Abstract
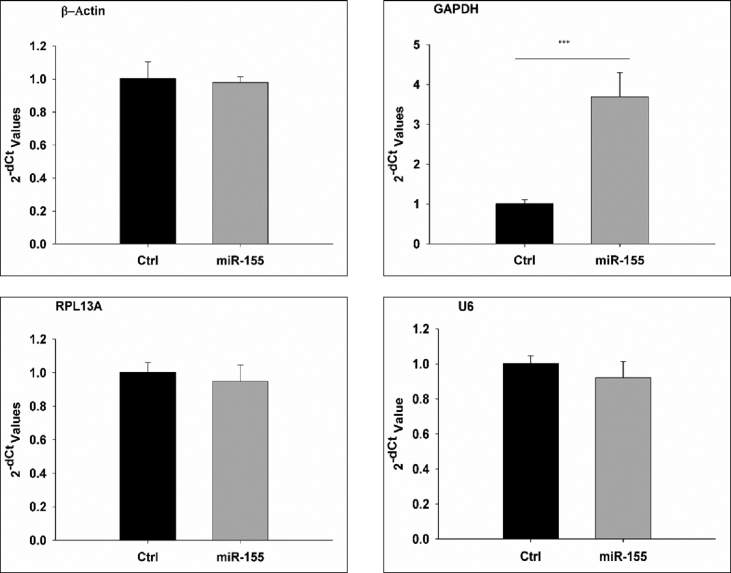
This data represents the effect of miR-155 on the expression of commonly used housekeeping genes, GAPDH, Beta Actin, RPL13A, and U6. The human miR-155 and control RNA were transfected to A549 cells by electroporation. Expression of these genes was compared in both groups by real-time PCR. The significant up-regulation in the expression of GAPDH was observed in the miR-155 transfected samples as compared to control while no major change was observed in the expression of the other three genes.
Keywords: MiR-155, Glucose metabolism, Housekeeping genes, microRNA electroporation
Specifications table
| Subject area | Biology |
|---|---|
| More specific subject area | Molecular biology |
| Type of data | Tables and figures |
| How data was acquired | Real-time PCR data acquired and analyzed by CFX manager software |
| Data format | Analyzed |
| Experimental factors | MiR-155 (30nM), time (24 hour post-transfection) |
| Experimental features | A549 cells were transfected with microRNA, and differential gene expression was analyzed |
| Data source location | Vallabhbhai Patel Chest Institute, University of Delhi, Delhi, India |
| Data accessibility | Data presented in current article only |
| Related research article | Kim S, Lee E, Jung J, Lee JW, Kim HJ, Kim J et al. microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene. 2018 May; 37 (22):2982–91. [1] |
Value of the data
|
1. Data
We report the relative gene-expression profile of four genes, Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), Beta-actin (β-Actin), Ribosomal Protein L13a (RPL13A) and U6 snRNA, in response to a microRNA (miR-155). The candidate cells, A549 cell line, were electroporated with miR-155 and Negative control RNA. The total RNA was isolated 24-h post-transfection and cDNA was prepared. We compared the expression profile of each gene at the transcriptomic level by quantitative-Polymerase Chain Reaction (qPCR) method. The qPCR efficiency, represented in Table 1, of each primer, was assessed by making the standard curve. The expression profiling of all four genes in control and miR-155 transfected samples shows a significant (p < 0.001) difference in the expression of gene GAPDH as compared to other test genes (Table 2). The data presented in Table 2 is Ct value data of qPCR performed on the cDNA sample obtained from A549 cells transfected with miR-155 and the cell transfected with negative control RNA. The Ct value representing in Table 2 is the average of technical replicates for each sample. Fig. 1 shows the upregulation in the expression of gene GAPDH in the miR-155 transfected group as compared to the control group while no critical change observed in the expression of other genes. The experiment performed on three biological and three technical replicates of each sample using primers in Table 3.
Table 1.
qPCR efficiency of target genes. 10 fold serial dilution of the target template of each primer was prepared, and real-time PCR was performed to calculate the qPCR efficiency.
| Target Gene | qPCR Efficiency |
|---|---|
| GAPDH | 95% |
| β-Actin | 98% |
| RPL13A | 93% |
| U6 snRNA | High efficiency claimed by manufacturer (ThermoScientific) |
Table 2.
Ct value data.
| Gene | Ct Value (Control) | Ct Value (miR-155 transfected) | p-value (based on ) |
|---|---|---|---|
| GAPDH | 20.18 | 18.19 | 0.0008 |
| 20.48 | 18.61 | ||
| 20.31 | 18.56 | ||
| β-Actin | 25.53 | 25.71 | 0.3609 |
| 25.82 | 25.76 | ||
| 25.69 | 25.66 | ||
| U6 | 22.29 | 22.33 | 0.2315 |
| 22.42 | 22.61 | ||
| 22.37 | 22.51 | ||
| RPL13A | 19.35 | 19.20 | 0.1246 |
| 19.21 | 19.50 | ||
| 19.19 | 19.30 |
Fig. 1.
The value of genes compared in the miR-155 transfected and control group (ctrl). (***p value < 0.001).
Table 3.
Primer sequences.
| Primer | Sequence | Reference |
|---|---|---|
| GAPDH (Glyceraldehyde 3-phosphate dehydrogenase) | Forward:TGCACCACCAACTGCTTAGC | [2] |
| Reverse: GGCATGGACTGTGGTCATGAG | ||
| RPL13A | Forward: GCCCTACGACAAGAAAAAGCG | [3] |
| Ribosomal Protein L13a | Reverse: TACTTCCAGCCAACCTCGTGA | |
| Beta Actin | Forward: GTCTGCCTTGGTAGTGGATAATG | [4] |
| Reverse: TCGAGGACGCCCTATCATGG | ||
| U6 snRNA | Predesigned primer probe set ((assay ID 001973, Thermo scientific) |
2. Experimental design, material and methods
2.1. Culture of human alveolar epithelial cell lineA549
Human alveolar epithelial cell line (A549) was maintained in Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 units/ml streptomycin in tissue culture flasks at 37 °C with 5% CO2.
2.2. microRNA transfection
A549 cells pellet was washed with 1XPBS and resuspended in electroporation buffer (Catalog # 1652676, Bio-Rad) with final cell number 1 × 106 cells/ml. The final concentration of microRNA (hsa-miR-155–5p mimic (Catalog # 4464066, Assay ID MC12601, Thermo scientific), Negative control microRNA (Catalog# 4464077, Thermo Scientific)) in electroporation buffer was 30nM. Electroporation was performed in 0.2ml Cuvette at 150V for 10 ms in a single pulse. Cells were seeded in six well plate after electroporation.
2.3. RNA isolation, cDNA preparation and real-time PCR
Total RNA was isolated using miRVANA RNA isolation kit (Catalog# AM1560, Thermo scientific) as per manufacturer's instructions. RNA quantification was done by qubit 3.0 fluorometer (Catalog #Q33216, life technologies). Total RNA (1μg) was reverse transcribed using high capacity reverse transcription kit (Catalog# 4368814, Thermo scientific) with oligo dT primer. PCR reaction (20μl) for the genes GAPDH, Beta Actin and RPL13A was performed using 2 μl of cDNA, 10 μlSSo Fast Eva green (Catalog# 1725203, Bio-Rad) and 0.25pmol primer mix (forward and reverse) on CFX96 Touch real-time PCR system (Catalog# 1855195, Bio-Rad) with temperature conditions as follow: Initial denaturation at 98 °C for 2min followed by 30 cycles of 98 °C for 5 sec and 60 °C for 5 sec.
The reverse transcription and PCR reaction of U6 was performed using predesigned assay (assay ID 001973, Thermo Scientific) as per the manufacturer instruction. Briefly, 10ng total RNA was reverse transcribed using RT primer with TaqMan reverse transcription kit (Catalog# 4366596, Applied Biosystems). PCR reaction was performed using a specific Taq Man primer-probe set by PCR master mix (Catalog# 4324018, Applied Biosystems).
The Ct value data was collected by CFX manager software (Bio-rad).
calculations were as follow:
2.4. Primer efficiency calculation
PCR reaction was performed for each gene of Table 1, using 2 μl cDNA. The PCR product of each gene was run on the agarose gel, DNA band was excised from the agarose gel and purified using gel extraction kit (Catalog# 28115, Qiagen) as per the manufacturer's instructions. The 10-fold serial dilution of all purified DNA samples was prepared and amplified by real-time PCR using the specific primer to generate a standard curve. Primer efficiency was calculated from the slop using formula.
2.5. Statistical analysis
Unpaired T-test was applied on the value of the data obtained from the samples transfected with negative control and miR-155.
Acknowledgment
We thank Vallabhbhai Patel Chest Institute for financial support.
Footnotes
Transparency document associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2019.103945.
Contributor Information
Madhu Khanna, Email: madhukhanna@hotmail.com.
Sanjesh Saini, Email: sanjeshsaini@gmail.com.
Malini Shariff, Email: malini.shariff@gmail.com.
Larance Ronsard, Email: laraphds@gmail.com.
Jitender K. Singh, Email: jitenderkumar.singh30@gmail.com.
Harish Kumar, Email: pushkar.harish@gmail.com.
Transparency document
The following is the transparency document related to this article:
References
- 1.Kim S., Lee E., Jung J., Lee J.W., Kim H.J., Kim J., ju Yoo H., Lee H.J., Chae S.Y., Jeon S.M., Son B.H., Gong G., Sharan S.K., Chang S. microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene. 2018;37:2982–2991. doi: 10.1038/s41388-018-0124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hruz T., Wyss M., Docquier M., Pfaffl M.W., Masanetz S., Borghi L., Verbrugghe P., Kalaydjieva L., Bleuler S., Laule O., Descombes P., Gruissem W., Zimmermann P. RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics. 2011;12:156. doi: 10.1186/1471-2164-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thibeaux R., Avé P., Bernier M., Morcelet M., Frileux P., Guillén N., Labruyère E. The parasite Entamoeba histolytica exploits the activities of human matrix metalloproteinases to invade colonic tissue. Nat. Commun. 2014;5 doi: 10.1038/ncomms6142. [DOI] [PubMed] [Google Scholar]
- 4.Lin J., Ji A., Qiu G., Feng H., Li J., Li S., Zou Y., Cui Y., Song C., He H., Lu Y. FBW7 is associated with prognosis, inhibits malignancies and enhances temozolomide sensitivity in glioblastoma cells. Cancer Sci. 2018;109:1001–1011. doi: 10.1111/cas.13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.