emission of 460 nm using a Tecan Infinite M1000 microplate reader.
HDAC activity assays were performed in duplicates at each concentration. The fluorescent intensity data were analysed using the computer software, Graphpad Prism. In the absence of the compound, the fluorescent intensity (Ft) in each data set was defined as 100% activity. In the absence of HDAC, the fluorescent intensity (Fb) in each data set was defined as 0% activity. The percent activity in the presence of each compound was calculated according to the following equation: % activity = (F-Fb)/(Ft-Fb), where F = the fluorescent intensity in the presence of the compound. The values of % activity versus a series of compound concentrations were then plotted using non-linear regression analysis of Sigmoidal dose–response curve generated with the equation Y = B+(T-B)/1 + 10((LogEC50-X)×Hill Slope), where Y = percent activity, B = minimum percent activity, T = maximum percent activity, X = logarithm of compound and Hill Slope = slope factor or Hill coefficient. The IC50 value was determined by the concentration causing a half-maximal percent activity.
2.2.2. Cytotoxicity assay using xCELLigence system
Established primary Choroid plexus carcinoma (CCHE-45) were cultured as previously described [41], in RPMI-1640 (Lonza, BE-12-702F) supplemented with 10% Fetal Bovine Serum (FBS, Gibco, 12483–020) and 100 U/ml penicillin and 100 µg/ml streptomycin (Lonza, DE17-602F). Cells were maintained at 37 °C in a 5% CO2 humidified atmosphere.
Cytotoxicity assay by the xCELLigence system was performed using RTCA xCELLigence DP system (ACEA Biosciences, Inc., San Diego, CA, USA) as per manufacturer’s instructions https://www.aceabio.com/products/rtca-dp/, https://www.aceabio.com/products/rtca-sp65–68. In brief, after setting up the instruments and blanking the wells with media, CCHE-45 cells were seeded into the 96 well E-plate (ACEA Biosciences) with a density of 20,000 cells/well. Attachment and growth of the cells were monitored every 1 h. Approximately 24 h after seeding, when the cells were in the log growth phase, the cells were exposed to a range of test compound 10a concentrations (3.125, 6.25, 12.5, 25, 50, 100, 200, 400) µM for 96 h. Controls received either medium alone, or medium + DMSO with a final concentration below of 0.5%. For each concentration, duplicates were tested. Experiments were conducted in three biological repeats. Results were analyzed using the RTCA software (Version 2.0). Data was exported and average IC50 value was analyzed.
2.2.3. Western blot analysis
CCHE-45 cells were treated with 100 µM of test compound for 24 h. HL60 cell line (a kind gift from Professor Azza Kamel from the National Cancer Institute, Cairo University); and is cultured in RPMI-1640 (Lonza, BE-12-702F) supplemented with 10% Fetal Bovine Serum (FBS, Gibco, 12483–020) and 100 U/ml penicillin and 100 µg/ml streptomycin (Lonza, DE17-602F), has also been treated with different concentrations of 10a in twofold serial dilutions for 24 h. Proteins were extracted from CCHE-45 cells using RIPA Lysis and Extraction Buffer (ThermoScientific, cat. No: 89901) containing 100x Halt Protease and Phosphate Inhibitor Cocktail (ThermoScientific, cat. No. 78444). Followed by protein quantification using Bradford Reagent (Pierce Coomassie Plus (Bradford) Assay Reagent) (ThermoScientific, cat. No. 23238). A total of 50 µg proteins were loaded on a 10% SDS page using 30% Acrylamide/Bisacrylamide 19:1(Serva, cat. No. 10679.01) then transferred by semidry transfer method using Bio-Rad Trans-Blot Turbo transfer system on PVDF Transfer Membrane (ThermoScientific, cat. No. 88518). The membranes were incubated overnight at 4 °C in the primary antibodies Mouse monoclonal Anti-Beta Actin antibody (1:100, abcam, ab8224) and Mouse monoclonal Anti-acetylated α Tubulin (1:200, Santa Cruz Biotechnology, sc-23950). Membranes were washed 3 times using TBST then incubated for an hour at room temperature in secondary antibody Anti-Mouse IgG, HRP-linked Antibody (1:1000, Cell Signaling, cat. No. 7076). Detection take place by Clarity Western ECL Substrate (Bio-Rad, Cat. No. 170–5061) using ChemiDoc MP imaging system (Bio-Rad). Densitometry analysis was performed using ImageJ software.
2.3. Docking studies
Molecular docking was performed using Biovia’s Discovery studio 4.0 software using the Dock ligands (CDOCKER) protocol which is an implementation of the CDOCKER algorithm. The X-ray crystal structure of the kinase domain of HDAC6 in complex with its propionic acid derivative inhibitor (PDB entry 5G0H) was recovered from RSCB protein data bank. The protein structure was prepared using protein preparation protocol of Biovia’s discovery studio 4.0. The amino acid residues were ionised using role-based technique and the missing residues and hydrogen atoms were added and minimised. The protein structure was typed by CHARMM. Synthesised compounds were prepared from ligands prepare tool which fix bad valences, adds hydrogen, and generates a 3 D coordinates using catalyst. Docking was performed using (CDOCKER) protocol. Top hits were set to 10 and pose cluster radius was set to 0.5 Å, while other docking parameters were kept as default. The best docking poses are analyzed according to docking score and interactions with key amino acids of the receptor using CDOCKER_ENERGY scoring function.
3. Results and discussion
3.1. Chemistry
The target product (benzimidazol-2-yl)thio-N-hydroxyacetamide derivatives (6a-d) were synthesised as shown in Scheme 1 starting from 2-mercaptobenzimidazole (1) to be alkylated with ethyl chloroacetate (2) in ethanolic suspension of potassium carbonate (K2CO3) at 60 °C to afford the corresponding ester (3) in 34% yield after stirring for 4 h. The resulting thioacetate ester (3) without further crystallisation, underwent an alkylation process that involves alkylation of benzimidazole-NH with 1.0 equivalent of benzyl chloride (4a), 4-bromobenzyl bromide (4b), cyclohexylmethyl bromide (4c) and cyclohexylethyl bromide (4d). The alkylation reaction was conducted in DMF in the presence of 1.5 equivalent of K2CO3 at 100 °C for 1 h to afford the respective ethyl 1-substituted(benzimidazole-2-yl)thioacetate esters (5a-d) in reasonable yields (40%-65%). The ester products (5a,c,d) were isolated as oil products while 4-bromobenzyl derivative (5b) was crystallised from methanol as golden yellow needles. The resulting esters (5a-d) were used for the next step without further chromatographic purification or crystallisation.
Scheme 1.
Synthesis of (benzimidazol-2-yl)thio-N-hydroxyacetamide derivatives (6).
Finally, The hydroxylamine hydrochloride (10 equivalent) was neutralised with 1 M NaOMe (14 equivalent) in methanolic solution and the produced hydroxylamine was promptly added to the respective ethyl 1-substituted(benzimidazole-2-yl)thio acetate esters (5a-d) (1.0 equivalent) dissolved in methanol. The reaction was considered complete after 10 min of stirring under reflux via observing disappearance of the ester spot on thin layer chromatography (TLC). The hydroxamic acid products (6b, 6c, 6d) were obtained in low yields while the benzyl derivative (6a) had the highest yield of 22%.
The structures of N-hydroxyacetamide derivatives (6a-d) were confirmed by IR, 1H-NMR, 13 C-NMR, Mass spectrometry and microanalysis. Furthermore, 1H-NMR confirmed the generation of S-acetate ester regioisomer not N-acetate ester. In 1H-NMR spectrum, Compound 6b as a representative figure to the whole series (6a-d) showed a singlet proton peak at chemical shift 4.14 ppm for methylene proton of S-CH2CONHOH while the reported69 value for methylene protons of N-CH2CONHOH is 4.98 ppm which verify the successful synthesis of the target regioisomers (6a-d).
On the other hand, we used different reaction conditions for preparation of the second series of target HDAC6 inhibitors (benzimidazol-2-yl)thio-N-hydroxybutanamide derivatives (10a-d) starting from 2-mercaptobenzimidazole (1) as shown in Scheme 2. Methyl 4-chlorobutyrate (7) was used to alkylate 2-mercaptobenzimidazole at position 2, potassium hydroxide was used as strong base to enhance the formation of thiolate salt in DMF as polar aprotic solvent to potentiate the efficacy of alkylation process with 4-carbon chain alkylating agent by stirring overnight at room temperature. The starting ester, methyl 4-((1H-benzo[d]imidazol-2-yl)thio) butanoate (8) was obtained in 76% yield and in pure needle crystals. Because the reaction condition was very regioselective, there were no any by-products require further purification. Interestingly, the resulting ester (8) gave positive hits on searching SciFinder for its preparation. But upon searching the references contents, we did not find the methyl ester of the prepared compound (8) but only the ethyl ester70. The ethyl ester counterpart of (8) was prepared using triethylamine as a mild base in DMF by stirring for 12 h at 80 °C and it was purified by column chromatography to infer that upon heating for such long period of time, multiple alkylation might have happened with these reaction conditions.
Scheme 2.
Synthesis of (benzimidazol-2-yl)thio-N-hydroxybutanamide derivatives (10).
The prepared ester (8) was heated with 1.0 equivalent of benzyl chloride (4a), 4-bromobenzyl bromide (4b), cyclohexylmethyl bromide (4c) and cyclohexylethyl bromide (4d) in the presence of 1.5 equivalent of K2CO3 in DMF at 100 °C for 2 h to afford the corresponding N-benzyl, N-(4-bromobenzyl), N-(cyclohexylmethyl) and N-(cyclohexylethyl) esters (9a-d) respectively, in quantitative yields.
The oil products of methyl 4-((1-benzyl/cycloalkyl-1H-benzo[d]imidazol-2-yl)thio)butanoates (9a-d) were used to prepare the corresponding N-hydroxybutanamide derivatives (10a-d) upon its treatment with hydroxylamine in methanol at room temperature after stirring for 2 h as described previously for synthesis of N-hydroxyacetamide derivatives (6a-b). The target compounds were obtained in yields (25%–35%).
The structures of the final N-hydroxybutanamide derivatives (10a-d) were confirmed by IR, 1H-NMR, 13 C-NMR, mass spectrometry and microanalysis. In 1HNMR, appearance of triplet proton peak of methylene group of S-CH2CH2CH2CONHOH for compound 10c at chemical shift 3.35 ppm while the reported value69 for methylene protons of N-CH2CH2CH2CONHOH is 4.28 ppm, confirmed the synthesis of target regioisomers (10a-d).
3.2. Biological activity
3.2.1. Human HDAC inhibition activity
Activity of the synthesised benzimidazole-based hydroxamic acid derivatives (6a-d) and (10a-d) against HDACs isoforms via in vitro enzymatic assay was investigated. The study revealed that the derivatives of one-carbon linker (6a-d) performed weak inhibiting activity against several HDAC isoforms including HDAC6 (Table 1). Among the investigated derivatives of three-carbon linker (10a-d), 1-benzylbenzimidazolyl-N-hydroxybutanamide 10a showed excellent inhibiting activity at 10 µM (92%) against HDAC6. Moreover, 10a exhibited impressive preferential activity against HDAC6 when compared to the inhibiting activity of the same derivative against all the other isoforms that represent class I (HDAC1, HDAC2, HDAC3, HDAC8), class IIa (HDAC4, HDAC5, HDAC7, HDAC9), class IIb (HDAC10) and class IV (HDAC11) (Table 1). There was no need to measure the corresponding IC50 values71 of 10a against other HDAC isoforms due to the weak inhibiting activity that didn’t reach 75% against any isoform (Table 1). The % inhibition was enough to highlight 10a that showed significant preference against HDAC6. Moreover, the other N-hydroxybutanamide derivatives were with insignificant activity against other HDAC isoforms as shown in (Table 1). The % inhibition results promoted 10a among all the synthesised derivatives to measure its HDAC6 IC50 value that specified the corresponding potency at 510 nM. In accordance, the nanomolar potency of 10a could primarily define it as new-preferential HDAC6 lead inhibitor 34,72–74.
Table 1.
In vitro inhibition activity of test compounds (6a-d) and (10a-d) against human HDACs.
| Compound | %inhibition of HDACs at 10µM of test compounda |
IC50±SEb | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HDAC1 | HDAC2 | HDAC3 | HDAC4 | HDAC5 | HDAC6 | HDAC7 | HDAC8 | HDAC9 | HDAC10 | HDAC11 | HDAC6 | |
| 6a | 6 | 5 | NDc | ND | ND | 2 | ND | 14 | ND | ND | ND | ND |
| 6b | 18 | 6 | ND | ND | ND | 6 | ND | 12 | ND | ND | ND | ND |
| 6c | 13 | 7 | ND | ND | ND | 2 | ND | 14 | ND | ND | ND | ND |
| 6d | 14 | 3 | ND | ND | ND | 2 | ND | 1 | ND | ND | ND | ND |
| 10a | 36 | 19 | 37 | 1 | 4 | 92 | 2 | 56 | 5 | 28 | 6 | 510 ± 0.015nM |
| 10b | 9 | 9 | ND | ND | ND | 12 | ND | 1 | ND | ND | ND | ND |
| 10c | 9 | 8 | ND | ND | ND | 14 | ND | 12 | ND | ND | ND | ND |
| 10d | 11 | 8 | ND | ND | ND | 35 | ND | 10 | ND | ND | ND | ND |
| SAHA (1µM) | 98d | 85 | 94 | ND | ND | 97 | ND | ND | ND | 97 | ND | ND |
| TSA (10µM) | ND | ND | ND | 64 | 64 | 100e | 78 | 89 | 60 | ND | 49 | 5.0 ± 0.00015 nM |
Mean value of two replicates of %inhibition of HDAC at 10 µM of test compound.
Mean value of two replicates of the concentration of test compound required to produce 50% inhibition of HDAC6 in nM±standard error.
Not determined.
Mean value of two replicates of % inhibition of HDAC1 at 3 µM of SAHA.
Mean value of two replicates of % inhibition of HDAC6 at 1 µM of TSA.
3.2.2. Cytotoxic activity assay against choroid plexus carcinoma (CCHE-45) cells
Choroid plexus carcinoma cell line CCHE-45 was then used to test the effect of the test HDAC6 inhibitor 10a on cell proliferation. CCHE-45 cell line is characterised by constitutive formation of aggresomes41, which are inclusion bodies for highly misfolded proteins formed by the collapse of the intermediate filament vimentin. Protein aggregates that are unable to be degraded by the proteasome are shuttled along the microtubules to the aggresomes assisted by dynein motor proteins and HDAC675. Due to the essential role of HDAC6 in aggresomes formation, CCHE-45 cell line was chosen to test the cytotoxic activity of HDAC6 inhibitor 10a. xCELLigence system was used to allow real-time monitoring of cell response to the drug; without any need for toxic labeling. The monitoring of cell proliferation for 96 h (Figure 4) identified the concentration of 400 µM or 200 µM of 10a led to a quick and dramatic reduction in cell index (CI), while 100 µM reduced CI to half the maximum CI after 24 h from addition of the drug with a mean IC50 of 112.67 ± 11.06 µM (Table 2).
Figure 4.
Real-time cell analysis of the cytotoxic effect of (10a) on CCHE-45. Cells were seeded into the E-plate then 24 h later, treated with a range of concentrations of (10a) for 96 h. Graph is showing CCHE-45 cell response profiles designated as cell index for the different concentrations of (10a) over 96 h. Graph is a representative of three independent experiments. Concentrations are 400 µM (blue line), 200 µM (orange line), 100 µM (gray line), 50 µM (yellow line), 25 µM (light blue line), 12.5 µM (green line), 6.25 µM (dark blue line), 3.125 µM (brown Line).
Table 2.
In vitro cytotoxic activity of (10a) against CCHE-45.
| Compound | CCHE-45 IC50 (µM)±SEMa |
|---|---|
| 10a | 112.67 ± 11.06 |
| Tubacin | 20.00 ± 10.18 |
Mean value of three replicates of the required concentration of test compound to produce 50% inhibition of CCHE-45cells ± standard error of the mean.
When (CCHE-45 IC50 = 112.76 µM) of 10a was compared to that of Tubacin; a standard HDAC6 selective inhibitor (CCHE-45 IC50 = 20 µM) (Table 2), it showed that the new lead inhibitor has just 5 times lower potency than the standard inhibitor. However, when compared to their enzymatic activity, where Tubacin has originally an HDAC6 IC50 = 4 nM55 which is 127.5 times more potent than the lead inhibitor 10a (HDAC6 IC50 = 510 nM) to infer that the new lead inhibitor carries remarkable efficacy and interesting cytotoxic profile. Accordingly, it is highly anticipated that the future structural optimisation of the lead inhibitor 10a might result in a selective HDAC6 inhibitor with a superior potency against CCHE-45 cells, compared to Tubacin.
3.2.3. Cell-based assay of acetylated α-tubulin
Western Blot analysis of acetylated α-tubulin was used to detect and semi-quantify the upregulation of acetylated α-tubulin that accumulates upon inhibition of HDAC6 by 10a treatment. A significant increase in acetylated α-tubulin was observed in CCHE-45 cells at 24 h after treatment with 10a (100 µM), which is statistically equivalent to the levels of acetylated α-tubulin induced by Tubacin addition (20 µM) (Figure 5(A)). This increase in acetylated α-tubulin was consistent through three biological replicates. Interestingly, the on-target mechanism of antiproliferative activity of 10a against brain cancer cells CCHE-45 as HDAC6 inhibitor was confirmed via exhibition of insignificant difference between the test inhibitor 10a and Tubacin according to densitometry measurements (Figure 5(B)) at their corresponding CCHE-45 IC50 values, (Table 2). Moreover, these results excluded the possibility of off-target mechanism of action of 10a against brain cancer cells CCHE-45 and the new inhibitor doesn’t have multiple targets by which it caused cancer cell death. This observation might highlight the new lead compound 10a as safe inhibitor with controlled biological interference as antiproliferative agent.
Figure 5.
The effect of (10a) on the acetylation of α-tubulin. Treatment of CCHE-45 cells for 24 h with 100 µM of (10a) upregulated acetylated α-tubulin using Tubacin as standard HDAC6 inhibitor at 20 µM. (A) Representative Western Blot analysis of acetylated α-tubulin protein levels of three replicates showing the effect of (10a) on acetylation of α-tubulin. (B) Relative mean densitometry measurement value of protein abundance levels using ImageJ software. β-Actin was used as a loading control.
We have extended the work to determine the effect of 10a on HL60; an acute promyeloblastic leukemia cell line, as HDAC6 has been observed to be overexpressed in acute myeloid leukemia76,77. Treating HL60 with the test inhibitor 10a was done through the addition of a range of concentrations over 2-fold serial dilutions starting from 100 µM and ended at 6.25 µM. According to the results shown (Figure 6(A)), acetylated α-tubulin levels were positively correlated with the test compound concentration, with a significant upregulation of acetylated α-tubulin detected at 25 µM (Figure 6(B)). This suggests that HL60 might be more sensitive to 10a compared to CCHE-45; as much lower concentration was able to induce the acetylation of α-tubulin.
Figure 6.
The effect of (10a) on the acetylation of α-tubulin. Treatment of HL60 cells for 24 h with several concentrations of (10a); 6.25, 12.5, 25, 50 and 100 µM upregulated acetylated α-tubulin at concentration starting from 12.5 µM. (A) Representative Western Blot analysis of acetylated α-tubulin protein levels of three replicates showing the effect of (10a) on acetylation of α-tubulin. (B) Relative mean densitometric value of protein abundance levels using ImageJ software. GAPDH was used as a loading control.
Conclusively, the enzymatic, cytotoxic activity and cell-based assays’ results could define benzimidazole-based hydroxamic acid derivative 10a of the new class as preferential HDAC6 lead inhibitor according to Graham Patrick’s definition74 of the new lead compound in drug discovery studies that says “the lead is the compound that exhibits therapeutic usefulness and the level of activity is not crucial”. Thus, the newly generated lead 10a is therapeutically useful lead for future development to improve the potency besides the selectivity, and eventually elaborate potent antiproliferative agent against solid tumour of choroid plexus carcinoma in addition to acute promyeloblastic leukaemia. Noteworthy to mention is that the generated lead inhibitor 10a gained its importance from being the first preferential HDAC6 inhibitor with verified cytotoxic activity against brain cancer CCHE-45 as type of serious solid tumours having such interesting profile and verified on-target cytotoxic activity when compared to tubacin standard inhibitor.
3.3. Docking studies
A molecular docking study using discovery studio 4.0 http://www.3dsbiovia.com/events/webinars/discovery-studio-25/index.html was performed using the Dock ligands (CDOCKER) protocol78. It is used to visualise the binding modes and orientation of some representative examples of the synthesised compounds into the active binding site of HDAC6. The coordinates of the target protein structure were obtained from the crystal structure of HDAC6 (PDB entry 5G0H)51 in complex with the preferential HDAC6 ligand inhibitor Trichostatin A (TSA) that shows high similarity to the human orthologue79. Validation of docking algorithm was achieved by redocking of co-crystallised structure of the inhibitor inside the active binding site of HDAC6. The root mean square difference (RMSD) between the top docking pose and original crystallographic geometry was 0.44 Å. This provides sufficient confidence in drawing meaningful conclusions from the docking study. In addition, redocking of TSA retrieved the reported binding mode of the inhibitor into the X-ray crystal structure of the active binding site of HDAC6 (C-docker energy = −25.49) as depicted in (Figure 7(A,B)), where the carbonyl and hydroxyl oxygens of hydroxamate moiety complexes with the corresponding Zn2+ metal ion in a bidentate fashion at metal-coordinate bond distances 2.25 Å and 2.20 Å, respectively (Figure 7(A,B)). The unsaturated aliphatic linker of TSA was nearly planar and sandwiched between the aromatic side chains of Phe583 and Phe643. Whereas the carbonyl group of the hydroxamate was almost coplanar with the unsaturated aliphatic chain and appeared twisted by ∼30° towards the zinc metal ion. The hydroxyl oxygen of hydroxamate makes hydrogen bonding with His573. Phenyl radical of docked ligand inhibitor TSA interacted with Phe643 via π-π stacking while the unsaturated aliphatic chain of the linker showed Alkyl-π interaction with Phe583 and Phe643 (Figure 7(A,B)).
Figure 7.
(A) Redocking of TSA (was built as solid stick model) into active biding site of HDAC6 (PDB entry 5G0H) in 3 D style and atoms are assigned by colors; (blue nitrogen, red oxygen and gray carbon). π-π stacking between the benzene ring of TSA and benzene rings of active binding site amino acid residues presented as pink dotted line. Alkyl-π stacking between alkyl chain of TSA and benzene rings of amino acid residues presented as light pink dotted line. Zinc metal ion-TSA coordinate bond formation presented as grey-dotted line. (B) Docking solution of TSA into the active binding site of HDAC6 (PDB entry 5G0H) in 2D style (C) Docking solution of compound (10a) (was built as solid stick model) into the active binding site of HDAC6 (PDB entry 5G0H). Yellow sulphur atom and hydrogen bond formation between compound (10a) and the amino acid residues represented as green dotted line, the description of the rest of binding interaction representations are the same as A and B. (D) Docking solution of compound (10a) into the active binding site of HDAC6 (PDB entry 5G0H) in 2D style.
The binding mode of the synthesised compounds of benzimidazole-based derivatives was investigated to justify the nanomolar activity of compound 10a (IC50 = 510 nM) against HDAC6 among all the other derivatives (5a-d) and (10b-d) that showed remarkably low %inhibition against HDAC6 compared to 10a (Table 1). On the other hand, identifying the binding mode of some representative examples of low-active benzimidazole derivatives 6a and 10b will provide us reliable clues for future development and optimisation of the identified lead inhibitor 10a.
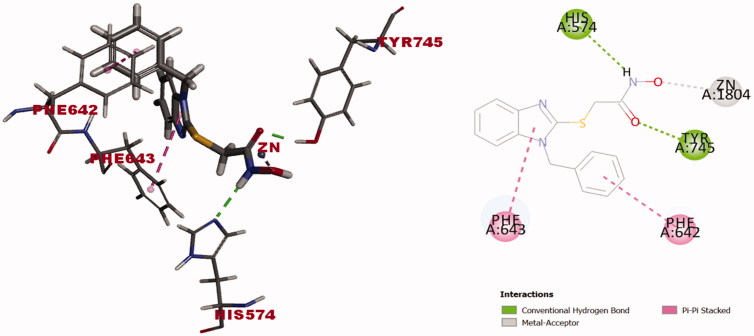
Docking of 1-benzylbenzimidazolyl-N-hydroxybutanamide 10a into the catalytic domain of HDAC6 (PDB entry 5G0H) revealed the (C-docker energy = −39.12 Kcal/mol) to be superior to that of ligand inhibitor, TSA (C-docker energy = −25.49 Kcal/mol). Binding interactions of 10a with the active binding site amino acid residues showed formation of three hydrogen bonds between hydroxamic acid and His573, Tyr745 and Gly582, π-π stacking between 1-benzyl radical and Phe643. Moreover, benzimidazole ring interacted with Phe642 via π-π stacking and with Leu712 via alkyl-π stacking (Figure 7(C,D)). Carbonyl and hydroxyl oxygens of hydroxamate moiety captured zinc metal ion in bidentate manner at respective bond distances 2.21 Å and 2.97 Å (Figure 7(C,D)) to imply the formation of metal-coordinate bond with lower strength due to the longer bond length80–82 when compared to TSA (Figure 7(A,B)). It is worthy to emphasise that the generated lead inhibitor 10a succeeded to interact with 3 amino acids; Phe643, Gly582, Leu712 out of 5 amino acids [51] that constitute the narrow hydrophobic channel of HDAC6 at which the acetylated lysine is set into the catalytic domain for deacetylation while the ligand inhibitor TSA interacted with only Phe643. In addition, 10a formed hydrogen bond as an extra type of interaction with Tyr745 that TSA in its bioactive conformer was not able to form (Figure 7(B–D)). Tyr745 is located next to zinc metal cation and thought to stabilise the transition state of intermediate to eventually release the product with deacetylated lysine residues51.
Compound 10b; the1–(4-bromobenzyl) counterpart of the lead inhibitor 10a that with 4-atoms-linker, was chosen to represent the derivatives of low inhibition activity (12%) against HDAC6 (Table 1). Docking solution of 10b into the catalytic domain of HDAC6 (PDB entry 5G0H) showed (C-docker energy = −36.34 Kcal/mol) to excel the binding affinity of ligand inhibitor TSA (C-docker energy = −25.49 Kcal/mol). Upon investigating the real reason(s) that lied behind the discrepancy in activity between 10b and 1-benzyl derivative 10a against HDAC6, it was found that the orientation of the docked compound 10b flipped around the S-alkyl chain axis (Figure 8) when compared to 10a (Figure 7(C)). This flip oriented the 4-bromobenzyl fragment upward in the hydrophobic channel to interact with Leu712 via alkyl-π stacking and be away from Phe643. Benzimidazole ring was then oriented downward to interact with Phe642 via π-π stacking. This might be attributed to the para substitution of 1-benzyl ring with bromine that led to steric clash with Phe643 forced the 4-bromobenzyl to flip and stay closer to Phe642. 4-Bromobenzyl exhibited π-stacking with Phe642 and Leu712 via bromine substituent and benzyl ring respectively (Figure 8) and lost the ability to interact with the hydrophobic channel amino acid Phe643. The hydroxamate moiety captured the metal cation in bidentate manner and formed two hydrogen bonds with His573 and Tyr745 (Figure 8). Thus, significant weaker inhibition activity of 10b might be attributed to the loss of π-π stacking interaction with Phe643.
Figure 8.
Docking solution of (10b) (was built as solid stick model) into catalytic domain of HDAC6 (PDB entry 5G0H) in 3D style in the left side and 2D style in the right side. Atoms are assigned by colours; (blue nitrogen, red oxygen, grey carbon, yellow sulphur and maroon bromine). Alkyl-π stacking presented as light pink dotted line. Zinc metal cation-hydroxamate coordinate bond formation presented as grey-dotted line. Hydrogen bond formation between compound (10b) and the amino acid residues represented as green dotted line.
Finally, we docked the 1-benzylbenzimidazole-N-hydroxyactamide derivative 6a; the one-carbon linker counter part of 10a that showed insignificant inhibiting activity (2%) against HDAC6 (Table 1) compared to 10a (92%). The docking solution of 6a into the active binding site of HDAC6 (PDB entry 5G0H) showed comparable binding affinity (C-docker energy = -28.63 Kcal/mol) to ligand inhibitor TSA (C-docker energy = −25.49 Kcal/mol) but lower than that of 10a (-39.12 Kcal/mol) and 10b (−36.34 Kcal/mol) to reflect significant alteration in the binding interaction forces with the hydrophobic channel amino acid residues of the catalytic domain. The orientation of the docked derivative 6a left an impression that the shortness of the linker made the compound struggle to take the pose that fulfills all the required binding interactions for successful inhibition to the enzyme. This could be justified by the resulting best pose of 6a that flipped on the S-alkyl chain axis to let benzimidazole ring catch the opportunity to interact with Phe643 via π- π stacking and it gives the hydroxamate moiety that built on short alkyl chain, the chance to face the zinc metal ion (Figure 9). The docked compound 6a also swung a little to get the hydroxamate residue closer to zinc metal ion to capture it via formation of metal-coordinate bond by the hydroxyl oxygen in a monodentate fashion (Figure 9). Thus, test compound’s flipping led the1-benzyl group to interact with Phe642 via π- π stacking and lost the interaction with Leu712 as one of the hydrophobic channel amino acid residues when compared to N-hydroxybutanamide 10b case. Two further hydrogen bonds are formed between the hydroxamate moiety and His574 and Tyr745 amino acid residues (Figure 9). Conclusively, the insignificant activity of 1-benzylbenzimidazole-N-hydroxyactamide 6a is definitely attributed to shortness of the linker between the 1-benzylbenzimidazole hydrophobic cap and the hydroxamate moiety.
Figure 9.
Docking solution of (6a) (was built as solid stick model) into catalytic domain of HDAC6 (PDB entry 5G0H) in 3 D style in the left side and 2D style in the right side. Atoms are assigned by colours; (blue nitrogen, red oxygen, yellow sulphur and grey carbon). π-π stacking presented as pink dotted line. Zinc metal cation-hydroxamate coordinate bond formation presented as grey-dotted line. Hydrogen bond formation between compound (6a) and the amino acid residues represented as green dotted line.
Based on the results of the above investigation that involved docking the compound 10a of the most inhibition activity (92%) against HDAC6 (IC50 = 510 nM) and two representative examples of insignificant inhibiting activity 10b (12%) and 6a (2%), it was obvious that the length of the linker, interaction of the 1-(4-unsubstituted)benzyl hydrophobic cap of benzimidazole ring with the narrow hydrophobic channel amino acid residues; Phe643 and Leu712 via π-stacking and bidentate capture of the hydroxamate moiety to zinc metal ion by the aid of carbonyl and hydroxyl oxygens are critical determinants that verified the inhibiting activity pattern of the test inhibitors generated from such benzimidazole skeleton. This also justifies the weak inhibiting activity of 1-cyclohexyl derivatives 6c,d and 10c,d that lack the interaction with the hydrophobic channel amino acids of HDAC6 via π-π stacking which is considered a critical type of interaction for successful inhibition of HDAC6.
After exhibiting the binding mode of the generated lead inhibitor; 1-benzylbenzimidazole-N-hydroxybutanamide 10a into the catalytic domain of HDAC6 and comparing it to that of the ligand inhibitor TSA, it was shown that the test inhibitor 10a was with superior binding affinity due to more interaction types with the narrow hydrophobic channel amino acid residues.
This raises the question: why the lead inhibitor10athat showed better binding mode and affinity to the ligand inhibitor TSA was much less potent in biological evaluation. The lead inhibitor 10a carries hydrophobic cap that is large enough to maintain the preferential and inhibiting activity against HDAC6 compared to HDAC1-5 and HDAC7-11 (Table 1) but is not enough to excel the selectivity. Moreover, the two critical factors that determine the activity and selectivity of the HDAC inhibitors are the hydrophobic cap and the linker64,83,84. Those are the two fragments that extensively varied in most of the designed HDAC inhibitors to either generate new inhibitor or to optimise the discovered lead inhibitor for improvement of the relative potency12,13,31. Since then, it might be that the three-carbon length of the linker of 10a was not enough to capture the zinc metal ion in HDAC6 catalytic domain as strong as TSA due to formation of metal-coordinate bond at longer distance (Figure 7(B,D)).
Definitely, the interesting results of the docking study could provide us with an excellent framework for setting up the future directions towards optimisation of the identified lead inhibitor 10a. The suggested directions involve: (i) longer carbon linker from five-carbon to seven-carbon to enhance the potency against the enzyme, (ii) bigger size of 1-arylmethyl residue via trying with binuclear arylmethyl residues instead of benzyl residue that is expected to change the profile of 10a from preferential to selective inhibitor and (iii) pyridylmethyl and quinolinyl methyl via trying with heterocycle substitute to benzyl residue that might enhance the affinity and the corresponding potency by nitrogen heteroatom.
Conclusion
The present study introduced molecular-, structural-based design and identification of new class of benzimidazole-based hydroxamic acid that involves a lead inhibitor with HDAC6 preferential inhibiting activity (HDAC6 IC50 = 510 nM) and on-target cytotoxic mechanism of action against CCHE-45 children brain cancer cells at (CCHE-45 IC50 = 112.76 µM). The generated lead inhibitor though low potency, it showed moderated cytotoxic activity against the CCHE-45 cells with interesting profile when compared to Tubacin as standard inhibitor that verified the better efficacy and the superior activity is anticipated upon enhancing the potency. The preferential inhibitor gave better activity against acute leukaemia cells HL60 at 25 µM according to Western analysis. The new class is feasible to synthesise with laboratory friendly reaction conditions and accessible to develop and modify to enhance both the potency and selectivity. Docking studies gave clues for the appropriate modifications to develop the structural features of the lead in order to change the profile from preferential to selective inhibitor. Referring to docking results, it has been recommended that introducing bigger size of arylmethyl and heterocycle substituents at position 1 of the lead scaffold will be in the favor of improving the selectivity and the longer carbons of the linker at position 2 might play a key role in enhancing the potency. For future study, these recommendations aim to enhance the level of activity and selectivity without compromising the therapeutic usefulness exhibited by the original lead inhibitor and ensure the on-target activity of the same inhibitor that may define it as safe lead inhibitor.
Supplementary Material
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Burns M, [Google Scholar]; Armstrong SA, Gutierrez A. Chapter 64 - Pathobiology of acute lymphoblastic leukemia In: Hoffman R, Benz EJ, Silberstein LE, et al., eds. Hematology. 7th ed Amsterdam, Netherlands: Elsevier; 2018:1005–1019.e1011. [Google Scholar]
- 2.Haupt S, Berger M, Goldberg Z, Haupt Y. Apoptosis - the p53 network. J Cell Sci 2003;116:4077. [DOI] [PubMed] [Google Scholar]
- 3.Riolo MT, Cooper ZA, Holloway MP, et al. Histone deacetylase 6 (HDAC6) deacetylates survivin for its nuclear export in breast cancer. J Biol Chem 2012;287:10885–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perri F, Longo F, Giuliano M, et al. Epigenetic control of gene expression: Potential implications for cancer treatment. Crit Rev Oncol/Hematol 2017;111:166–72. [DOI] [PubMed] [Google Scholar]
- 5.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis 2010;31:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 2014;124:30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schnekenburger M, Florean C, Dicato M, Diederich M. Epigenetic alterations as a universal feature of cancer hallmarks and a promising target for personalized treatments. Curr Top Med Chem 2016;16:745–76. [DOI] [PubMed] [Google Scholar]
- 8.Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol 2007;1:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biel M, Wascholowski V, Giannis A. Epigenetics–an epicenter of gene regulation: histones and histone-modifying enzymes. Angewandte Chemie 2005;44:3186–216. [DOI] [PubMed] [Google Scholar]
- 10.Schafer S, Jung M. Chromatin modifications as targets for new anticancer drugs. Arch Pharm. 2005;338:347–57. [DOI] [PubMed] [Google Scholar]
- 11.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature 1997;389:349–52. [DOI] [PubMed] [Google Scholar]
- 12.Lee HY, Fan SJ, Huang FI, et al. 5-Aroylindoles act as selective histone deacetylase 6 inhibitors ameliorating Alzheimer's disease phenotypes. J Med Chem 2018;61:7087–102. [DOI] [PubMed] [Google Scholar]
- 13.Chao SW, Chen LC, Yu CC, et al. Discovery of aliphatic-chain hydroxamates containing indole derivatives with potent class I histone deacetylase inhibitory activities. Eur J Med Chem 2018;143:792–805. [DOI] [PubMed] [Google Scholar]
- 14.Seidel C, Schnekenburger M, Dicato M, Diederich M. Histone deacetylase 6 in health and disease. Epigenomics 2015;7:103–18. [DOI] [PubMed] [Google Scholar]
- 15.Cosenza M [Google Scholar]; , Pozzi S. The Therapeutic Strategy of HDAC6 Inhibitors in Lymphoproliferative Disease. Int J Mol Sci 2018;19:e2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar A, Chauhan S. How much successful are the medicinal chemists in modulation of SIRT1: A critical review. Eur J Med Chem 2016;119:45–69. [DOI] [PubMed] [Google Scholar]
- 17.Giannini G, Cabri W, Fattorusso C, Rodriquez M. Histone deacetylase inhibitors in the treatment of cancer: overview and perspectives. Future Med Chem 2012;4:1439–60. [DOI] [PubMed] [Google Scholar]
- 18.Wickstrom SA, Masoumi KC, Khochbin S, et al. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. The EMBO J 2010;29:131–44.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang C, Pan J, Hawke DH, et al. NudC deacetylation regulates mitotic progression. PLoS One 2013;8:e73841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montgomery RL, Davis CA, Potthoff MJ, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Development 2007;21:1790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W, Fan LX, Zhou X, et al. HDAC6 regulates epidermal growth factor receptor (EGFR) endocytic trafficking and degradation in renal epithelial cells. PLoS One 2012;7:e49418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pugacheva EN, Jablonski SA, Hartman TR, et al. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 2007;129:1351–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene 2007;26:5420–32. [DOI] [PubMed] [Google Scholar]
- 24.Cohen HY, Lavu S, Bitterman KJ, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Molecul Cell 2004;13:627–38. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Holloway MP, Ma L, et al. Acetylation directs survivin nuclear localization to repress STAT3 oncogenic activity. J Biol Chem 2010;285:36129–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gryder BE, Sodji QH, Oyelere AK. Targeted cancer therapy: giving histone deacetylase inhibitors all they need to succeed. Future Med Chem 2012;4:505–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reid T, Valone F, Lipera W, et al. Phase II trial of the histone deacetylase inhibitor pivaloyloxymethyl butyrate (Pivanex, AN-9) in advanced non-small cell lung cancer. Lung Cancer 2004;45:381–6. [DOI] [PubMed] [Google Scholar]
- 28.Rivieccio MA, Brochier C, Willis DE, et al. HDAC6 is a target for protection and regeneration following injury in the nervous system. Proc Natl Acad Sci USA 2009;106:19599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Best JD, Carey N. Epigenetic therapies for non-oncology indications. Drug Discovery Today 2010;15:1008–14. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Kwon S, Yamaguchi T, et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol 2008;28:1688–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leonhardt M, Sellmer A, Kramer OH, et al. Design and biological evaluation of tetrahydro-beta-carboline derivatives as highly potent histone deacetylase 6 (HDAC6) inhibitors. Eur J Med Chem 2018;152:329–57. [DOI] [PubMed] [Google Scholar]
- 32.Ingham OJ, Paranal RM, Smith WB, et al. Development of a Potent and Selective HDAC8 Inhibitor. ACS Med Chem Lett 2016;7:929–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner FF, Zhang YL, Fass DM, et al. Kinetically selective inhibitors of histone deacetylase 2 (HDAC2) as cognition enhancers. Chem Sci 2015;6:804–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Negmeldin AT, Knoff JR, Pflum MKH. The structural requirements of histone deacetylase inhibitors: C4-modified SAHA analogs display dual HDAC6/HDAC8 selectivity. Eur J Med Chem 2018;143:1790–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frumm SM, Fan ZP, Ross KN, et al. Selective HDAC1/HDAC2 inhibitors induce neuroblastoma differentiation. Chem Biol 2013;20:713–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dallavalle S, Pisano C, Zunino F. Development and therapeutic impact of HDAC6-selective inhibitors. Biochem Pharmacol 2012;84:756–65. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen TK, Hildmann C, Dickmanns A, et al. Crystal structure of a bacterial class 2 histone deacetylase homologue. J Mol Biol 2005;354:107–20. [DOI] [PubMed] [Google Scholar]
- 38.Krennhrubec K, Marshall BL, Hedglin M, et al. Design and evaluation of 'Linkerless' hydroxamic acids as selective HDAC8 inhibitors. Bioorg Med Chem Lett 2007;17:2874–8. [DOI] [PubMed] [Google Scholar]
- 39.Santo L, Hideshima T, Kung AL, et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood 2012;119:2579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rey M, Irondelle M, Waharte F, et al. HDAC6 is required for invadopodia activity and invasion by breast tumor cells. Eur J Cell Biol 2011;90:128–35. [DOI] [PubMed] [Google Scholar]
- 41.Nassar M, Samaha H, Ghabriel M, et al. LC3A silencing hinders aggresome vimentin cage clearance in primary choroid plexus carcinoma. Sci Rep 2017;7:8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergman JA, Woan K, Perez-Villarroel P, et al. Selective histone deacetylase 6 inhibitors bearing substituted urea linkers inhibit melanoma cell growth. J Med Chem 2012;55:9891–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aldana-Masangkay GI, Rodriguez-Gonzalez A, Lin T, et al. Tubacin suppresses proliferation and induces apoptosis of acute lymphoblastic leukemia cells. Leuk Lymp 2011;52:1544–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li T, Zhang C, Hassan S, et al. Histone deacetylase 6 in cancer. J Hematol Oncol 2018;11:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selenica ML, Benner L, Housley SB, et al. Histone deacetylase 6 inhibition improves memory and reduces total tau levels in a mouse model of tau deposition. Alzheimer's Res Ther 2014;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parmigiani RB, Xu WS, Venta-Perez G, et al. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proc Natl Acad Sci USA 2008;105:9633–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim C, Choi H, Jung ES, et al. HDAC6 inhibitor blocks amyloid beta-induced impairment of mitochondrial transport in hippocampal neurons. PLoS One 2012;7:e42983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greer JM, McCombe PA. The role of epigenetic mechanisms and processes in autoimmune disorders. Biol targets Ther 2012;6:307–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003;299:1057–61. [DOI] [PubMed] [Google Scholar]
- 50.de Zoeten EF, Wang L, Butler K, et al. Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3(+) T-regulatory cells. Mol Cell Biol 2011;31:2066–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyake Y, Keusch JJ, Wang L, et al. Structural insights into HDAC6 tubulin deacetylation and its selective inhibition. Nature Chem Biol 2016;12:748–54. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Shin D, Kwon SH. Histone deacetylase 6 plays a role as a distinct regulator of diverse cellular processes. FEBS J 2013;280:775–93. [DOI] [PubMed] [Google Scholar]
- 53.Aldana-Masangkay GI, Sakamoto KM. The role of HDAC6 in cancer. J Biomed Biotechnol 2011;2011:875824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bobrowska A, Paganetti P, Matthias P, Bates GP. Hdac6 knock-out increases tubulin acetylation but does not modify disease progression in the R6/2 mouse model of Huntington's disease. PLoS One 2011;6:e20696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haggarty SJ, Koeller KM, Wong JC, et al. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA 2003;100:4389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Butler KV, Kalin J, Brochier C, et al. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J Am Chem Soc 2010;132:10842–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalin JH, Bergman JA. Development and therapeutic implications of selective histone deacetylase 6 inhibitors. J Med Chem 2013;56:6297–313. [DOI] [PubMed] [Google Scholar]
- 58.Cosenza M, Civallero M, Marcheselli L, et al. Ricolinostat, a selective HDAC6 inhibitor, shows anti-lymphoma cell activity alone and in combination with bendamustine. Apoptosis 2017;22:827–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vogl DT, Raje N, Jagannath S, et al. Ricolinostat, the first selective histone deacetylase 6 inhibitor, in combination with bortezomib and dexamethasone for relapsed or refractory multiple myeloma. Clin Cancer Res 2017;23:3307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yee AJ, Bensinger WI, Supko JG, et al. Ricolinostat plus lenalidomide, and dexamethasone in relapsed or refractory multiple myeloma: a multicentre phase 1b trial. Lancet Oncol 2016;17:1569–78. [DOI] [PubMed] [Google Scholar]
- 61.Lee DH, Won HR, Ryu HW, et al. The HDAC6 inhibitor ACY1215 enhances the anticancer activity of oxaliplatin in colorectal cancer cells. Int J Oncol 2018;53:844–54. [DOI] [PubMed] [Google Scholar]
- 62.Wang L, Xiang S, Williams KA, et al. Depletion of HDAC6 enhances cisplatin-induced DNA damage and apoptosis in non-small cell lung cancer cells. PLoS One 2012;7:e44265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Butler KV, Kozikowski AP. Chemical origins of isoform selectivity in histone deacetylase inhibitors. Curr Pharmaceut Design 2008;14:505–28. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki T. Explorative study on isoform-selective histone deacetylase inhibitors. Chem Pharm Bull 2009;57:897–906. [DOI] [PubMed] [Google Scholar]
- 65.Marlina S, Shu M-H, AbuBakar S, Zandi K. Development of a Real-Time Cell Analysing (RTCA) method as a fast and accurate screen for the selection of chikungunya virus replication inhibitors. Parasites Vectors 2015;8:579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Freedman JD, Hagel J, Scott EM, et al. Oncolytic adenovirus expressing bispecific antibody targets T-cell cytotoxicity in cancer biopsies. EMBO Mol Med 2017;9:1067–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soto-Pantoja DR, Wilson AS, Clear KY, et al. Unfolded protein response signaling impacts macrophage polarity to modulate breast cancer cell clearance and melanoma immune checkpoint therapy responsiveness. Oncotarget 2017;8:80545–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Türker Şener L, Albeniz G, Dinç B, Albeniz I. iCELLigence real-time cell analysis system for examining the cytotoxicity of drugs to cancer cell lines. Exp Ther Med 2017;14:1866–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adiguzel E, Yilmaz F, Emirik M, Ozil M. Synthesis and characterization of two new hydroxamic acids derivatives and their metal complexes. An investigation on the keto/enol, E/Z and hydroxamate/hydroximate forms. J Mol Struct 2017;1127:403–12. [Google Scholar]
- 70.Tsuchiya TN ; , Mizuno HS, Matsumoto Y, et al. US Pat. 2005/0267148A1; 2005.
- 71.Milik SN, Abdel-Aziz AK, Lasheen DS, et al. Surmounting the resistance against EGFR inhibitors through the development of thieno[2,3-d]pyrimidine-based dual EGFR/HER2 inhibitors. Eur J Med Chem 2018;155:316–36. [DOI] [PubMed] [Google Scholar]
- 72.Sawyer JS, Anderson BD, Beight DW, et al. Synthesis and activity of new aryl- and heteroaryl-substituted pyrazole inhibitors of the transforming growth factor-beta type I receptor kinase domain. J Med Chem 2003;46:3953–6. [DOI] [PubMed] [Google Scholar]
- 73.Congreve M, Chessari G, Tisi D, Woodhead AJ. Recent developments in fragment-based drug discovery. J Med Chem 2008;51:3661–80. [DOI] [PubMed] [Google Scholar]
- 74.Patrick GL . [Google Scholar]; .An introduction to medicinal chemistry. Oxford, England: Oxford University Press;2013. [Google Scholar]
- 75.Kawaguchi Y, Kovacs JJ, McLaurin A, et al. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 2003;115:727–38. [DOI] [PubMed] [Google Scholar]
- 76.Hackanson B, Rimmele L, Benkißer M, et al. HDAC6 as a target for antileukemic drugs in acute myeloid leukemia. Leukemia Res 2012;36:1055–62. [DOI] [PubMed] [Google Scholar]
- 77.Bradbury CA, Khanim FL, Hayden R, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia 2005;19:1751–9. [DOI] [PubMed] [Google Scholar]
- 78.Lu SH, Wu JW, Liu HL, et al. The discovery of potential acetylcholinesterase inhibitors: a combination of pharmacophore modeling, virtual screening, and molecular docking studies. J Biomed Sci 2011;18:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ouyang H, Ali YO, Ravichandran M, et al. Protein aggregates are recruited to aggresome by histone deacetylase 6 via unanchored ubiquitin C termini. J Biol Chem 2012;287:2317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harris SE, Orpen AG, Bruno IJ, Taylor R. Factors affecting d-block metal-ligand bond lengths: toward an automated library of molecular geometry for metal complexes. J Chem Inform Model 2005;45:1727–48. [DOI] [PubMed] [Google Scholar]
- 81.Mason R, [Google Scholar]; Randaccio L. The dependence of metal–ligand bond lengths on the nature of ligand groups: centroids of overlap density in metal–ligand bonds. J Chem Soc 1971;1150–4. [Google Scholar]
- 82.Nimmermark A, Öhrström L, Reedijk J. Metal-ligand bond lengths and strengths: are they correlated? A Detailed CSD Analysis. Z Kristallogr Cryst Mater 2013;228:311–17. [Google Scholar]
- 83.Somoza JR, Skene RJ, Katz BA, et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure 2004;12:1325–34. [DOI] [PubMed] [Google Scholar]
- 84.Wang DF, Wiest O, Helquist P, et al. On the function of the 14 A long internal cavity of histone deacetylase-like protein: implications for the design of histone deacetylase inhibitors. J Med Chem 2004;47:3409–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.