Abstract
Honeybee populations in Côte d’Ivoire have been previously identified as belonging to one subspecies, Apis mellifera scutellata, but other studies have since reported a mixed population consisting of A. m. adansonii and A. m. jemenitica. The population structure and the geographic distribution of honeybees in Côte d’Ivoire remain unclear. This study aimed to profile the population structure of honeybees and their biogeography in Côte d’Ivoire. A total of 33 honeybee colonies were sampled from 15 localities to investigate the maternal ancestry of indigenous honeybee populations using the DraI COI-COII mtDNA test. The results revealed that the honeybee population in Côte d’Ivoire is composed of African haplotypes, all belonging to the AI sublineage. Haplotypes A1 and A4 were recorded with five new sequence variants, including three types of haplotype A1 and two types of haplotype A4. The A1e variant was the most frequent in the A. m. adansonii distributional area. The distribution of the haplotype variants was correlated with the climate pattern in Côte d’Ivoire. This is the first study in Côte d’Ivoire that gives insights into the biogeography and mitotype structure of the local honeybee populations.
Keywords: Apis mellifera, genetic diversity, haplotypes, Côte d’Ivoire, population structure
1. Introduction
Based on morphometrics and multivariate analyses [1], the honeybee species Apis mellifera L. has been split into four lineages. Lineage A is spread from the north to the south of Africa, lineages M and C are distributed in Europe and lineage O in the Middle East [1,2,3]. Recently, the DraI mitochondrial DNA test was used in the identification of maternal honeybee populations [3,4,5,6,7,8,9]. Using this method, four mitochondrial DNA (mtDNA) lineages have been identified, including lineages A, M, C, and Y [2,7,10]. Honeybees from the M and C lineages have been more extensively studied for their taxonomy, biodiversity, and geographic distribution than their counterparts, the A and Y lineages. The genetic diversity of honeybees in the African continent is still understudied [3,11,12], making genetic conservation programs difficult in Africa [12]. To gain further insights into the genetic diversity of indigenous honeybee populations, more studies are needed to improve the availability of reference data in different African regions. This information could contribute towards monitoring the endemic populations, improving honeybee services, and designing strategies for ecological conservation of the local population. Several studies have been done to discriminate the honeybee population in Africa [1,3,13,14,15,16,17,18,19,20,21,22]. Using morphometric traits, eleven A. mellifera L. subspecies have been taxonomically recognized in the African continent, i.e., A. m. lamarckii [14], A. m. jemenitica [15], A. m. intermissa [16], A. m. sahariensis [17], A. m. unicolor [18], A. m. scutellata [19], A. m. capensis [20], A. m. adansonii [18], A. m. monticola, A. m. litorea [21], and A. m. sinensis [22].
In West Africa, particularly in Côte d’Ivoire, the biodiversity of honeybee populations has been studied with sample sizes that are low for the size of the region (322,462 km2). Classification of local honeybee populations in Côte d’Ivoire has, therefore, been controversial. Based on the morphometric characterization, the honeybee populations in Côte d’Ivoire were initially claimed to be A. m. scutellata [19]. Later, Ruttner [1] recognized these populations as belonging to A. m. adansonii in the tropical dry and subequatorial climates. Similarly, according to the morphometric study carried out by Radloff et al. [13], honeybee populations from Côte d’Ivoire were identified as A. m. adansonii in the tropical dry and tropical humid climates, with some A. m. jemenitica and A. m. adansonii hybrid populations.
Mitochondrial DNA variation in the cytochrome oxidase subunit I-II (COI-COII) intergenic region has been used to distinguish lineages and to refine the classification of A. mellifera L. [2,23,24,25,26]. Mitochondrial DNA markers were employed to demonstrate that the honeybee populations from Côte d’Ivoire belong to lineage A [3,27]. However, lineage A consists of four sublineages, namely, AI (haplotype A1–A4, A6, A12, A13, A19, A24–A27, etc.), AII (haplotype A8, A10’ A9, etc.) [3], AIII (haplotype A20, A11, A14, A30, A33, etc.) [28], and Z (haplotype Z1–Z4, Z7) [29,30]. These sublineages are usually differentiated by the presence or absence of an additional DraI site (TTTAAA) and a deletion at the 3’ end of the P element [2,3]. Two forms of the P element, P0 and P1, are typical of lineage A. The P1 form is characterized by a 15-bp deletion at the 3’ end of the P element, whereas P0 does not exhibit any large deletion. The P0 form is carried by sublineages AI, AII, and Z [3,29,30], whereas P1 is carried by sublineage AIII [28]. Sublineage AII is differentiated from sublineage AI by the absence of the DraI site at the 5’ end of the first Q element, whereas sublineage Z has an additional DraI site in the middle of the first Q element. The classification into sublineages and the haplotype geographical distribution patterns of the honeybee populations of Côte d’Ivoire are yet to be clarified. Getting an accurate view of the genetic diversity could allow targeting of the sensitive honeybee populations and conservation of their biodiversity.
In this study, we provide for the first time the matriline structure and biogeographic distribution of the honeybee populations in Côte d’Ivoire using the highly polymorphic intergenic COI-COII region of the mtDNA.
2. Materials and Methods
2.1. Study Area
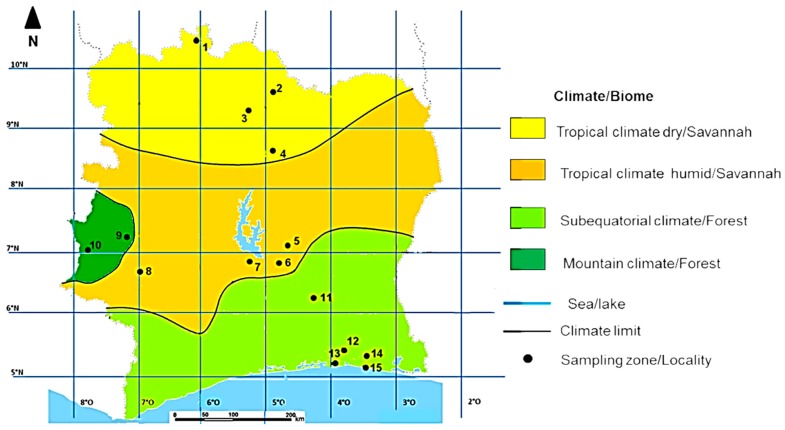
The study was conducted in Côte d’Ivoire, a West African country geographically located at 5°18′34″ N and 4°00′45″ W, between the Sahara Desert and the Atlantic Ocean. The country has four climate types from the north to the south (Figure 1), i.e., tropical dry climate, tropical humid climate, subequatorial climate, and a mountain climate that is found on the western side (Table 1).
Figure 1.
Map of honeybee sampling sites along with different climate and biome types in Côte d’Ivoire. Each sampling locality is presented with a black dot. The 15 sampling localities were spread over the 4 climate types (tropical dry and humid climates, subequatorial dry, and mountain climates) and the 2 biome types (Savannah and Forest). The numbers in the map represent a site with correspondence names and coordinates shown in Table 1.
Table 1.
Geo-coordinates, climatic conditions and landscape type of the sampling sites and colonies.
| Climate Type | Total Number of Colonies (N) | Sampling Localities (Map Code) | Number of Colonies per Sampling Location | Latitude | Longitude | Altitude | Mean Annual Temperature (°C) | Mean Relative Humidity (%) | Mean Annual Precipitation (mm) | Biome Type |
|---|---|---|---|---|---|---|---|---|---|---|
| Tropical dry climate | 10 | Tengrela (1) | 2 | 10°2′ N | 6°42′ W | 382 | 27.6 | 60.7 | 841 | Savannah |
| Ferkéssedougou (2) | 2 | 9°6′ N | 5°19′ W | 370 | 27 | 68.0 | 951 | |||
| Korhogo (3) | 3 | 9°36′ N | 5°63′ W | 342 | 27 | 65.4 | 951 | |||
| Tafiére (4) | 3 | 8°4′ N | 5°17′ W | 385 | 26.6 | 65.0 | 1100 | |||
| Tropical humid climate | 10 | Didievi (5) | 3 | 7°4′ N | 4°3′ W | 116 | 27.2 | 74.1 | 1090.6 | |
| Yamoussoukrou (6) | 4 | 6°54′ N | 5°21′ W | 213 | 26 | 76.5 | 1118 | |||
| Bouaflé (7) | 2 | 6°59′ N | 5°45′ W | 205 | 26.6 | 75.8 | 1242 | |||
| Duekoué (8) | 1 | 6°45′ N | 7°21′ W | 234 | 25.4 | 78.2 | 1572 | |||
| Mountain climate | 3 | Man (9) | 1 | 7°23′ N | 7°31′ W | 339 | 25 | 78 | 1930 | Forest |
| Danané (10) | 2 | 7°16′ N | 8°09′ W | 354 | 24.9 | 77.2 | 1930 | |||
| Subequatorial climate | 10 | Dimbokro (11) | 4 | 6°39′ N | 4°42′ W | 92 | 27 | 75 | 1176 | |
| Alepé (12) | 1 | 5°3′ N | 3°36′ W | 89 | 26.5 | 82.7 | 1544 | |||
| Dabou (13) | 2 | 5°2′ N | 4°23′ W | 20 | 26.4 | 83.5 | 1743 | |||
| Abobo (14) | 1 | 5°25′ N | 4°01′ W | 108 | 26.5 | 82.7 | 1544 | |||
| Bingerville (15) | 2 | 5°21′ N | 4°53′ W | 59 | 26.5 | 90.4 | 1230 |
2.2. Collection of Honeybees
Between May and July, 2015, sixty adult worker honeybees were collected from each of 33 colonies from 15 localities in Côte d’Ivoire (Table 1). Ten colonies were sampled from the tropical dry, tropical humid, and subequatorial climates each, and three colonies were sampled from the mountain climate (Table 1). Samples were taken from feral swarms collected by the traditional beekeepers in the countryside. All samples were preserved in 90% ethanol and were transferred under cool conditions to the Institute of Apicultural Research (IAR), Chinese Academy of Agricultural Sciences, Beijing, China, for their molecular characterization. The low sample size collected in Côte d’Ivoire was mainly due to the quasi absence of beekeeping in several regions. Moreover, in the selected regions, beekeepers (only 18) held less than five hives.
2.3. DNA Extraction, Sequencing, and DraI Test
One worker honeybee from each sampled colony was subjected to DNA extraction. The DNA was extracted from the individual honeybee thoraces by using an E.Z.N.A® Tissue DNA Kit (Omega Bio-Tek, Doraville, GA, USA) according to the manufacturer’s instructions.
PCR amplification of the COI-COII intergenic region was carried out according to a protocol detailed previously [4]. The size of the amplified DNA amplicon was determined by running 10 μL of the PCR-amplified products for 20 min on 1.0% agarose gel using gel electrophoresis. After gel migration, it was visualized and photographed using a UV-equipped gel documentation system (Bio-Rad laboratories, 6000; Biorad, Hercules, CA, USA). About 20 μL of PCR products from each sample were sent to Sangon Biotech (Beijing, China) for purification and direct Sanger sequencing in both directions. To confirm the result quality of the sequenced DNA, we repeated the same analysis independently with two more individuals from each colony, yielding a total of three sequences per colony.
To conduct the restriction fragment length polymorphism (RFLP) analysis, we used DraI (Boehringer Manheim) to digest 500 ng of the PCR products from each colony individually. The digested products were then run on 2.0% Metaphor agarose gels prepared in 1 × TBE at 10 V/cm to carry out the electrophoretic analysis. The nucleotide bands were visualized under a UV transilluminator [31].
2.4. Data Analysis
Before data analysis, we used the CodonCode aligner (www.codoncode.com) to clean the sequences. Then, the mtDNA sequences were aligned using MEGA 5.04 software [32]. Comparisons of the sequences were conducted with the Basic Local Alignment Search Tool (BLAST®) by searching the most relevant DNA sequences available on the GenBank® web portal (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Newly described sequences were submitted to the GenBank NCBI database under the accession numbers MF984182, MF984186, MH152663, MH152664, and MH152665. Similarity among the COI-COII haplotypes was investigated using the PopART version 1.6 (Population Analysis with Reticulate Trees) software (http://popart.otago.ac.nz). The classical phylogenetic tree, including lineages M, C, and Y, and sub-lineages AI, AII, AIII, Z, was built. We trimmed all nucleotide sequences to the same length. Then, the data was aligned on the online service of www.ebi.ac.uk. Using the software Jalview version 2.10.3 [33], the parsimony tree of mtDNA COI-COII was generated by average distance with the percentage identity method (PID).
3. Results
Honeybee populations from Côte d’Ivoire were composed of two mtDNA sequences, P0Q and P0QQ, corresponding to the sequence lengths of 545 and 737 bp, respectively. P0Q and P0QQ types were described by the haplotype A1 [2] and A4 respectively. While P0Q was the most recorded with an overall presence in 31 out of 33 sampled honeybee colonies, P0QQ was found in 2 out of 33. Among the 33 colonies examined, we recorded four variants of haplotype A1, i.e., A1e [34], A1s, A1q, and A1r, and two variants of haplotype A4, i.e., A4o and A4r. In this study, the haplotype variants A1s, A1q, A1r, A4o, and A4r were newly described. The sequence A1e was the most dominant (23 out of the 33) in the samples.
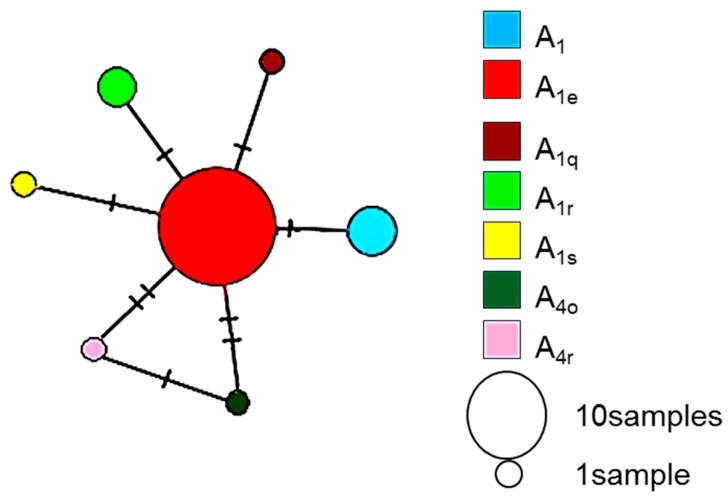
The sequences A1, A1e, A1s, A1q, A1r, A4o, and A4r comprised one haplogroup by network analysis (Figure 2). The sequence A1e represents the internal component of the haplogroup. The sequences A1, A1s, A1q, and A1r diverged from the dominant sequence A1e by only one base (Figure 3) and the sequences A4o and A4r were separated from A1e by two bases.
Figure 2.
Haplotype network displaying the relationship between the mtDNA sequences of Apis mellifera L. samples collected from different localities in Cote d’Ivoire. Circle size represents the number of haplotype copies recorded in the dataset. Each hatch mark represents a nucleotide change.
Figure 3.
Pattern of P0 sequences section from 1 to 68bp (a) and Q sequences section of the COI–COII intergenic region of A1, A1e, A1s, A1q, A1r, A4o, A4r from 69 to 218 bp (b) and from 339 to 468 bp (c). Asterisk (*) mark indicates 2nd Q section of A4o and A4r from 459 to 589 bp. Letters in red correspond to mutation sites.
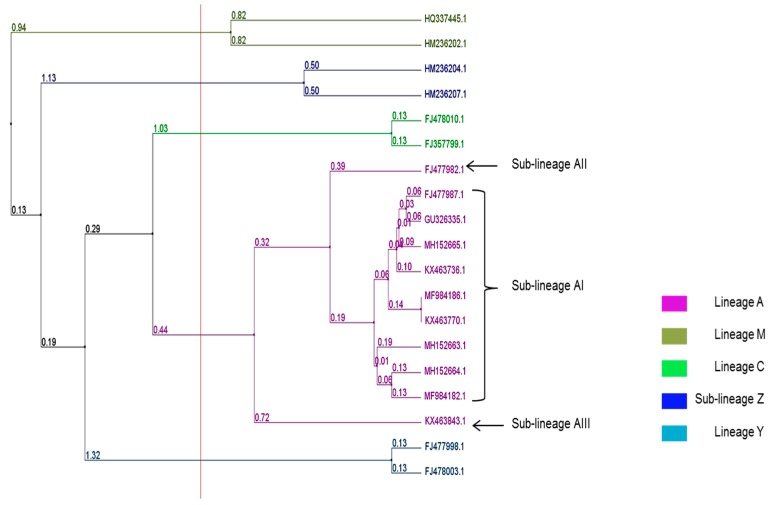
Our results clearly show that Côte d’Ivoire honeybee colonies belong to the A lineage and the AI sublineage. The P0Q sequence pattern of A1e, A1s, A1q, and A1r and the P0QQ sequence pattern of A4o and A4r from the collected data were genetically close to the sequence pattern of the honeybee A. m. scutellata mtDNA of haplotype A4 recorded in South Africa [5] and A. m. adansonii matrilines of haplotype A1 recorded in Zambia [3] (Figure 4).
Figure 4.
Phylogenetic tree of the mtDNA COI-COII sequence A1s (MH152665), A1q (MF984182), A1r (MF984186), A4o (MH152663), A4r (MH152664), A1 (KX463736), A1e (GU326335) of honeybee populations in Côte d’Ivoire. Additionally, the observed mtDNA sequence recorded in the lineage Y (FJ478003 = Y2d, FJ477998 = Y1a), M (HQ337445 = M12, HM236202 = M63), C (FJ478010 = C1, FJ357799 = C1b), and A with the sublineages AI (KX463770 = A6, FJ477987 = A4), AII (FJ477982 = A9), AIII (KX463843 = A20) and Z (HM236204 = Z1, HM236207 = Z3). Distances are indicated near the node.
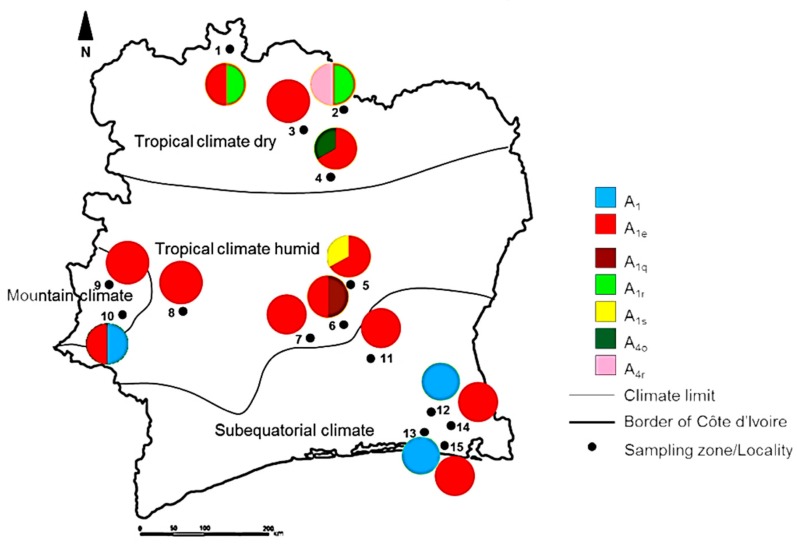
The sequence A1e was distributed across all climatic types (i.e., dry tropical, humid tropical, mountain, and subequatorial climates) in both savannah and forest biotypes (Figure 5) with a frequency of six in the dry tropical climate, eight in the humid tropical climate, one in the mountain climate, and eight in the subequatorial climate. We found that A1r was only distributed in the dry tropical climate in the savannah, and A1q and A1s were recorded only in the humid tropical climate, in the savannah biome. Haplotype A1 was recorded in the subequatorial climate in the forest biotype, while the haplotype A4 and its variants A4o and A4r were predominant in the northern part of the country in the tropical dry climate of the savannah biotype. With the exception of A1e, there was a clear co-segregation of haplotype variants and climatic types.
Figure 5.
Map of Côte d’Ivoire showing the distribution of COI-COII according to the sample locations (1 to 15). Pie-charts indicate the frequencies of sequence at each location based on our collected samples.
4. Discussion
Among the sequences of haplotype A1 and A4 (i.e., A1e, A1s, A1q, A1r, A4o, and A4r) recorded in this study from the Côte d’Ivoire, three variants of haplotype A1, namely A1s, A1q, and A1r, and two variants of haplotype A4, namely A4o and A4r, were not yet recorded and are described in this study. The haplotype A1 and its variant A1e were previously reported by Chávez-Galarza et al. [2] from Iberia and by Szalanski and Magnus [34] from Washington and Iron Counties, Utah, USA, in NCBI GenBank using mtDNA markers. All of them belonged to the evolutionary lineage A, particularly to the AI sublineage, indicating that honeybees from Côte d’Ivoire belong exclusively to the African lineage [3,13]. The hybridization with A. m. jemenitica from lineage Y, as reported earlier, may occur, between A. m. adansonii queen and A. m. jemenitica drone. Therefore, haplotypes of Y lineage ancestry (A. m. jemenitica) could not be found in our samples. The absence of Y lineage in our study does not mean that they are not present at all in Côte d’Ivoire. The rarity of A. m. jemenitica could be due to an inadequate environment, mainly due to low altitudes (ranging from 20 to 385 m) within the country. This result corroborates well with the previous work of El-Niweiri and Moritz [35] in Sudan, who reported that the populations of A. m. jemenitica are scarce at altitudes below 500 m.
The absence of honeybees from the M and C mtDNA lineages could be explained by the fact that these lineages are not endemic to West Africa [1,3]. In fact, the importation program of honeybees from the M lineage and Z sublineage initiated by the Government of Côte d’Ivoire in 1980 to improve the production of honey in the country had failed. The unsuccessful breeding of European honeybees in Côte d’Ivoire was due to the aggressiveness of the endemic honeybees against the imported honeybees and also to their poor adaptation to the new environment [36]. These factors significantly reduced the number of worker honeybees imported until the end of the honeybee importation program in Côte d’Ivoire in 1983. Therefore, the probability that these M and C lineages, and Z sublineage, were collected in our samples was very low.
The haplotype variants A4 occurred in the tropical dry climate in the ecological region of A. m. scutellata [35,36]. Haplotype A1 and its variants (A1e, A1s, A1q, and A1r) were recorded in the distributional area of A. m. adansonii [1,15,35]. The geographical distribution of the sequences recorded in this study is marked by a co-segregation of haplotypes and climate types. The genetic diversity of honeybee populations found in this study, therefore, might be a result of a long period of ecological adaptation to the environment. Thus, it is crucial to conserve the diversity of honeybee populations in Côte d’Ivoire.
5. Conclusions
According to our analyses of the intergenic COI-COII region of mtDNA, honeybee populations in Côte d’Ivoire exhibited the occurrence of two haplotypes, A1 and A4, with different variants i.e., A1e, A1s, A1q, A1r, A4o, and A4r. We did not find any local evidence of the introgression of imported honeybees from M and C lineages and Z sublineage. The haplotypes A1 and A4 occurred in the ecological regions of A. m. adansonii and A. m. scutellata, respectively. However, we were unable to fully infer the taxonomy of our honeybee colonies. Therefore, we recommend that future research should combine both morphometric and nuclear DNA analyses such as microsatellite markers for the accurate identification of the local honeybees in Côte d’Ivoire. Nevertheless, a larger sampling area is needed to be prospected to get a deeper insight into the biogeography of A. mellifera L. in Côte d’Ivoire.
Acknowledgments
We are thankful to the Nangui Abrogoua University (Abidjan, Côte d’Ivoire) for helping us to collect honeybees throughout the country and to the Institute of Apicultural Research (Beijing, China) for their assistance and cooperation in molecular genetic analyses of honeybees. We thank Komivi Dossa for his critical comments on the manuscript. We wish to thank Mr. Yao Konan Aline and Mr. Iritie Bruno for providing us the financial support and facility for data collection in the field.
Author Contributions
Conceptualization, investigation, K.A.S.C.; methodology, W.S. and C.C.; data curation, formal analysis, writing—original draft, K.A.S.C., C.C., and W.S.; visualization, writing—review and editing, K.A.S.C., M.Z.M., Y.K., W.S., and C.-S.M.; supervision, W.S. and C.-S.M.; all authors have read and approved the final version of the manuscript.
Funding
We are grateful to the Chinese Government for the financial support to the first author under China Scholarship Council (CSC). This study was financially supported by the Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2018-IAR) and Earmarked Fund for Modern Agro-industry Technology Research System (CARDS-45-KXJ1).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ruttner F. Biogeography and Taxonomy of Honeybees. Springer; New York, NY, USA: 1988. [Google Scholar]
- 2.Chávez-Galarza J., Garnery L., Henriques D., Neves C.J., Loucif-Ayad W., Jonhston J.S., Pinto M.A. Mitochondrial DNA variation of Apis mellifera iberiensis: Further insights from a large-scale study using sequence data of the tRNAleu-cox2 intergenic region. Apidologie. 2017;48:533–544. doi: 10.1007/s13592-017-0498-2. [DOI] [Google Scholar]
- 3.Franck P., Garnery L., Loiseau A., Oldroyd B.P., Hepburn H.R., Solignac M., Cornuet J.M. Genetic diversity of the honeybee in Africa: Microsatellite and mitochondrial data. Heredity. 2001;86:420–430. doi: 10.1046/j.1365-2540.2001.00842.x. [DOI] [PubMed] [Google Scholar]
- 4.Garnery L., Cornuet J.M., Solignac M. Evolutionary history of the honey bee Apis mellifera inferred from mitochondrial DNA analysis. Mol. Ecol. 1992;1:145–154. doi: 10.1111/j.1365-294X.1992.tb00170.x. [DOI] [PubMed] [Google Scholar]
- 5.Clarke K.E., Rinderer T.E., Franck P., Quezada-Euán J.J.G., Oldroyd B.P. The Africanization of honey bees (Apis mellifera L.) of the Yucatan: A study of a massive hybridization event across time. Evolution. 2002;56:1462–1474. doi: 10.1111/j.0014-3820.2002.tb01458.x. [DOI] [PubMed] [Google Scholar]
- 6.Jensen A.B., Palmer K.A., Boomsma J.J., Pedersen B.V. Varying degrees of Apis mellifera ligustica introgression in protected populations of the black honey bee, Apis mellifera mellifera, in northwest Europe. Mol. Ecol. 2005;14:93–106. doi: 10.1111/j.1365-294X.2004.02399.x. [DOI] [PubMed] [Google Scholar]
- 7.Franck P., Garnery L., Solignac M., Cornuet J.M. Molecular confirrmation of a fourth lineage in honeybees from Middle-East. Apidologie. 2000;31:167–180. doi: 10.1051/apido:2000114. [DOI] [Google Scholar]
- 8.Cánovas F., De La Rúa P., Serrano J., Galián J. Geographical patterns of mitochondrial DNA variation in Apis mellifera iberiensis (Hymenoptera: Apidae) J. Zool. Syst. Evol. Res. 2008;46:24–30. doi: 10.1111/j.1439-0469.2007.00435.x. [DOI] [Google Scholar]
- 9.De La Rúa P., Galián J., Pedersen B.V., Serrano J. Molecular characterization and population structure of Apis mellifera from Madeira and the Azores. Apidologie. 2006;38:699–708. doi: 10.1051/apido:2006044. [DOI] [Google Scholar]
- 10.Palmer M.R., Smith D.R., Kaftanoglu O. Turkish honeybees: Genetic variation and evidence for a fourth lineage of Apis mellifera mt DNA. J. Hered. 2000;91:42–46. doi: 10.1093/jhered/91.1.42. [DOI] [PubMed] [Google Scholar]
- 11.Whitfield C.W., Behura S.K., Stewart I., Berlocher H., Clark A.G., Johnston J.S., Sheppard W.S., Smith D.R., Suarez A.V., Weaver D., et al. Thrice out of Africa: Ancient and recent expansions of the honey bee, Apis mellifera. Science. 2006;314:642–645. doi: 10.1126/science.1132772. [DOI] [PubMed] [Google Scholar]
- 12.Dietemann V., Pirk C., Crewe R. Is there a need for conservation of honeybees in Africa? Apidologie. 2009;40:285–295. doi: 10.1051/apido/2009013. [DOI] [Google Scholar]
- 13.Radloff S.E., Hepburn H.R., Fuchs S. Ecological and morphological differentiation of the Apis mellifera linnaeus honeybees (Hymenoptera: Apidae) of West Africa. Afr. Entomol. 1998;6:17–23. [Google Scholar]
- 14.Cockerell T.D.A. New rocky mountain bees, and other notes. Can. Entomol. 1906;38:160–166. doi: 10.4039/Ent38160-5. [DOI] [Google Scholar]
- 15.Ruttner F. Les races d’abeille de l’Afrique; Proceedings of the XXV Congrès International d’Apiculture; Grenoble, France. 4 September 1976; pp. 347–367. [Google Scholar]
- 16.Maa T.C. An inquiry into the systematics of the tribus Apidini or honeybees (Hymenoptera) Treubia. 1953;21:525–640. [Google Scholar]
- 17.Baldensperger P.J. Variétés d’abeilles en Afrique du nord; Proceedings of the 5th Congrès International d’Entomologie; Paris, France. 18 July 1932. [Google Scholar]
- 18.Latreille P.A. Notice des espèces d’abeilles vivant en grande société, ou abeilles proprement dites, et description d’espèces nouvelles. Ann. Mus. Natl. Hist. Nat. 1804;5:161–178. [Google Scholar]
- 19.Lepeletier A. Histoire Naturelle des Insectes. Suites à Buffon. Hymenopte Áres; Roret, Paris: 1836. [Google Scholar]
- 20.Eschscholtz J.F. Entomographien. Reimer; Berlin, Germany: 1822. [Google Scholar]
- 21.Smith F.G. The races of honeybees in Africa. Bee World. 1961;42:255–260. doi: 10.1080/0005772X.1961.11096896. [DOI] [Google Scholar]
- 22.Meixner M.D., Leta M.A., Koeniger N., Fuchs S. The honey bees of Ethiopia represent a new subspecies of Apis mellifera—Apis mellifera simensis n. ssp. Apidologie. 2011;42:425–437. doi: 10.1007/s13592-011-0007-y. [DOI] [Google Scholar]
- 23.Cornuet J.M., Garnery L. Mitochondrial DNA variability in honey bees and its phylogeographic implications. Apidologie. 1991;22:627–642. doi: 10.1051/apido:19910606. [DOI] [Google Scholar]
- 24.Nikolenko A.G., Poskryakov A.V. Polymorphism of locus COI-COII of mitochondrial DNA in the honeybee Apis mellifera L. from the southern Ural Region. Russ. J. Genet. 2002;38:364–368. doi: 10.1023/A:1015289900666. [DOI] [PubMed] [Google Scholar]
- 25.Tan K., Warrit N., Smith D.R. Mitochondrial DNA diversity of the Chinese Apis cerana. Apidologie. 2007;38:238–246. doi: 10.1051/apido:2007008. [DOI] [Google Scholar]
- 26.Achou M., Loucif-Ayad W., Legout H., Hmidan H., Alburaki M., Garnery L. An insightful molecular analysis reveals foreign honeybees among Algerian honeybee populations (Apis mellifera L.) J. Data Min. Genomics Proteomics. 2015;6:166. doi: 10.4172/2153-0602.1000166. [DOI] [Google Scholar]
- 27.Arias M.C., Sheppard W.S. Molecular phylogenetics of honey bee subspecies (Apis mellifera L.) inferred from mitochondrial DNA sequence. Mol. Phylogenet. Evol. 1996;5:557–566. doi: 10.1006/mpev.1996.0050. [DOI] [PubMed] [Google Scholar]
- 28.De la Rúa P.J., Serrano J., Galián J. Mitochondrial DNA variability in the Canary Islands honeybees (Apis mellifera L.) Mol. Ecol. 1998;7:1543–1547. doi: 10.1046/j.1365-294x.1998.00468.x. [DOI] [PubMed] [Google Scholar]
- 29.Alburaki M., Moulin S., Legout H., Alburaki A., Garnery L. Mitochondrial structure of Eastern honeybee populations from Syria, Lebanon and Iraq. Apidologie. 2011;42:628. doi: 10.1007/s13592-011-0062-4. [DOI] [Google Scholar]
- 30.Alburaki M., Bertrand B., Legout H., Moulin S., Alburaki A., Sheppard W.S., Garnery L. A fifth major genetic group among honeybees revealed in Syria. BMC Genet. 2013;14:117. doi: 10.1186/1471-2156-14-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J., Russell D. Molecular cloning: A laboratory manual (3-volume set) Immunology. 2001;49:895–909. [Google Scholar]
- 32.Kumar S., Nei M., Dudley J., Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterhouse A.M., Proter J.B., Martin D.M.A. Jalview version 2-a multiple sequence alignement editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szalanski A.L., Magnus M.R. Mitochondrial DNA characterization of Africanized honey bee (Apis mellifera L.) populations from the USA. J. Apic. Res. 2010;49:177–185. doi: 10.3896/IBRA.1.49.2.06. [DOI] [Google Scholar]
- 35.El-Niweiri M.A., Moritz R.F. Mitochondrial discrimination of honeybees (Apis mellifera) of Sudan. Apidologie. 2008;39:566–573. doi: 10.1051/apido:2008039. [DOI] [Google Scholar]
- 36.Fletcher D.J. The African bee, Apis mellifera adansonii, in Africa. Annu. Rev. Entomol. 1978;23:151–171. doi: 10.1146/annurev.en.23.010178.001055. [DOI] [Google Scholar]