Summary
In seeds and other parts of cultivated, tetraploid cotton (Gossypium hirsutum L.), multicellular groups of cells lysigenously form dark glands containing toxic terpenoids such as gossypol that defend the plant against pests and pathogens. Using RNA‐seq analysis of embryos from near‐isogenic glanded (Gl 2 Gl 2 Gl 3 Gl 3) versus glandless (gl 2 gl 2 gl 3 gl 3) plants, we identified 33 genes that expressed exclusively or at higher levels in embryos just prior to gland formation in glanded plants. Virus‐induced gene silencing against three gene pairs led to significant reductions in the number of glands in the leaves, and significantly lower levels of gossypol and related terpenoids. These genes encode transcription factors and have been designated the ‘Cotton Gland Formation’ (CGF) genes. No sequence differences were found between glanded and glandless cotton for CGF1 and CGF2 gene pairs. The glandless cotton has a transposon insertion within the coding sequence of the GoPGF (synonym CGF3) gene of the A subgenome and extensive mutations in the promoter of D subgenome homeolog. Overexpression of GoPGF (synonym CGF3) led to a dramatic increase in gossypol and related terpenoids in cultured cells, whereas CRISPR/Cas9 knockout of GoPGF (synonym CGF3) genes resulted in glandless phenotype. Taken collectively, the results show that the GoPGF (synonym CGF3) gene plays a critical role in the formation of glands in the cotton plant. Seed‐specific silencing of CGF genes, either individually or in combination, could eliminate glands, thus gossypol, from the cottonseed to render it safe as food or feed for monogastrics.
Keywords: cotton (Gossypium hirsutum), glanding, RNA‐seq, comparative transcriptomics, VIGS, CRISPR/Cas9, transcription factors, cottonseed, gossypol, protein source, nutrition security
Introduction
Pigmented glands are one of the major characteristics of the tribe Gossypieae, belonging to family Malvaceae, that includes Gossypium L. and seven other genera (Fryxell, 1968). Most of the commercially grown cotton plants have dark glands in the subepidermal tissues of the aerial parts and in the cortex of roots that produce and store terpenoids such as gossypol (Stanford and Viehoever, 1918; Tian et al., 2018). While glands in the seed kernel and flower petals predominantly contain gossypol, those present in other parts of the plant contain additional terpenoids derived from the same biosynthetic pathway. Presence of these terpenoids serves a protective function against various insect pests and some pathogens (Hedin et al., 1992a; Lukefahr and Martin, 1966; Maxwell et al., 1965; Stipanovic et al., 1978a,b, 1999).
Research on the basis of gland formation in the cotton plant began following the discovery of ‘Hopi Moencopi’, a genotype cultivated well into the early 20th century by the native Hopi peoples of Central Arizona (Fulton, 1938; McMichael, 1954, 1959, 1960). The bolls of this plant were reported to have variable number of pigment glands (Fulton, 1938). Since then, research conducted by several investigators implicated the roles of six genes in gland formation, however, only two major genes (Gl 2 and Gl 3) are believed to be involved in gland formation (Endrizzi et al., 1985; Gutierrez et al., 1972). In the tetraploid G. hirsutum, alleles Gl 2, Gl 2, Gl 3 and Gl 3 result in the glanded phenotype, whereas gl 2, gl 2, gl 3 and gl 3 are responsible for the glandless phenotype. Different combinations of dominant (Gl) and recessive (gl) alleles produced lesser number of glands with varying distribution in different parts of the plant at different stages of development (Gutierrez et al., 1972; Lee, 1965; McCarty et al., 1996; McMichael, 1960; Scheffler and Romano, 2008, 2012). Lee (1965) reported that Gl 2 has approximately twice the expressivity of Gl 3, with Gl 2 originating from the ancestor that contributed to the A subgenome and Gl 3 belonging to the D subgenome. Gl 2 and Gl 3 were localized to the A12 and D12 chromosomes of G. hirsutum respectively (Lee, 1965; Percy et al., 2015; Samora et al., 1994). During the course of the current investigation, Ma et al. (2016) published a report describing the identification of a gene named GoPGF (Gossypium Pigment Gland Formation) through map‐based cloning approach using a glandless (dominant) G. barbadense mutant (Gl 2 e) that was originally created in Egypt by mutagenizing radiation (Afifi et al., 1966). This was followed by sequence analysis of the two homeologs of this gene in a recessive glandless mutant of G. hirsutum. The origins of this glandless mutant were not described. The causative mutations in this glandless cotton were presumed to be insertions of a single nucleotide into the coding sequences of each of the two GoPGF homeologs resulting in premature translation termination. The authors designated the GoPGF gene on chromosome A12 as the Gl 2 gene and its homeolog on chromosome D12 as the Gl 3 gene.
In this study that began in May, 2015, we utilized a more direct, RNA‐seq based approach to identify the genes that are involved in gland formation in cotton. In the developing cotton embryo, gland formation begins around 15 days post‐anthesis (dpa) (Reeves and Beasley, 1935; Scheffler et al., 2014). We performed RNA‐seq analysis to identify differentially expressed genes in 14‐, 16‐ and 32‐dpa embryos from glanded (STV GL; GVS4) and glandless (STV gl; GVS5) cotton (Stoneville 7A; Gossypium hirsutum L.) (Scheffler and Romano, 2012). The genes that were expressed at significantly higher levels in the 14‐dpa embryos of the glanded plants, and thus deemed to have possible regulatory functions in gland development, were subjected to virus‐induced gene silencing (VIGS) to ascertain whether these played a role in gland formation. These analyses resulted in the identification of three genes that played critical roles in gland formation. VIGS targeting of any of these three genes not only resulted in inhibition of gland formation, but also in the reduction in gossypol and related terpenoids in the leaves of treated plants. These have been designated Cotton Gland Formation (CGF) genes. We have determined the genomic sequence of these genes in the A and D subgenomes of glanded and glandless cotton plants. Only the GoPGF (synonym CGF3) gene homeologs show differences between the glanded and glandless cotton plants. Furthermore, GoPGF (synonym CGF3) knockout lines showed complete absence of glands. We discuss the implications of suppressing the expression of CGF genes in a seed‐specific manner to obtain cotton plants that produce seeds with significant reduction in the number of glands (and thus gossypol) so that the immense protein resource available in cottonseed can be safely used either as feed for monogastric animals or directly as food for humans to improve their nutrition security.
Results
Comparative transcriptome analysis of developing embryos from glanded and glandless cotton plants reveals several genes associated with gland formation
Cotton embryos at 14‐, 16‐ and 32‐dpa were used for transcriptome analysis; these encompassed a stage preceding visible gland formation to one of active gland‐filling. Embryos from near‐isogenic lines of Stoneville 7A referred to as Stoneville 7A Glanded (STV GL, GVS4) and Stoneville 7A glandless (STV gl, GVS5) were compared with the aim of finding differentially expressed genes. As shown in Figure S1a, no glands were observed in 14‐dpa embryos of GVS4, however, at 16‐dpa some glands can be seen in the embryos from this line (shown with arrows in Figure S1b). Gossypol, the major storage terpenoid of seed‐glands, can be detected in the embryos of glanded cotton plants around 24‐dpa and later (Scheffler et al., 2014). No glands were detected in line GVS5 embryos at any stage of development (Figures S1c and d). RNA was isolated from three replicate samples of 14‐, 16‐ and 32‐dpa embryos each from GVS4 and GVS5. RNA‐seq was performed on these three different developmental stages from two different glanding types to give six different tissues and a total 377 million quality‐filtered paired‐end reads were obtained. Out of these, 273 million unique reads (72.13%) were mapped to the reference genome (Zhang et al., 2015), and 22.82% of them mapped more than one time. Overall, 94.95% of reads were mapped to the reference genome. Only the uniquely mapped reads were used to measure transcript abundance. Tissue‐wise data for the mapped reads are given in Table S1.
To ascertain transcript abundance, only the uniquely mapped reads were quantified using HTSeq‐count program to obtain read count values for all the annotated 70,478 genes in G. hirsutum (Zhang et al., 2015). Of these, 57,510 genes were expressed in at least one of the samples analysed, which were further considered for downstream analysis. At least, 30 million unique reads were counted for each tissue (every replicate had 10 million or more read counts). DESeq2 program was used to identify differentially expressed genes (log fold change ≥2 and FDR <0.05). Figure 1 shows the number of genes that are differentially expressed in the glanded vs. glandless embryos at different time points. At 14 dpa, a small number of genes were differentially expressed, with only 33 genes expressed at higher levels in the glanded embryos compared to glandless embryos (Table S2). Seven genes were expressed at lower levels in the glanded embryos. At 16 dpa, 178 genes were expressed at higher levels and 73 genes at lower levels in the glanded embryos (Data S1). At 32 dpa, 894 genes were expressed at higher levels and 240 genes at lower levels in the glanded embryos (Data S1). Table S2 shows the 33 genes that were expressed at significantly higher levels in the 14‐dpa embryos of glanded cotton plant. This stage precedes 1–2 days before the glands become visible, therefore we focused on this stage of development to identify the genes that would presumably be responsible for and involved in gland formation.
Figure 1.
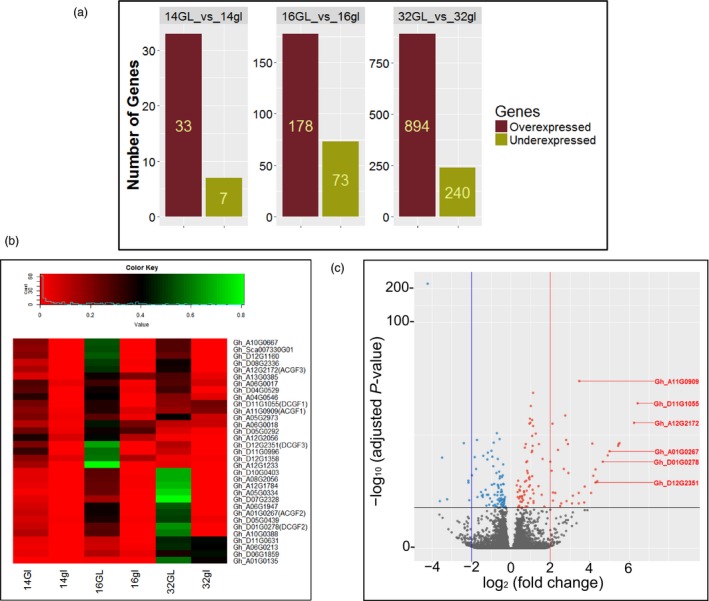
Differentially expressed genes in developing embryos from glanded (GL) and glandless (gl) cotton plants. (a) Differentially expressed genes identified in pairwise comparisons between GL and gl embryos at 14‐, 16‐ and 32‐days post‐anthesis (dpa). (b) Heatmap visualization of expression of the 33 genes that were expressed at higher levels in the 14‐dpa glanded embryos during the course of embryo development. (c) Volcano plot showing gene expression differences between 14‐dpa embryos from glanded and glandless cotton. Genes with absolute fold change ≥2 (red line) and P < 0.05 are indicated as red dots. The CGF gene homeologs are shown in red coloured letters.
Virus‐induced gene silencing validates the involvement of three genes in gland formation
Virus‐induced gene silencing is a rapid and simple method to transiently silence a target gene in the young emerging leaves of a plant. Therefore, we used this method to silence individual genes in cotton seedlings in order to determine their role in gland development. The subset of genes that were predicted to encode proteins of regulatory function were subjected to VIGS, based on the assumption that one or more of these might play an important role in gland formation. In cases where both of the homeologs were found to be expressed at higher levels in the glanded embryos, a single VIGS construct was used to target both homeologs for silencing – a viable approach given the high degree of sequence similarity (over 95%) between the two (Table S2). In all our VIGS experiments, the gene‐silencing efficacy was confirmed by targeting GhCLA gene that results in albino leaf phenotype (Figure S2). Target sequences, ranging in size from 357 to 634 bp, were amplified using a set of primers listed in Table S3. Of the ten genes targeted in this manner, we observed negative effects on gland formation after silencing of three gene pairs, designated Cotton Gland Formation (CGF) genes. We observed a dramatic reduction in the number of glands in response to silencing of both the Gh_A11G0909/Gh_D11G1055 gene pair (CGF1; 78% reduction) and the Gh_A12G2172/Gh_D12G2351 [GoPGF (synonym CGF3); 90% reduction] (Figures 2a, and S3). The reduction in gland numbers in the newly emerging leaves was observed starting at 2‐weeks post‐infiltration. At 21 days post‐infiltration, the leaves were scanned and the gland number was quantified. Figure 2a shows the representative images of the leaves from the plantlets that had undergone VIGS treatment as compared to an empty vector control. The CGF1 and GoPGF (synonym CGF3) gene pairs both encode basic Helix‐Loop‐Helix (bHLH) transcription factors. VIGS silencing of another gene pair, Gh_A01G0267/Gh_D01G0278 (CGF2), which encodes NAC‐family transcription factors, did not show such a dramatic reduction in the number of glands compared to the lines that were silenced at the CGF1 and GoPGF (synonym CGF3) loci. However, the visual and microscopic appearance of the glands in CGF2‐targeted leaves were qualitatively different from glanded cotton in terms of colour intensity and structure, as though their development was adversely affected (Figure 2a). No effects on gland number or appearance were observed with the remaining seven VIGS constructs and therefore those respective genes were not investigated further.
Figure 2.
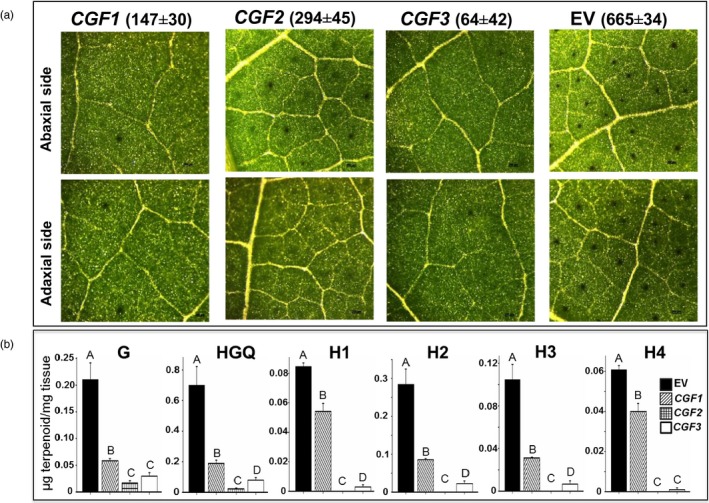
Effect of virus‐induced gene silencing (VIGS) of the CGF1,CGF2 and GoPGF (synonym CGF3) genes on gland formation. (a) Microscopic images of leaves (magnification: X42). Mean gland number per unit area (6.45 cm2) ±SE shown above the respective image. (b) terpenoid levels in the leaves. EV: empty vector control; G: gossypol; HGQ: hemigossypolon; H: heliocides. The values indicated by bars within a group are significantly different at P ≤ 0.05 if labelled with different letters.
Unlike the glands in cottonseed that contain mainly gossypol, the glands in leaves contain not only gossypol, but also hemigossypolon and heliocides that are derived from the same biosynthesis pathway. Thus, reduced number of functional glands would be expected to result in lower amounts of these terpenoids in the leaves of cotton plants that have undergone VIGS against the CGF genes. Therefore, we conducted HPLC analysis to measure the levels of these terpenoids in the leaves. Indeed a significant reduction in the level of gossypol and related terpenoids (hemigossypolon and heliocides) was observed in the leaves of plants that were subjected to VIGS‐mediated silencing of CGF1, CGF2 or GoPGF (synonym CGF3) genes (Figure 2b). Since the terpenoids are usually produced and stored in the glands, the reduced levels of these compounds likely result from fewer glands or fewer functional glands. Thus, based on the results from RNA‐seq analysis and VIGS experiments, we identified three transcription factors and their homeologs that play an important, positive role in the formation of glands in the cotton plant. The CGF gene homeologs of the A subgenome will be referred to as ACGF and those of the D subgenome as DCGF in the remaining text, figures and tables in this report.
qRT‐PCR validates the involvement of three CGF gene pairs in gland formation
Transcript abundance for the CGF genes (and the respective homeologs) in glanded (GVS4) and glandless (GVS5) embryos at different developmental stages is depicted as normalized read counts in Figure 3a. To validate the RNA‐seq expression profile of these genes, qRT‐PCR was performed using the same set of RNA samples that was used to perform RNA‐seq analysis. PCR efficiencies of the three CGF gene pairs and internal control histone gene were determined following the protocol described by Livak and Schmittgen (2008). The efficiencies for each of the CGF homeologs and histone gene were in the range of 2.0 to 2.2 confirming the validity of qPCR results (Figure S4). Results obtained from qRT‐PCR analysis confirmed the expression profiles of the three CGF genes that were observed with the RNA‐seq analysis (Figure 3b). Expression of these CGF genes in the glandless embryos was indeed substantially lower than that of the glanded embryos at 14‐ and 16‐dpa stages of development.
Figure 3.
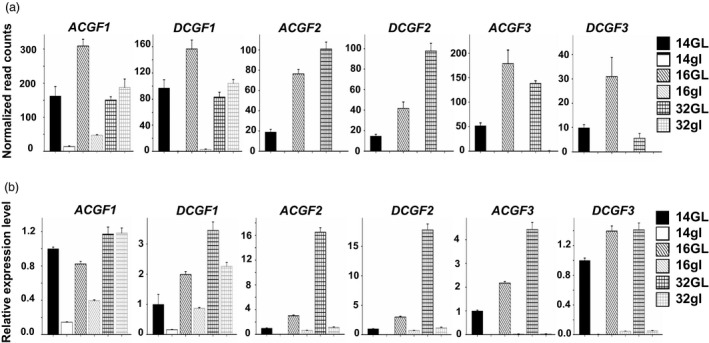
Expression levels for the three CGF genes in A and D subgenomes at 14‐, 16‐ and 32‐days post‐anthesis embryos from glanded (GL) and glandless (gl) cotton plants. (a) Mean normalized read counts of three biological replicates, based on RNA‐seq analysis; (b) qRT‐PCR results showing relative expression levels.
Sequencing of CGF genes reveals the underlying cause of glandless phenotype
The results for the expression profile of CGF genes show that these genes have little or no activity in the glandless embryos, especially at 14‐dpa stage. In order to understand the reasons for these differences, we sequenced each of these genes and their homeologs from glanded and glandless cotton plants. Large PCR fragments were amplified from the genomic DNA of glanded and glandless cotton plants using specific primers (Table S4) that can differentiate between the A and D subgenome homeologs of each of the CGF genes. These amplicons included approx. 2 kb of promoter region (4.2 kb in case of DCGF3), UTRs, introns (if present), exons and terminator. No sequence differences were found between glanded and glandless cotton plants in either CGF1 or CGF2 genes or in their respective homeologs (Figures S5–S8). Major sequence differences between the glanded and glandless cotton plants were observed in both GoPGF (synonym CGF3) gene homeologs (Figure 4). The glandless line (GVS5) showed a 5.1 kb transposon insertion between 362 and 363 bp of the coding sequence of the ACGF3 gene (Gh_A12G2172; Figures 4a,b and S9). In addition, there were two SNPs and a 2‐bp deletion in the promoter sequence, and two SNPs in the coding sequence of Gh_A12G2172 gene in the glandless GVS5. The coding sequence of the DCGF3 gene (Gh_D12G2351) of the glandless mutant (GVS5) has two SNPs (one synonymous and one nonsynonymous) compared to the wild‐type glanded cotton (GVS4). In addition, the terminator sequence of the DCGF3 from the glandless mutant line (GVS5) has one base pair deletion. However, the significant differences in the DCGF3 gene between glanded and glandless cotton were in the promoter (Figures 4c,d and S10). The ~4.2 kb promoter region of this gene in the glandless mutant (GVS5) had fifteen SNPs, two deletions (1 and 49 bp long) and two insertions (1 and 3 bp) compared to the glanded cotton (GVS4). Interestingly, the CGF3 gene that we have identified and sequenced from the glanded cotton, GVS4, is the same gene designated as GoPGF by Ma et al. (2016). However, the underlying cause of mutation responsible for glandless phenotype in each case is different, as discussed later.
Figure 4.
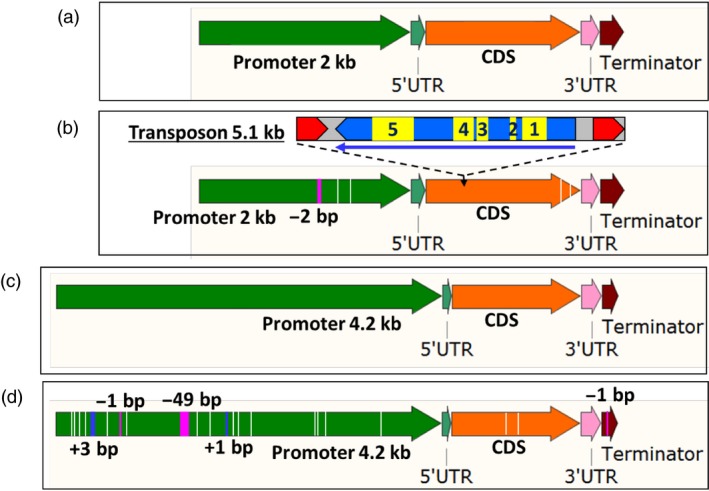
Illustration showing differences between glanded and glandless cotton for GoPGF (synonym CGF3) gene in A‐ and D‐subgenome. (a) ACGF3 in glanded (GVS4) cotton. (b) ACGF3 in glandless (GVS5) cotton showing four SNPs (white lines), one deletion (pink line) and a transposon insertion. The Copia‐like, retrotransposon is 5.1 kb in size. Red arrows represent direct repeats. The long thin arrow indicates direction and size of an open reading frame. Functional domains are: 1: gag‐polypeptide of LTR copia‐type, 2: GAG‐pre‐integrase domain, 3: Integrase core domain, 4: Reverse transcriptase (RNA‐dependent DNA polymerase), 5: Ty1/Copia family of RNase HI in long‐term repeat retro elements. (c) DCGF3 in glanded (GVS4) cotton. (d) DCGF3 in glandless (GVS5) cotton showing 17 SNPs (white lines), three deletions (pink lines) and two insertions (blue lines).
Both the A‐ and D‐subgenome homeologs of GoPGF (synonym CGF3) gene are expressed in the embryos of glanded cotton
There are some genes in an allotetraploid such as G. hirsutum in which one homeolog for a particular gene is expressed, whereas the other remains silent in a given tissue (Adams et al., 2003; Grover et al., 2012). RNA‐seq results showed that while both the A‐ and D‐subgenome homeologs of the GoPGF (synonym CGF3) gene were expressed in the developing embryos of the glanded cotton, the DCGF3 was less active (Figure 3a). In order to further confirm whether both the homeologs of the GoPGF (synonym CGF3) gene are expressed in the embryos of glanded cotton, a PCR amplicon was generated with a primer set which can amplify both A and D‐subgenome homeologs using the cDNA from 14‐dpa embryos. Direct sequencing of this amplicon clearly showed the expected SNPs (Figure S11) thus confirming the results from RNA‐seq analysis and qRT‐PCR showing that both the GoPGF (synonym CGF3) homeologs are expressed in the embryos of glanded cotton.
Promoter sequence analysis and activity evaluation of the D subgenome GoPGF (synonym CGF3) homeolog reveals the basis for its inactivity in glandless cotton
As described earlier, the GoPGF (synonym CGF3) homeologs in both the A and D subgenomes show no expression in the embryos of glandless GVS5 (Figure 3). The undetectable level of expression of the GoPGF (synonym CGF3) gene in the A subgenome is likely due the insertion of the 5.1 kb transposon (Figure 4b). As we had done for the other CGF genes and their homeologs, at first, we amplified only the ~2 kb of the promoter (2009 bp), 5’‐UTR (97 bp), the coding sequence, 3’‐UTR and 182 bp of the terminator region of the DCGF3 gene from glanded and glandless cotton. We observed four SNPs in the promoter region and two SNPs in the coding sequence between glanded and glandless cotton. To investigate whether these SNPs in the promoter region were responsible for the lack of transcripts in the glandless GVS5, we assembled promoter::gusA constructs using ~2.1 kb long promoter (including 5’‐UTR) sequences from the DCGF3 gene of GVS4 and GVS5. Agrobacterium tumefaciens cells containing the reporter gene construct were used to transform hypocotyl segments of cotton seedlings. Callus tissue growing on hypocotyl segments following transformation were examined histochemically for GUS activity, 30 days after transformation. The results presented in Figure S12 show clearly that the D subgenome GoPGF (synonym CGF3) gene promoter sequences (~2.1 kb) from the glanded and glandless cotton were equally active. It is possible that the ~2.1 kb sequence does not fully represent the entire promoter region of this gene and that important regulatory elements reside further upstream. We therefore isolated a longer, ~4.2 kb of the promoter region of the DCGF3 gene from glanded (GVS4) and glandless (GVS5) cotton. As described earlier, the ~4.2 kb promoter region of this gene in the glandless mutant (GVS5) showed significant mutations, including fifteen SNPs, two deletions (1 and 49 bp long) and two insertions (1 and 3 bp) compared to the glanded cotton (GVS4). In order to examine whether these sequence differences in the glandless cotton were responsible for the lack of expression of the DCGF3 gene, we assembled reporter gene constructs as described above. Callus tissues growing from the transformed cotyledon, hypocotyl and petiole explants were examined histochemically for GUS activity, 5 weeks after Agrobacterium‐mediated transformation with each of the constructs. The results from this analysis are shown in Figure S13. While the tissue transformed with a construct wherein the gusA gene was under the control of DCGF3 promoter from glanded cotton showed strong GUS activity, the callus originating from explants following transformation with glandless DCGF3 promoter construct showed drastic reduction in reporter gene activity. The results suggest that the lack of DCGF3 transcripts in the glandless (GVS5) cotton is due to the attenuation of the activity of its heavily mutated promoter.
Sequencing of GoPGF (synonym CGF3) gene from four additional glandless lines reveals the nature of mutations
As mentioned above, the underlying cause for the glandless trait in the recessive, G. hirsutum mutant as proposed by Ma et al. (2016) was the insertion of a single nucleotide in the coding sequence of each of the two GoPGF homeologs. However, the GoPGF (synonym CGF3) gene identified in this study showed a 5.1 kb, copia‐like, retrotransposon in the A subgenome homeolog and several SNPs, insertions and deletions in the promoter of the D subgenome homeolog of the glandless mutant GVS5 (Figures 4, S8 and S9). In order to further explore the genetic basis of the glandless phenotype in cotton germplasm, four additional glandless cotton lines that have been developed by other breeders [Acala GLS, NM‐13P1088, NM‐13P1115 and NM‐13P1117 strains (Bowman et al., 2006; Zhang et al., 2014)] were examined for allelic variation in the GoPGF (synonym CGF3) gene pair by PCR amplification and sequencing. The results showed that the glandless Acala GLS and NM‐13P1088 had the same transposon insertion in the ACGF3 gene that we had discovered in the GVS5 line. These results agree with the pedigree information available for these lines (Bowman et al., 2006). The glandless source of GVS5 is STV 7A gl which is believed to have C6‐5 in its pedigree, and one of C6‐5 parents is Hopi Moencopi. Bowman et al. (2006) traces Acala GLS back to Hopi Moencopi and NM‐13P1088 has Acala GLS as its glandless parent. As mentioned earlier, Hopi Moencopi is a glandless source discovered and described in the mid‐twentieth century (Fulton, 1938; McMichael, 1954, 1959, 1960). The other two glandless cotton lines (NM‐13P1115 and NM‐13P1117) had a total of three SNPs in the coding region of ACGF3 gene, including two synonymous and one nonsynonymous, at residue 43, which alters an alanine to valine. Thus, these two lines have the same dominant mutation Gl e 2 obtained through irradiation to create the Egyptian glandless cotton cultivar ‘Bahtim 110’, as reported previously (Kohel and Lee, 1984; Ma et al., 2016). Their pedigrees show that the glandless parent for both NM‐13P1115 and NM‐13P1117 was an experimental line that had Bahtim 110 as one of its parents.
CRISPR/Cas9‐mediated knockout of CGF2 genes reduces gland density and terpenoids in the leaves of mutants, and knockout of GoPGF (synonym CGF3) genes results in glandless phenotype
We conducted additional experiments using the CRISPR/Cas9 system to knockout CGF2 and GoPGF (synonym CGF3) genes in order to validate their role in gland formation. In each case, both A and D homeologs were targeted for knockout. Since CGF1 homeologs are active in the 32‐dpa embryos of both glanded and glandless plants (Figure 3), and therefore possibly also involved in other activities, so these were not targeted for CRISPR/Cas9‐mediated knockout. Four lines from targeting of the CGF2 gene (LCT236 construct) and nine lines from targeting of the GoPGF (synonym CGF3) gene (LCT237 and LCT238 constructs) were recovered. Detailed biochemical and molecular analyses were performed on two lines in each case. The leaves obtained from the regenerated plants (T0 generation) were examined for their terpenoid content. Results presented in Table 1 show significant reduction in terpenoid levels in the leaf tissues of the CGF2 mutants (236‐8 and 236‐10). In line with our observations in VIGS experiments, the number of glands was substantially reduced in various parts of the mutant lines (Figure 5). The glands that were present were smaller and appeared abnormal as shown in the high magnification images presented in Figure S14. The mutations obtained in these two CGF2‐targeted lines are shown in Figure S15. Virtually no gossypol was detected in the leaves of GoPGF (synonym CGF3) knockout plants (237‐3 and 237‐4; Table 1) and all parts of the plants examined were devoid of glands (Figure 5). The mutations observed in these two GoPGF (synonym CGF3)‐targeted lines are shown in Figure S16. The trait created by CRISPR‐Cas9‐mediated knockout of CGF2 and GoPGF (synonym CGF3) genes is heritable in the T1 generation as illustrated in Figure S17. These results confirm that CGF2 and GoPGF (synonym CGF3) genes play important roles in the development of glands in the cotton plant. Furthermore, a completely glandless phenotype observed in the GoPGF (synonym CGF3) knockout mutants validates the primacy of this gene as a key regulator of gland development.
Table 1.
Terpenoid values in the leaves of mutant lines generated by CRISPR/Cas9 mediated knockout of CGF2 (236‐8 and 236‐10) and GoPGF (synonym CGF3) (237‐3 and 237‐4) genes in comparison to wild‐type control
| Terpenoids (μg terpenoid/mg tissue) | ||||||
|---|---|---|---|---|---|---|
| HGQ | G | H1 | H2 | H3 | H4 | |
| Wild‐type | 0.33 | 0.18 | 0.07 | 0.28 | 0.11 | 0.04 |
| Line 236‐8 | 0.01 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 |
| Line 236‐10 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 |
| Line 237‐3 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Line 237‐4 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
G, gossypol; HGQ, hemigossypolon; H, heliocides.
Figure 5.

Mutant lines showing the effect of CRISPR/Cas9‐mediated knockout of CGF2 (236‐10) and GoPGF (synonym CGF3) (237‐3) genes on gland formation in T0 plants in comparison to glanding pattern seen in a wild‐type, control cotton plant. (a,f,k) leaf image (adaxial), illuminated from underside. (b,g,l) leaf image (abaxial), illuminated from above. (c,h,m) leaf petiole. (d,i,n) unopened floral bud. (e,j,o) developing cotton boll.
Overexpression of A subgenome GoPGF (synonym CGF3) in cotton callus tissue significantly increases the terpenoid levels
While glands are present in most parts of the cotton plants, these have never been observed in callus cultures. We wanted to examine the impact of overexpressing GoPGF (synonym CGF3) gene under the control of a constitutive promoter, such as the CaMV 35S promoter. Therefore, we assembled an overexpression vector using the ACGF3 coding sequence driven by this promoter and transformed cotton seedling explants using the Agrobacterium method. Individual transgenic events (in the form of small, kanamycin‐resistant calli) developing on the explants were excised and further cultured as per our laboratory protocol. When observed after 4 months, a majority of these events had turned unusually dark brown, while a few events remained light pale‐green colour similar to what transgenic callus lines, transformed with any other gene, usually appear at this stage. We suspected that the dark‐coloured events were expressing the transgenic ACGF3 gene, whereas the lighter‐coloured ones were not. In order to examine this possibility, qRT‐PCR was performed on these two types of culture lines. Results presented in Figure S18 show that the dark‐coloured culture lines indeed showed higher level transcription of GoPGF (synonym CGF3) gene compared to the lighter‐coloured lines that showed activities similar to the nontransgenic control cultures. This molecular analysis was followed by an additional biochemical analysis in which we examined the two types of culture lines for their terpenoid content. Terpenoids that are usually found in glands, such as gossypol, were detected at significantly higher levels in the dark‐coloured cultures compared to the light‐coloured ones and the nontransgenic callus cultures (Figure 6). In addition to gossypol, some other terpenoids were found either exclusively (hemigossypol, desoxyhemigossypol, hemigossylic acid lactone, methoxyhemigossypol and desoxymethoxyhemigossypol) or at significantly higher levels (methoxygossypol and dimethoxygossypol) in the dark‐coloured culture lines.
Figure 6.
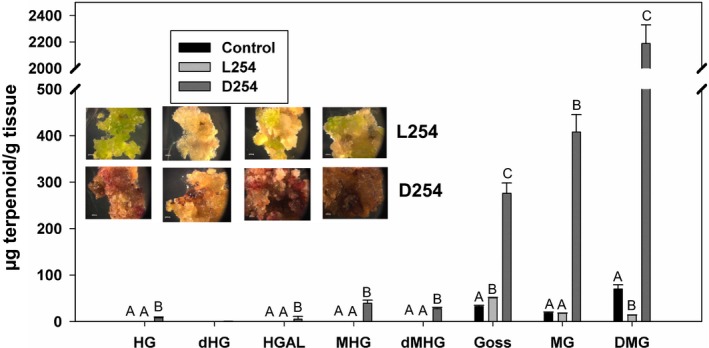
Terpenoid levels in cotton callus cultures, obtained following transformation with A subgenome GoPGF (synonym CGF3) overexpression construct. L254: light‐coloured callus lines; D254 dark‐coloured callus lines; Control: nontransgenic callus. HG: Hemigossypol,; dHG: Desoxyhemigossypol; HGAL: Hemigossylic acid lactone; MHG: Methoxyhemigossypol; dMHG: Desoxymethoxy‐hemigossypol; Goss: Gossypol; MG: Methoxygossypol; DMG: Dimethoxygossypol. The values indicated by bars within a group are significantly different at P ≤ 0.05 if labelled with different letters.
Discussion
Studies on cotton embryo development and gland formation by Reeves and Beasley (1935) and Scheffler et al. (2014) indicate that gland formation starts around 15 dpa. A similar timing of gland formation was observed in our study. No glands were observed in the embryos at the 14‐dpa stage in the greenhouse‐grown, glanded cotton (STV GL) GVS4 (Figure S1a), however, at 16‐dpa stage, the glands were clearly visible under a microscope in the embryos from this glanded line (Figure S1b). No glands were observed in the glandless (STV gl) GVS5 at any stage of embryo development. On the basis of this information, we conducted transcriptome analyses on embryos at 14‐, 16‐, 32‐dpa stage of development obtained from glanded, GVS4 and glandless, GVS5 near‐isogenic cotton lines. RNA‐seq analysis revealed that 33 genes were expressed at higher levels in the glanded embryos at 14 dpa compared to their counterparts from the glandless plants. Since gossypol biosynthesis does not begin in the embryos before 20 dpa (Scheffler et al., 2014), genes involved in terpenoid biosynthesis such as the one encoding δ‐cadinene synthase were not active in the embryos of the glanded plants at 14 dpa. Because we found no visible glands in the embryos at 14 dpa, we hypothesized that comparative transcriptomics at this time‐point would reveal the identity of the genes that play an important role in initiating gland formation. The later stages of embryo development are likely to reveal the genes that are involved in gland maturation and biosynthesis of secondary metabolites, including gossypol (Huchelmann et al., 2017).
RNA‐seq proved to be a rather straightforward and useful technique in identifying a number of genes that were either solely expressed or more highly expressed in the embryos (14 dpa) of glanded cotton compared to those in the glandless cotton. VIGS was used against ten different genes that were predicted to encode proteins with regulatory functions to ascertain their involvement in gland formation. VIGS targeting of three different genes and their homeologs (designated CGF) significantly reduced the number of glands, and the terpenoids that are stored within, in the young emerging leaves of a cotton plantlet. Further, qRT‐PCR results on each of these genes validated the RNA‐seq analysis in terms of relative expression levels for the homeologs of the three CGF genes.
Sequencing of the respective homeologs of CGF1 and CGF2 did not show any differences between the glanded and glandless cotton. However, the ACGF3 gene in the glandless cotton had a 5.1 kb transposon insertion within its coding sequence, thus accounting for its silencing (Figures 4 and S9). The D subgenome homeolog of GoPGF (synonym CGF3) gene in the glandless cotton showed two SNPs in the coding sequence and one SNP in the terminator between glanded and glandless cotton. However, the ~4.2 kb upstream regulatory sequence showed some major differences in the glandless cotton, including fifteen SNPs, two deletions (1 and 49 bp long) and two insertions (1 and 3 bp), compared to the glanded cotton (Figures 4 and S10). Comparative promoter activity analysis of this region between glanded and glandless cotton showed that the heavily mutated, DCGF3 gene promoter from the glandless cotton was substantially weakened.
No sequence differences between the glanded and glandless cotton were observed for the CGF1 and CGF2 genes and their respective homeologs. However, the fact that VIGS‐mediated downregulation of these genes did have a negative impact on the gland numbers and terpenoid levels, the respective encoded proteins probably do play an important role in gland formation. Particularly, the importance of CGF2 in gland development is supported by the fact that both VIGS and CRISPR/Cas9‐mediated knockout of this gene not only had a negative effect on gland numbers, the glands that were visible appeared abnormal and the terpenoid content of the leaves was greatly reduced. Of the three CGF gene pairs, GoPGF (synonym CGF3) genes seem to play the most critical role in gland development. Validation for this notion comes from the following results: 1) complete absence of GoPGF (synonym CGF3) transcripts in the glandless embryos at all stages of development, 2) significant reduction in leaf glands and terpenoids by VIGS treatment, 3) totally glandless phenotype and absence of terpenoids in the knockout lines created by CRISPR/Cas9‐mediated mutations. The two GoPGF (synonym CGF3) homeologs were localized on A12 and D12 chromosomes of G. hirsutum.
As mentioned earlier, a report by Ma et al. (2016) describes the identification of a gene, named GoPGF, using a glandless (dominant) mutant (G. barbadense, Hai‐1) that was derived from a mutant (Gl 2 e) originally created in Egypt by mutagenizing radiation (Afifi et al., 1966). The authors proposed that one amino acid change from alanine to valine at residue 43 in the protein as a result of substitution of ‘C’ to ‘T’ at base 128 in the coding sequence of the A subgenome GoPGF was the underlying cause for this dominant mutation. These authors followed this work by sequence analysis of the two homeologs of this gene in a recessive glandless mutant of G. hirsutum of unknown origin. They found that this glandless cotton had a single nucleotide insertion in the coding sequence of each of the two GoPGF homeologs resulting in premature translation termination (insertion of a ‘T’ between 735 and 736 bp in A subgenome homeolog and insertion of an ‘A’ between 916 and 917 in the D subgenome homeolog), thus accounting for the glandless trait. In our investigation of GoPGF (synonym CGF3) gene(s) of GVS5, the cause of mutation is entirely different. The basis for the silencing of the A subgenome GoPGF (synonym CGF3) is likely due to the insertion of a 5.1 kb transposon, whereas the D subgenome GoPGF (synonym CGF3) gene promoter of the glandless cotton has undergone extensive mutations, thus silencing the gene activity.
The ACGF3 is localized on chromosome A12 while its homeolog the DCGF3 is present on chromosome D12. Here, we have provided substantial evidence that these two homeologs are the main genes controlling the development of glands in cotton plants and, based on chromosomal location, correspond to the Gl 2 and Gl 3 loci described by several geneticists previously (Lee, 1965; Percy et al., 2015). The results from RNA‐seq analysis also show that while both homeologs of the GoPGF (synonym CGF3) gene are expressed in the developing embryos of glanded cotton, the A subgenome homeolog is more active, thus providing confirmation for the earlier contention that Gl 2 is expressed at higher level compared to Gl 3 gene in glanded cotton (Lee, 1965). Hovav et al. (2015) conducted global transcriptome analysis on developing seeds of G. hirsutum (TM1) at 10, 20, 30 and 40 dpa and found that about 20% of the genes showed homeolog expression bias. This group also observed that the ACGF3 homeolog in TM1 had higher level of expression than DCGF3 in the seeds at 30‐ and 40‐dpa.
While cotton is grown for its fibre, the plant produces ~1.6 X more seed by weight. In addition to the oil, cottonseed also contains ~23% protein. Thus, global cottonseed production (~45 million metric tons, MMT) containing ~10 MMT of protein can potentially meet the basic protein requirements of ~550 million people (Rathore et al., 2017). However, because of the presence of toxic gossypol in the seed glands (Gadelha et al., 2014; Risco and Chase, 1997), this abundant resource cannot be used for food or even as feed for monogastric animals. Whole cottonseed and cottonseed meal are used simply as feed for older cattle that are highly inefficient in converting feed protein into meat protein (Rathore et al., 2017). Gossypol‐free cottonseed meal can be a new source of protein for the more efficient aquaculture species and poultry, or can even be used as human food. The identification of the three CGF genes that play a direct or indirect role in gland formation provides us with the tools to suppress gland formation by silencing any one or more of these genes. Thus, strict tissue‐specific silencing of the CGF gene(s) in the seed kernel should eliminate or significantly reduce its gossypol content. Tissue‐specific silencing of a gene represents a powerful approach to examine the effects of silencing a gene in a particular tissue, and the trait created by these methods is stable and heritable (Houmard et al., 2007; Liu et al., 2002; Palle et al., 2013; Rathore et al., 2012; Schmidt et al., 2011; Sunilkumar et al., 2006). Strict tissue specificity of such gene silencing is critical because the terpenoid contents of the glands in the rest of the cotton plant provide protection against various pests and pathogens (Hedin et al., 1992b; Stipanovic et al., 1999). The expression profile of the three CGF genes in the embryos of glanded and glandless cotton at various stages of development suggests that CGF2 and GoPGF (synonym CGF3) can be safely targeted for silencing as these two genes are not transcribed in the embryos of glandless cotton and thus not necessary for normal embryo development (Figure 3). To ensure complete elimination of gossypol from the cottonseed, it may be advisable to target one of these two CGF genes in combination with δ‐cadinene synthase gene for silencing. Silencing of δ‐cadinene synthase, that catalyses a key step in the biosynthesis of gossypol, has been used successfully to significantly reduce gossypol in the cottonseed by 98% (Sunilkumar et al., 2006). The combined targeting of two different types of genes should ensure complete elimination of seed gossypol in case some glands do develop despite silencing of CGF2 and GoPGF (synonym CGF3) genes. There are several gene‐silencing technologies available such as RNAi (Hebert et al., 2008; Smith et al., 2000; Sunilkumar et al., 2006), CRISPR interference (CRISPRi) (Larson et al., 2013; Zhao et al., 2014) and C2c2 (CRISPR‐Cas13a)‐mediated destruction of specific transcripts (Abudayyeh et al., 2016). Any such gene silencing technologies in conjunction with a seed‐specific promoter can be used to eliminate the glands and thus gossypol from the cottonseed only. While elimination or significant reduction in gossypol from the cottonseed is a highly desirable goal, its commercial success may require modification of cultivation and especially seed storage practices to address the possibility of increased predation, especially by rodents.
While the CaMV35S promoter is typically considered to be too strong to drive a gene encoding a regulatory protein, our results on the callus cultures overexpressing the ACGF3 gene point to an intriguing possibility of increasing the number of glands in the foliage and floral tissue by driving the expression of this gene under the control of its own promoter or another suitable promoter. It is also possible to increase the expression of the GoPGF (synonym CGF3) genes using some form of CRISPR/Cas9 technology to enhance the activity of the respective native promoters. In this regard, it will be important to understand the molecular basis underlying higher gland density in some of the cotton genotypes. Thus, we believe that seed‐specific silencing of CGF2/CGF3 genes and/or δ‐cadinene synthase genes, whereas overexpressing the CGF3 gene in other organs, by modification of native promoters or transgenic overexpression, can provide a cotton plant that produces gossypol‐free seeds, while having greater number of glands (and therefore higher levels of gossypol and related terpenoids) in rest of the plant for more robust defence against pests and pathogens, albeit at a slightly higher metabolic cost. There is an increasing need for such a ‘natural’ defence mechanism against pests because more and more insect species are developing resistance to various forms of Bt‐cotton. The cost of refining oil from such gossypol‐free cottonseed will be lower, and the meal can be used as a source of protein for the more efficient monogastric animals (poultry, swine and aquaculture species) and even as food, thus enhancing nutrition security in the cotton‐producing parts of the world.
Experimental procedures
Plant materials, RNA isolation, library preparation and RNA‐sequencing
Near‐isogenic lines of tetraploid cotton (Gossypium hirsutum L.) cultivar Stoneville 7A, designated Stoneville 7A glanded (STV GL; GVS4; Gl 2 Gl 2 Gl 3 Gl 3) and Stoneville 7A glandless (STV gl; GVS5; gl 2 gl 2 gl 3 gl 3) (Scheffler and Romano, 2012) were used for comparative RNA‐seq analysis to identify the genes that are involved in gland formation. Fully opened flowers were tagged on the greenhouse‐grown plants of GVS4 and GVS5. Bolls at 14‐, 16‐, 32‐dpa were collected and embryos were carefully dissected from the developing seeds under a stereo‐microscope. A glanded cultivar, Coker 312 was used to conduct VIGS and CRISPR/Cas9 experiments to validate the function of the candidate genes.
Total RNA was extracted from three independent biological replicates of each embryo sample (100–200 mg) using the Spectrum Plant Total RNA Kit (Sigma‐Aldrich, St. Louis, MO) following manufacturer's instructions. After on‐column DNase I treatment to remove the DNA from samples, RNA was eluted with nuclease‐free water. RNA quantity was measured using micro spectrophotometer (Nano‐Drop Technologies, Inc., Thermo Fisher Scientific Inc., Waltham, MA), and its quality was assessed with Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA). Only the samples with RNA integrity number (RIN) above 8.0 were used for the analysis.
Library preparation and RNA‐seq were performed by Texas A&M AgriLife Genomics and Bioinformatics Services. Poly‐A enriched mRNA from each replicate sample was used for the library preparation, 125‐bp paired‐end sequencing was performed using Illumina HiSeq 2500. Sequence cluster identification, quality pre‐filtering, base calling and uncertainty assessment were done in real time using Illumina HCS 2.2.58 and RTA 1.18.64 software (Illumina Inc., San Diego, CA) with default parameter settings.
Bioinformatics analysis
RNA‐seq data were further processed using Trimmomatic software to filter out the low‐quality reads (Bolger et al., 2014) using LEADING:20 TRAILING:20 SLIDINGWINDOW:5:20 MINLEN:100 as parameters. Filtered reads were then mapped to the G. hirsutum (Texas Marker‐1) reference genome (Zhang et al., 2015) using HISAT2 program (Kim et al., 2015) and gene annotation in GFF3 format (NBI_Gossypium_hirsutum_v1.1.gene.gff3) (Yu et al., 2014). The allotetraploid cotton G. hirsutum L. acc. Texas Marker‐1 (TM‐1) is widely used as a genetic standard and its genome was sequenced in 2015 (Zhang et al., 2015). The output from the HISAT2 program was then analysed to quantify the reads per gene using the HTSeq‐count program (Anders et al., 2015). The differentially expressed genes were identified using DESeq2 (Love et al., 2014). The False Discovery Rate was set to ≤0.05 and the log fold change value to ≥2 to determine differentially expressed genes.
CRISPR/Cas9‐mediated knockout of CGF2 and GoPGF (synonym CGF3) genes
CGF2 and GoPGF (synonym CGF3) genes were targeted for knockout using the CRISPR/Cas9 system. The guide sequences used to target CGF2 and GoPGF (synonym CGF3) are listed in Table S5. Selected lines showing the mutant phenotype (absence of glands or malformed glands) were analysed by sequencing the amplicon, encompassing the target sites, generated from the genomic DNA isolated from the leaves of T0 plants. Primer sequences are provided in Table S6. A complete description of the CRISPR/Cas9‐mediated knockout and mutation analysis is provided in the Supplementary Methods.
A detailed description of procedures, including VIGS, terpenoid analysis, qRT‐PCR, sequencing, promoter activity assay, etc. is provided in Supplementary Methods (Data S2).
Supporting information
Figure S1 Microscopic images of developing embryos of Stoneville 7A glanded (GL; GVS4) and glandless (gl; GVS5), near‐isogenic lines, used for comparative RNA‐seq analysis.
Figure S2 Virus‐induced gene silencing (VIGS) in cotton.
Figure S3 Leaves from plants that had undergone virus‐induced gene silencing against CGF genes showing the effects on gland formation.
Figure S4 Figure S4 Real‐time PCR standard curves representing PCR efficiency for each of the CGF homeologs and Histone gene.
Figure S5 Alignment of A subgenome CGF1 gene sequences from GVS4 (glanded) and GVS5 (glandless, recessive mutant) near‐isogenic lines.
Figure S6 Alignment of D subgenome CGF1 gene sequences from GVS4 (glanded) and GVS5 (glandless, recessive mutant) near‐isogenic lines.
Figure S7 Alignment of A subgenome CGF2 gene sequences from GVS4 (glanded) and GVS5 (glandless, recessive mutant) near‐isogenic lines.
Figure S8 Alignment of D subgenome CGF2 gene sequences from GVS4 (glanded) and GVS5 (glandless, recessive mutant) near‐isogenic lines.
Figure S9 Alignment of A subgenome GoPGF (synonym CGF3) gene sequences from GVS4 (glanded) and GVS5 (glandless, recessive mutant) near‐isogenic lines.
Figure S10 Alignment of D subgenome GoPGF (synonym CGF3) gene sequences from GVS4 (glanded) and GVS5 (glandless, recessive mutant) near‐isogenic lines.
Figure S11 Sequencing results showing four different SNPs that differentiate A and D subgenome GoPGF (synonym CGF3) genes.
Figure S12 Promoter (2.05 kb) activity evaluation of the D subgenome GoPGF (synonym CGF3) gene from glanded and glandless cotton using gusA as the reporter gene.
Figure S13 Promoter (~4.2 kb) activity evaluation of the D subgenome GoPGF (synonym CGF3) gene from glanded and glandless cotton using gusA as the reporter gene.
Figure S14 Effect of CRISPR/Cas9‐mediated knockout of CGF2 and GoPGF (synonym CGF3) genes on gland formation in T0 plants in comparison to glanding pattern seen in a wild‐type, control cotton plant.
Figure S15 Mutations observed in two CGF2 knockout lines, (a) 236‐8 and (b) 236‐10. The two target sites are highlighted yellow and PAM sequences red.
Figure S16 Mutations observed in two GoPGF (synonym CGF3) knockout lines, (a) 237‐3 and (b) 237‐4.
Figure S17 Effect of CRISPR/Cas9‐mediated knockout of CGF2 and GoPGF (synonym CGF3) genes on gland formation observed in cottonseed kernels.
Figure S18 qRT‐PCR analysis of GoPGF (synonym CGF3) transcripts in cotton callus cultures obtained following transformation with ACGF3 overexpression construct. L254: light‐coloured callus lines; D254 dark‐coloured callus lines; Control: nontransgenic callus.
Table S1 RNA‐seq reads for glanded (GL; GVS4) and glandless (gl; GVS5) embryos at 14‐, 16‐ and 32‐days post‐anthesis and their mapping to the reference genome.
Table S2 Genes that expressed at higher levels in the glanded embryos (STV GL; GVS4; Gl2Gl2Gl3Gl3) in comparison to those in the glandless (STV gl; GVS5; gl2gl2gl3gl3) embryos at 14‐days post‐anthesis stage of development based on RNA‐seq analysis. Genes encoding putative transcription factors were tested for their role in gland formation using virus‐induced gene silencing (VIGS).
Table S3 Primers used to amplify segments of the coding sequence of the target gene for cloning into TRV2 binary vector to conduct VIGS experiments.
Table S4 Primers used to amplify and isolate CGF genes from A and D subgenomes of glanded (GVS4) and glandless (GVS5) cotton plants.
Table S5 Guide sequences used to target CGF2 and GoPGF (synonym CGF3) genes.
Table S6 Primers used for amplicon sequencing of regenerated plants targeted with LCT236, LCT237 and LCT238 constructs.
Data S1 Differentially expressed genes between glanded and glandless embryos at 16‐ and 32 days post anthesis.
Data S2 Supplementary methods.
Acknowledgements
The base vectors for assembling the transformation constructs were kindly provided by Dr. Daniel Voytas, University of Minnesota. The two glandless lines, NM13P1115 and NM13P1118, were developed by Dr. Jinfa Zhang, New Mexico State University. We thank Drs. Wayne Smith and David Stelly for critical reading of the manuscript. This research was supported by funds from Cotton Inc. and Texas AgriLife Research. The authors declare no conflict of interest.
Data availability
RNA‐seq data from developing embryos at different stages of development (14‐, 16‐ and 32‐dpa) from both glanded (GVS4), and glandless (GVS5) cotton are available in the NCBI SRA archive (accession # PRJNA448612).
References
- Abudayyeh, O.O. , Gootenberg, J.S. , Konermann, S. , Joung, J. , Slaymaker, I.M. , Cox, D.B.T. , Shmakov, S. et al (2016) C2c2 is a single‐component programmable RNA‐guided RNA‐targeting CRISPR effector. Science, 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K.L. , Cronn, R. , Percifield, R. and Wendel, J.F. (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ‐specific reciprocal silencing. Proc. Natl Acad. Sci. USA, 100, 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afifi, A. , Bary, A. , Kamel, S. and Heikal, I. (1966) Bahtim 110, a new strain of Egyptian cotton free from gossypol. Emp. Cot. Grow. Rew. 43, 112–120. [Google Scholar]
- Anders, S. , Pyl, P.T. and Huber, W. (2015) HTSeq—a Python framework to work with high‐throughput sequencing data. Bioinformatics, 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, D.T. , Gutierrez, O.A. , Percy, R.G. , Calhoun, D.S. and May, O.L. (2006) Pedigrees of upland and Pima cotton cultivars released between 1970 and 2005. Mississippi Agricultural & Forestry Experiment Station Bulletin 1155, December 2006.
- Endrizzi, J. , Turcotte, E. and Kohel, R. (1985) Genetics, cytology, and evolution of Gossypium In Advances in Genetics (Caspari E.W. and John G.S. eds), pp. 271–375. Amsterdam, Netherlands: Elsevier. [Google Scholar]
- Fryxell, P.A. (1968) A redefinition of the tribe Gossypieae. Bot. Gaz. 129, 296–308. [Google Scholar]
- Fulton, H. (1938) Hopi cotton: a variable species. J. Agr. Res. 56, 333–336. [Google Scholar]
- Gadelha, I.C.N. , Fonseca, N.B.S. , Oloris, S.C.S. , Melo, M.M. and Soto‐Blanco, B. (2014) Gossypol toxicity from cottonseed products. Sci. World J. 2014, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover, C. , Gallagher, J. , Szadkowski, E. , Yoo, M. , Flagel, L. and Wendel, J. (2012) Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 196, 966–971. [DOI] [PubMed] [Google Scholar]
- Gutierrez, M. , Vrdoljak, J. and Ricciardi, A. (1972) Development of gossypol‐glandless strains of cotton. In Induced Mutations and Plant Improvement, pp. 397–404. Vienna, Austria: International Atomic Energy Agency. [Google Scholar]
- Hebert, C.G. , Valdes, J.J. and Bentley, W.E. (2008) Beyond silencing—engineering applications of RNA interference and antisense technology for altering cellular phenotype. Curr. Opin. Biotechnol. 19, 500–505. [DOI] [PubMed] [Google Scholar]
- Hedin, P.A. , Jenkins, J.N. and Parrott, W.L. (1992a) Evaluation of flavonoids in Gossypium arboreum (L.) cottons as potential source of resistance to tobacco budworm. J. Chem. Ecol. 18, 105–114. [DOI] [PubMed] [Google Scholar]
- Hedin, P.A. , Parrott, W.L. and Jenkins, J.N. (1992b) Relationships of glands, cotton square terpenoid aldehydes, and other allelochemicals to larval growth of Heliothis virescens (Lepidoptera: Noctuidae). J. Econ. Entomol. 85, 359–364. [Google Scholar]
- Houmard, N.M. , Mainville, J.L. , Bonin, C.P. , Huang, S. , Luethy, M.H. and Malvar, T.M. (2007) High‐lysine corn generated by endosperm‐specific suppression of lysine catabolism using RNAi. Plant Biotechnol. J. 5, 605–614. [DOI] [PubMed] [Google Scholar]
- Hovav, R. , Faigenboim‐Doron, A. , Kadmon, N. , Hu, G. , Zhang, X. , Gallagher, J.P. and Wendel, J.F. (2015) A transcriptome profile for developing seed of polyploid cotton. Plant Genome 8, 1–15. [DOI] [PubMed] [Google Scholar]
- Huchelmann, A. , Boutry, M. and Hachez, C. (2017) Plant glandular trichomes: natural cell factories of high biotechnological interest. Plant Physiol. 175, 6–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Langmead, B. and Salzberg, S.L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat. Meth. 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohel, R. and Lee, J. (1984) Genetic analysis of Egyptian glandless cotton 1. Crop Sci. 24, 1119–1121. [Google Scholar]
- Larson, M.H. , Gilbert, L.A. , Wang, X. , Lim, W.A. , Weissman, J.S. and Qi, L.S. (2013) CRISPR interference (CRISPRi) for sequence‐specific control of gene expression. Nat. Protoc. 8, 2180–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.A. (1965) THE genomic allocation of the principal foliar‐gland loci in gossypium hirsutum and gossypium barbadense. Evolution, 19, 182–188. [Google Scholar]
- Liu, Q. , Singh, S.P. and Green, A.G. (2002) High‐stearic and high‐oleic cottonseed oils produced by hairpin RNA‐mediated post‐transcriptional gene silencing. Plant Physiol. 129, 1732–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2008) Analyzing real‐time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukefahr, M.J. and Martin, D.F. (1966) Cotton‐plant pigments as a source of resistance to the bollworm and tobacco Budworm12. J. Econ. Entomol. 59, 176–179. [Google Scholar]
- Ma, D. , Hu, Y. , Yang, C. , Liu, B. , Fang, L. , Wan, Q. , Liang, W. et al (2016) Genetic basis for glandular trichome formation in cotton. Nat. Commun. 7, 10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, F.G. , Lafever, H.N. and Jenkins, J.N. (1965) Blister beetles on glandless cotton. J. Econ. Entomol. 58, 792–793. [Google Scholar]
- McCarty, J.C. , Hedin, P.A. and Stipanovic, R.D. (1996) Cotton Gossypium spp. plant gossypol contents of selected GL2 and GL3 alleles. J. Agric. Food Chem. 44, 613–616. [Google Scholar]
- McMichael, S. (1954) Glandless boll in upland cotton and its use in the study of natural crossing. Agron. J. 46, 527–528. [Google Scholar]
- McMichael, S.C. (1959) Hopi cotton, a source of cottonseed free of gossypol pigments 1. Agron. J. 51, 630. [Google Scholar]
- McMichael, S.C. (1960) Combined effects of glandless genes gl2 and gl3 on pigment glands in the cotton plant. Agron. J. 52, 385–386. [Google Scholar]
- Palle, S.R. , Campbell, L.M. , Pandeya, D. , Puckhaber, L. , Tollack, L.K. , Marcel, S. , Sundaram, S. et al (2013) RNAi‐mediated ultra‐low gossypol cottonseed trait: performance of transgenic lines under field conditions. Plant Biotechnol. J. 11, 296–304. [DOI] [PubMed] [Google Scholar]
- Percy, R. , Hendon, B. , Bechere, E. and Auld, D. (2015) Qualitative genetics and utilization of mutants In Cotton (Fang D.D. and Percy R.G., eds), pp. 155–186. Madison, WI: American Society of Agronomy, Inc., Crop Science Society of America, Inc., and Soil Science Society of America, Inc. [Google Scholar]
- Rathore, K.S. , Sundaram, S. , Sunilkumar, G. , Campbell, L.M. , Puckhaber, L. , Marcel, S. , Palle, S.R. et al (2012) Ultra‐low gossypol cottonseed: generational stability of the seed‐specific, RNAi‐mediated phenotype and resumption of terpenoid profile following seed germination. Plant Biotechnol. J. 10, 174–183. [DOI] [PubMed] [Google Scholar]
- Rathore, K.S. , Wedegaertner, T.C. and Hake, K. (2017) Ultra‐low gossypol cottonseed (ULGCS) as a feed for non‐ruminants to enhance human nutrition security In Broadening Horizons. Feedipedia ‐ Animal Feed Resources Information System ‐ INRA CIRAD AFZ and FAO. [Google Scholar]
- Reeves, R. and Beasley, J. (1935) The development of the cotton embryo. J. Agric. Res. 51, 935–944. [Google Scholar]
- Risco, C. and Chase, C. Jr . (1997) Gossypol In Handbook of Plant and Fungal Toxicants (Felix D'Mello J.P., ed), pp. 87–98. Boca Raton, FL: CRC Press. [Google Scholar]
- Samora, P.J. , Stelly, D.M. and Kohel, R.J. (1994) Localization and Mapping of the Le1, and Gl2 Loci of Cotton (Gossypium hirsutum L.). J. Hered. 85, 152–157. [Google Scholar]
- Scheffler, J.A. and Romano, G.B. (2008) Modifying gossypol in cotton (Gossypium hirsutum L.): a cost effective Method for small seed samples. J. Cotton Sci. 12, 202–209. [Google Scholar]
- Scheffler, J.A. and Romano, G.B. (2012) Registration of GVS1, GVS2, and GVS3 upland cotton lines with varying gland densities and two near‐isogenic lines, GVS4 and GVS5. J. Plant Reg. 6, 190–194. [Google Scholar]
- Scheffler, J.A. , Taliercio, E.W. , Tonos, J.L. and Romano, G.B. (2014) Microscopic methods to evaluate gland initiation and development in cotton ovules. J. Cotton Sci. 18, 420–429. [Google Scholar]
- Schmidt, M.A. , Barbazuk, W.B. , Sandford, M. , May, G. , Song, Z. , Zhou, W. , Nikolau, B.J. et al (2011) Silencing of soybean seed storage proteins results in a rebalanced protein composition preserving seed protein content without major collateral changes in the metabolome and transcriptome. Plant Physiol. 156, 330–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, N.A. , Singh, S.P. , Wang, M.‐B. , Stoutjesdijk, P.A. , Green, A.G. and Waterhouse, P.M. (2000) Gene expression: total silencing by intron‐spliced hairpin RNAs. Nature, 407, 319. [DOI] [PubMed] [Google Scholar]
- Stanford, E.E. and Viehoever, A. (1918) Chemistry and histology of the glands of the cotton plant, with notes on the occurrence of similar glands in related plants. J. Agric. Res. 13, 419–435. [Google Scholar]
- Stipanovic, R.D. , Bell, A.A. , O'Brien, D.H. and Lukefahr, M.J. (1978a) Heliocide H1. A new insecticidal C25 terpenoid from cotton (Gossypium hirsutum). J. Agric. Food Chem. 26, 115–118. [Google Scholar]
- Stipanovic, R.D. , Bell, A.A. , O'Brien, D.H. and Lukefahr, M.J. (1978b) Heliocide H3 aninsecticidal terpenoid from Gossypium hirsutum. Phytochemistry, 17, 151–152. [Google Scholar]
- Stipanovic, R. , Bell, A. and Benedict, C. (1999) Cotton pest resistance: the role of pigment gland constituents In Biologically Active Natural Products: Agrochemicals (Cutler H.G. and Cutler S., eds), pp. 211–220. Boca Raton, FL: CRC Press. [Google Scholar]
- Sunilkumar, G. , Campbell, L.M. , Puckhaber, L. , Stipanovic, R.D. and Rathore, K.S. (2006) Engineering cottonseed for use in human nutrition by tissue‐specific reduction of toxic gossypol. Proc. Natl Acad. Sci. USA, 103, 18054–18059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, X. , Ruan, J.‐X. , Huang, J.‐Q. , Yang, C.‐Q. , Fang, X. , Chen, Z.‐W. , Hong, H. et al (2018) Characterization of gossypol biosynthetic pathway. Proc. Natl Acad. Sci. USA, 115, E5410–E5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas, D.F. , Atkins, P. and Baltes, N.J. (2015) Engineering plant genomes using CRISPR/Cas systems. 2015/0167000 A1
- Yu, J. , Jung, S. , Cheng, C.‐H. , Ficklin, S.P. , Lee, T. , Zheng, P. , Jones, D. et al (2014) CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42, D1229–D1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Idowu, O.J. , Flynn, R. , Wedegaertner, T. and Hughs, S.E. (2014) Genetic variation and selection within glandless cotton germplasm. Euphytica, 198, 59–67. [Google Scholar]
- Zhang, T. , Hu, Y. , Jiang, W. , Fang, L. , Guan, X. , Chen, J. , Zhang, J. et al (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM‐1) provides a resource for fiber improvement. Nat. Biotech. 33, 531–537. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Dai, Z. , Liang, Y. , Yin, M. , Ma, K. , He, M. , Ouyang, H. et al (2014) Sequence‐specific inhibition of microRNA via CRISPR/CRISPRi system. Sci. Rep. 4, 3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Microscopic images of developing embryos of Stoneville 7A glanded (GL; GVS4) and glandless (gl; GVS5), near‐isogenic lines, used for comparative RNA‐seq analysis.
Figure S2 Virus‐induced gene silencing (VIGS) in cotton.
Figure S3 Leaves from plants that had undergone virus‐induced gene silencing against CGF genes showing the effects on gland formation.
Figure S4 Figure S4 Real‐time PCR standard curves representing PCR efficiency for each of the CGF homeologs and Histone gene.
Figure S5 Alignment of A subgenome CGF1 gene sequences from GVS4 (glanded) and GVS5 (glandless, recessive mutant) near‐isogenic lines.
Figure S6 Alignment of D subgenome CGF1 gene sequences from GVS4 (glanded) and GVS5 (glandless, recessive mutant) near‐isogenic lines.
Figure S7 Alignment of A subgenome CGF2 gene sequences from GVS4 (glanded) and GVS5 (glandless, recessive mutant) near‐isogenic lines.
Figure S8 Alignment of D subgenome CGF2 gene sequences from GVS4 (glanded) and GVS5 (glandless, recessive mutant) near‐isogenic lines.
Figure S9 Alignment of A subgenome GoPGF (synonym CGF3) gene sequences from GVS4 (glanded) and GVS5 (glandless, recessive mutant) near‐isogenic lines.
Figure S10 Alignment of D subgenome GoPGF (synonym CGF3) gene sequences from GVS4 (glanded) and GVS5 (glandless, recessive mutant) near‐isogenic lines.
Figure S11 Sequencing results showing four different SNPs that differentiate A and D subgenome GoPGF (synonym CGF3) genes.
Figure S12 Promoter (2.05 kb) activity evaluation of the D subgenome GoPGF (synonym CGF3) gene from glanded and glandless cotton using gusA as the reporter gene.
Figure S13 Promoter (~4.2 kb) activity evaluation of the D subgenome GoPGF (synonym CGF3) gene from glanded and glandless cotton using gusA as the reporter gene.
Figure S14 Effect of CRISPR/Cas9‐mediated knockout of CGF2 and GoPGF (synonym CGF3) genes on gland formation in T0 plants in comparison to glanding pattern seen in a wild‐type, control cotton plant.
Figure S15 Mutations observed in two CGF2 knockout lines, (a) 236‐8 and (b) 236‐10. The two target sites are highlighted yellow and PAM sequences red.
Figure S16 Mutations observed in two GoPGF (synonym CGF3) knockout lines, (a) 237‐3 and (b) 237‐4.
Figure S17 Effect of CRISPR/Cas9‐mediated knockout of CGF2 and GoPGF (synonym CGF3) genes on gland formation observed in cottonseed kernels.
Figure S18 qRT‐PCR analysis of GoPGF (synonym CGF3) transcripts in cotton callus cultures obtained following transformation with ACGF3 overexpression construct. L254: light‐coloured callus lines; D254 dark‐coloured callus lines; Control: nontransgenic callus.
Table S1 RNA‐seq reads for glanded (GL; GVS4) and glandless (gl; GVS5) embryos at 14‐, 16‐ and 32‐days post‐anthesis and their mapping to the reference genome.
Table S2 Genes that expressed at higher levels in the glanded embryos (STV GL; GVS4; Gl2Gl2Gl3Gl3) in comparison to those in the glandless (STV gl; GVS5; gl2gl2gl3gl3) embryos at 14‐days post‐anthesis stage of development based on RNA‐seq analysis. Genes encoding putative transcription factors were tested for their role in gland formation using virus‐induced gene silencing (VIGS).
Table S3 Primers used to amplify segments of the coding sequence of the target gene for cloning into TRV2 binary vector to conduct VIGS experiments.
Table S4 Primers used to amplify and isolate CGF genes from A and D subgenomes of glanded (GVS4) and glandless (GVS5) cotton plants.
Table S5 Guide sequences used to target CGF2 and GoPGF (synonym CGF3) genes.
Table S6 Primers used for amplicon sequencing of regenerated plants targeted with LCT236, LCT237 and LCT238 constructs.
Data S1 Differentially expressed genes between glanded and glandless embryos at 16‐ and 32 days post anthesis.
Data S2 Supplementary methods.
Data Availability Statement
RNA‐seq data from developing embryos at different stages of development (14‐, 16‐ and 32‐dpa) from both glanded (GVS4), and glandless (GVS5) cotton are available in the NCBI SRA archive (accession # PRJNA448612).


