Abstract
Root morphology is essential for plant survival. NO3− is not only a nutrient, but also a signal substance affecting root growth in plants. However, the mechanism of NO3−-mediated root growth in rice remains unclear. In this study, we investigated the effect of OsNRT2.1 on root elongation and nitrate signaling-mediated auxin transport using OsNRT2.1 overexpression lines. We observed that the overexpression of OsNRT2.1 increased the total root length in rice, including the seminal root length, total adventitious root length, and total lateral root length in seminal roots and adventitious roots under 0.5-mM NO3− conditions, but not under 0.5-mM NH4+ conditions. Compared with wild type (WT), the 15NO3− influx rate of OsNRT2.1 transgenic lines increased by 24.3%, and the expressions of auxin transporter genes (OsPIN1a/b/c and OsPIN2) also increased significantly under 0.5-mM NO3− conditions. There were no significant differences in root length, ß-glucuronidase (GUS) activity, and the expressions of OsPIN1a/b/c and OsPIN2 in the pDR5::GUS transgenic line between 0.5-mM NO3− and 0.5-mM NH4+ treatments together with N-1-naphthylphalamic acid (NPA) treatment. When exogenous NPA was added to 0.5-mM NO3− nutrient solution, there were no significant differences in the total root length and expressions of OsPIN1a/b/c and OsPIN2 between transgenic plants and WT, although the 15NO3− influx rate of OsNRT2.1 transgenic lines increased by 25.2%. These results indicated that OsNRT2.1 is involved in the pathway of nitrate-dependent root elongation by regulating auxin transport to roots; i.e., overexpressing OsNRT2.1 promotes an effect on root growth upon NO3− treatment that requires active polar auxin transport.
Keywords: OsNRT2.1, NO3−, auxin transport, root length, rice
1. Introduction
Nitrogen (N) is an essential macronutrient for plant growth and crop productivity [1]. Plant roots can absorb various forms of nitrogen, including nitrate (NO3−), ammonium (NH4+), and organic molecules, which are mainly amino acids [2]. Traditionally, rice is cultivated under flooding conditions. NH4+ is the primary form in the paddy fields, and rice prefers NH4+ to NO3− [3]. NH4+ is absorbed into plants by ammonium transporters (AMTs) [1,2]. Excessive NH4+ in soil is considered to be toxic to rice [1]. However, the concentration of NH4+ is generally lower than that of NO3−, and NH4+ is converted to nitrite and NO3− in well-drained soil. NO3− is usually the most abundant nitrogen source in aerobic soil; however, this anionic form is soluble in soil water and easy to migrate in soil [4,5]. NO3− is very important for rice to improve nitrogen use efficiency and grain yield. NO3− is not only a nutrient, but also a signal substance affecting plant growth and development in plants, including inducing the expressions of auxin-related genes [6,7,8,9,10], breaking seed dormancy [11,12], regulating leaf growth [13,14], modulating flowering time [15], and regulating root architecture [16,17,18,19,20,21]. Auxin plays a central role in modulating every step in root growth [22,23,24,25,26,27]. Many studies have shown that the regulation of root system architecture by NO3− involves the interaction between NO3− and the auxin signaling pathway [16,28,29,30,31].
NO3− is absorbed by roots through NO3− transporters and then transported to the whole plant, or it combines with carbon to produce amino acids before redistribution [1,32]. The concentration of NO3− in soil fluctuates greatly in time and space, and it can reach a range of 1 to 10 mM from few µM after fertilization or nitrification [32,33]. The NO3− uptake system in higher plants is composed of the low-affinity transport system (LATS) and the high-affinity transport system (HATS) [1,2,34,35,36]. Rice grows in submerged soils with a low concentration of nitrate; NO3− uptake by roots is dominated by high-affinity transport in rice [1,2]. As high-affinity nitrate transporters, NRT2s include five members in rice [37]. Among them, OsNRT2.1, OsNRT2.2, and OsNRT2.3a need a partner protein OsNAR2.1 to absorb and transport NO3− in rice [37,38].
NRT1.1 participates in NO3− uptake by roots under high and low concentrations of NO3− [39,40]. NRT1.1 plays a major role in regulating NO3− in root system architecture, because it modulates root growth in response to NO3− [10,41,42,43]. Nitrate modulates root development by regulating NRT1.1 [10,19,42,43,44,45]. NRT2.1 is a high-affinity nitrate transporter [42,46,47], and plays an important role in regulating root development under low concentrations of NO3− in Arabidopsis thaliana [17,18]. In the previous study, we have reported that the knockdown of OsNAR2.1 inhibits lateral root formation by reducing auxin transport from shoots to roots under low concentrations of NO3−, and OsNAR2.1 might be involved in both NO3− uptake and NO3− signaling [30].
Auxin transport is mediated by auxin influx carriers (AUX1/LAX family) and efflux carriers (PINs and ABCB/PGPs) in plants [48,49,50,51]. Moreover, PIN proteins regulate auxin gradients during the growth of lateral roots (LRs) [52]. PIN1 is localized in the basal (root apex-facing) side of the root vasculature [53,54], PIN2 is expressed in the basal side of the cortical cells and in the apical (shoot apex-facing) side of the epidermal and root cap cells [55,56], PIN3 is distributed in the columella cells in roots in an apolar manner, PIN4 is localized in the basal side of cells in the central root meristem with less pronounced polarity in the quiescent center, and PIN7 is localized in the basal side of the stele cells and in columella cells in an apolar manner [52,57,58].
Our previous results showed that the overexpression of OsNRT2.1 significantly improves NO3− uptake and rice growth under conditions with a low concentration of NO3− [47,59]. In this study, we investigated the effect of OsNRT2.1 on root elongation through nitrate signaling-mediated auxin transport by testing root development, the effects of N-1-naphthylphalamic acid (NPA) on nitrate uptake, and the expression of OsPINs in OsNRT2.1 overexpression lines.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
OsNRT2.1 transgenic lines (OE1, OE2, and OE3) and the pDR5::GUS transgenic line have been described in Luo et al. [59] and Chen et al. [60], respectively. We got the F1 generation seeds of the crossing line of OE3/DR5::GUS OsNRT2.1 transgenic lines.
Plants were grown in the greenhouse under natural light at day/night temperatures of 30 °C/22 °C and 60% relative humidity. The seeds of wild type (WT) and transgenic lines were surface-sterilized with 10% (v/v) hydrogen peroxide solution for 30 min, thoroughly rinsed, and washed six times with deionized water. Seeds of uniform size were germinated on the top cover of a 1-L pot with 40 holes per top cover and one seed per hole. The containers were filled with a quarter concentration of nutrient solution with 0.25 mM of NH4NO3, 0.5 mM of NH4+ or 0.5 mM of NO3−. Ten-day-old seedlings grown in 0.5 mM of NH4+ or 0.5 mM of NO3− nutrient solution were transplanted into holes on the top cover of a 7-L pot containing the corresponding nutrient solution, respectively.
The complete nutrient solution of the International Rice Research Institute (IRRI) included 1.0 mM of MgSO4·7H2O, 1.0 mM of CaCl2, 0.5 mM of Na2SiO3, 0.35 mM of K2SO4, 0.3 mM of KH2PO4, 20.0 μM of Fe-EDTA (ethylene diaminetetra acetic acid tetrasodium salt), 20.0 μM of H3BO3, 9.0 μM of MnCl2, 0.77 μM of ZnSO4, 0.39 μM of (NH4)6Mo7O24, and 0.32 of μM CuSO4, with pH 5.5. The nutrient solution was replaced once every day. NH4+ and NO3− supplied in the nutrient solution were (NH4)2SO4 and Ca(NO3)2, respectively. Ca2+ in NH4+ nutrient solution was supplemented with CaCl2, and the same concentration of Ca2+ was added under different treatments. Nitrification inhibitor dicyandiamide was added to each pot to prevent the oxidation of nutrient solution.
2.2. qRT-PCR Analysis
Total RNA was extracted using TRIzol reagent (Vazyme Biotech Co, Ltd., Nanjing, China). DNase I-treated total RNAs were used for reverse transcription (RT) with HiScript II Q Select RT SuperMix for qPCR (+gDNA wiper) kit (Vazyme Biotech Co). Quantitative assays were performed in triplicate using the 2 × T5 Fast qPCR Mix (SYBRGreenI) kit (Vazyme Biotech Co). The primers for qRT-PCR are shown in Table S1.
2.3. Assay of the Concentration of Total Nitrogen
About 0.05 g of crushed dry samples were digested with H2SO4-H2O2 at 280 ℃. After cooling, the digested samples were diluted to 100 mL in distilled water. The concentration of total N was measured using the Kjeldahl method [61].
2.4. N-1-Naphthylphalamic Acid Treatment
Three-day-old normal grown seedlings were transferred to nutrient solutions containing 1 μM of auxin efflux inhibitor N-1-naphthylphalamic acid (NPA) from 100 mM of NPA that were dissolved in dimethyl sulfoxide(DMSO)and control containing the same amount of DMSO. Sampling was performed after seven days of treatment.
2.5. Determination of 15NO3− Influx Rate in Roots
The influx rates of 15NO3− in roots were assayed as described previously [62]. Plant seedlings were transferred to 0.5 mM of 15NO3− (atom% 15N: 15NO3−, 99%) nutrient solution with or without 1 μM of NPA for 5 min, and then, the seedlings were transferred to 0.1 mM of CaSO4 solution for 1 min before sampling. The concentration of 15N was analyzed by isotope ratio mass spectrometry (DELTA V Advantage Isotope Ratio Mass Spectrometer, Thermo Fisher Scientific, Waltham, MA, USA).
2.6. Analysis of ß-Glucuronidase Activity
For ß-Glucuronidase (GUS) staining, crown roots were immersed in GUS staining solution (1 mg/mL of X-glucuronide in 100 mM of sodium phosphate, 0.5 mM of ferrocyanide, 0.5 mM of ferricyanide, and 0.1% Triton X-100, pH 7.2), and then incubated at 37 °C in the dark.
For GUS activity assay, 5 μL of extract was added to 450 μL of GUS extraction buffer containing 1 mM of 4-methylumbelliferyl β-d-glucuronide, and incubated at 37 °C. Thereafter, 20 μL of reaction mixture was added into a 180-μL stop solution (1 M of sodium carbonate) for 10 min. Then, the fluorescence values were measured at 365 nm by a Fluorolite 1000 fluorometer (DYNEX technologies, USA).
2.7. Root Scanning
The root parameters were measured as previously described [30]. The length of seminal and adventitious roots was measured with a ruler, and the LR density was calculated as the ratio of the root length to the number of roots. The total root length and LR length were measured using the WinRhizo scanner-based image analysis system (Regent Instruments, Montreal, QC, Canada). The total root length represents the sum of the seminal roots, adventitious roots, and all the lateral roots. The total adventitious roots represent the sum of all the adventitious root lengths in every plant. The total LR length in seminal roots is the sum of the lateral root length of the seminal roots in every plant and the total LR length in adventitious roots is the sum of the lateral root length in the adventitious roots of every plant.
2.8. Statistical Analysis
The data of the experiments were analyzed by one-way ANOVA and Tukey’s test at p < 0.05 to determine the statistically significant differences among different treatments. All the statistical evaluations were performed using SPSS version 20.0 statistical software (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Effect of OsNRT2.1 Overexpression on Root Growth under 0.25-mM NH4NO3 Conditions
In previous studies, three genetically stable transgenic rice lines were obtained by transgenic technology [59]. There were no significant differences in grain length, grain width, and 1000-grain weight between OsNRT2.1 transgenic lines and WT (Figure S1A–C). The overexpression of OsNRT2.1 had no significant effect on grain size in rice. Moreover, the concentration of total N in the seeds of OsNRT2.1 transgenic lines was analyzed, and no significant difference was found between them and WT (Figure S1D).
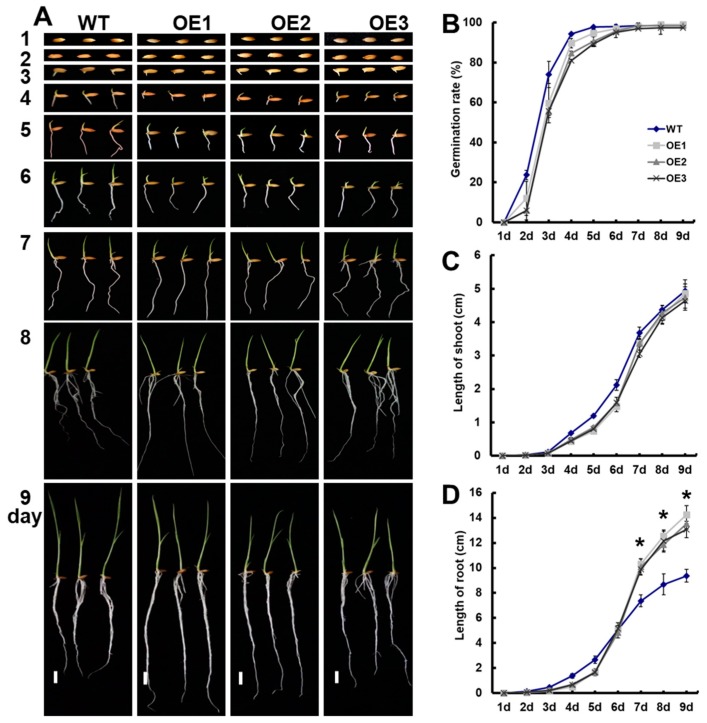
We also performed germination experiments and examined the growth of seedlings under 0.25-mM NH4NO3 conditions (Figure 1A). There were no significant differences in the final seed germination rate (Figure 1B) and shoot length between transgenic lines and WT on the ninth day (Figure 1C). Since the seventh day, the root length of transgenic lines was significantly longer than that of WT (Figure 1D).
Figure 1.
Overexpression of OsNRT2.1 affects root growth at the seedling stage under 0.25-mM NH4NO3 conditions. Rice seedlings were grown in a quarter concentration of nutrient solution containing 0.25 mM of NH4NO3 from the beginning. The nutrient solution was replaced daily. (A) Seed germination in wild type (WT) and transgenic plants. Scale bars = 1 cm. (B) Seed germination rate, (C) shoot length, and (D) root length of WT and transgenic plants. Error bars: SE (n = 10). Significant differences between transgenic lines and WT are indicated by asterisks (p < 0.05, one-way ANOVA).
The roots of transgenic lines on the ninth day were scanned and analyzed (Figure 2). Compared with WT, the total root length of transgenic plants under the 0.25-mM NH4NO3 conditions increased by 42.2% (Figure 3B), including a 45.3% increase in the seminal root length (Figure 3C), a 37.7% increase in the total adventitious root length (Figure 3D), a 48.7% increase in the total lateral root length in seminal roots (Figure 3G), and a 41.7% increase in the total lateral root length in adventitious roots (Figure 3H). However, there were no significant differences in the lateral root density in seminal roots and the lateral root density in adventitious roots between the transgenic lines and WT, respectively (Figure 3E,F).
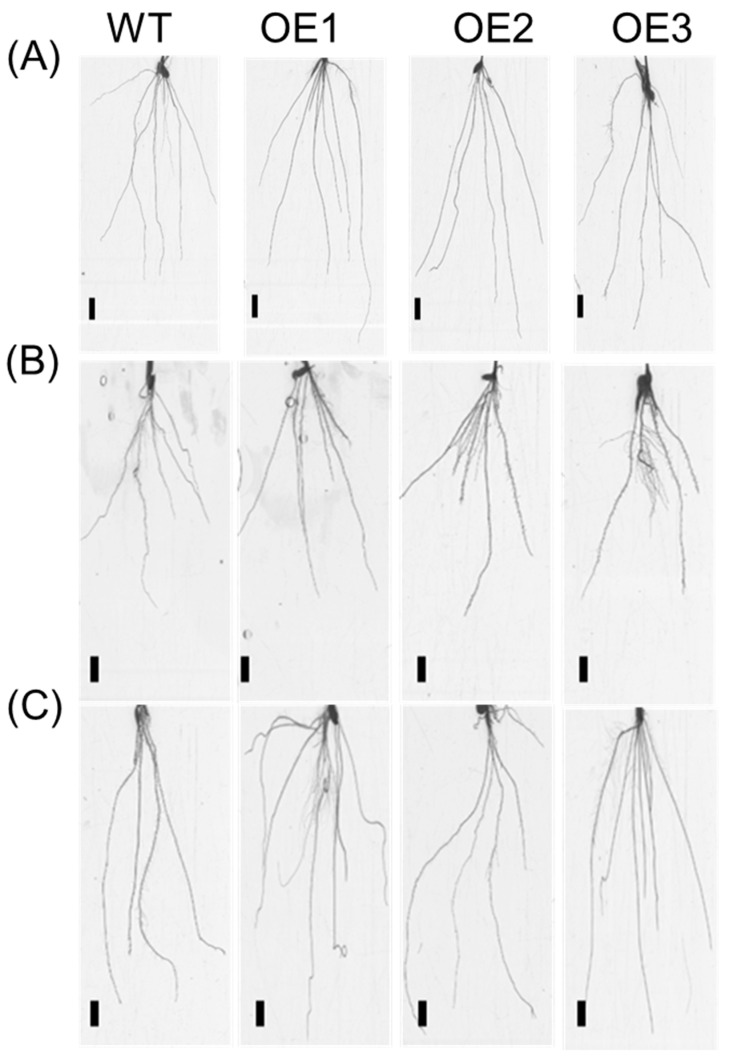
Figure 2.
Root morphology of WT and OsNRT2.1 transgenic lines (A) under 0.25-mM NH4NO3 conditions, (B) under 0.5-mM NH4+ conditions, (C) under 0.5-mM NO3− conditions. WT and transgenic lines on the ninth day were shown in Figure 1 (bar = 1 cm).
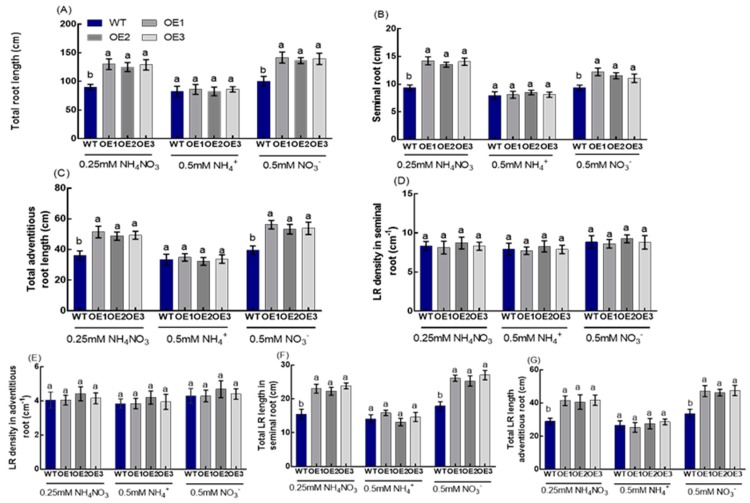
Figure 3.
Root morphology of WT and OsNRT2.1 transgenic lines under 0.25-mM NH4NO3, 0.5-mM NH4+, or 0.5-mM NO3− conditions. WT and transgenic plants on the 10th day were shown in Figure S2. (A) Total root length, (B) seminal root length, (C) total adventitious root length, (D) LR density in seminal roots, (E) lateral root (LR) density in adventitious roots, (F) total LR length in seminal roots, and (G) total LR length in adventitious roots. Total root length is the sum of the seminal root, adventitious roots, and all lateral roots. Error bars: SE (n = 5). Significant differences between transgenic lines and WT are indicated by different letters (p < 0.05, one-way ANOVA).
In addition, we examined the growth of OsNRT2.1 transgenic seedlings under 0.5-mM NH4+ (Figure S2A) or NO3− (Figure S2D) conditions. Under 0.5-mM NH4+ conditions, there were no significant differences in shoot length and root length between the transgenic lines and WT (Figure S2B,C). Under 0.5-mM NO3− conditions, the shoot length of transgenic lines was significantly longer than that of WT from the 25th day (Figure S2E), and the root length of transgenic lines was significantly longer than that of WT from the 10th day (Figure S2F).
The roots of transgenic plants were scanned and analyzed under 0.5-mM NH4+ (Figure 2B and Figure 3A) or NO3− (Figure 2C and Figure 3) conditions on the 10th day. Under 0.5-mM NH4+ conditions, there was no significant difference in the total root length between the transgenic lines and WT (Figure 3A), including seminal root length (Figure 3B), total adventitious root length (Figure 3C), lateral root density in seminal roots (Figure 3D), lateral root density in adventitious roots (Figure 3E), total lateral root length in seminal roots (Figure 3F), and total lateral root length in adventitious roots (Figure 3G).
Under 0.5-mM NO3− conditions, the total root length of transgenic plants increased by 38.6% compared with that of WT (Figure 3A), including a 24.0% increase in seminal root length (Figure 3B), a 36.4% increase in total adventitious root length (Figure 3C), a 46.0% increase in total lateral root length in seminal roots (Figure 3F), and a 39.7% increase in total lateral root length in adventitious roots (Figure 3G). However, there were no significant differences in lateral root density in seminal roots and lateral root density in adventitious roots between the transgenic lines and WT, respectively (Figure 3D,E).
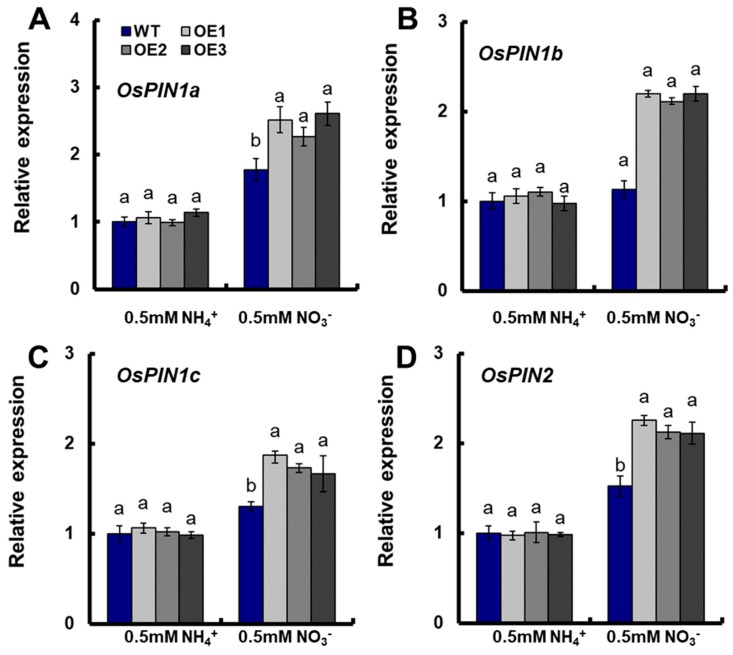
3.2. Effect of OsNRT2.1 Overexpression on the Expressions of OsPINs under 0.5-mM NH4+ or NO3− Conditions
The expression of auxin transporter OsPINs in roots were also assayed in this study. There were no significant differences in the expression of OsPIN1a, OsPIN1b, OsPIN1c, and OsPIN2 between the transgenic plants and WT under 0.5-mM NH4+ conditions (Figure 4A–D). Meanwhile, the expression of OsPIN1a, OsPIN1b, OsPIN1c, and OsPIN2 in transgenic lines increased significantly compared with those in WT under 0.5-mM NO3− conditions (Figure 4A–D).
Figure 4.
The expressions of OsPINs in the roots of WT and OsNRT2.1 transgenic lines under 0.5-mM NH4+ or 0.5 mM NO3− conditions. WT and transgenic plants on the 10th day were shown in Figure 3. Real-time quantitative RT-PCR analysis of the expressions of (A) OsPIN1a, (B) OsPIN1b, (C) OsPIN1c, and (D) OsPIN2 in WT and OsNRT2.1 transgenic lines. RNA was extracted from roots. Error bars: SE (n = 5). Significant differences between transgenic lines and WT are indicated by different letters (p < 0.05, one-way ANOVA).
3.3. Effect of NPA on Root Growth under 0.5-mM NH4+ and NO3− Conditions
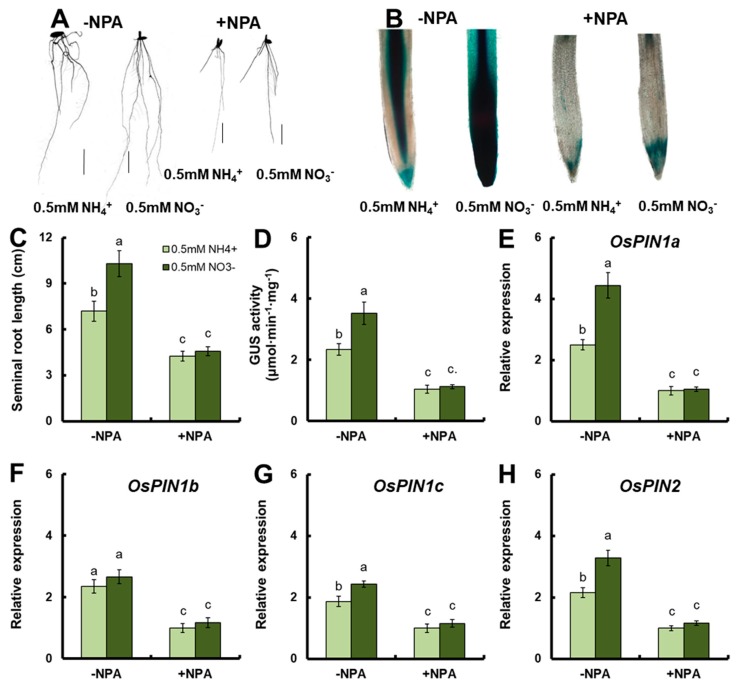
In order to investigate the effect of auxin transport on nitrate-mediated root growth, the rice seedlings of the pDR5::GUS transgenic line were treated with NPA, which is an auxin transport inhibitor. Exogenous NPA treatment significantly reduced the seminal root length under 0.5-mM NH4+ or NO3− conditions, respectively (Figure 5A,C). The seminal root length under 0.5-mM NO3− conditions was significantly longer than that under 0.5-mM NH4+ conditions without NPA treatment (Figure 6A,C). There was no significant difference in rice root length between 0.5-mM NO3− and 0.5-mM NH4+ treatments with NPA treatment (Figure 5A,C).
Figure 5.
Effects of N-1-naphthylphalamic acid (NPA) on root growth under 0.5-mM NH4+ or 0.5 mM NO3− conditions. Three-day-old normal grown pDR5::GUS transgenic seedlings were transferred to nutrient solutions containing 1 μM of NPA from 100 mM of NPA that was dissolved in DMSO (dimethyl sulfoxide), and the control containing the same amount of DMSO (dimethyl sulfoxide). Sampling was performed after seven days of treatment. (A) Root morphology of rice plants under 0.5-mM NH4+ or 0.5-mM NO3− conditions with or without NPA treatment. (bar = 1 cm). (B) ß-Glucuronidase (GUS) expression in the root tips of pDR5::GUS seedlings under 0.5-mM NH4+ or 0.5 mM NO3− conditions with or without NPA treatment. Root tips were stained for 2 h at 37 °C in the dark. (C) Seminal root length. (D) GUS activity of roots. Real-time quantitative RT-PCR analysis of the expressions of (E) OsPIN1a, (F) OsPIN1b, (G) OsPIN1c, and (H) OsPIN2 in pDR5::GUS seedlings under 0.5-mM NH4+ or 0.5-mM NO3− conditions with or without NPA treatment. RNA was extracted from roots. Error bars: SE (n = 5). Significant differences between transgenic lines and WT are indicated by different letters (p < 0.05, one-way ANOVA).
Figure 6.
Root morphology of WT and OsNRT2.1 transgenic lines with NPA treatment under 0.5 mM of NO3−. Three-day-old normal grown WT and transgenic seedlings were transferred to nutrient solutions containing 1 μM of NPA from 100 mM of NPA that was dissolved in DMSO and the control containing the same amount of DMSO. Sampling was performed after seven days of treatment. (A) Root morphology (bar = 1 cm), (B) total root length, (C) seminal root length, (D) total adventitious root length, (E) LR density in seminal roots, (F) LR density in adventitious roots, (G) total LR length in seminal roots, and (H) total LR length in adventitious roots. Total root length is the sum of the seminal roots, adventitious roots, and all lateral roots. Error bars: SE (n = 5). Significant differences between transgenic lines and WT are indicated by different letters (p < 0.05, one-way ANOVA).
ß-Glucuronidase staining showed that the color of seminal roots of pDR5::GUS transgenic seedlings without NPA treatment was significantly darker under 0.5-mM NO3− treatment than under 0.5-mM NH4+ treatment (Figure 5B). The color of pDR5::GUS transgenic seedlings with NPA treatment was very light under 0.5-mM NH4+ or NO3− treatment (Figure 5B). The GUS activity of pDR5::GUS transgenic seedlings was further analyzed to confirm the result of GUS staining. NPA could significantly inhibit the GUS activity under 0.5-mM NH4+ or NO3− conditions (Figure 5D). The GUS activity under 0.5-mM NO3− was significantly higher than that under 0.5-mM NH4+ without NPA treatment, and there was no significant difference in the GUS activity between 0.5-mM NH4+ and 0.5 mM-NO3− with NPA treatment (Figure 5D). The same results were found for OE3/DR5::GUS transgenic seedlings: the seminal root tip color of the OE3/DR5::GUS transgenic line without NPA treatment was significantly stronger under 0.5-mM NO3− treatment than under 0.5-mM NH4+ treatment (Figure S5B). The color of pDR5::GUS transgenic seedlings with NPA treatment was very light under 0.5-mM NH4+ or NO3− treatment (Figure S5B).
The expressions of OsPIN1a, OsPIN1c, and OsPIN2 under 0.5-mM NO3− was significantly higher than those under 0.5-mM NH4+ without NPA treatment (Figure 5E–H). The expressions of OsPIN1a, OsPIN1b, OsPIN1c, and OsPIN2 were significantly inhibited under 0.5-mM NH4+ or NO3− conditions with NPA treatment; however, there were no significant differences in the expressions of OsPIN1a, OsPIN1b, OsPIN1c, and OsPIN2 under 0.5-mM NH4+ conditions and under 0.5-mM NO3− with NPA treatment (Figure 5E–H).
3.4. NPA Inhibits the Effect of OsNRT2.1 on Root Growth under 0.5-mM NO3− Conditions
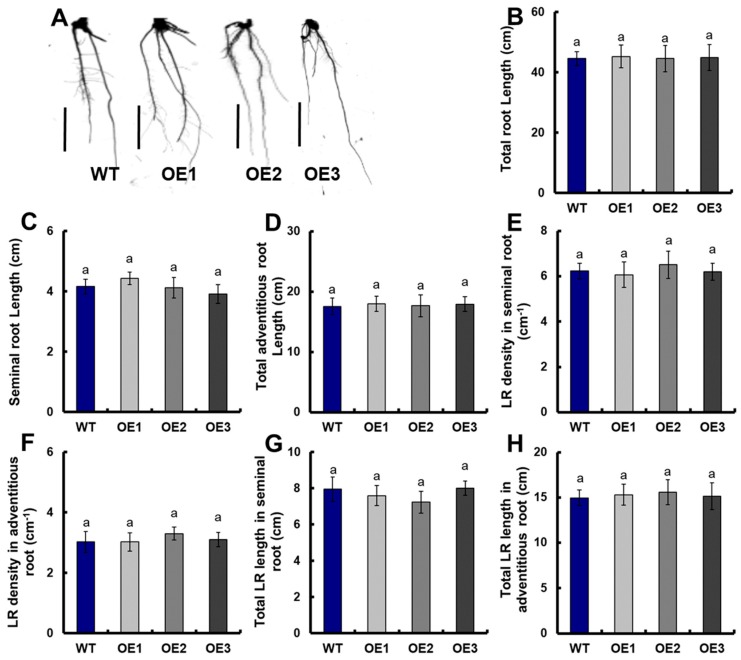
Moreover, NPA was added to OsNRT2.1 transgenic lines under 0.5-mM NO3− conditions (Figure 6A). There were no significant differences in the total root length between transgenic plants and WT (Figure 7B), including the seminal root length (Figure 6C), total adventitious root length (Figure 6D), lateral root density in seminal roots (Figure 6E), lateral root density in adventitious roots (Figure 6F), total lateral root length in seminal roots (Figure 6G), and total lateral root length in adventitious roots (Figure 6H).
Figure 7.
15NO3− influx rate in roots of WT and OsNRT2.1 transgenic lines under 0.5-mM 15NO3− conditions with NPA treatment. WT and transgenic plants under 0.5-mM NO3− with 1 μM of NPA treatment are shown. The plants were transferred to a quarter concentration of nutrient solution containing 0.5 mM of 15NO3− and 1 μM of NPA for 5 min. Error bars: SE (n = 5). Significant differences between transgenic lines and WT are indicated by different letters (p < 0.05, one-way ANOVA).
Similarly, there were no significant differences in the expressions of OsPIN1a, OsPIN1b, OsPIN1c, and OsPIN2 between OsNRT2.1 transgenic lines and WT under 0.5-mM NO3− conditions with NPA treatment (Figure S3).
3.5. NPA Inhibits the Effect of OsNRT2.1 on 15NO3− Influx Rate in Roots under 0.5-mM NO3− Conditions
The short-term NO3− uptake in roots of the transgenic lines and WT was analyzed by exposing the seedlings to 0.5 mM of 15NO3− for 5 min, so as to determine the effect of OsNRT2.1 overexpression on root NO3− influx to the whole plants. Compared with WT, the 15NO3− influx rate of OsNRT2.1 transgenic lines increased by 24.3% (Figure S4 [59] and Table S2).
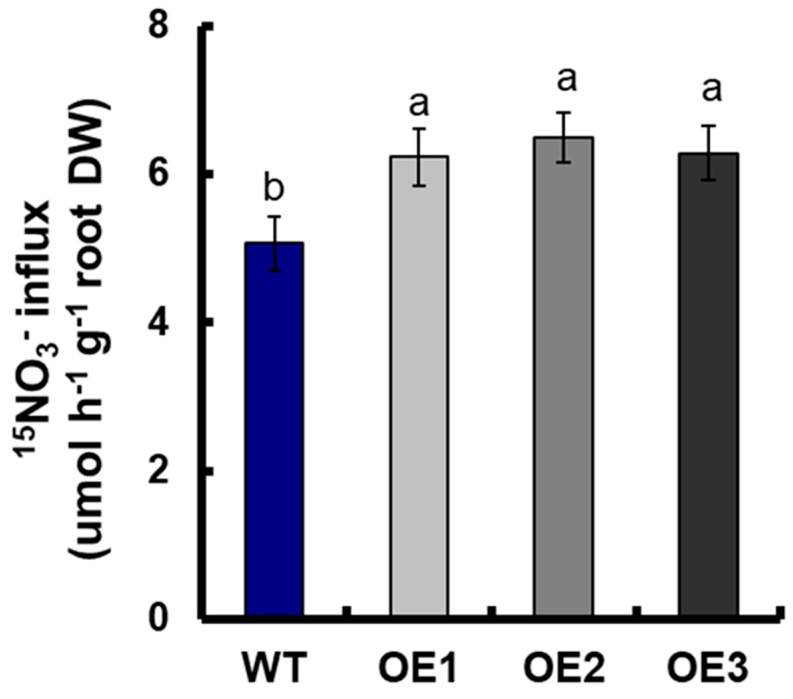
The 15NO3− influx rate of the transgenic lines under 0.5-mM 15NO3− conditions with NPA treatment was also analyzed. The results showed that WT and all the rice overexpression lines decreased the 15NO3− influx rate with NPA treatment [Table S2]. These data suggested that the NRT2.1-related import of NO3− may be partially auxin-dependent. However, compared with WT, the 15NO3− influx rate of OsNRT2.1 transgenic lines under 0.5-mM 15NO3− conditions with NPA treatment was higher by 25.2% (Figure 7 and Table S2).
4. Discussion
In plants, NO3− is not only a nutrient, but also a signal substance affecting root growth [16,17,18,19,20,21,30,31,63,64]. However, the mechanism of NO3−-mediated root growth in rice remains unclear.
We observed that the overexpression of OsNRT2.1 increased the total root length in rice, including the seminal root length, total adventitious root length, and total lateral root length in seminal roots and adventitious roots under 0.25-mM NH4NO3 conditions (Figure 1 and Figure 2). OsNRT2.1 overexpression lines have longer seminal roots under NO3−, while their respective LR number was not increased (Figure 3). That would indicate there are more LRs in seminal roots following NO3− treatment, which was in agreement with other plant species [16,65]. Further analysis showed that the total root length of transgenic plants increased by 38.6% compared with WT under 0.5-mM NO3− conditions (Figure 3A), including a 24.0% increase in seminal root length (Figure 3B), a 36.4% increase in total adventitious root length (Figure 3C), a 46.0% increase in total lateral root length in the seminal roots (Figure 3F), and a 39.7% increase in total lateral root length in the adventitious roots (Figure 3G). However, there was no significant difference between the transgenic lines and WT under 0.5-mM NH4+ conditions (Figure S2, Figure 2 and Figure S3). The regulation of NO3− on plant root growth is not exerted by the direct perception of external NO3−, but rather depends on the amount of NO3− absorbed by plants [16,30,65]. The short-term NO3− uptake in roots of the transgenic lines and WT was analyzed by exposing the plants to 0.5 mM of 15NO3− for 5 min, in order to determine the effect of OsNRT2.1 overexpression on root NO3− influx into the whole plants. Compared with WT, the 15NO3− influx rate of OsNRT2.1 transgenic lines increased by 24.3% (Figure S4 [59] and Table S2). Overall, these results suggested that the overexpression of OsNRT2.1 might promote root elongation by increasing NO3− uptake in rice.
Many studies have shown that the regulation of root system architecture by NO3− involves a strong interaction between NO3− and the auxin-signaling pathway [16,28,29,30,31]. Auxin plays a central role in modulating every step of root growth [22,23,24,25,26,27]. There were no significant differences in the expressions of OsPIN1a, OsPIN1b, OsPIN1c, and OsPIN2 between transgenic plants and WT under 0.5-mM NH4+ conditions (Figure 4A–D). However, the expressions of OsPIN1a, OsPIN1b, OsPIN1c, and OsPIN2 in transgenic lines increased significantly compared with WT under 0.5-mM NO3− conditions (Figure 4A–D). An analysis of gene structure showed that the distribution of OsPIN1a, OsPIN1b, and OsPIN1c in rice is similar to that of AtPIN1 [66]. AtPIN1 is highly conserved and plays an important role in auxin polar transport [48]. AtPIN1 mediates the vertical transport of IAA from shoot to root along the embryonic apical-basal axis [49,50]. OsPIN1a and OsPIN1c are expressed in the lateral root cap region, OsPIN1b is expressed in the root cap, and OsPIN1b and OsPIN1c are expressed in the meristem [66]. OsPIN1a is an auxin efflux protein that regulates the negative phototropism in rice roots [53]. OsPIN1b regulates root growth and seminal root elongation; the adventitious roots of ospin1b RNAi transgenic plants are significantly reduced compared with WT, and the seminal roots of ospin1b T-DNA mutants are shorter than those of WT plants [54,67]. OsPIN2 is expressed in the lateral root cap region, root epidermal, and outer cortex cells [55,66,68]. The overexpression of OsPIN2 results in reduced auxin levels in the rice root tip [60]. Wang et al. [55] recently reported that OsPIN2 plays an important role in mediating root gravitropic responses and is essential for plants to produce normal root growth angle in rice. OsPIN2 is an auxin efflux carrier protein, and may regulate the flow of auxin from the root tip to the root elongation zone and the distribution of auxin in seminal root tips, thereby modulating the seminal root elongation and lateral root formation in rice [56]. Therefore, OsNRT2.1 might be involved in the auxin transport pathway, regulating root growth and development under NO3− supplied conditions.
In order to investigate the effect of auxin transport on nitrate-mediated root growth, the pDR5::GUS transgenic seedlings were treated with NPA, which is an auxin transport inhibitor. The pDR5::GUS reporter system is a sensitive and simple system for monitoring auxin response and distribution in plant cells [57,60,69]. The seminal root length of pDR5::GUS transgenic seedlings under 0.5-mM NO3− conditions was significantly longer than that under 0.5-mM NH4+ conditions without NPA treatment (Figure 5A,C Figure S5B). Sun et al. [51] showed that the auxin levels in LRs and RT were higher under NO3− conditions than those under NH4+ conditions, indicating that auxin distribution in roots may be regulated by NO3− supply, which can increase the polar auxin transport to improve the seminal root length. Exogenous NPA treatment could significantly reduce the seminal root length under 0.5-mM NH4+ or NO3− conditions, respectively (Figure 5A,C and Figure S5A). There was no significant difference in root length under 0.5-mM NO3− conditions and under 0.5-mM NH4+ conditions with NPA treatment (Figure 5A,C and Figure S5A). Without NPA treatment, the expressions of OsPIN1a, OsPIN1c, and OsPIN2 under 0.5-mM NO3− conditions was significantly higher than those under 0.5-mM NH4+ conditions (Figure 5E–H). In addition, the expressions of OsPIN5a, OsPIN5b, OsPIN5c, OsPIN8, OsPIN9, OsPIN10a, and OsPIN10b were also increased under 0.5-mM NO3− conditions [51]. The expressions of OsPIN1a, OsPIN1b, OsPIN1c, and OsPIN2 was significantly inhibited under 0.5-mM NH4+ or NO3− conditions with NPA treatment (Figure 5E–H). Exogenous NPA treatment could significantly inhibit the GUS activity under 0.5-mM NH4+ or NO3− conditions (Figure 5D). The GUS activity under 0.5-mM NO3− conditions was significantly higher than that under 0.5-mM NH4+ conditions without NPA treatment, and there was no significant difference in GUS activity under 0.5-mM NH4+ conditions and under 0.5-mM NO3− conditions with NPA treatment (Figure 5B,D and Figure S5B). However, the DR5 signal was increased in the root tip of the “Nanguang” variety of seedlings under NH4+ conditions with the addition of NPA compared with that without NPA [70]. This could be attributed to the difference between rice varieties. These results indicated that nitrate could be used as a signal substance to induce the expressions of OsPIN1a/b/c and OsPIN2, regulate auxin transport to roots, and promote root elongation. However, nitrate could not induce root growth when auxin transport was inhibited.
Plants adjust their growth and development in response to changing environmental conditions by sensing external signals and integrating them into plant hormone-signaling pathways such as the auxin-signaling pathway [29,71,72,73,74,75,76]. The root growth of the auxin-insensitive mutant axr4 cannot be promoted by adding NO3− [16]. Different environmental signals and endogenous signals can regulate auxin distribution by affecting polar transport [66,77]. When NPA was added to 0.5 mM of NO3− nutrient solution, there was no significant difference in the total root length between transgenic plants and WT (Figure 6). Similarly, there was no significant difference in the expressions of OsPIN1a/b/c and OsPIN2 between OsNRT2.1 transgenic plants and WT under 0.5-mM NO3− conditions with NPA treatment (Figure S3), although the 15NO3− influx rate of OsNRT2.1 transgenic lines under 0.5-mM 15NO3− conditions with NPA treatment increased by 25.2% (Figure 7 and Table S2). These results indicated that OsNRT2.1 promoted root elongation by regulating auxin transport to roots, when auxin transport was inhibited; only increasing NO3− uptake could not promote root growth in rice.
However, the overexpression of OsNRT2.1 increased NO3− influx rate, induced the expressions of OsPINs in turn with the increase of the nitrate influx, and up-regulated auxin transport to roots, resulting in enhanced root development. Based on the above results in overexpression lines, the seedlings showed greater growth and yield than WT under low concentrations of nitrate in hydroponic solutions (Figure S2) and low concentrations of nitrogen fertilizer in the field [59]. Thus, this study provides clear evidence for the super phenotype of overexpression lines from the perspective of the root.
5. Conclusions
NO3− could be used as a signal substance to induce the expressions of OsPIN1a/b/c and OsPIN2, regulate auxin transport to the root system, and promote root elongation. We first report that the overexpression of OsNRT2.1 can show great root growth phenotypes under low NO3− conditions through the modulation of auxin transport, and root elongation may depend on auxin transport, but not the increase of nitrate influx.
Acknowledgments
The authors would like to thank Guohua Xu (Nanjing Agricultural University) for experimental guidance; and Shiwei Guo (Nanjing Agricultural University) for the root-scanning experiment. The English in this document has been checked by two native speakers of English, and for a certificate (Order NO:20190215MJ20190214022), please see: http://www.scimj.com/.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/4/290/s1. Table S1. Primers used for qRT-PCR, Table S2. The average data statistics of the 15NO3− influx rate in roots of WT and OsNRT2.1 transgenic lines under 0.5-mM 15NO3− conditions with NPA and without NPA, Figure S1. Rice seed size, Figure S2. Plant growth of WT and OsNRT2.1 transgenic lines under 0.5-mM NH4+ or NO3− conditions, Figure S3. The expressions of OsPINs in the roots of WT and OsNRT2.1 transgenic lines under 0.5-mM NO3− conditions with NPA treatment, Figure S4. 15NO3− influx rate in roots of WT and OsNRT2.1 transgenic lines under 0.5-mM 15NO3− conditions, Figure S5. Effects of NPA on root growth under 0.5-mM NH4+ or 0.5 mM NO3− condition.
Author Contributions
J.C. and X.F. conceived and designed the experiments; M.N., B.L., X.Y., B.L., and J.C. performed the experiments; J.C. and M.N. analyzed the data; J.C., M.N., and X.F. wrote and revised the paper. All the authors read and approved the final manuscript.
Funding
This study was financially supported by the China National Key Program for Research and Development (2016YFD0100700), Jiangsu Science Fund for Distinguished Young Scholars (Grant BK20160030), the Transgenic Project (Grant 2016ZX08001003-008), the Innovation project of basic scientific research in the central universities of the Ministry of Education (KYYJ201604, KJJQ201701), the Innovative Research Team Development Plan of the Ministry of Education of China (Grant No. IRT_17R56), and the Fundamental Research Funds for the Central Universities (Grant No. KYT201802).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Xu G., Fan X., Miller A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012;63:153–182. doi: 10.1146/annurev-arplant-042811-105532. [DOI] [PubMed] [Google Scholar]
- 2.Fan X., Naz M., Fan X., Xuan W., Miller A.J., Xu G. Plant nitrate transporters: From gene function to application. J. Exp. Bot. 2017;68:2463–2475. doi: 10.1093/jxb/erx011. [DOI] [PubMed] [Google Scholar]
- 3.Wang M.Y., Siddiqi M.Y., Ruth T.J., Glass A.D. Ammonium uptake by rice roots (II. Kinetics of 13NH4+ influx across the plasmalemma) Plant Physiol. 1993;103:1259–1267. doi: 10.1104/pp.103.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zarabi M., Jalali M. Leaching of nitrogen from calcareous soils in western Iran: A soil leaching column study. Environ. Monit. Assess. 2012;184:7607–7622. doi: 10.1007/s10661-012-2522-3. [DOI] [PubMed] [Google Scholar]
- 5.Jin Z., Zhu Y., Li X., Dong Y., An Z. Soil N retention and nitrate leaching in three types of dunes in the Mu Us desert of China. Sci. Rep. 2015;5:14222. doi: 10.1038/srep14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dechorgnat J., Patrit O., Krapp A., Fagard M., Daniel-Vedele F. Characterization of the Nrt2.6 gene in Arabidopsis thaliana: A link with plant response to biotic and abiotic stress. PLoS ONE. 2012;7:e42491. doi: 10.1371/journal.pone.0042491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho C.H., Lin S.H., Hu H.C., Tsay Y.F. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien J.A., Vega A., Bouguyon E., Krouk G., Gojon A., Coruzzi G., Gutiérrez R.A. Nitrate Transport, Sensing, and Responses in Plants. Mol. Plant. 2016;9:837–856. doi: 10.1016/j.molp.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y.Y., Hsu P.K., Tsay Y.F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012;17:458–467. doi: 10.1016/j.tplants.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Bouguyon E., Brun F., Meynard D., Kubeš M., Pervent M., Leran S., Lacombe B., Krouk G., Guiderdoni E., Zažímalová E., et al. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants. 2015;1:15015. doi: 10.1038/nplants.2015.15. [DOI] [PubMed] [Google Scholar]
- 11.Lboresi A., Gestin C., Leydecker M.T., Bedu M., Meyer C., Truong H.N. Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ. 2005;28:500–512. doi: 10.1111/j.1365-3040.2005.01292.x. [DOI] [PubMed] [Google Scholar]
- 12.Matakiadis T., Alboresi A., Jikumaru Y., Tatematsu K., Pichon O., Renou J.P., Kamiya Y., Nambara E., Truong H.N. The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 2009;149:949–960. doi: 10.1104/pp.108.126938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu P.K., Tsay Y.F. Two phloem nitrate transporters, NRT1.11 and NRT1.12, are important for redistributing xylem-borne nitrate to enhance plant growth. Plant Physiol. 2013;163:844–856. doi: 10.1104/pp.113.226563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahayu Y.S., Walch-Liu P., Neumann G., Römheld V., von Wirén N., Bangerth F. Root-derived cytokinins as long-distance signals for NO3—Induced stimulation of leaf growth. J. Exp. Bot. 2005;56:1143–1152. doi: 10.1093/jxb/eri107. [DOI] [PubMed] [Google Scholar]
- 15.Castro Marín I., Loef I., Bartetzko L., Searle I., Coupland G., Stitt M., Osuna D. Nitrate regulates floral induction in Arabidopsis, acting independently of light, gibberellin and autonomous pathways. Planta. 2011;233:539–552. doi: 10.1007/s00425-010-1316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H., Jennings A., Barlow P.W., Forde B.G. Dual pathways for regulation of root branching by nitrate. Proc. Nalt. Acad. Sci. USA. 1999;96:6529–6534. doi: 10.1073/pnas.96.11.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Little D.Y., Rao H., Oliva S., Daniel-Vedele F., Krapp A., Malamy J.E. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Nalt. Acad. Sci. USA. 2005;102:13693–13698. doi: 10.1073/pnas.0504219102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Remans T., Nacry P., Pervent M., Girin T., Tillard P., Lepetit M., Gojon A. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 2006;140:909–921. doi: 10.1104/pp.105.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouguyon E., Perrine-Walker F., Pervent M., Rochette J., Cuesta C., Benkova E., Martinière A., Bach L., Krouk G., Gojon A., et al. Nitrate Controls Root Development through Posttranscriptional Regulation of the NRT1.1/NPF6.3 Transporter/Sensor. Plant Physiol. 2016;172:1237–1248. doi: 10.1104/pp.16.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun C.H., Yu J.Q., Hu D.G. Nitrate: A Crucial Signal during Lateral Roots Development. Front. Plant Sci. 2017;8:485. doi: 10.3389/fpls.2017.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fredes I., Moreno S., Díaz F.P., Gutiérrez R.A. Nitrate signaling and the control of Arabidopsis growth and development. Curr. Opin. Plant Biol. 2018;47:112–118. doi: 10.1016/j.pbi.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Dubrovsky J.G., Sauer M., Napsucialy-Mendivil S., Ivanchenko M.G., Friml J., Shishkova S., Celenza J., Benková E. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Nalt. Acad. Sci. USA. 2008;105:8790–8794. doi: 10.1073/pnas.0712307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/S0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 24.Bhalerao R.P., Eklöf J., Ljung K., Marchant A., Bennett M., Sandberg G. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 2002;29:325–332. doi: 10.1046/j.0960-7412.2001.01217.x. [DOI] [PubMed] [Google Scholar]
- 25.Fukaki H., Okushima Y., Tasaka M. Auxin-mediated lateral root formation in higher plants. Int. Rev. Cytol. 2007;256:111–137. doi: 10.1016/S0074-7696(07)56004-3. [DOI] [PubMed] [Google Scholar]
- 26.Fukaki H., Tasaka M. Hormone interactions during lateral root formation. Plant Mol. Biol. 2009;69:437–449. doi: 10.1007/s11103-008-9417-2. [DOI] [PubMed] [Google Scholar]
- 27.Ljung K., Bhalerao R.P., Sandberg G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 2001;28:465–474. doi: 10.1046/j.1365-313X.2001.01173.x. [DOI] [PubMed] [Google Scholar]
- 28.Forde B.G. Local and long-range signaling pathways regulating plant responses to nitrate. Annu. Rev. Plant Biol. 2002;53:203–224. doi: 10.1146/annurev.arplant.53.100301.135256. [DOI] [PubMed] [Google Scholar]
- 29.Vidal E.A., Araus V., Lu C., Parry G., Green P.J., Coruzzi G.M., Gutiérrez R.A. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Nalt. Acad. Sci. USA. 2010;107:4477–4482. doi: 10.1073/pnas.0909571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang S., Chen S., Liang Z., Zhang C., Yan M., Chen J., Xu G., Fan X., Zhang Y. Knockdown of the partner protein OsNAR2.1 for high-affinity nitrate transport represses lateral root formation in a nitrate-dependent manner. Sci. Rep. 2015;5:18192. doi: 10.1038/srep18192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canales J., Contreras-López O., Álvarez J.M., Gutiérrez R.A. Nitrate induction of root hair density is mediated by TGA1/TGA4 and CPC transcription factors in Arabidopsis thaliana. Plant J. 2017;92:305–316. doi: 10.1111/tpj.13656. [DOI] [PubMed] [Google Scholar]
- 32.Miller A.J., Fan X., Orsel M., Smith S.J., Wells D.M. Nitrate transport and signalling. J. Exp. Bot. 2007;58:2297–2306. doi: 10.1093/jxb/erm066. [DOI] [PubMed] [Google Scholar]
- 33.Crawford N.M., Glass A.D.M. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 1998;3:389–395. doi: 10.1016/S1360-1385(98)01311-9. [DOI] [Google Scholar]
- 34.Gojon A., Krouk G., Perrine-Walker F., Laugier E. Nitrate transceptor(s) in plants. J. Exp. Bot. 2011;62:2299–2308. doi: 10.1093/jxb/erq419. [DOI] [PubMed] [Google Scholar]
- 35.Krapp A., David L.C., Chardin C., Girin T., Marmagne A., Leprince A.S., Chaillou S., Ferrario-Méry S., Meyer C., Daniel-Vedele F. Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 2014;65:789–798. doi: 10.1093/jxb/eru001. [DOI] [PubMed] [Google Scholar]
- 36.Lupini A., Mercati F., Araniti F., Miller A.J., Sunseri F., Abenavoli M.R. NAR2.1/NRT2.1 functional interaction with NO3(−) and H(+) fluxes in high-affinity nitrate transport in maize root regions. Plant Physiol. Biochem. 2016;102:107–114. doi: 10.1016/j.plaphy.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 37.Feng H., Yan M., Fan X., Li B., Shen Q., Miller A.J., Xu G. Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J. Exp. Bot. 2011;62:2319–2332. doi: 10.1093/jxb/erq403. [DOI] [PubMed] [Google Scholar]
- 38.Yan M., Fan X., Feng H., Miller A.J., Shen Q., Xu G. Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ. 2011;34:1360–1372. doi: 10.1111/j.1365-3040.2011.02335.x. [DOI] [PubMed] [Google Scholar]
- 39.Tsay Y.F., Schroeder J.I., Feldmann K.A., Crawford N.M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell. 1993;72:705–713. doi: 10.1016/0092-8674(93)90399-B. [DOI] [PubMed] [Google Scholar]
- 40.Liu K.H., Tsay Y.F. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 2003;22:1005–1013. doi: 10.1093/emboj/cdg118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remans T., Nacry P., Pervent M., Filleur S., Diatloff E., Mounier E., Tillard P., Forde B.G., Gojon A. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Nalt. Acad. Sci. USA. 2006;103:19206–19211. doi: 10.1073/pnas.0605275103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krouk G., Lacombe B., Bielach A., Perrine-Walker F., Malinska K., Mounier E., Hoyerova K., Tillard P., Leon S., Ljung K., et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell. 2010;18:927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 43.Mounier E., Pervent M., Ljung K., Gojon A., Nacry P. Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ. 2014;37:162–174. doi: 10.1111/pce.12143. [DOI] [PubMed] [Google Scholar]
- 44.Forde B.G. Nitrogen signalling pathways shaping root system architecture: An update. Curr. Opin. Plant Biol. 2014;21:30–36. doi: 10.1016/j.pbi.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 45.Cerezo M., Tillard P., Filleur S., Muños S., Daniel-Vedele F., Gojon A. Major alterations of the regulation of root NO(3)(−) uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol. 2001;127:262–271. doi: 10.1104/pp.127.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filleur S., Dorbe M.F., Cerezo M., Orsel M., Granier F., Gojon A., Daniel-Vedele F. An arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 2001;489:220–224. doi: 10.1016/S0014-5793(01)02096-8. [DOI] [PubMed] [Google Scholar]
- 47.Katayama H., Mori M., Kawamura Y., Tanaka T., Mori M., Hasegawa H. Production and characterization of transgenic rice plants carrying a high-affinity nitrate transporter gene (OsNRT2.1) Breed. Sci. 2009;59:237–243. doi: 10.1270/jsbbs.59.237. [DOI] [Google Scholar]
- 48.Friml J., Palme K. Polar auxin transport-old questions and new concepts? Plant Mol. Biol. 2002;49:273–284. doi: 10.1023/A:1015248926412. [DOI] [PubMed] [Google Scholar]
- 49.Wisniewska J., Xu J., Seifertova D., Brewer P.B., Ruzicka K., Blilou I., Rouquie D., Benkova E., Scheres B., Friml J. Polar PIN localization directs auxin flow in plants. Science. 2006;312:883. doi: 10.1126/science.1121356. [DOI] [PubMed] [Google Scholar]
- 50.Petrasek J., Friml J. Auxin transport routes in plant development. Development. 2009;136:2675–2688. doi: 10.1242/dev.030353. [DOI] [PubMed] [Google Scholar]
- 51.Sun H., Feng F.J., Zhao Q. Nitric oxide affects rice root growth by regulating auxin transport under nitrate supply. Front. Plant Sci. 2018;9:659. doi: 10.3389/fpls.2018.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vieten A., Sauer M., Brewer P.B., Friml J. Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 2007;12:160–168. doi: 10.1016/j.tplants.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 53.Xu H., Mo Y., Wang H., Zhang Y. OsPIN1a Gene Participates in Regulating Negative Phototropism of Rice Roots. Rice Sci. 2014;21:83–89. doi: 10.1016/S1672-6308(13)60168-7. [DOI] [Google Scholar]
- 54.Xu M., Zhu L., Shou H., Wu P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005;46:1674–1681. doi: 10.1093/pcp/pci183. [DOI] [PubMed] [Google Scholar]
- 55.Wang L., Guo M., Li Y., Ruan W., Mo X., Wu Z., Sturrock C.J., Yu H., Lu C., Peng J., et al. LARGE ROOT ANGLE1, encoding OsPIN2, is involved in root system architecture in rice. J. Exp. Bot. 2018;69:385–397. doi: 10.1093/jxb/erx427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inahashi H., Shelley I.J., Yamauchi T., Nishiuchi S., Takahashi-Nosaka M., Matsunami M., Ogawa A., Noda Y., Inukai Y. OsPIN2, which encodes a member of the auxin efflux carrier proteins, is involved in root elongation growth and lateral root formation patterns via the regulation of auxin distribution in rice. Physiol. Plant. 2018;164:216–225. doi: 10.1111/ppl.12707. [DOI] [PubMed] [Google Scholar]
- 57.Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. Aux ⁄ IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bao J., Chen F., Gu R., Wang G., Zhang F., Mi G. Lateral root development of two Arabidopsis auxin transport mutants, aux1-7 and eir1-1, in response to nitrate supplies. Plant Sci. 2007;173:417–425. doi: 10.1016/j.plantsci.2007.07.003. [DOI] [Google Scholar]
- 59.Luo B., Chen J., Zhu L., Liu S., Li B., Lu H., Ye G., Xu G., Fan X. Overexpression of a High-Affinity Nitrate Transporter OsNRT2.1 Increases Yield and Manganese Accumulation in Rice Under Alternating Wet and Dry Condition. Front. Plant Sci. 2018;9:1192. doi: 10.3389/fpls.2018.01192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y., Fan X., Song W., Zhang Y., Xu G. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 2012;10:139–149. doi: 10.1111/j.1467-7652.2011.00637.x. [DOI] [PubMed] [Google Scholar]
- 61.Chen J., Zhang Y., Tan Y., Zhang M., Zhu L., Xu G., Fan X. Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT2.1 expression with the OsNAR2.1 promoter. Plant Biotechnol. J. 2016;14:1705–1715. doi: 10.1111/pbi.12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen J., Fan X., Qian K., Zhang Y., Song M., Liu Y., Xu G., Fan X. pOsNAR2.1:OsNAR2.1 expression enhances nitrogen uptake efficiency and grain yield in transgenic rice plants. Plant Biotechnol. J. 2017;15:1273–1283. doi: 10.1111/pbi.12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang H.M., Forde B.G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279:407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]
- 64.Signora L., de Smet I., Foyer C.H., Zhang H. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 2001;28:655–662. doi: 10.1046/j.1365-313x.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- 65.Zhang H.M., Forde B.G. Regulation of Arabidopsis root development by nitrate availability. J. Exp. Bot. 2000;51:51–59. doi: 10.1093/jxb/51.342.51. [DOI] [PubMed] [Google Scholar]
- 66.Wang J.R., Hu H., Wang G.H., Li J., Chen J.Y., Wu P. Expression of PIN genes in rice (Oryza sativa L.): Tissue specificity and regulation by hormones. Mol. Plant. 2009;2:823–831. doi: 10.1093/mp/ssp023. [DOI] [PubMed] [Google Scholar]
- 67.Sun H., Tao J., Bi Y., Hou M., Lou J., Chen X., Zhang X., Luo L., Xie X., Yoneyama K., et al. OsPIN1b is Involved in Rice Seminal Root Elongation by Regulating Root Apical Meristem Activity in Response to Low Nitrogen and Phosphate. Sci. Rep. 2018;8:13014. doi: 10.1038/s41598-018-29784-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miyashita Y., Takasugi T., Ito Y. Identification and expression analysis of PIN genes in rice. Plant Sci. 2010;178:424–428. doi: 10.1016/j.plantsci.2010.02.018. [DOI] [Google Scholar]
- 69.Scarpella E., Rueb S., Meijer A.H. The RADICLELESS1 gene is required for vascular pattern formation in rice. Development. 2003;130:645–658. doi: 10.1242/dev.00243. [DOI] [PubMed] [Google Scholar]
- 70.Song W., Sun H., Li J., Gong X., Huang S., Zhu X., Zhang Y., Xu G. Auxin distribution is differentially affected by nitrate in roots of two rice cultivars differing in responsiveness to nitrogen. Ann. Bot. 2013;112:1383–1393. doi: 10.1093/aob/mct212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vidal E.A., Moyano T.C., Riveras E., Contreras-Lopez O., Gutierrez R.A. Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc. Nalt. Acad. Sci. USA. 2013;110:12840–12845. doi: 10.1073/pnas.1310937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Smet I., Vanneste S., Inzé D., Beeckman T. Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 2006;60:871–887. doi: 10.1007/s11103-005-4547-2. [DOI] [PubMed] [Google Scholar]
- 73.López-Bucio J., Cruz-Ramírez A., Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 2003;6:280–287. doi: 10.1016/S1369-5266(03)00035-9. [DOI] [PubMed] [Google Scholar]
- 74.Malamy J.E. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 2005;28:67–77. doi: 10.1111/j.1365-3040.2005.01306.x. [DOI] [PubMed] [Google Scholar]
- 75.Krouk G., Ruffel S., Gutiérrez R.A., Gojon A., Crawford N.M., Coruzzi G.M., Lacombe B. A framework integrating plant growth with hormones and nutrients. Trends Plant Sci. 2011;16:178–182. doi: 10.1016/j.tplants.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 76.Kazan K. Auxin and the integration of environmental signals into plant root development. Ann. Bot. 2013;112:1655–1665. doi: 10.1093/aob/mct229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vanneste S., Friml J. Auxin: A trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.