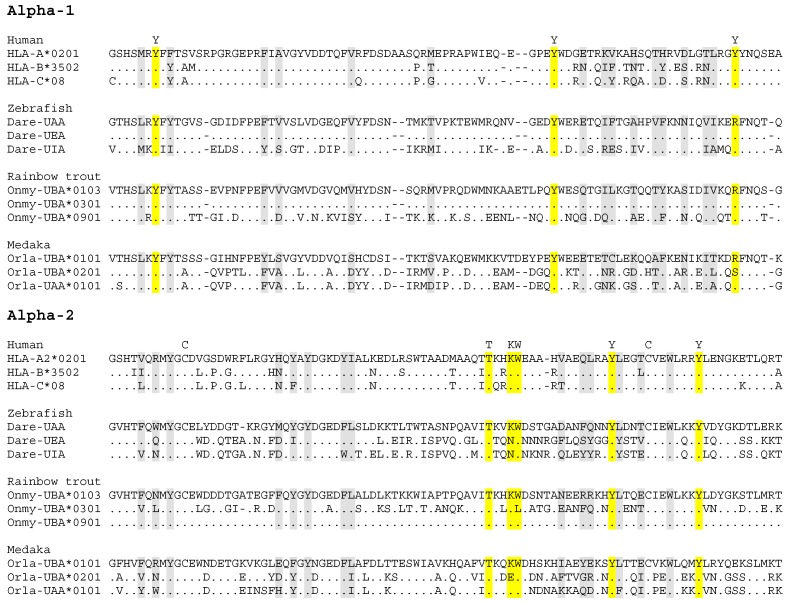
Figure 2.
Alignment of deduced classical MHC class I α1 + α2 domain amino acid sequences. This figure is dedicated to showing aspects of within species variation in the most important functional parts of the classical MHC class I molecules, namely the α1 and α2 domains. The three sequences shown each for zebrafish, rainbow trout and medaka were chosen because they reveal past recombination events. In zebrafish, the Dare-UAA and Dare-UEA sequences share an identical α1 sequence, but have very different α2 sequences, indicative of a recombination event involving the intron between the respective exons [11]. On the other hand, Dare-UIA has an α2 sequence which is very similar to Dare-UEA, but has a very different α1 sequence. Among investigated fish species, recombination events involving the intron between the α1 and α2 exons may have been the most abundant in rainbow trout, since many alleles show their recent traces (e.g., [46,113]). This is exemplified here by Onmy-UBA*0103 having an identical versus very different α1 sequence compared to Onmy-UBA*0301 and Onmy-UBA*0901, respectively, whereas the reverse is found for their α2 domains. Investigated medaka haplotypes have two intact classical MHC class I loci, which have consistently been designated UAA and UBA, although in some haplotypes the “UBA” sequence is quite similar to the UAA sequence (exemplified here by Orla-UBA*0201) and in other haplotypes the UBA sequence is highly divergent from the UAA sequence (exemplified here by Orla-UBA*0101); Nonaka and Nonaka, 2010 [116], explained this situation by interlocus recombination. The levels of divergence among the here depicted zebrafish, rainbow trout and medaka sequences, can be found between individuals of the same species (as allelic or haplotype variation), and are larger than found between three sequences of different human classical MHC class I loci HLA-A, -B and, C. Gray and yellow shading highlight residues that may form part of the peptide binding groove, with yellow shading and a letter indication above the alignment used for conserved residues that bind the peptide ligand termini [41,131]. The letter C above the alignment indicates conserved cysteines. GenBank accessions of the depicted sequences are: HLA-A2, P01892; HLA-B*3502, AAB96790; HLA-C*08, AVQ10002; Dare-UAA, Z46776; Dare-UEA, BC053140; Dare-UIA, KC626502; Onmy-UBA*0103, AF287483 (previous name Onmy-UBA*101); Onmy-UBA*0301, AF287492 (previous name Onmy-UBA*701); Onmy-UBA*0901, AF296366; Orla-UBA*0101, BAD93266; Orla-UBA*0201, AB450999; Orla-UAA*0101, BAD93265.