Abstract
Foodborne safety has become a global public health problem in both developed and developing countries. The rapid and precise monitoring and detection of foodborne pathogens has generated a strong interest by researchers in order to control and prevent human foodborne infections. Traditional methods for the detection of foodborne pathogens are often time-consuming, laborious, expensive, and unable to satisfy the demands of rapid food testing. Owing to the advantages of simplicity, real-time analysis, high sensitivity, miniaturization, rapid detection time, and low cost, electrochemical biosensing technology is more and more widely used in determination of foodborne pathogens. Here, we summarize recent developments in electrochemical biosensing technologies used to detect common foodborne pathogens. Additionally, we discuss research challenges and future prospects for this field of study.
Keywords: electrochemical biosensor, pathogen, food, detection
1. Introduction
In the 21st century, foodborne diseases are particularly problematic. The development of science and technology and economic progress has been unable to effectively control the spread of foodborne diseases, which are instead showing an upward trend [1,2]. Food safety-related poisoning incidents occur frequently around the world and the incidence of foodborne diseases is high, regardless of country. The diseases caused by foodborne pathogens can be divided into four categories. The first is food poisoning, which refers to acute or sub-acute diseases that occur after eating food contaminated with toxic or hazardous substances [3,4]; the second is allergic diseases associated with food [5,6,7,8]; the third kind includes infectious diseases (dysentery) [9,10], zoonotic diseases (foot-and-mouth disease) [11,12] and so on; the last is disease characterized by chronic toxicity, caused by long-term ingestion of a large amount of certain toxic and harmful substances [13,14]. There is no doubt that foodborne diseases have become a global public health problem affecting everyone. It is difficult to evaluate the global incidence of foodborne disease, however, according to CDC 2011 estimates, one in six Americans get foodborne disease, 128,000 are hospitalized, and 3000 die of foodborne diseases annually [15,16]. A great proportion of these cases are due to the contamination of food and drinking water [17]. There are many kinds of pathogens that are capable of producing toxins causing foodborne diseases [18,19], among them Escherichia coli, Vibrio cholerae, Bacillus cereus, Staphylococcus aureus and Clostridium perfringens are common [20,21].
Routine detection process of pathogens includes non-selective and selective enrichment culture, plate separation, pure steps, biochemical reaction and serological identification, which are cumbersome, time-consuming and laborious [22,23,24,25]. The traditional technique is unable to meet the need of food safety supervision and rapid diagnosis of food pathogens [26,27,28]. In recent years, some rapid detection techniques were established with the development of biotechnology, such as detecting certain bacteria, bacterial automatic identification system and point-of-care technologies [29,30]. However, these methods also still have some limitations. Most of these techniques still require such steps as purifying cultures of bacteria and enriching bacteria. Furthermore, there may be more than one pathogen and microorganism in the food [31,32,33], hence, how to detect multi-target microorganisms at the same time by separating and enriching the pathogens in the food samples has increasingly become the focus of food microbial testing [34,35]. Therefore, the development of rapid detection of foodborne diseases has no time to delay.
At present, biosensing technology has more applications due to its advantages of unique sensitivity, low detection limit, and simple operation. Compared with traditional analytical methods, biological sensing technology has irreplaceable advantages: the first one is real-time, which can make mutual interactions with biological macromolecules and analysis using the changes that occur every moment of the process; the second is speediness, as the process takes only 5–15 min, and a large number of samples can be measured in a short time; the third is specificity and detection of other non-specific molecules in the sample; the last is simplicity, such that large molecules do not need to be labeled. The emerging electrochemical biosensing technique has been developed and applied to the microbial analysis of foodborne pathogens in a much shorter time, with high sensitivity and selectivity comparable to the conventional methods, which makes the idea of rapid detection of foodborne pathogens possible [36,37,38].
2. The Principle of Electrochemical Biosensors
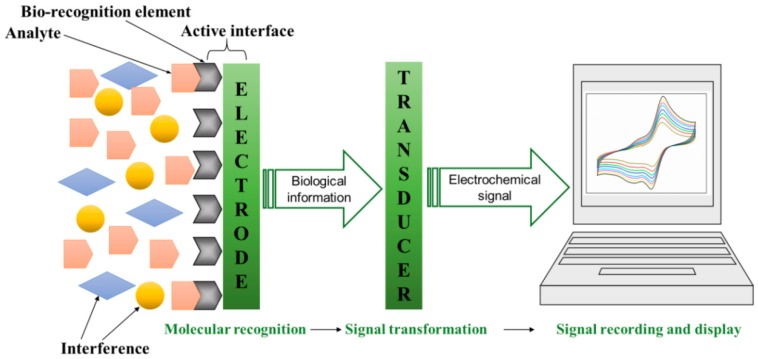
The bio-recognition element is the core component of the electrochemical biosensor which was fixed on the surface of the electrode by physical or chemical method. The biosensor can selectively identify the target molecule and capture it onto the electrode surface, owing to the specific recognition function of bio-recognition element with the substance to be tested. As the main body of the signal converter, the electrode can derive the identification signal generated on the surface of the electrode and convert it into an electrical signal, including current, voltage, and resistance, which can be measured and analyzed in order to achieve qualitative or quantitative analysis of the analysis target. The operating principle of electrochemical biosensor is shown in Figure 1.
Figure 1.
A schematic representation of the electrochemical biosensor. After the analyte contacts a recognition element on the surface of the biosensor, physical or chemical changes yield a reaction that is transformed into an electrochemical signal. This information can be further processed to determine the concentration of the pathogen and changes in the composition of the analyte.
Electrochemical biosensors can be classified into amperometric, impedimetric, potentiometric and conductometric biosensors according to the observed data type, such as current, impedance, potential and conductance, respectively [37,38]. Electrochemical biosensors were the first reported type of commercialized biosensors in the history of biosensor development. The preparation of electrochemical-active interference is the crux for the superior reported biosensors developed to date [39]. However, electrochemical biosensors certainly possess disadvantages similar to other biosensors. Among the limits of electrochemical biosensor, the immobilization of bio-recognition element without denaturation or random orientation is the most insurmountable. Hence, most of the biosensors take advantage of self-assembled monolayer (SAM) modified gold electrode surfaces because they could supply favorable substrates and binding sites for bio-recognition element via the chemical groups (such as salines, thiols, acid, disulphides, or amines) in the surface of electrode [40]. According to the number of publications about electrochemical biosensors over the recent years, we can also declare that electrochemical biosensing technology is one of the most promising techniques within the field of foodborne pathogen detection.
3. Detection of Foodborne Pathogens Using Electrochemical Biosensing Techniques
In recent years, an increasing number of researchers focused on the detection of foodborne pathogens using electrochemical biosensing techniques. Therefore, in this review we summarize recent developments of electrochemical biosensors used to detect common foodborne pathogens. The detection methods, materials used and performance of electrochemical biosensors for foodborne pathogens are shown in Table 1.
Table 1.
Biosensors for the detection of foodborne pathogens.
| Analyst | Detection Type | Materials | Performance | Reference |
|---|---|---|---|---|
| E. coli | Amperometric | screen-printed electrode | Rapid determination of four E. coli subspecies Assay time: approximately 2 min |
[46] |
| E. coli | Amperometric | DNA nanopyramids | Linear range: 1–102 CFU/mL LOD: 1.20 CFU/mL |
[47] |
| E. coli | Amperometric | G-quadruplex/hemin/Gold electrode | Linear range: 9.4–9.4 × 105 CFU/mL LOD: 8 CFU/mL |
[86] |
| E. coli | Impedimetric | rGO-CysCu/Gold electrode | Linear range: 100–108 CFU/mL LOD: 3.8 CFU/mL Assay time: > 1 h |
[48] |
| E. coli | Impedimetric | BSA-conjugated 3D Ag nanoflowers | Linear range: 3.0 × 102–3.0 × 108 CFU/mL LOD: 100 CFU/mL |
[49] |
| E. coli | Amperometric | T7lacZ phages/PAGE | 105 CFU/mL in 3 h and 102 CFU/mL after 7 h | [50] |
| E. coli | Amperometric | CdS@ZIF-8 particles | Linear range: 10–108 CFU/mL Assay time: < 3 h LOD: 3 CFU/mL (S/N=3) |
[51] |
| Vibrio cholerae | Amperometric | ALP/screen-printed electrodes | LOD: 105 cells/mL Assay time: < 55 min |
[55] |
| Vibrio cholerae | Amperometric | screen-printed electrodes | 8 CFU/mL in sea water, 80 CFU/mL sewer water and tap water Assay time: 55 min |
[56] |
| Vibrio cholera | Amperometric | Biotinylated-PAb/ SPE | LOD: 4 × 102 cells/mL Assay time: < 1 h |
[57] |
| Vibrio cholerae | Impedimetric | CeO2 nanowire-modified microelectrode | Linear range: 1.0 × 102–1.0 × 104 CFU/mL | [58] |
| B. cereus | Impedimetric | GNPs-sDNA-(nheA)/PGE | Sensitivity: 100 CFU/mL LOD: 9.4 × 10−12 mol/L |
[66] |
| B. cereus | Amperometric | GNPs-Chit-GCE | Linear range: 5.0 × 101 to 5.0 × 104 CFU/mL LOD: 10.0 CFU/mL (S/N = 3) |
[67] |
| B. cereus | Potentiometric | CPE/SIP | Linear range: 102–105 CFU/mL | [68] |
| S. aureus | Amperometric | HRP-MPA/gold electrode | LOD: 1.6 × 105 cells/mL | [77] |
| S. aureus | Amperometric | HRP-DTSP-/Screen-printed electrodes | Linear range: 1.3 × 103–7.6 × 104 cells/mL LOD: 3.7 × 102 cells/mL Assay time: approximately 30 min |
[78] |
| S. aureus | Amperometric | AP-MPA/gold electrode | Linear range: 4.4 × 105–1.8 × 107 cells/mL LOD: 1.7 × 105 cells/mL Assay time: approximately 25 min |
[79] |
| S. aureus | Impedimetric | Aptamer/rGO-AuNP/GCE | Linear range:10–106 CFU/mL LOD: 10 CFU/mL (S/N=3) Assay time: < 1 h |
[80] |
| S. aureus | Impedimetric | MPA/gold electrode | Linear range: 101–107 CFU/mL LOD: 10 CFU/mL |
[81] |
| S. aureus | Impedimetric | screen printed electrode | Linear range: 3.6 × 107–9.3 × 107 CFU/mL Assay time: approximately 30 min |
[82] |
| DNA of C. perfringens | Electrochemiluminescence | gold electrode (rolling circle amplification) | LOD: 10−15 M Assay time: approximately 1 h |
[85] |
| DNA of C. perfringens | Amperometric | SA/ADH/Fe3O4 nanocomposites | Linear range: 10−12–10−6 M Assay time: same as PCR |
[64] |
| C. perfringens | Impedimetric | CeO2/chitosan/GCE | Linear range: 1.0 × 10−14–1.0 × 10−7 mol/L LOD: 7.06 × 10−15 mol/L |
[87] |
CPE: carbon paste electrode; SIP: spore-imprinted polymer; HRP: horseradish peroxidase; MPA: 3-mercaptopropionic acid; ADH: alcohol dehydrogenase.
3.1. Escherichia coli
Escherichia coli (E. coli) was discovered by Escherich in 1885, and had been considered a non-pathogenic bacterium and a normal part of gut flora for a long period of time [41]. Around the middle of the 20th century, it was recognized that some special serotypes of E. coli were pathogenic to humans and animals, especially to infants and young animals, and often cause severe diarrhea and sepsis [42]. Human are likely to be infected with E. coli by drinking contaminated water or eating unripe foods (especially beef, burgers and roast beef). In addition, a person whose hygiene is poor may be infected by human transmission, or by eating food contaminated with feces [43,44]. Therefore, detection of E. coli in our diet is vital for our health.
The reports of electrochemical biosensors for detection of E. coli are plentiful in foodborne pathogens [45]. As early as 2003, R. Mikkelsen et al. [46] have published screen-printed sensor arrays for the rapid determination of four E. coli subspecies (E. coli B, E. coli Neotype, E. coli JM105 and E. coli HB101). DNA biosensors are an effective means for detection of E. coli. For example, DNA nanopyramids were used by Leong et al. [47] to anchor E. coli lipopolysaccharides, lysate, and whole bacteria. Huang et al. constructed a simple, label-free, and low-cost electrochemical biosensor for highly sensitive detection of E. coli, based on rolling circle amplification (RCA) coupled with peroxidase-mimicking DNA enzyme amplification. The E. coli could specifically bind to the G-quadruplex units in an aptamer-primer probe, which leads to the formation of numerous G-quadruplex oligomers on electrode. Owing to the K+ and hemin on the electrode, the G-quadruplex/hemin complexes were able to generate extremely strong catalytic activity toward H2O2, and then strong electrochemical response could be detected. Recently, Ranjbar et al. prepared polyanilinated amino-functionalized metal–organic frameworks (MOFs) to link amine-modified DNA aptamer by glutaraldehyde (GA). The fabricated biocomposite was used to capture E. coli O157:H7 and methylene blue (MB) as electrochemical indicators in differential pulse voltammetry detection.
Label-free electrochemical biosensors also were developed for detection of E. coli. Using graphene wrapped copper(II)-assisted cysteine hierarchical structure (rGO-CysCu), Malhotra et al. [48] fabricated an immune-electrode which realized that E. coli O157: H7 cells could be differentiated from the non-pathogenic E. coli and other bacterial cells. Another label-free electrochemical biosensor was developed by Wang et al. [49] based on a novel 3D Ag nanoflower. The [Fe(CN)6]3−/4− was used as the redox probe to detect the resistance changes when E. coli O157:H7 was captured by the biosensor.
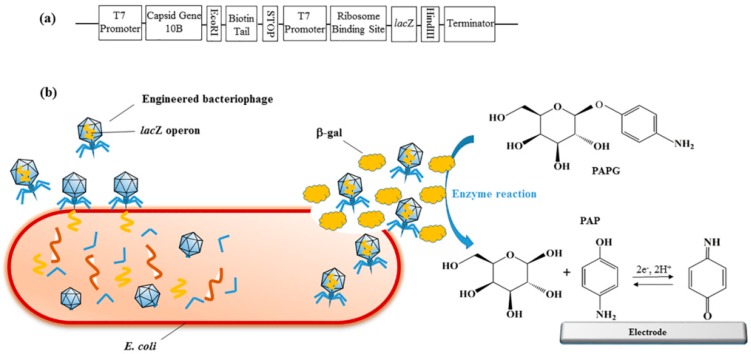
Furthermore, as shown in Figure 2, Nugen et al. [50] tactfully used T7 bacteriophages engineered with lacZ operon to infect E. coli and trigger the overexpression of beta-galactosidase (β-gal). The β-gal would catalyze the 4-aminophenyl-β-galactopyranoside (PAPG) as a substrate and release the electroactive species paminophenol, which could be detected by electrochemical method. Tan et al. [51] introduced amino groups by decorating the surfaces of CdS@ZIF-8 muti-core-shell particles through polyethyleneimine, in order to absorb the anti-E. coli O157:H7 antibody. The Cd(II) ions would release from CdS@ZIF-8 after the target was captured, and then the current could be detected by differential pulse voltammetry.
Figure 2.
Scheme representation of electrochemical detection of E. coli using engineered phage. (a) The designed construct of genome of T7lacZ phage. (b) Specific capture and infection of E. coli by T7lacZ phage resulted in the release and overexpression of enzyme β-gal. PAPG was catalyzed by β-gal into an electroactive species PAP that can be quantified by electrochemical device. Reprinted with permission from [50]. Copyright (2017) American Chemical Society.
3.2. Vibrio cholerae
Vibrio cholerae is the pathogen of human cholera which is one of the ancient and widespread epidemic diseases. Vibrio cholerae has caused many pandemics in the world, mainly characterized by severe vomiting, diarrhea, water loss, and high mortality [52,53,54]. Therefore, it belongs to international quarantine classifications of infectious diseases.
The first Vibrio cholerae electrochemical biosensor was developed by Rao et al. [55] based on disposable screen-printed electrodes (SPE) to adsorb the polyclonal antibodies (PAb) of Vibrio cholerae. When bacterial cells bound to the surface of electrode, the antibodies conjugated to alkaline phosphatase (ALP), as the enzyme tracer catalyzed 1-naphtyl phosphate as its substrate, and then gave an electroactive product which could be detected via an amperometric method. This amperometric biosenor was further applied to study spiked water samples detecting as few as 8 CFU mL−1 in sea water and 80 CFU mL−1 in tap water through an enrichment step [56]. A similar amperometric biosensor for the detection of Vibrio cholerae was described by Doblin et al. using a biotinylated PAb, immobilized on neutravidine modified surface of SPE [57]. A one-step label-free biosensor for V. cholerae detection was developed using antibodies covalently immobilized on a CeO2 nanowire-modified microelectrode to capture the targets. The resulting biosensor was detected by impedance analysis with [Fe(CN)6]3−/4− as the redox probe [58].
3.3. Bacillus cereus
Bacillus cereus (B. cereus), a species of the genus Bacillus, has close contact with humans and can cause food poisoning [59]. Many foods, especially leftovers that have been improperly refrigerated, can cause this type of diarrhea [60,61]. The symptoms caused by Bacillus cereus are abdominal pain, vomiting and diarrhea which are very similar to that caused by Clostridium perfringens [62,63]. Moreover, it is more difficult to distinguish from other short-term symptoms caused by deterioration (such as those caused by Staphylococcus aureus) [64,65]. So, developing an accurate detection method of Bacillus cereus in our food is quite significant.
A B. cereus electrochemical biosensor based on DNA-based Au nanoparticle modified pencil graphite electrode (PGE) was developed by Soleimanian-Zad et al. [66]. The target was captured by the sensing element comprisinggold nano-particles (GNPs) self-assembled with single-stranded DNA of nheA gene immobilized with thiol linker on the GNPs-modified PGE. The researchers also detected the bacteria in milk and infant formula, which showed that the biosensor was suitable for food safety and quality control applications [13]. Liang et al. [67] published a novel B. cereus electrochemical sensor using monoclonal antibodies of B. cereus immobilized on double-layer gold nanoparticles to capture the target, and chitosan was used to link the sensing element with GCE. The sensor displayed a fast detection response, long-term stability and high sensitivity to bacterial contamination. A label-free electrochemical biosensor for Bacillus anthracis spores was fabricated by Amine et al. [68] using pyrrole to modify the electrode and [Fe(CN)6]3−/4− as redox probe.
3.4. Staphylococcus aureus
Staphylococcus aureus (S. aureus) is a typical gram-positive bacterium which could lead to serious purulent infection in human beings, causing pneumonia, pseudomembranous colitis, pericarditis, and even systemic infections such as sepsis [69,70,71]. Food poisoning caused by Staphylococcus aureus enterotoxin accounts for between 33% and 45% of all bacterial food poisoning in the United States and Canada, respectively [65,72]. There are also numerous poisoning incidents in China [73,74,75,76].
M. Pingarro’n et al. [77] developed an amperometric biosensor for the quantification of S. aureus based on rabbit immunoglobulin (RbIgG) immobilized onto the 3-mercaptopropionic acid (MPA) modified electrode. Using the competitive effect between protein A-bearing S. aureus cells and anti-RbIgG labeled with horseradish peroxidase (HRP), the prepared biosensor realized the detection of S. aureus in semi-skimmed milk. Subsequently, the research group reported other two electrochemical biosensors for S. aureus detection. One is an improvement of previous work which used covalent immobilization for anti-RbIgG at SAM modified gold electrodes by 3, 3′- Dithiodipropionic acid di (N-succinimidyl ester) (DTSP) [78]. Another work took advantage of the MPA-SAM gold electrode modified by RbIgG and tyrosinase [79].
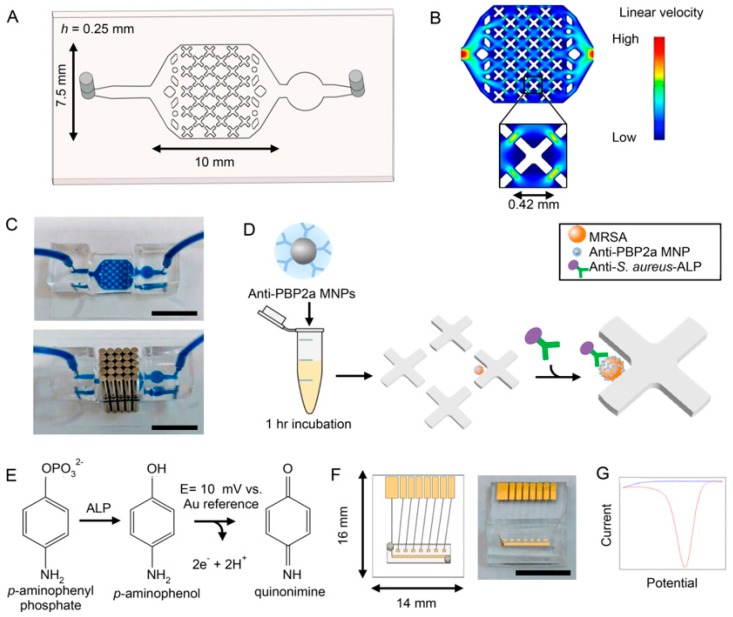
Wei et al. [80] reported an electrochemical sensor for S. aureus detection using single-stranded DNA as aptamer linked to reduced graphene oxide-gold nanoparticles (rGO-AuNP) nanocomposite by impedance spectroscopy. Mansour et al. [81] also detected S. aureus by impedance spectroscopy through monitoring the change of resistance before and after the S. aureus, recognized by anti-S. aureus, immobilized on gold electrode using ferri-/ferrocyanide as redox probe. The developed biosensor was further used to detect stressed and resuscitated pathogens. Recently, a low-cost screen-printed electrode was applied to build an S. aureus biosensor by Connolly et al. [82] using impedance spectroscopy. The targets were incubated in chambers containing the electrodes, and the results analyzed through a novel approach. Impedance spectroscopy provides a label-free method; however, its detection limit is still not low enough compared to other electrochemical biosensors. Methicillin-resistant S. aureus collected from patient nasal swabs was captured and detected using a microfluidic device and antibody-functionalized magnetic nanoparticles. As displayed in Figure 3, the identification of S. aureus is realized by the use of a strain-specific antibody functionalized with alkaline phosphatase for electrochemical detection [83].
Figure 3.
Bacterial capture and electrochemical detection. (A) Schematic of bacterial capture device fabricated in PDMS. (B) Flow profile of capture device simulated on COMSOL Multiphysics. X-shaped features create areas of reduced flow velocity. (C) Photograph of bacterial capture device filled with dye in the absence, and presence of, an array of external magnets (above and below images, respectively). Scale is 10 mm. (D) Filtered nasal swab specimen is incubated with anti-PBP2a MNPs for 1 h. The solution is then processed with the device, where magnetically-labeled bacteria are captured in areas of low flow velocity. After wash steps, anti-S. aureus antibodies functionalized with ALP are introduced into the device and washed. (E) The substrate p-APP is introduced to the device, where it is converted to electrochemically active p-AP by ALP. p-AP is oxidized to quinonimine at a potential of 10 mV against a gold reference electrode. (F) Schematic (left) and photograph (right) of electrochemical detector chip. A PDMS channel allows simple transfer of electrochemical readout solution from the capture device. Detection utilizes on-chip working and reference gold electrodes and an external Pt counter electrode. Scale is 10 mm. (G) Differential pulse voltammogram displaying signal from p-APP (blue) and p-AP (red). The measured current correlates to number of captured bacteria. Reprinted with permission from [83]. Copyright (2019) American Chemical Society.
3.5. Clostridium Perfringens
Clostridium perfringens (C. perfringens) is the most common type of Clostridium in clinically genital gangrene pathogens. C. perfringens can break down sugar in muscle and connective tissue and then release a large amount of gas, which results in severe emphysema of the tissue and affects the supply of blood, ultimately causing a large area of tissue necrosis. The bacterial was named of C. perfringens also due to the bacteria can form a capsule in the body [84].
The detection of clostridium perfringens by electrochemical method is mainly owing to its DNA. Pu et al. [85] published an electrochemiluminescence sensor for detection of DNA of C. perfringens using RCA, like the work of Huang et al. [86]. This research team reported another Clostridium perfringens DNA biosensor based on screen-printed electrodes in the same year [64]. They used the stable hairpin form of the initial molecular beacon, which will open after incubating with target DNA, and then the streptavidin aptamer is reactivated. The electrochemical signal of DPV could be detected by “sandwich” reaction. Recently, Wang et al. [87] described an electrochemical biosensor for the detection of DNA of C. perfringens based on CeO2/chitosan-modified electrodes by monitoring the changes of impedance.
3.6. Simultaneous Detection of Multiple Foodborne Pathogens
There seems to be a trend of developing the electrochemical biosensor for the simultaneous and multiple detection of biologically pathogens [88]. A multi-junction sensor was constructed for potential multiplexed detection of E. coli and S. aureus based on a 2 × 2 junction array formed with gold tungsten wires on single walled carbon nanotube and polyethylenimine. The detection time is rapid and the LODs for E. coli and S. aureus were 10 μL and 100 μL, respectively [89]. Li et al. [57] developed a sandwich-type electrochemical biosensor based on Au/GCP for simultaneous ultrasensitive detection of E. coli O157:H7 and Vibrio cholerae O1. The detection antibodies specific for E. coli O157:H7 and Vibrio cholerae O1 were labeled by CdS and PbS nanoparticles via C60@AuNPs as nanocarriers and HCR amplification, respectively. The antibodies used for capture pathogens were linked to streptavidin-coated magnetic beads (MB@SA). The prepared biosensor displayed excellent performance and this method could be expanded readily for detecting other pathogenic bacteria and would be of great value for future applications in food safety. Furthermore, Ai et al. [90] built an efficient electrochemical disinfection for E. coli and S. aureus in drinking water based on ferrocene–PAMAM–multi-walled carbon nanotubes–chitosan nanocomposite modified pyrolytic graphite electrode. When applying a potential of 0.4 V for 10 min, almost all pathogens were killed, demonstrating that they provided a valid electrochemical method for the disinfection of pathogens.
4. Conclusions and Perspective
Although some traditional methods for detection of foodborne pathogens are sensitive, most of them are also time-consuming (a few days to a week), which limit their practical application. Therefore, developing new methods to detect foodborne pathogens is necessary. Electrochemical biosensing technology has been maturely applied to the rapid determination of pathogens through exploration and development.
Electrochemical biosensors based on nucleic acid or aptamer displayed high sensitivity and low detection limit, however the stability and accuracy should be improved. The electrochemical biosensor based on the combination between antigen and antibody is a big family of biosensors used for the detection of pathogens. These biosensors have high accuracy, but the detection limit is not low enough, especially the biosensors based on the sandwiched principle. The tendency for electrochemical biosensors of pathogens is that multiple pathogens were detected simultaneously. In summary, there is room for further improvement for the detection methods for food pathogens. A rapid, sensitive and low-cost detection method for foodborne pathogens has a huge market prospect. Given the demand and preponderance of electrochemical sensing, there is still a great chance for further developments in the detection of food pathogens in the near future.
Author Contributions
Conceptualization, Z.Z.; validation, J.Z. and X.D.; writing—original draft preparation, Z.Z.; writing—review and editing, J.Z. and X.D.; supervision, X.D.; project administration, Z.Z. and X.D.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant nos. 31801200 and 31801137), Shandong Provincial Natural Science Foundation (ZR2017BC052), Shandong Key Laboratory of Animal Resistance Biology Open fund (2017KF06) and Postdoctoral Science Foundation of China (2017M612334).
Conflicts of Interest
The authors have declared that no competing interest exists.
References
- 1.Lawrence F. The Poison Squad: One Chemist’s Single-Minded Crusade for Food Safety at the Turn of the Twentieth Century. Nature. 2018;562:334–335. doi: 10.1038/d41586-018-07038-0. [DOI] [Google Scholar]
- 2.Cep M. The Poison Squad: One Chemist’s Single-Minded Crusade for Food Safety at the Turn of the Twentieth Century. Science. 2018;361:971. [Google Scholar]
- 3.Inoue H., Suzuki T., Hyodo M., Miyake M. Evaluation of multinomial logistic regression models for predicting causative pathogens of food poisoning cases. J. Vet. Med. Sci. 2018;80:1223–1227. doi: 10.1292/jvms.17-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang L., Huang T. Food poisoning associated with ingestion of wild wasp broods in the upstream region of the Lancang river valley, Yunnan province, China. Toxicon. 2018;145:1–5. doi: 10.1016/j.toxicon.2018.02.045. [DOI] [PubMed] [Google Scholar]
- 5.Markevych I., Standl M., Lehmann I., von Berg A., Heinrich J. Food diversity during the first year of life and allergic diseases until 15 years. J. Allergy Clin. Immun. 2017;140:1751–1754. doi: 10.1016/j.jaci.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Rosa J.S., Hernandez J.D., Sherr J.A., Smith B.M., Brown K.D., Farhadian B., Mahony T., McGhee S.A., Lewis D.B., Thienemann M., et al. Allergic Diseases and Immune-Mediated Food Disorders in Pediatric Acute-Onset Neuropsychiatric Syndrome. Pediatr. Allergy Immunol. Pulmonol. Impact. 2018;31:158–165. doi: 10.1089/ped.2018.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch A.G., Pollak J., Glass T.A., Poulsen M.N., Bailey-Davis L., Mowery J., Schwartz B.S. Early-life antibiotic use and subsequent diagnosis of food allergy and allergic diseases. Clin. Exp. Allergy. 2017;47:236–244. doi: 10.1111/cea.12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hose A.J., Pekkanen J., Roduit C., Riedler J., Dalphin J., Von Mutius E., Ege M.J. Food introduction styles in the first year of life and risk of allergic diseases in the PASTURE birth cohort. Allergy. 2018;73:80. [Google Scholar]
- 9.Berhe H.W., Makinde O.D., Theuri D.M. Co-dynamics of measles and dysentery diarrhea diseases with optimal control and cost-effectiveness analysis. Appl. Math. Comput. 2019;347:903–921. doi: 10.1016/j.amc.2018.11.049. [DOI] [Google Scholar]
- 10.Oliver S.P. Foodborne Pathogens and Disease Celebrates Its Fifteen Year Anniversary. Foodborne Pathog. Dis. 2018;15:1–2. doi: 10.1089/fpd.2017.28999.edi. [DOI] [PubMed] [Google Scholar]
- 11.Zhu D., Zhao X.Y., Yao Y., Dai F.F., He H., Li R.Q., Jin R.H., Liang L.C., Li N. A new factor influencing pathogen detection by molecular assay in children with both mild and severe hand, foot, and mouth disease. Diagn. Microbiol. Infect. Dis. 2013;76:162–167. doi: 10.1016/j.diagmicrobio.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riddle M.S., Murray J.A., Cash B.D., Pimentel M., Porter C.K. Pathogen-Specific Risk of Celiac Disease Following Bacterial Causes of Foodborne Illness: A Retrospective Cohort Study. Digest. Dis. Sci. 2013;58:3242–3245. doi: 10.1007/s10620-013-2733-7. [DOI] [PubMed] [Google Scholar]
- 13.Yu J., Wu J.Q., Zhang Y.Y., Guo L.H., Cong X.Y., Du Y.J., Li J., Sun W.B., Shi J.L., Peng J., et al. Concurrent highly pathogenic porcine reproductive and respiratory syndrome virus infection accelerates Haemophilus parasuis infection in conventional pigs. Vet. Microbiol. 2012;158:316–321. doi: 10.1016/j.vetmic.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Q.L., Sun L., Lian J.J., Gao X.L., Zhao L., Ding M.Y., Li J., Liang Y.C. The phospholipase C (FgPLC1) is involved in regulation of development, pathogenicity, and stress responses in Fusarium graminearum. Fungal Genet. Biol. 2016;97:1–9. doi: 10.1016/j.fgb.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Zheng S., Wu X., Zhang L., Xin C., Liu Y., Shi J., Peng Z., Xu S., Fu F., Yu J., et al. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound. Emerg. Dis. 2017;64:1337–1341. doi: 10.1111/tbed.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao T.L., Ren Q. New records of Ochrolechia and Placopsis from the Hengduan Mountains, China. Mycotaxon. 2012;122:461–466. doi: 10.5248/122.461. [DOI] [Google Scholar]
- 17.Yang H.T., Zou S.S., Zhai L.J., Wang Y., Zhang F.M., An L.G., Yang G.W. Pathogen invasion changes the intestinal microbiota composition and induces innate immune responses in the zebrafish intestine. Fish Shellfish Immunol. 2017;71:35–42. doi: 10.1016/j.fsi.2017.09.075. [DOI] [PubMed] [Google Scholar]
- 18.He C.Q., Liu Y.X., Wang H.M., Hou P.L., He H.B., Ding N.Z. New genetic mechanism, origin and population dynamic of bovine ephemeral fever virus. Vet. Microbiol. 2016;182:50–56. doi: 10.1016/j.vetmic.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 19.Ammar A.M., Attia A.M., Abd El-Aziz N.K., Abd El Hamid M.I., El-Demerdash A.S. Class 1 integron and associated gene cassettes mediating multiple-drug resistance in some food borne pathogens. Int. Food Res. J. 2016;23:332–339. [Google Scholar]
- 20.Oliver S.P. My, How Time Flies: Foodborne Pathogens and Disease to Begin Its Seventh Year with Publication Frequency to Increase in 2010. Foodborne Pathog. Dis. 2010;7:1. doi: 10.1089/fpd.2010.9999. [DOI] [PubMed] [Google Scholar]
- 21.Huang J.Y., Henao O.L., Griffin P.M., Vugia D.J., Cronquist A.B., Hurd S., Tobin-D’Angelo M., Ryan P., Smith K., Lathrop S., et al. Infection with Pathogens Transmitted Commonly Through Food and the Effect of Increasing Use of Culture-Independent Diagnostic Tests on Surveillance—Foodborne Diseases Active Surveillance Network, 10 US Sites, 2012-2015. Morb. Mortal. Wkly. 2016;65:368–371. doi: 10.15585/mmwr.mm6514a2. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y.Y., Qi C.C., Shan S.J., Zhang F.M., Li H., An L.G., Yang G.W. Characterization of common carp (Cyprinus carpio L.) interferon regulatory factor 5 (IRF5) and its expression in response to viral and bacterial challenges. BMC Vet. Res. 2016;12 doi: 10.1186/s12917-016-0750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Zhang F.M., Guo H.Y., Zhu Y.Y., Yuan J.D., Yang G.W., An L.G. Molecular characterization of hepcidin gene in common carp (Cyprinus carpio L.) and its expression pattern responding to bacterial challenge. Fish Shellfish Immunol. 2013;35:1030–1038. doi: 10.1016/j.fsi.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Hou P.L., Wang H.M., Zhao G.M., He C.Q., He H.B. Rapid detection of infectious bovine Rhinotracheitis virus using recombinase polymerase amplification assays. BMC Vet. Res. 2017:13. doi: 10.1186/s12917-017-1284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng S., Wu X., Shi J., Peng Z., Gao M., Xin C., Liu Y., Wang S., Xu S., Han H., et al. Rapid specific and visible detection of porcine circovirus type 3 using loop-mediated isothermal amplification (LAMP) Transbound. Emerg. Dis. 2018;65:597–601. doi: 10.1111/tbed.12835. [DOI] [PubMed] [Google Scholar]
- 26.Zheng S., Shi J., Wu X., Peng Z., Xin C., Zhang L., Liu Y., Gao M., Xu S., Han H., et al. Presence of Torque teno sus virus 1 and 2 in porcine circovirus 3-positive pigs. Transbound. Emerg. Dis. 2018;65:327–330. doi: 10.1111/tbed.12792. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X.Y., Lou M.F., Shen W., Fu R.S., Wang D.H. A Maternal Low-Fiber Diet Predisposes Offspring to Improved Metabolic Phenotypes in Adulthood in an Herbivorous Rodent. Physiol. Biochem. Zool. 2017;90:75–84. doi: 10.1086/688978. [DOI] [PubMed] [Google Scholar]
- 28.Cam D., Oktem H.A. Development of rapid dipstick assay for food pathogens, Salmonella, by optimized parameters. J. Food Sci. Technol. 2019;56:140–148. doi: 10.1007/s13197-018-3467-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan F., Chen M., Leng B.Y., Wang B.S. An efficient autofluorescence method for screening Limonium bicolor mutants for abnormal salt gland density and salt secretion. S. Afr. J. Bot. 2013;88:110–117. doi: 10.1016/j.sajb.2013.06.007. [DOI] [Google Scholar]
- 30.Choi J.R., Yong K.W., Choi J.Y., Cowie A.C. Emerging Point-of-care Technologies for Food Safety Analysis. Sensor. 2019;19:817. doi: 10.3390/s19040817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii S., Segawa T., Okabe S. Simultaneous Quantification of Multiple Food- and Waterborne Pathogens by Use of Microfluidic Quantitative PCR. Appl Environ. Microb. 2013;79:2891–2898. doi: 10.1128/AEM.00205-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y.L., Ravindranath S., Irudayaraj J. Separation and detection of multiple pathogens in a food matrix by magnetic SERS nanoprobes. Anal. Bioanal. Chem. 2011;399:1271–1278. doi: 10.1007/s00216-010-4453-6. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi M., Natori T., Kubota-Hayashi S., Miyata M., Ohkusu K., Kawamoto K., Kurazono H., Makino S., Ezaki T. A New Protocol to Detect Multiple Foodborne Pathogens with PCR Dipstick DNA Chromatography after a Six-Hour Enrichment Culture in a Broad-Range Food Pathogen Enrichment Broth. Biomed. Res. Int. 2013 doi: 10.1155/2013/295050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J.Z., Fu Y.F., Ju L.W., Miao X.Y., Shen Y.J., He L., Wang W.J., Jin J.L., Shao L.Y., Sampath R., et al. Detection and identification of viral pathogens in patients with hand, foot, and mouth disease by multilocus PCR, reverse-transcription PCR and electrospray ionization mass spectrometry. J. Clin. Virol. 2014;59:115–119. doi: 10.1016/j.jcv.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y., Gao Y., Wang T., Dong Q.G., Li J.W., Niu C. Detection of 12 Common Food-Borne Bacterial Pathogens by TaqMan Real-Time PCR Using a Single Set of Reaction Conditions. Front. Microbiol. 2019:10. doi: 10.3389/fmicb.2019.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L., Liang B., Shi J.G., Li F., Mascini M., Liu A.H. A selective and sensitive D-xylose electrochemical biosensor based on xylose dehydrogenase displayed on the surface of bacteria and multi-walled carbon nanotubes modified electrode. Biosens. Bioelectron. 2012;33:100–105. doi: 10.1016/j.bios.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 37.Reta N., Saint C.P., Michelmore A., Prieto-Simon B., Voelcker N.H. Nanostructured Electrochemical Biosensors for Label-Free Detection of Water- and Food-Borne Pathogens. ACS Appl. Mater. Interface. 2018;10:6055–6072. doi: 10.1021/acsami.7b13943. [DOI] [PubMed] [Google Scholar]
- 38.Viswanathan S., Rani C., Ho J.A.A. Electrochemical immunosensor for multiplexed detection of food-borne pathogens using nanocrystal bioconjugates and MWCNT screen-printed electrode. Talanta. 2012;94:315–319. doi: 10.1016/j.talanta.2012.03.049. [DOI] [PubMed] [Google Scholar]
- 39.Camacho C., Chico B., Cao R., Matias J.C., Hernandez J., Palchetti I., Simpson B.K., Mascini M., Villalonga R. Novel enzyme biosensor for hydrogen peroxide via supramolecular associations. Biosens. Bioelectron. 2009;24:2028–2033. doi: 10.1016/j.bios.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 40.Smith S.R., Seenath R., Kulak M.R., Lipkowski J. Characterization of a Self-Assembled Monolayer of 1-Thio-beta-D-Glucose with Electrochemical Surface Enhanced Raman Spectroscopy Using a Nanoparticle Modified Gold Electrode. Langmuir. 2015;31:10076–10086. doi: 10.1021/acs.langmuir.5b02767. [DOI] [PubMed] [Google Scholar]
- 41.Kralj J.M., Hochbaum D.R., Douglass A.D., Cohen A.E. Electrical Spiking in Escherichia coli Probed with a Fluorescent Voltage-Indicating Protein. Science. 2011;333:345–348. doi: 10.1126/science.1204763. [DOI] [PubMed] [Google Scholar]
- 42.Liu G., Lao R.J., Xu L., Xu Q., Li L.Y., Zhang M., Shen H., Mathur S., Fan C.H., Song S.P. Detection of Single-Nucleotide Polymorphism on uidA Gene of Escherichia coli by a Multiplexed Electrochemical DNA Biosensor with Oligonucleotide-Incorporated Nonfouling Surface. Sensor. 2011;11:8018–8027. doi: 10.3390/s110808018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo S.Y., Tay M.Y.F., Aung K.T., Seow K.L.G., Ng L.C., Purbojati R.W., Drautz-Moses D.I., Schuster S.C., Schlundt J. Phenotypic and genotypic characterization of antimicrobial resistant Escherichia coli isolated from ready-to-eat food in Singapore using disk diffusion, broth microdilution and whole genome sequencing methods. Food Control. 2019;99:89–97. doi: 10.1016/j.foodcont.2018.12.043. [DOI] [Google Scholar]
- 44.Zhang S.H., Yang G.Z., Huang Y.B., Zhang J.M., Cui L.H., Wu Q.P. Prevalence and Characterization of Atypical Enteropathogenic Escherichia coli Isolated from Retail Foods in China. J. Food Prot. 2018;81:1761–1767. doi: 10.4315/0362-028X.JFP-18-188. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z., Liu J., Fan J., Wang Z.Y., Li L. Detection of catechol using an electrochemical biosensor based on engineered Escherichia coli cells that surface-display laccase. Anal. Chim. Acta. 2018;1009:65–72. doi: 10.1016/j.aca.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Ertl P., Wagner M., Corton E., Mikkelsen S.R. Rapid identification of viable Escherichia coli subspecies with an electrochemical screen-printed biosensor array. Biosens. Bioelectron. 2003;18:907–916. doi: 10.1016/S0956-5663(02)00206-3. [DOI] [PubMed] [Google Scholar]
- 47.Giovanni M., Setyawati M.I., Tay C.Y., Qian H., Kuan W.S., Leong D.T. Electrochemical Quantification ofEscherichia coliwith DNA Nanostructure. Adv. Funct. Mater. 2015;25:3840–3846. doi: 10.1002/adfm.201500940. [DOI] [Google Scholar]
- 48.Pandey C.M., Tiwari I., Singh V.N., Sood K.N., Sumana G., Malhotra B.D. Highly sensitive electrochemical immunosensor based on graphene-wrapped copper oxide-cysteine hierarchical structure for detection of pathogenic bacteria. Sens. Actuators B Chem. 2017;238:1060–1069. doi: 10.1016/j.snb.2016.07.121. [DOI] [Google Scholar]
- 49.Huang H., Liu M., Wang X., Zhang W., Yang D.P., Cui L., Wang X. Label-Free 3D Ag Nanoflower-Based Electrochemical Immunosensor for the Detection of Escherichia coli O157:H7 Pathogens. Nanoscale Res. Lett. 2016;11:507. doi: 10.1186/s11671-016-1711-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang D., Chen J., Nugen S.R. Electrochemical Detection of Escherichia coli from Aqueous Samples Using Engineered Phages. Anal. Chem. 2017;89:1650–1657. doi: 10.1021/acs.analchem.6b03752. [DOI] [PubMed] [Google Scholar]
- 51.Zhong M., Yang L., Yang H., Cheng C., Deng W., Tan Y., Xie Q., Yao S. An electrochemical immunobiosensor for ultrasensitive detection of Escherichia coli O157:H7 using CdS quantum dots-encapsulated metal-organic frameworks as signal-amplifying tags. Biosens. Bioelectron. 2019;126:493–500. doi: 10.1016/j.bios.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 52.Zhang F., Huang Y.H., Liu S.Z., Zhang L., Li B.T., Zhao X.X., Fu Y., Liu J.J., Zhang X.X. Pseudomonas reactans, a Bacterial Strain Isolated From the Intestinal Flora of Blattella germanica With Anti-Beauveria bassiana Activity. Environ. Entomol. 2013;42:453–459. doi: 10.1603/EN12347. [DOI] [PubMed] [Google Scholar]
- 53.Huang Y.H., Wang X.J., Zhang F., Huo X.B., Fu R.S., Liu J.J., Sun W.B., Kang D.M., Jing X. The Identification of a Bacterial Strain BGI-1 Isolated From the Intestinal Flora of Blattella Germanica, and Its Anti-Entomopathogenic Fungi Activity. J. Econ. Entomol. 2013;106:43–49. doi: 10.1603/EC12120. [DOI] [PubMed] [Google Scholar]
- 54.Li L., Yang H.J., Liu D.C., He H.B., Wang C.F., Zhong J.F., Gao Y.D., Zeng Y.J. Analysis of Biofilms Formation and Associated Genes Detection in Staphylococcus Isolates from Bovine Mastitis. Int. J. Appl. Res. Vet. Med. 2012;10:62–68. doi: 10.5897/AJB11.081. [DOI] [Google Scholar]
- 55.Yuan F., Leng B.Y., Wang B.S. Progress in Studying Salt Secretion from the Salt Glands in Recretohalophytes: How Do Plants Secrete Salt? Front. Plant Sci. 2016:7. doi: 10.3389/fpls.2016.00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma M.K., Goel A.K., Singh L., Rao V.K. Immunological Biosensor for Detection of Vibrio cholerae O1in Environmental Water Samples. World J. Microbiol. Biotechnol. 2006;22:1155–1159. doi: 10.1007/s11274-006-9156-y. [DOI] [Google Scholar]
- 57.Laczka O., Labbate M., Doblin M. Application of an ELISA-type amperometric assay to the detection of Vibrio species with screen-printed electrodes. Anal. Methods-UK. 2014;6:2020–2023. doi: 10.1039/C3AY42169D. [DOI] [Google Scholar]
- 58.Tam P.D., Thang C.X. Label-free electrochemical immunosensor based on cerium oxide nanowires for Vibrio cholerae O1 detection. Mat. Sci. Eng. C-Mater. 2016;58:953–959. doi: 10.1016/j.msec.2015.09.027. [DOI] [PubMed] [Google Scholar]
- 59.Nwaru B.I., Virtanen S.M. Allergenic Food Introduction and Childhood Risk of Allergic or Autoimmune Disease. JAMA-J. Am. Med. Assoc. 2017;317:86. doi: 10.1001/jama.2016.18329. [DOI] [PubMed] [Google Scholar]
- 60.Xie W., Zhou J. Aberrant regulation of autophagy in mammalian diseases. Biol. Lett. 2018:14. doi: 10.1098/rsbl.2017.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Z.B., Qi X.Y., Wang Z.L., Li P.H., Wu C.X., Zhang H., Zhao Y.X. Overexpression of TsGOLS2, a galactinol synthase, in Arabidopsis thaliana enhances tolerance to high salinity and osmotic stresses. Plant Physiol. Biochem. 2013;69:82–89. doi: 10.1016/j.plaphy.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 62.Hou P.L., Zhao G.M., He C.Q., Wang H.M., He H.B. Biopanning of polypeptides binding to bovine ephemeral fever virus G(1) protein from phage display peptide library. BMC Vet. Res. 2018:14. doi: 10.1186/s12917-017-1315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang C., Wang J., Zhang X., Zhang L., Zhang H., Wang L., Wood L.G., Wang G. Do Fast Foods Relate To Asthma Or Other Allergic Diseases? Am. J. Resp. Crit. Care. 2017;195:A3005. [Google Scholar]
- 64.Jiang D., Liu F., Zhang L., Liu L., Liu C., Pu X. An electrochemical strategy with molecular beacon and hemin/G-quadruplex for the detection of Clostridium perfringens DNA on screen-printed electrodes. RSC Adv. 2014;4:57064–57070. doi: 10.1039/C4RA09834J. [DOI] [Google Scholar]
- 65.Liang J.W., Tian F.L., Lan Z.R., Huang B., Zhuang W.Z. Selection characterization on overlapping reading frame of multiple-protein-encoding P gene in Newcastle disease virus. Vet. Microbiol. 2010;144:257–263. doi: 10.1016/j.vetmic.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 66.Izadi Z., Sheikh-Zeinoddin M., Ensafi A.A., Soleimanian-Zad S. Fabrication of an electrochemical DNA-based biosensor for Bacillus cereus detection in milk and infant formula. Biosens. Bioelectron. 2016;80:582–589. doi: 10.1016/j.bios.2016.02.032. [DOI] [PubMed] [Google Scholar]
- 67.Zhu Y.Y., Xing W.X., Shan S.J., Zhang S.Q., Li Y.Q., Li T., An L., Yang G.W. Characterization and immune response expression of the Rig-I-like receptor mda5 in common carp Cyprinus carpio. J. Fish Biol. 2016;88:2188–2202. doi: 10.1111/jfb.12981. [DOI] [PubMed] [Google Scholar]
- 68.Ait Lahcen A., Arduini F., Lista F., Amine A. Label-free electrochemical sensor based on spore-imprinted polymer for Bacillus cereus spore detection. Sens. Actuators B Chem. 2018;276:114–120. doi: 10.1016/j.snb.2018.08.031. [DOI] [Google Scholar]
- 69.Wang X.G., Huang J.M., Feng M.Y., Ju Z.H., Wang C.F., Yang G.W., Yuan J.D., Zhong J.F. Regulatory mutations in the A2M gene are involved in the mastitis susceptibility in dairy cows. Anim. Genet. 2014;45:28–37. doi: 10.1111/age.12099. [DOI] [PubMed] [Google Scholar]
- 70.Wang N., Liu T.T., Xu J.S., Jiang B. The leaf-mining genus Antispila Hubner, 1825 feeding on Vitaceae in Shandong Peninsula, China with one new species (Lepidoptera, Heliozelidae) ZooKeys. 2018;744:49–65. doi: 10.3897/zookeys.744.22421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang F., Yang H.J., He H.B., Wang C.F., Gao Y.D., Zhong Q.F., Wang X.H., Zeng Y.J. Study on the Hemolysin Phenotype and the Genetype Distribution of Staphyloccocus aureus Caused Bovine Mastitis in Shandong Dairy Farms. Int. J. Appl. Res. Vet. Med. 2011;9:416–421. [Google Scholar]
- 72.Liu M., Xie S.B., Zhou J. Use of animal models for the imaging and quantification of angiogenesis. Exp. Anim. 2018;67:1–6. doi: 10.1538/expanim.17-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ren Q. A new species and new records of the lichen genus Pertusaria from China. Mycotaxon. 2015;130:689–693. doi: 10.5248/130.689. [DOI] [Google Scholar]
- 74.Chang J., Zhang E.L., Liu E.F., Sun W.W., Langdon P.G., Shulmeister J. A 2500-year climate and environmental record inferred from subfossil chironomids from Lugu Lake, southwestern China. Hydrobiologia. 2018;811:193–206. doi: 10.1007/s10750-017-3488-5. [DOI] [Google Scholar]
- 75.Lu M., Zhang Y.Y., Tang S.K., Pan J.B., Yu Y.K., Han J., Li Y.Y., Du X.H., Nan Z.J., Sun Q.P. AtCNGC2 is involved in jasmonic acid-induced calcium mobilization. J. Exp. Bot. 2016;67:809–819. doi: 10.1093/jxb/erv500. [DOI] [PubMed] [Google Scholar]
- 76.Wang X.Y., Zhang L.L., Joshi Y., Wang H.Y., Hur J.S. New species and new records of the lichen genus Porpidia (Lecideaceae) from western China. Lichenologist. 2012;44:619–624. doi: 10.1017/S0024282912000242. [DOI] [Google Scholar]
- 77.Escamilla-Gómez V., Campuzano S., Pedrero M., Pingarrón J.M. Development of an Amperometric Immunosensor for the Quantification of Staphylococcus aureus Using Self-Assembled Monolayer-Modified Electrodes as Immobilization Platforms. Electroanalysis. 2007;19:1476–1482. doi: 10.1002/elan.200703893. [DOI] [Google Scholar]
- 78.Escamillagomez V., Campuzano S., Pedrero M., Pingarron J. Electrochemical immunosensor designs for the determination of Staphylococcus aureus using 3,3-dithiodipropionic acid di(N-succinimidyl ester)-modified gold electrodes. Talanta. 2008;77:876–881. doi: 10.1016/j.talanta.2008.07.045. [DOI] [Google Scholar]
- 79.Escamilla-Gomez V., Campuzano S., Pedrero M., Pingarron J.M. Immunosensor for the determination of Staphylococcus aureus using a tyrosinase-mercaptopropionic acid modified electrode as an amperometric transducer. Anal. Bioanal. Chem. 2008;391:837–845. doi: 10.1007/s00216-007-1810-1. [DOI] [PubMed] [Google Scholar]
- 80.Jia F., Duan N., Wu S., Ma X., Xia Y., Wang Z., Wei X. Impedimetric aptasensor for Staphylococcus aureus based on nanocomposite prepared from reduced graphene oxide and gold nanoparticles. Microchim. Acta. 2014;181:967–974. doi: 10.1007/s00604-014-1195-8. [DOI] [Google Scholar]
- 81.Bekir K., Barhoumi H., Braiek M., Chrouda A., Zine N., Abid N., Maaref A., Bakhrouf A., Ouada H.B., Jaffrezic-Renault N., et al. Electrochemical impedance immunosensor for rapid detection of stressed pathogenic Staphylococcus aureus bacteria. Environ. Sci. Pollut. Res. Int. 2015;22:15796–15803. doi: 10.1007/s11356-015-4761-7. [DOI] [PubMed] [Google Scholar]
- 82.Ward A.C., Hannah A.J., Kendrick S.L., Tucker N.P., MacGregor G., Connolly P. Identification and characterisation of Staphylococcus aureus on low cost screen printed carbon electrodes using impedance spectroscopy. Biosens. Bioelectron. 2018;110:65–70. doi: 10.1016/j.bios.2018.03.048. [DOI] [PubMed] [Google Scholar]
- 83.Nemr C.R., Smith S.J., Liu W., Mepham A.H., Mohamadi R.M., Labib M., Kelley S.O. Nanoparticle-Mediated Capture and Electrochemical Detection of Methicillin-Resistant Staphylococcus aureus. Anal. Chem. 2019;91:2847–2853. doi: 10.1021/acs.analchem.8b04792. [DOI] [PubMed] [Google Scholar]
- 84.Saitoh Y., Suzuki H., Tani K., Nishikawa K., Irie K., Ogura Y., Tamura A., Tsukita S., Fujiyoshi Y. Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science. 2015;347:775–778. doi: 10.1126/science.1261833. [DOI] [PubMed] [Google Scholar]
- 85.Jiang D., Liu F., Liu C., Liu L., Li Y., Pu X. Induction of an electrochemiluminescence sensor for DNA detection of Clostridium perfringens based on rolling circle amplification. Anal. Methods. 2014;6:1558–1562. doi: 10.1039/C3AY41961D. [DOI] [Google Scholar]
- 86.Zhao S.S., Jiang Y.X., Zhao Y., Huang S.J., Yuan M., Zhao Y.X., Guo Y. CASEIN KINASE1-LIKE PROTEIN2 Regulates Actin Filament Stability and Stomatal Closure via Phosphorylation of Actin Depolymerizing Factor. Plant Cell. 2016;28:1422–1439. doi: 10.1105/tpc.16.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qian X., Qu Q., Li L., Ran X., Zuo L., Huang R., Wang Q. Ultrasensitive Electrochemical Detection of Clostridium perfringens DNA Based Morphology-Dependent DNA Adsorption Properties of CeO(2) Nanorods in Dairy Products. Sensor. 2018;18:1878. doi: 10.3390/s18061878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tian F., Lyu J., Shi J.Y., Tan F., Yang M. A polymeric microfluidic device integrated with nanoporous alumina membranes for simultaneous detection of multiple foodborne pathogens. Sens. Actuator B-Chem. 2016;225:312–318. doi: 10.1016/j.snb.2015.11.059. [DOI] [Google Scholar]
- 89.Yamada K., Choi W., Lee I., Cho B.K., Jun S. Rapid detection of multiple foodborne pathogens using a nanoparticle-functionalized multi-junction biosensor. Biosens. Bioelectron. 2016;77:137–143. doi: 10.1016/j.bios.2015.09.030. [DOI] [PubMed] [Google Scholar]
- 90.Shang K., Qiao Z., Sun B., Fan X., Ai S. An efficient electrochemical disinfection of E. coli and S. aureus in drinking water using ferrocene–PAMAM–multiwalled carbon nanotubes–chitosan nanocomposite modified pyrolytic graphite electrode. J. Solid State Electrochem. 2013;17:1685–1691. doi: 10.1007/s10008-013-2031-5. [DOI] [Google Scholar]