Abstract
Comparison of the secondary and three-dimensional structures of the hammerhead and pistol ribozymes reveals many close similarities, so in this work we have asked if they are mechanistically identical. We have determined a new crystal structure of the pistol ribozyme and have shown that G40 acts as general base in the cleavage reaction. The conformation in the active site ensures an in-line attack of the O2′ nucleophile, and the conformation at the scissile phosphate and the position of the general base are closely similar to those in the hammerhead ribozyme. However, the two ribozymes differ in the nature of the general acid. 2′-Amino substitution experiments indicate that the general acid of the hammerhead ribozyme is the O2′ of G8, while that of the pistol ribozyme is a hydrated metal ion. The two ribozymes are related but mechanistically distinct.
Introduction
The nucleolytic ribozymes are a group of catalytic RNA species that carry out site-specific cleavage reactions by nucleophilic attack of a 2′-hydroxyl group on the adjacent phosphodiester linkage.1 In some cases the reverse ligation reaction can also be catalyzed. These reactions can be accelerated by 106-fold in the context of the ribozyme. This class of ribozymes currently comprises nine different species, of which four have been identified in the last five years as a result of bioinformatic analysis in the Breaker laboratory.2,3 This expanding group now provides an opportunity to discern common catalytic strategies and to contrast different mechanisms and thus to extract some general principles of RNA catalysis. As an example, we proposed that the hairpin and VS ribozymes would have essentially the same chemical mechanism despite unrelated structures,4 and this is supported by all available evidence.5−7
The common catalytic strategy that links the nucleolytic ribozymes is the use of general acid–base catalysis,8 deprotonating the nucleophile and protonating the oxyanion leaving group. This probably makes the largest contribution to the overall catalytic rate enhancement; interference with the function of either acid or base typically lowers the reaction rate by 3 or 4 orders of magnitude. However, the identity of the catalytic functional groups varies from ribozyme to ribozyme. The majority use nucleobases in proton transfer and most use guanine N1 as the general base in the cleavage reaction. The hairpin, VS, and twister ribozymes all use adenine and guanine nucleobases as general acid and base, respectively.4,9−12 The other potential catalytic participants are hydrated metal ions and 2′-hydroxyl groups. The best evidence indicates that the HDV ribozyme uses metal-ion-bound water as general base13 and a cytosine nucleobase as the general acid,14 and the TS ribozyme likely uses a similar mechanism.15 Exceptionally the GlmS ribozyme uses the amine group of bound glucosamine 6-phosphate as general acid.16
Like many nucleolytic ribozymes, the hammerhead uses a guanine nucleobase (G12) as general base,17 but the structure of the active site reveals no nucleobase close to the O5′ leaving group.18 Thomas and Perrin19 have shown that the reduced cleavage activity of a G8 O2′H variant can be restored by 5′-phosphorothiolate substitution at the scissile phosphate. The thiolate leaving group does not require protonation, so rescue indicates that G8 O2′ participates in general acid catalysis. 5′-Phosphorothiolate substitution also restores the activity reduced in the absence of divalent cations. These data allow two straightforward interpretations:
Mechanism 1. G8 O2′ acts as the general acid, activated by a lowered pKa arising from an interaction with a metal ion.
Mechanism 2. A hydrated divalent cation acts as the general acid, and the G8 O2′ is required for the correct positioning of the cation.
Thomas and Perrin19 proposed that G8 O2′ was the general acid on the basis of changes in apparent pKa values observed under different conditions. However, this was called into question by recent data showing that additional factors influence apparent pKa values20 and by a transition-state analogue hammerhead structure in which a divalent cation appears to be better placed to act as general acid.21
The pistol ribozyme is one of the new nucleolytic ribozymes.3,22 Examination of the secondary structure and location of conserved nucleotides suggests a strong similarity with the hammerhead ribozyme (Figure 1). The hammerhead ribozyme (Figure 1A) has a three-way junction structure with a tertiary interaction between stems I and II23 that stabilizes the folded structure24 and is required for high activity. Recently a class of hammerhead ribozymes has been described in which stems I and II interact through a pseudoknot3 (Figure 1B). A pseudoknot is also a key element in the structure of the pistol ribozyme (Figure 1C). If the conventional depiction of the pistol ribozyme is rotated so that the position of cleavage is located equivalently to those for the hammerhead structures (Figure 1D), it becomes apparent that the overall secondary structures, the positions of the conserved nucleotides, and the scissile phosphate are very similar in the two ribozymes. Furthermore, rates of cleavage for the two ribozymes have similar dependence of pH, rising monotonically with pH to a plateau above pH 8.22,25,26 This clearly raises the question of how similar are the catalytic mechanisms of the pistol and hammerhead ribozymes.
Figure 1.
Comparison of the secondary structures of the pistol and hammerhead ribozymes. A generalized depiction of the secondary structure is shown, which is not indicative of the true number of base pairs formed in a given helix. Highly conserved nucleotides are colored red. The cleavage sites are arrowed. Long-range interactions are indicated by broken lines. (A) Conventional hammerhead structure, with a loop–receptor interaction between helices II and I. The nucleotides proposed to be involved in general acid (G8) and general base (G12) catalysis are shown bold (also in part B). (B) Hammerhead variant in which there is a long-range pseudoknot interaction (PS) between helices I and II. (C) Pistol ribozyme in its conventional depiction.3,22 (D) Pistol ribozyme rotated anticlockwise by 90° showing the pseudoknot interaction (PS). Comparison with (B) shows the strong similarity with the hammerhead variant.
In this paper we present structural and mechanistic data on the pistol ribozyme and a comparison with the hammerhead ribozyme. We show that a guanine nucleobase in pistol acts as the general base in the cleavage reaction and is located in an equivalent position to G12 in the hammerhead ribozyme. However, in other respects the two ribozymes differ. 2′-Amino substitution results in significantly different pH dependence for the two ribozymes, consistent with the hammerhead ribozyme using mechanism 1 with G8 O2′ as the general acid, while the pistol ribozyme uses mechanism 2 where a divalent cation acts as the general acid. Thus, the hammerhead and pistol ribozymes are not mechanistically identical.
Experimental Section
Chemical Synthesis of RNA
Oligonucleotides were synthesized using t-BDMS phosphoramidite chemistry27 as described in Wilson et al.,28 implemented on an Applied Biosystems 394DNA/RNA synthesizer. RNA was synthesized using UltraMILD ribonucleotide phosphoramidites with 2′O-tert-butyldimethylsilyl (t-BDMS) protection29,30 (Link Technologies). 2′-Amino-G and 2′-amino-A phosphoramidites were synthesized according to the literature.31,32 Due to limited solubility in acetonitrile, 2′-amino-A phosphoramidite was dissolved in anhydrous 3:1 methylene chloride/acetonitrile prior to use.
Oligoribonucleotides containing 5-bromocytidine (ChemGenes Corp.) were deprotected using anhydrous 2 M ammonia in methanol (Sigma-Aldrich) for 36 h. Oligonucleotides containing 2′-amino-G or 2′-amino-A were deprotected using 3:1 ammonia/ethanol at 55 °C for 17 h. Unmodified sequences and oligonucleotides incorporating other modifications were deprotected either in 3:1 ammonia/ethanol solution at room temperature for 3 h or in 1:1 ammonia/methylamine at 60 °C for 20 min according to the manufacturer’s recommendations and evaporated to dryness. All oligonucleotides were redissolved in 115 μL of anhydrous DMSO and 125 μL of triethylamine trihydrofluoride (Aldrich) to remove t-BDMS groups and agitated at 65 °C in the dark for 2.5 h prior to butanol precipitation. Full-length RNA was then purified as described previously.12
Ribozyme Kinetics
Pistol ribozymes used for kinetic analysis were assembled from two oligonucleotides termed ribozyme and substrate derived from the env-25 ribozyme sequence3 (all sequences written 5′ to 3′): ribozyme (47 nt), CGUGGUUAGGGCCACGUUAAAUAGUUGCUUAAGCCCUAAGCGUUGAU; substrate (15 nt), CGAUCAGGUGCAAGG.
The underlined nucleotides in the substrate sequence hybridize with the ribozyme strand to form helices P3 and P2. Additional nucleotides were added at the 5′ and 3′ ends to improve separation of products on acrylamide gels for analysis. Note that the construct used for kinetics has four Watson–Crick base pairs in P2 whereas the ribozyme used for crystallography has one less. The two constructs also differ in the sequence of the conserved loop comprising nucleotides 30–33 (UAAG compared to CAGG). These changes have only minor effects on activity (Table 1).
Table 1. Rates of Cleavage for the Pistol Ribozyme and Modified Variantsa.
| ribozyme | rate (min–1) | sd | fold decrease |
|---|---|---|---|
| unmodified | 9.8 | 0.6 | 1 |
| Sequence Variants and Alignment | |||
| ΔG25 | 0.29 | 0.01 | 34 |
| ΔU26 | 11 | 1 | 0.89 |
| U30C | 2.2 | 0.4 | 4.5 |
| A32G | 7.3 | 0.6 | 1.3 |
| Modification of Putative General Base and Scissile P Phosphorothioate Derivatives | |||
| G40 6S | 12.3 | 0.4 | 0.79 |
| G40I | 1.23 | 0.02 | 8.0 |
| U+1 PS Rp | 0.00022 | 0.00002 | 45000 |
| U+1 PS Sp | 5.6 | 0.1 | 1.8 |
| U+1 PS Rp + G40I | 0.0039 | 0.0001 | 2500 |
| U+1 PS Sp + G40I | 0.7 | 0.2 | 14 |
| Replacement of 1 mM MgCl2 with 1 mM MnCl2 | |||
| unmodified | 120b | 0.082 | |
| G40I | 15b | 0.65 | |
| U+1 PS Rp | 0.033 | 0.005 | 300 |
| U+1 PS Rp + G40I | 0.39 | 0.03 | 25 |
| 2′OH Variants at Position 32 | |||
| 2′OMe-A32 | 0.00054 | 0.00002 | 18000 |
| dA32 | 0.15 | 0.01 | 65 |
| dA32 + G42I | 0.18 | 0.01 | 54 |
| A32 2′NH2 | 0.102 | 0.001 | 96 |
| Putative Mg2+ Binding Pocket | |||
| G–1I | 8.0 | 0.7 | 1.2 |
| G33 N7C | 0.00077 | 0.00005 | 13000 |
| G33 PS | 9.4 | 0.7 | 1.0 |
| C35 PS | 5.5 | 0.5 | 1.8 |
| G42AP | 0.014 | 0.002 | 700 |
| G42AP + G–1I | 0.012 | 0.002 | 820 |
| G42AP + G40I | 0.0108 | 0.0005 | 910 |
| G42I | 8.5 | 0.2 | 1.2 |
| hexammine CoCl3c | 0.025 | 0.002 | 390 |
All data collected under standard conditions except where noted. PS = phosphorothioate.
Extrapolation based on the difference in measured rates for Mg2+ and Mn2+ for the unmodified ribozyme at pH 5.5 as shown in Figure 5A.
Activity of unmodified ribozyme in 1 mM hexammine cobalt chloride in the absence of Mg2+.
Hammerhead ribozymes based on a sequence from Schistosoma mansoni, were also assembled from two oligonucleotides: ribozyme (42 nt), GCAGGUACAUACAGCUGAUGAGUCCCAAAUAGGACGAAACGC; substrate (20 nt), CGCGUCCUGUAUUCCACUGC. One nucleotide was added at the 5′ end of the substrate to improve gel electrophoresis analysis.
Cleavage kinetics were studied under single-turnover conditions. Ribozyme and radioactively [5′-32P]-labeled substrate were annealed in 50 mM NaCl, 0.1 mM EDTA (pH 7.0) by heating to 80 °C and then cooling to 25 °C at 0.2 °C per second in a thermal cycler. Annealed ribozyme was equilibrated to 25 °C and the reaction initiated by adding an equal volume of equilibrated 2× reaction buffer. The final reaction contained 1 μM ribozyme and 20 nM substrate. Standard reaction conditions were 50 mM TAPS (pH 8.0), 1.0 mM MgCl2, 2.0 M NaCl, 0.1 mM EDTA. For the study of the pH dependence of rates, standard conditions were used except that 50 mM of the following buffers was used to achieve the desired pH: MES, pH 5.0–6.5; MOPS, pH 7.0–7.5; TAPS, pH 8.0–9.0; CHES, pH 9.25–9.5. Slow reactions requiring long incubations were carried out under mineral oil to prevent evaporation. 2 μL aliquots were removed at intervals, and the reaction was terminated by addition to 13 μL of a mixture containing 95% (v/v) formamide, 50 mM EDTA, and electrophoresis dyes. Substrate and product were separated by electrophoresis in 20% polyacrylamide gels containing 7 M urea and quantified by phosphorimaging. Progress curves were fitted by nonlinear regression analysis to single exponential functions using KaleidaGraph (Abelbeck Software).
Apparent pKa values for ionizable functional groups were determined from cleavage rate data as a function of reaction pH fitted to equations for either a single ionizable group,
| 1 |
or two ionizable groups,
| 2 |
where kcleave is the intrinsic rate of cleavage when acid and/or base is in the active state. To fit data for the pistol ribozyme with a 2′-NH2 modification of A32, kcleave was assumed to depend on the ionization state of the amino group so that eq 2 was modified such that
| 3 |
Crystallography
The construct that gave the best crystals was hybridized from two oligonucleotides with sequences GGCCGUUCGGGCGGCCAUAAACAGCCCUCAGGCC-(BrC)-GAAGCGUGGCGUUCC and GGAACGCCG-(dG)-UGGGA. The sequence was adapted from that of Alistipes putredinis.3 The numbering in the .pdb file commences with 2 for the first G so that the numbering of the active core of the ribozyme is consistent with that of previously determined structures. 5-Br-C was substituted at C36 to allow anomalous data collection, and 2′-deoxy-G was substituted at −1G to inactivate ribozyme activity. A mixture containing equimolar quantities of the two oligonucleotides was annealed by incubation at 65 °C for 5 min and slowly cooled to 4 °C. The concentration of the RNA was adjusted to 0.1–0.2 mM before mixing with an equal volume of 0.01 M MgSO4 heptahydrate, 0.05 M Na cacodylate trihydrate (pH 6.0), 1.8 M Li2SO4 monohydrate. This was sealed with 0.5 mL of 1.8 M Li2SO4 monohydrate solution at 4 °C in hanging drop mode and crystals of 50 μm × 50 μm × 100 μm grew in 2 weeks. Crystals were flash frozen and stored in liquid nitrogen.
The 3.1 Å single-wavelength anomalous dispersion (SAD) data set was acquired from Diamond beamline I24. Phases were acquired from the SAD data by locating the bromine atoms with Autosol in the PHENIX suite.33 The initial model was generated automatically by PHENIX Autobuild Wizard.34 The model was adjusted manually using Coot35 and subjected to several rounds of adjustment and optimization using Coot, REFMAC, and phenix.refine. The structure was finally refined to Rfactor = 18.34%, Rfree = 20.44%. The statistics are shown in Table S1. The structure has been deposited with the PDB with accession code 6R47.
Results
Structure of the Pistol Ribozyme
We have solved the structure of the pistol ribozyme at 3.1 Å resolution, with sequence adapted from that of Alistipes putredinis.3 The structure is consistent with the two previously determined structures36,37 with minor differences in the positions of G40 and G42 in the vicinity of the cleavage site that are discussed below. The pistol ribozyme comprises four double-stranded segments, P1, P2, P3, and PS, each of which adopts a standard A-form helix (Figures 2A–C and S1). The global structure of the ribozyme can be described as the combination of a pseudoknot (P1, PS, and P3) and a three-way junction comprising P2, PS, and P3. Helices P1, PS, and P3 are coaxial, but there is a pronounced curvature along the length of this so that the axes of P1 and P3 are inclined at an angle of 100°. The core of the three-way junction contains most of the highly conserved nucleotides (Figure S2), and we shall see below it contains key functional nucleotides that cluster around the scissile phosphate. It is thus the active site of the ribozyme. The three-way junction of the hammerhead ribozyme has the same helical architecture with helices II and III coaxially stacked (Figure 2D), but with negligible curvature, and the highly conserved nucleotides clustered at the junction.
Figure 2.
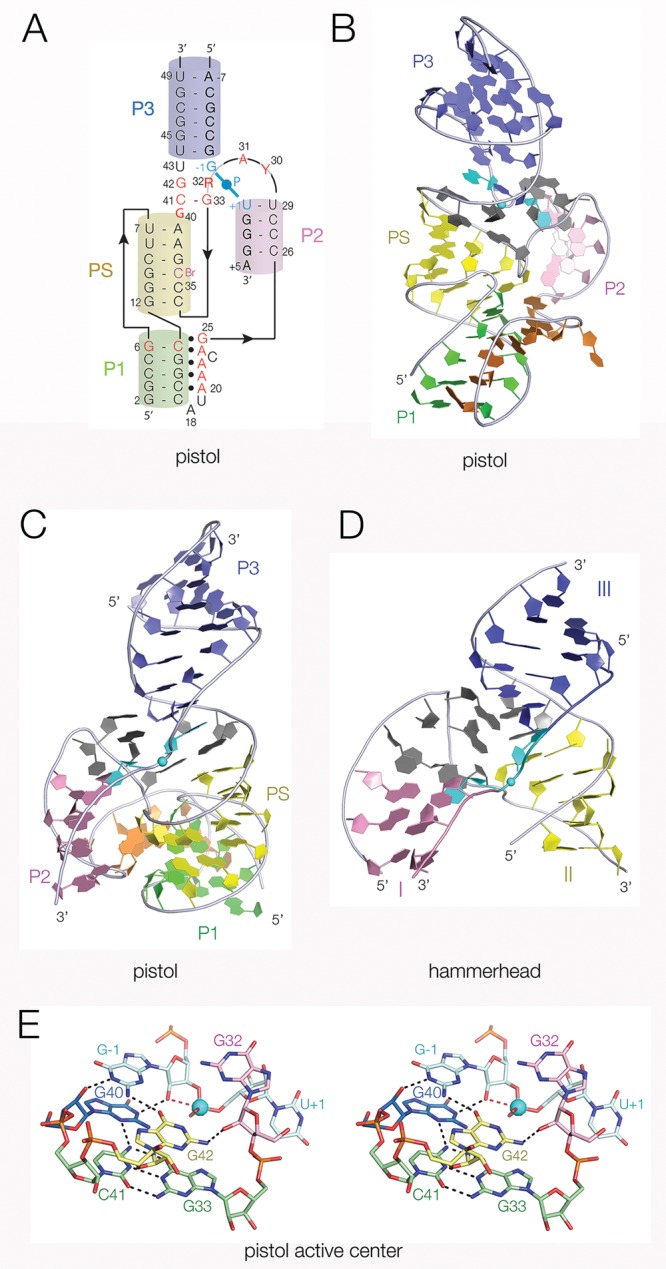
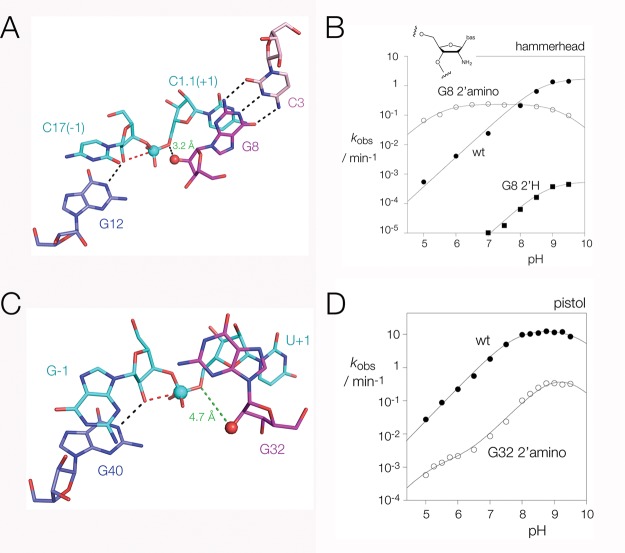
Crystal structure of the pistol ribozyme and comparison with the hammerhead ribozyme. We have solved a new crystal structure of the pistol ribozyme at 3.1 Å resolution. (A) Scheme showing the secondary structure as it relates to the three-dimensional structure in the crystal. The helices are colored: P1 green, P2 pink, P3 blue, and pseudoknot PS yellow. Highly conserved nucleotides are colored red. The scissile phosphate and the flanking nucleotides (−1 and +1 positions) are colored cyan. (B, C) Views of the crystal structure of pistol seen from opposite sides, rotated around the vertical axis. The scissile phosphate is shown as the cyan sphere, and the flanking nucleotides are also colored cyan. A stereoscopic view of the complete pistol structure with electron density is shown in Figure S1. The positions of conserved nucleotides in the structure are shown in stereoscopic view in Figure S2. (D) Structure of the core of the hammerhead ribozyme determined by Scott and co-workers,18,43 taken from PDB code 3ZP8. Corresponding helices have been colored in the same manner as those of the pistol ribozyme, and the structure may be directly compared with that in part C. (E) Parallel-eye stereoscopic view of the active center of the pistol ribozyme. The scissile phosphate is highlighted as the cyan sphere. In this and subsequent figures the direction of nucleophilic attack is shown by the broken red line.
The pistol ribozyme has a variable length section that links the 3′ end of P1 to the 5′ end of P2 and makes a number of interactions in the minor groove of P1, particularly at the conserved A3 sequence (A20–A22) plus A24. These make a series of A-minor hydrogen bonds predominantly between adenine ring N atoms and 2′O atoms of the P1 helix. G25 is stacked on A24 and forms a base triple with the conserved G6:C13 base pair at the end of P1. A revised alignment of pistol ribozyme sequences shows that A24 and G25 are more highly conserved than has been recognized previously (Figure S3) and deletion of G25 significantly impairs cleavage (Table 1).
Central Core of the Ribozyme, In-Line Attack, and Catalytic Role of a Guanine Nucleobase G40
The central core of the pistol ribozyme comprises three strands: (a) that linking the PS and P3 helices contains G40, C41, and G42 and runs continuously through this region in a quasi-helical manner, (b) the strand linking helices P3 and P2 containing G–1 and U+1 that flank the scissile phosphate, and (c) the 180° turn connecting P2 and PS. The secondary structure of the hammerhead ribozyme is very similar, the major difference being that a longer sequence makes the 180° turn.
An approximately in-line geometry between the O2′ nucleophile, the scissile phosphate, and the O5′ leaving group is required for the SN2 cleavage reaction. Comparison of the structure of the junction between helices P3 and P2 of the pistol ribozyme and that between helices III and I of the hammerhead ribozyme18 reveals a significant similarity (Figure 3A,B), with the nucleobases flanking the scissile phosphate splayed apart to create the in-line geometry required for catalysis. In the pistol ribozyme G–1 is stacked between the first base pair of P3 and G40 (Figure 3A) and held in place by hydrogen-bonding with G40 and G42 (Figure 2E). U+1 contributes to the first (noncanonical) base pair of P2 and the O2′–P–O5′ angle is 166°, close to the optimal angle of 180°. In the hammerhead ribozyme C-1 is stacked between the first base pair of helix III and G12, held in place by hydrogen bonding with G5 and A13 (Figure S4). C+1 is part of the first base pair of helix I, and the O2′–P–O5′ angle is 157° (Figure 3B). G12 acts as a general base in the hammerhead cleavage reaction17 and G40 occupies a very similar position in the pistol ribozyme. Note that the pistol structure has a deoxyribose at position −1 to prevent cleavage and the position of the O2′ has been modeled. Nevertheless, it is clear that G40 is well positioned to act as a general base as has been proposed previously.36
Figure 3.

pH dependence of the pistol ribozyme cleavage reaction, and the conformation around the scissile phosphate. (A) Crystal structure of the pistol ribozyme around the scissile phosphate. The nucleotides flanking the scissile phosphate are splayed apart to generate an in-line conformation. G–1 is stacked on G–2 at the base of P3, while U+1 is base paired with U29 and stacked on the end of P2. G40 N1 is hydrogen bonded to the G–1 O2′ nucleophile (this was modeled as the structure was determined with G–1 O2′H to prevent activity), and the broken red line shows the direction of nucleophilic attack. Electron density (2Fo – Fc) contoured at 2σ is shown for nucleotides G–1, U+1, and G40. (B) Structure of the hammerhead ribozyme (PDB code 3ZP8) around the scissile phosphate.18,43 The viewpoint has been chosen to show the close similarity between the pistol and hammerhead structures. The conformations of the nucleotides flanking the scissile phosphate are very similar, and the juxtaposition of the general bases (G12 for the hammerhead) is almost identical. (C) Plot of cleavage rate as a function of reaction pH for the natural ribozyme sequence (filled circles) and for ribozyme with G40 replaced by inosine (G40I; open circles). The data have been scaled together for comparison, with the unmodified pistol corresponding to the left axis, and the G40I ribozyme using the right axis. These data were measured under single turnover conditions in the presence of 1 mM Mg2+ and 2 M Na+ ions. The data have been fitted to eq 2 (lines), giving apparent pKa values of 7.8 and 9.8 for the unmodified ribozyme and 8.5 and 8.5 for the G40I ribozyme.
To investigate the role of G40, we first measured the rate of cleavage by the pistol ribozyme as a function of pH in the presence of 1 mM Mg2+ and 2 M Na+ ions (Figure 3C). The resulting profile is a narrow bell-shape, corresponding to titration of two groups with apparent pKa values of 7.8 and 9.7. We repeated the titration for a G40I (I = inosine; this has a pKa 0.8 unit below that of guanine) variant. This resulted in the higher apparent pKa being lowered to 8.5, consistent with a direct role of this nucleobase in proton transfer in the reaction. If G40 is acting as a general base, then substitution of a nucleotide with a lower pKa will result in a greater fraction being in the deprotonated active state. On this basis, we calculate that an inosine substitution at G40 should lead to a 6-fold increase in the cleavage activity, yet the activity of G40I is 8-fold lower. This difference indicates that the G40I substitution results in significant perturbation to activity. Since G40 lies adjacent to the G–1 O2′, our data are consistent with the hypothesis that it serves as a general base to deprotonate the nucleophile of the cleavage reaction, performing the same function as G12 in the hammerhead. This leaves the question of what might serve as a general acid, which we discuss below.
Structural Comparison of the Central Cores of the Pistol and Hammerhead Ribozymes
In order to identify other functional groups that might contribute to catalysis, it is helpful to compare the core of the pistol ribozyme with that of the more-extensively studied hammerhead. The strand linking PS and P3 in the pistol ribozyme has three nucleotides, but there are only two main planes of nucleotide interactions because G40 forms a triple base interaction with the G33-C41 Watson–Crick base pair that stacks onto the end of PS. Above this plane (in our figures P3 lies at the top) G42 is stacked on the G33-C41 base pair and coplanar with G–1. G–1 N2 donates a hydrogen bond to G42 O6, and G42 N2 is hydrogen bonded to G32 O2′. G–1 N1 donates a hydrogen bond to G40 O2′ (Figure 2E). In contrast, the three nucleotides of the equivalent strand in the hammerhead junction form a helical structure. G12 is held in position to activate the nucleophile by a Hoogsteen-sugar edge base pair with A9 that is stacked on helix II. A13 is stacked on A9 and bonds to C-1 and is thus similarly positioned to G42 in the pistol structure. A14 is then stacked between A13 and the first base pair of helix III. These structural elements serve to position the general base but are too far from the O5′ leaving group to act as a general acid.
The junction sequence between P2 and PS of the pistol ribozyme forms a loop that interacts with P3 on the minor groove side (Figure S5). A31 lies at the apex of the turn between P2 and PS and is stacked on G32. Hydrogen bonds between A31 N3 and U43 O2′ plus G32 N7 and C30 O2′ stabilize the narrow minor groove at the turn. C30 N3 is 2.9 Å from the proS O of G33 and well positioned to form a hydrogen bond that would also stabilize the turn. Position 30 is an invariant pyrimidine and most frequently a U, and there is a hydrogen bond between U30 N3 and the proS O of G33 in the other pistol structures.36,37 Our crystals were obtained at pH = 6, so it therefore seems likely that the pKa of C30 is raised such that protonated C30+ donates a hydrogen bond in our structure. This hypothesis is supported by the observation that C30 is less active then U30 at pH = 8 (Table 1). The last nucleotide in the sequence (G33) serves to position G40 on the opposite strand as described above.
The loop of the hammerhead ribozyme is more elaborate but shares common features. The three nucleotides U4-G5-A6 lie at the apex of the loop and form an identical loop structure to that described for the pistol ribozyme (Figure S5), although there is greater interaction between the loop and the adjacent helix in the hammerhead. The final nucleotide (A9) serves to position G12 to act as general base and thus has a similar structural role to that of G33 in the pistol ribozyme. C3 and G8 form an additional base pair at the end of helix II that further stabilizes the loop, and U7, which is not conserved, is stacked on the base pair and tilts the loop toward helix III, thus promoting their interaction.
G8 O2′ of the hammerhead is 3.2 Å from O5′ (Figure 4A) and either acts as the general acid in the cleavage reaction or helps position a catalytic Mg2+ ion.18,19,38 The equivalent nucleotide in the pistol ribozyme is A or G32, yet participation by the nucleobase in catalysis is unlikely given that the ribozyme is equally active with either A32 or G32 (Table 1) and atomic mutagenesis of the ring nitrogen atoms of A32 has little effect on activity.39 A32 O2′H substitution results in an 80-fold loss of activity, indicating that the O2′ has an important function (Table 1). However, G32 O2′ is 4.7 Å from O5′ (Figure 4C), and thus some structural alteration would be required for this to act as the general acid.
Figure 4.
Role of the 2′-hydroxyl groups of G8 and G32 in the hammerhead and pistol ribozymes. In these experiments the pH dependence of cleavage has been measured for ribozymes with a specific 2′-amino substitution. (A) Crystal structure of the hammerhead ribozyme around the scissile phosphate showing the position and orientation of G8. Note that the nucleotide is oriented so that the 2′-hydroxyl group is directed toward the O5′ leaving group with an O2′-O5′ distance of 3.2 Å. (B) Plot of cleavage rate as a function of reaction pH for the hammerhead ribozyme. This has been performed for the unmodified ribozyme (filled circles), G8 2′NH2 ribozyme (open circles), and G8 O2′H ribozyme (filled squares) in 2 M Na+ ions. Equivalent data have been obtained in the presence of 1 mM Mg2+ and 2 M Na+ ions, shown in Figure S6. The data for the unmodified and G8 O2′H ribozymes were fitted using eq 1, giving apparent pKa values of 8.7 and 8.8, respectively. The data for the G8 2′NH2 ribozyme were fitted using eq 2, giving apparent pKa values of 5.5 and 9.3. (C) Crystal structure of the pistol ribozyme around the scissile phosphate showing the position and orientation of G32. Comparison with the structure of the hammerhead ribozyme (part A) shows that G32 is rotated by 90° such that the 2′-hydroxyl group is not directed toward the O5′ leaving group, with an O2′–O5′ distance of 4.7 Å. (D) Plot of cleavage rate as a function of reaction pH for the pistol ribozyme. This has been performed for the unmodified ribozyme (filled circles) and the A32 2′NH2 ribozyme (open circles) in 1 mM Mg2+ and 2 M Na+ ions. The data for the unmodified ribozyme are the same as those in Figure 3C. The data for the A32 2′NH2 ribozyme were fitted using eqs 2 and 3, giving an apparent pKa values for the 2′ amino group of 5.2.
Investigation of the Mechanistic Role of G8 O2′ in the Hammerhead Ribozyme
We have explored the potential roles of the putative catalytic O2′ groups of the pistol and hammerhead ribozymes by atomic mutation coupled with pH titration of cleavage rates. This involves synthesizing variant ribozymes in which the O2′ in question is replaced by an amino group. The pKa of 2′NH2-ribose has been measured as 6.2 40 and is thus substantially lower than that of an OH group; it will deprotonate (and thus be unavailable as a general acid) at significantly lower pH. This was performed first for the hammerhead ribozyme, using the Schistosoma mansoni form with a tertiary contact between the terminal loop of helix II and an internal receptor in helix I.18,23 The dependence of cleavage rate on pH is shown in Figure 4B. In the absence of divalent cations the unmodified ribozyme exhibits a log–linear increase in reaction rate across the pH range up to pH = 9, consistent with a titration of the general base, G12, and with the participation of a general acid of high pKa that is fully protonated over the observable pH range. A G8 O2′H variant gave a similar profile, with rates 1000-fold lower. However, the pH profile of the G8 2′NH2 variant was quite different with a plateau between pH 6 and 8, and progressively lower rates to lower (<6) and higher (>8) pH. Fitting the data to a two-ionization model gave apparent pKa values of 5.5 and 9.3. We assign these to the titration of the 2′NH2 group and G12 respectively. The plateau at intermediate pH is due to the decline in the proportion of acid in the active protonated state with increasing pH being offset by a corresponding increase in the proportion of base in the active deprotonated state. At extreme pH values the rate decreases because either the acid (at low pH) or the base (at high pH) is fully in the active state, so the rate is determined by the titration of the other species. These data provide good evidence for the direct participation of the G8 O2′ in general acid–base catalysis by the hammerhead ribozyme.
Mechanistic Difference between the Pistol and Hammerhead Ribozymes
The equivalent substitution was made in the pistol ribozyme by synthesis of an A32 2′NH2 variant, and the rate of cleavage was measured as a function of pH (Figure 4D). Cleavage by the unmodified ribozyme increased log–linearly with pH values up to pH = 8. Cleavage rates by the A32 2′NH2 pistol ribozyme were 100-fold lower, but the dependence on pH was closely similar to that of the unmodified ribozyme. This is quite different from the effect of the corresponding modification of the hammerhead ribozyme. There is a deviation from linearity at low pH corresponding to a pKa = 5.2 indicating that the 2′NH2 does influence the activity of the ribozyme. However, this is consistent with a group that influences the rate rather than a group that participates directly in proton transfer in the transition state.20 The results show that there is a mechanistic difference between the two ribozymes, and in contrast to the hammerhead ribozyme, our data do not support a direct role for the A32 O2′ in general acid catalysis.
Potential Role of Metal Ions in the Pistol Ribozyme Cleavage Reaction
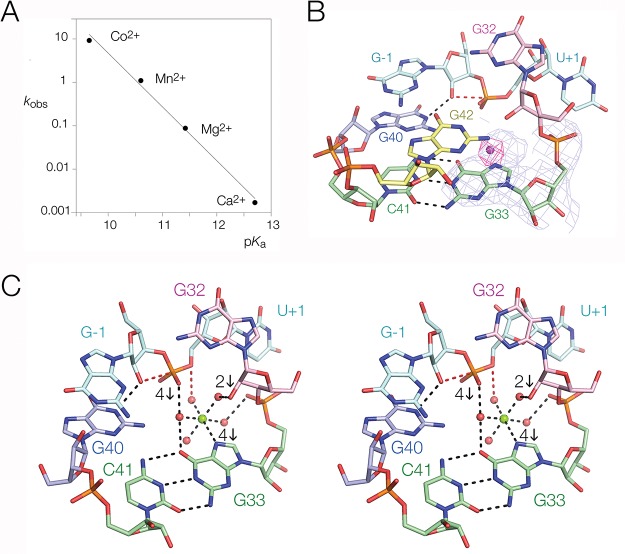
The results of the 2′-amino substitution argue against the direct participation of the A32 O2′ as general acid in the pistol ribozyme, and there are no other functionalities within the active center that offer an alternative. We have therefore examined the role of divalent metal ions in the cleavage reaction. We have measured rates of cleavage in the presence of Ca2+, Mg2+, Mn2+, and Co2+ ions. The rates increase 4 orders of magnitude across the range from Ca2+ to Co2+, and there is a log–linear dependence of cleavage rate on the pKa of the hydrated metal ions (Figure 5A). This indicates an important role of the hydrated metal ion in the chemistry of the cleavage reaction. This result is consistent with an increase in reactivity of inner sphere water molecules acting as a general acid as their pKa is reduced. Participation of an inner sphere water molecule in proton transfer is also consistent with the 390-fold reduction in activity observed when the functionally inert hexammine cobalt chloride is substituted in place of magnesium chloride (Table 1).
Figure 5.
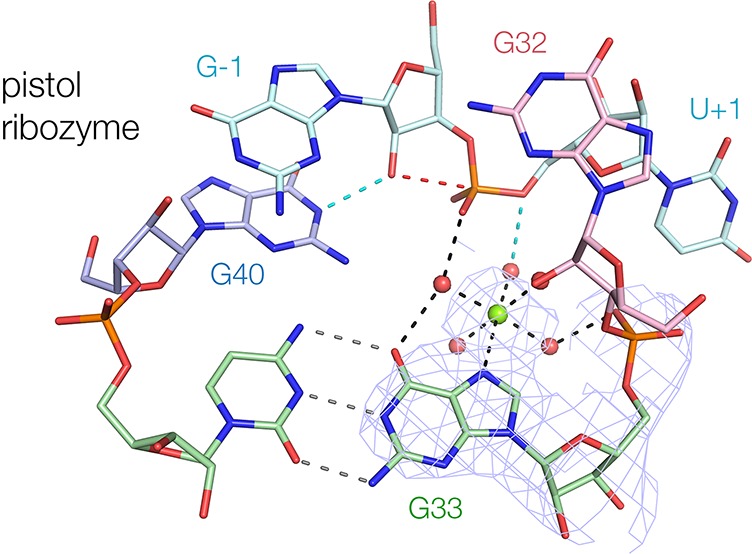
Role of a bound metal ion in the pistol ribozyme. (A) The rate of ribozyme cleavage was measured in 50 mM MES (pH 6.0), 2 M NaCl, 0.1 mM EDTA in the presence of 1 mM of chosen divalent metal ions. The observed rate of cleavage (kobs) is plotted as a function of hydrated metal ion pKa. (B) Metal ion binding site close to the active center of the pistol ribozyme. The light blue mesh shows the 2Fo – Fc electron density contoured at 2σ in the vicinity of G33. The crystals were soaked with 1 mM MnCl2 and excited with X-rays close to the peak of anomalous scatter for Mn2+. The magenta mesh shows the anomalous peak of electron density contoured at 7σ. A stereoscopic view is shown in Figure S7. (C) A divalent metal ion has been modeled into the active center of the pistol ribozyme in the position indicated by the Mn2+ ion anomalous scatter. This is directly bonded to G33 N7, and the five inner sphere water molecules have been placed to be most consistent with the atomic mutagenesis data. Those data are summarized by the downward arrows, with the numbers indicating the order of magnitude reduction in cleavage activity resulting from mutation. The inner sphere water molecule apical to G33 N7 lies within hydrogen bonding distance of the O5′ leaving group (colored red) and thus is well positioned to act as a general acid. A parallel-eye stereoscopic view is shown. G42 is not displayed in this view for ease of clarity.
Possible Metal Ion in the Active Site of the Pistol Ribozyme
Examination of the electron density map for the pistol ribozyme revealed unassigned density within the region of the active center (Figures 5B and S7). While the resolution of the structure is insufficient to permit unambiguous assignment of this density, it is consistent with a hydrated Mg2+ ion. We soaked Mn2+ ions into the crystals and observed strong anomalous scatter from the same position (Figures 5B and S7). The center of the density is almost in plane with G33 and 2.4 Å from G33 N7. While consistent with a direct metal–N bond, direct bonding of Mg2+ to N7 of guanine is highly unusual and frequently misassigned.41 Nevertheless, the position of the Mn2+ is unambiguous. In addition, atomic substitution G33 N7C lowers cleavage activity by nearly 4 orders of magnitude (see following section). Ren and co-workers36 also assigned a bound metal ion at the corresponding position in their structure of the pistol ribozyme.
Response of Cleavage Activity to Atomic Mutation in the Active Center
We wished to identify other functional groups of the ribozyme that could bind to and orient the inner sphere water molecules of the Mg2+ ion. In addition we wished to learn if other functional groups in the core of the ribozyme could participate in the catalytic mechanism. We therefore performed atomic mutagenesis on functional groups lining the cation binding pocket (Table 1). Significant change in cleavage rates resulted from some of these alterations, and the magnitudes have been mapped onto the structure in Figure 5C.
The most impaired variant studied is the Rp stereoisomer of a phosphorothioate substitution at the scissile phosphate. The proR nonbridging oxygen is 3.7 Å from the Mg2+ ion, so it is plausible that it accepts a hydrogen bond from an inner sphere water. Harris et al.22 had previously shown that one phosphorothioate stereoisomer was significantly impaired and activity was not restored by addition of Mn2+ ions. We now extend this observation with resolved stereoisomers. The Sp stereoisomer exhibits negligible impairment. By contrast the rate of cleavage of the Rp stereoisomer is 45 000-fold lower than that of the unmodified substrate under standard conditions (Table 1). Although it is 150-fold more active in Mn2+ ions, much of this increase can be attributed to the greater reactivity of Mn2+ ion-bound water molecules, a 13-fold effect at pH 5.5 (Figure 5A).
In contrast to the other two structures, G40 N2 donates a hydrogen bond to the proR O of the scissile phosphate in the structure of Nguyen et al.37 We sought to test this by comparing the cleavage activity of the phosphorothioate stereoisomers by a G40I ribozyme (Table 1). Like the unmodified substrate, the Sp stereoisomer is cleaved 8-fold slower by the G40I ribozyme. Surprisingly, the Rp stereoisomer is cleaved 20× more rapidly by the G40I ribozyme. Although the increase in activity is somewhat greater than expected for an inosine substitution based on the difference in pKa of the nucleotides, this result is consistent with the inosine substitution being nonperturbing in the context of the Rp phosphorothioate whereas it is perturbing in the presence of the proR O. We conclude that G40 N2 donates a hydrogen bond to the proR O. However, the absence of this hydrogen bond makes it difficult to assess the extent of rescue of the Rp phosphorothioate by Mn2+ ions. This problem is avoided by a comparison of the cleavage rates of the Rp stereoisomer by the G40I ribozyme, which is 100-fold more active in the presence of Mn2+ ions compared to Mg2+ ions. Again, much of this increase can be attributed to differences in pKa of ion-bound water molecules, and the rate is 38-fold lower than the estimated cleavage rate of the unmodified substrate by the G40I ribozyme in the presence of Mn2+ ions. Therefore, rescue of the Rp phosphorothioate by Mn2+ ions is far from complete and the data are more consistent with an outer sphere interaction between a bound Mg2+ ion and the proR O of the scissile phosphate.
Modification of A32 2′OH and G42 2-aminopurine substitution also resulted in significantly reduced cleavage rates. However, of these two positions only A32 O2′ is close enough to the metal ion to act as an outer sphere ligand. We have constructed a model in which a pentahydrated Mg2+ ion is bound to G33 N7 with the inner sphere water molecules positioned to interact with the proR O of the scissile phosphate and A32 O2′, Figure 5C. In this model one of the inner sphere water molecules lies within hydrogen bonding distance of the O5′ leaving group and is thus well positioned to act as a general acid.
Reorganization of the Active Site Is Required To Attain the Transition State
Several of the modifications listed in Table 1 are not readily explained by the available structures of the pistol ribozyme. In each structure G32 2′OH accepts a hydrogen bond from G42 N2 and an A32 O2′H modification leads to an 80-fold decrease in activity. Yet a G42I substitution, which lacks the N2 donor, suffers no significant impairment. Similarly, in our structure G–1 N2 donates a hydrogen bond to G42 O6, and G42 2-aminopurine, which lacks the O6 acceptor, has 1000-fold lower activity. However, a G–1I substitution, that lacks the N2 donor has unaltered activity. It is notable that G42 is not coplanar with G–1 in the pistol ribozyme structure of Ren et al.36 but is intermediate between G40 and G42, and G42 O6 potentially accepts a hydrogen bond from either nucleotide, suggesting a degree of structural flexibility in this area. Thus, neither of the hydrogen bonds made by G42 in our structure appears to be necessary for activity. Furthermore, our model for Mg2+ binding suggests that two of the inner sphere water molecules are able to bond to the phosphates of nucleotides 33 and 35, but phosphorothioate substitutions at these positions have little effect. These observations suggest that G42 and possibly the Mg2+ ion alter position as the transition state is attained.
Discussion
We have presented evidence that the pistol ribozyme implements all four recognized catalytic strategies. In-line orientation is achieved by stacking the nucleobases G–1 and U+1 on helices P3 and P2, respectively. U+1 base pairs with U29 and is thus part of P2. G–1 is stabilized by hydrogen bonding with G40 O2′. In the crystal G–1 also donates a hydrogen bond to G42 O6; however this bond is not necessary for full activity.
Atomic mutagenesis of G40 and the scissile phosphate supports the hypothesis that the proR nonbridging O accepts hydrogen bonds from G40 N2 and from an inner-sphere water molecule of a Mg2+ ion. These provide structural stabilization of the general base and acid and the phosphorane transition state. Furthermore, the proximity between the phosphate and the Mg2+ ion will also provide a degree of electrostatic stabilization. The Rp phosphorothioate substitution was the most deleterious substitution investigated, indicating the importance of the proR O to the activity of the ribozyme.
Like the other nucleolytic ribozymes, the pistol ribozyme obtains significant rate enhancement from general acid–base catalysis. It has been assumed that G40 would act as the general base due to its proximity to the nucleophile,36,37 and our G40I substitution data support this. If the substitution was nonperturbing, an increase in rate would be expected since inosine has a lower pKa than guanosine. However, a decrease in activity was observed that we attribute to the loss of the hydrogen bond between G40 N2 and the proR nonbridging oxygen atom. When the substrate contained an Rp phosphorothioate substitution, weakening the hydrogen bond, G40I substitution resulted in a 20-fold increase in activity.
The pistol ribozyme employs guanine as a general base in common with most nucleolytic ribozymes. However, its use of a hydrated divalent cation as general acid is novel. The evidence for this is structural and mechanistic. The only nucleotide close enough to the leaving group to act as a general acid is that at position 32, which can be either an A or a G in the sequence alignment. This suggests that the nucleobase is unlikely to act as a general acid, confirmed by Neuner et al.39 with a series of deaza-adenine substitutions. The 2′OH of nucleotide 32 appears better positioned to act as a proton donor to the leaving group. However, 2′-amino substitution does not significantly change the pH dependence of cleavage rate (Figure 4D), although there is a slight increase in activity at low pH due to protonation of the amino group.
This leaves a divalent cation as the best candidate for general acid. In support of this hypothesis we have demonstrated a log–linear relationship between cleavage activity and metal ion pKa. By itself this does not prove the metal ion acts directly since an equivalent relationship has been found for the hammerhead.42 However, since activation of a hydroxyl group has been ruled out, this is strong evidence for a divalent cation acting as the general acid. In addition, there is a bound metal ion in the crystal structure in proximity to the leaving group, that may be occupied by a Mn2+ ion. Mutagenesis of functional groups lining the metal ion binding pocket identified several that exhibited significant loss of activity. In addition to the Rp phosphorothioate, the results of modifications to the A32 2′OH are interesting. Deoxy and amino substitutions lowered activity 2 orders of magnitude, consistent with the loss of a significant bond. Addition of a methyl group gave an 18 000-fold loss of activity, likely due to steric occlusion of the metal ion binding. On the basis of the crystallographic and the mutagenesis data, we have modeled a hydrated Mg2+ ion in the pocket, showing that a hydrated metal ion occupying this position would be well placed to act as a general acid.
Not all the mutagenesis data are consistent with the crystal structure, in particular the network of bonds holding G42 in position. We propose that there is some reorganization of the active site in the transition state involving a repositioning of G42. Nevertheless our conclusions regarding the pistol ribozyme mechanism are reliable since they are predominantly based on kinetic data that reflect the transition state.
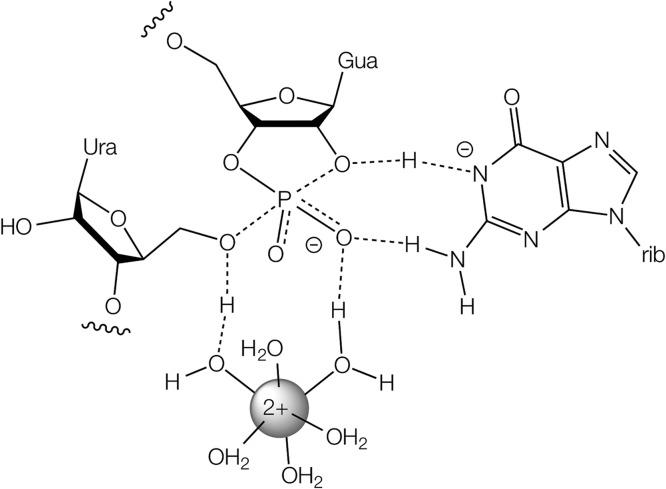
The catalytic strategies used by the pistol ribozyme are depicted in Figure 6. Most nucleolytic ribozymes use a more reactive functional group with a low pKa as a general acid, where the advantage of high reactivity is partially offset by the low abundance of the protonated active state. We propose that the pistol ribozyme adopts the inverse strategy wherein it utilizes a fully protonated general acid with relatively low reactivity. The mechanism involves the transfer of two protons, and there are two apparent pKa values in the data in Figure 3. We have assigned the higher pKa to G40, but this leaves the question of what is responsible for the lower pKa. Hydrated Mg2+ has a pKa of 11.4, so this is unlikely to be the source. It is more likely to be a nucleobase with a significantly shifted pKa, and since the apparent pKa increases when the structure is perturbed (e.g., G40I and A32 2′NH2), we propose that the nucleobase is G or U. One plausible explanation is that the nucleobase has a lowered pKa through proximity to the metal ion and that perturbation of the structure results in the pKa relaxing back toward its normal value.
Figure 6.
Proposed catalytic mechanism for the pistol ribozyme.
The role of G12 as the general base in the hammerhead ribozyme is well established.17 Here we have focused on the general acid since there remained ambiguity between two possible general acids, a hydrated divalent cation and G8 2′OH. G8 2′NH2 substitution results in a ribozyme that is highly active in the absence of divalent cations with a pH dependence of cleavage consistent with the 2′NH2 group participating in catalysis. This clearly establishes that the ribozyme uses mechanism 1 in the absence of divalent cations, in which G8 2′O acts as the general acid. Addition of 1 mM Mg2+ has little effect on the activity of the G8 2′NH2 hammerhead ribozyme, but this increases the activity of the G8 2′OH and G8 O2′H ribozymes by 10-fold and 100-fold, respectively. The best explanation for these data is that the G8 O2′ acts as the general acid and the Mg2+ ion activates it by lowering its pKa. If the ribozyme used the Mg2+ ion as general acid with the G8 O2′ helping to position it (mechanism 2), then addition of Mg2+ ion would be expected to increase the activity of G8 O2′ by at least as much as it does the activity of G8 O2′H since G8 O2′ would be expected to position the Mg2+ ion better for catalysis. Nevertheless, the G8 O2′H data show that Mg2+ ions can act as general acid, so this alternative mechanism could become dominant when more reactive cations with lower pKa values are used.
It is clear that the hammerhead and pistol ribozymes share a lot of similarity in terms of both structure and catalytic mechanism. However, they differ markedly in their general acid catalysis. Here the key nucleotide is G8 in the hammerhead, and its equivalent in pistol is A or G32. While the pH dependence of cleavage is similar for the two ribozymes, 2′OH to NH2 mutants of G8 or A32 exhibit very different pH profiles (Figure 4B and Figure 4D), reflecting a clear mechanistic difference. In addition the conformations of G8 and G32 are very different in the two ribozymes. In the hammerhead ribozyme G8 is base paired with C3, thereby directing the G8 O2′ toward the O5′ of the scissile phosphate (Figure 4A). The O2′–O5′ distance is 3.2 Å, and thus the 2′ hydroxyl group is well positioned to act directly as general acid in the cleavage reaction. By contrast in the pistol ribozyme G32 is not base paired and is rotated by 90° so that the O2′–O5′ distance is lengthened to 4.7 Å (Figure 4C). The O2′ (but not the nucleobase) of G32 is important in the cleavage reaction, but our structural and mechanistic evidence indicates that it acts as a ligand for a bound metal ion, an inner-coordinated water molecule of which is the general acid in the cleavage reaction.
With respect to the general acid component of catalysis, the pistol and hammerhead ribozymes appear to be mechanistic mirror images. The hammerhead ribozyme uses the 2′-hydroxyl of G8. However, the pKa will normally be very high, and computational studies suggest that this is probably activated by binding a metal ion.38 Our evidence for the pistol ribozyme indicates that a water molecule directly coordinated to a bound metal ion acts as the general acid but that an important ligand for this metal ion is the 2′-hydroxyl of G or A32. It is conceivable that the two ribozymes share a common ancestor, where the role of the metal ion has diversified between directly acting as the general acid or activating a 2′-hydroxyl group to take this role.
Acknowledgments
We thank Saira Ashraf for expert RNA synthesis and acknowledge financial support from Cancer Research UK (Program Grant A18604 to D.M.J.L.) and the U.S. National Institutes of Health (Grant GM131568 to J.A.P.). We thank Diamond Light Source for synchrotron time and the Wellcome Trust for the in-house diffractometer.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.9b02141.
Seven supplementary figures and one supplementary table (PDF)
Author Present Address
§ Y.L.: State Key Laboratory of Medicinal Chemical Biology, College of Pharmacy, Nankai University, Haihe Education Park, 38 Tongyan Road, Tianjin 300353, People’s Republic of China.
The authors declare no competing financial interest.
Supplementary Material
References
- Lilley D. M. J. How RNA acts as a nuclease: some mechanistic comparisons in the nucleolytic ribozymes. Biochem. Soc. Trans. 2017, 45, 683–691. 10.1042/BST20160158. [DOI] [PubMed] [Google Scholar]
- Roth A.; Weinberg Z.; Chen A. G.; Kim P. B.; Ames T. D.; Breaker R. R. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 2014, 10 (1), 56–60. 10.1038/nchembio.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z.; Kim P. B.; Chen T. H.; Li S.; Harris K. A.; Lunse C. E.; Breaker R. R. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat. Chem. Biol. 2015, 11 (8), 606–610. 10.1038/nchembio.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T. J.; McLeod A. C.; Lilley D. M. J. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 2007, 26 (10), 2489–2500. 10.1038/sj.emboj.7601698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T. J.; Li N.-S.; Lu J.; Frederiksen J. K.; Piccirilli J. A.; Lilley D. M. J. Nucleobase-mediated general acid-base catalysis in the Varkud satellite ribozyme. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 11751–11756. 10.1073/pnas.1004255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T. J.; Lilley D. M. J. Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid-base catalysis ? A critical assessment of available experimental data. RNA 2011, 17 (2), 213–221. 10.1261/rna.2473711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kath-Schorr S.; Wilson T. J.; Li N. S.; Lu J.; Piccirilli J. A.; Lilley D. M. J. General acid-base catalysis mediated by nucleobases in the hairpin ribozyme. J. Am. Chem. Soc. 2012, 134 (40), 16717–24. 10.1021/ja3067429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua P. C. Mechanistic considerations for general acid-base catalysis by RNA: revisiting the mechanism of the hairpin ribozyme. Biochemistry 2003, 42 (8), 2259–2265. 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- Lafontaine D. A.; Wilson T. J.; Norman D. G.; Lilley D. M. J. The A730 loop is an important component of the active site of the VS ribozyme. J. Mol. Biol. 2001, 312 (4), 663–674. 10.1006/jmbi.2001.4996. [DOI] [PubMed] [Google Scholar]
- Pinard R.; Hampel K. J.; Heckman J. E.; Lambert D.; Chan P. A.; Major F.; Burke J. M. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 2001, 20 (22), 6434–6442. 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood V. D.; Collins R. A. Identification of the catalytic subdomain of the VS ribozyme and evidence for remarkable sequence tolerance in the active site loop. J. Mol. Biol. 2002, 320 (3), 443–454. 10.1016/S0022-2836(02)00521-1. [DOI] [PubMed] [Google Scholar]
- Wilson T. J.; Liu Y.; Domnick C.; Kath-Schorr S.; Lilley D. M. J. The novel chemical mechanism of the twister ribozyme. J. Am. Chem. Soc. 2016, 138 (19), 6151–6162. 10.1021/jacs.5b11791. [DOI] [PubMed] [Google Scholar]
- Nakano S.; Chadalavada D. M.; Bevilacqua P. C. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science 2000, 287, 1493–1497. 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- Das S. R.; Piccirilli J. A. General acid catalysis by the hepatitis delta virus ribozyme. Nat. Chem. Biol. 2005, 1 (1), 45–52. 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Wilson T. J.; Lilley D. M. J. The structure of a nucleolytic ribozyme that employs a catalytic metal ion. Nat. Chem. Biol. 2017, 13, 508–513. 10.1038/nchembio.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W. C.; Nahvi A.; Roth A.; Collins J. A.; Breaker R. R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 2004, 428 (6980), 281–286. 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Han J.; Burke J. M. Model for general acid-base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site. Biochemistry 2005, 44 (21), 7864–7870. 10.1021/bi047941z. [DOI] [PubMed] [Google Scholar]
- Martick M.; Scott W. G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 2006, 126 (2), 309–320. 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. M.; Perrin D. M. Probing general acid catalysis in the hammerhead ribozyme. J. Am. Chem. Soc. 2009, 131 (3), 1135–1143. 10.1021/ja807790e. [DOI] [PubMed] [Google Scholar]
- Frankel E. A.; Bevilacqua P. C. Complexity in pH-dependent ribozyme kinetics: Dark pKa shifts and wavy rate-pH profiles. Biochemistry 2018, 57 (5), 483–488. 10.1021/acs.biochem.7b00784. [DOI] [PubMed] [Google Scholar]
- Mir A.; Golden B. L. Two active site divalent Ions in the crystal structure of the hammerhead ribozyme bound to a transition state analogue. Biochemistry 2016, 55 (4), 633–636. 10.1021/acs.biochem.5b01139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K. A.; Lunse C. E.; Li S.; Brewer K. I.; Breaker R. R. Biochemical analysis of pistol self-cleaving ribozymes. RNA 2015, 21 (11), 1852–1858. 10.1261/rna.052514.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A.; Lescoute A.; Westhof E.; Jayasena S. D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Mol. Biol. 2003, 10 (9), 708–712. 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Penedo J. C.; Wilson T. J.; Jayasena S. D.; Khvorova A.; Lilley D. M. J. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA 2004, 10, 880–888. 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis E. A.; Bartel D. P. The hammerhead cleavage reaction in monovalent cations. RNA 2001, 7 (4), 546–552. 10.1017/S1355838201002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny M. D.; Jucker F. M.; Kellogg E.; Khvorova A.; Jayasena S. D.; Pardi A. Fast cleavage kinetics of a natural hammerhead ribozyme. J. Am. Chem. Soc. 2004, 126 (35), 10848–10849. 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- Beaucage S. L.; Caruthers M. H. Deoxynucleoside phosphoramidites - a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981, 22, 1859–1862. 10.1016/S0040-4039(01)90461-7. [DOI] [Google Scholar]
- Wilson T. J.; Zhao Z.-Y.; Maxwell K.; Kontogiannis L.; Lilley D. M. J. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry 2001, 40, 2291–2302. 10.1021/bi002644p. [DOI] [PubMed] [Google Scholar]
- Hakimelahi G. H.; Proba Z. A.; Ogilvie K. K. High yield selective 3′-silylation of ribonucleosides. Tetrahedron Lett. 1981, 22 (52), 5243–5246. 10.1016/S0040-4039(01)92470-0. [DOI] [Google Scholar]
- Perreault J.-P.; Wu T.; Cousineau B.; Ogilvie K. K.; Cedergren R. Mixed deoxyribo- and ribooligonucleotides with catalytic activity. Nature 1990, 344, 565–567. 10.1038/344565a0. [DOI] [PubMed] [Google Scholar]
- Dai Q.; Deb S. K.; Hougland J. L.; Piccirilli J. A. Improved synthesis of 2′-amino-2′-deoxyguanosine and its phosphoramidite. Bioorg. Med. Chem. 2006, 14 (3), 705–713. 10.1016/j.bmc.2005.08.050. [DOI] [PubMed] [Google Scholar]
- Karpeisky A.; Sweedler D.; Haeberli P.; Read J.; Jarvis K.; Beigelman L. Scaleable and efficient synthesis of 2′-deoxy-2′-N-phthaloyl nucleoside phosphoramidites for oligonucleotide synthesis. Bioorg. Med. Chem. Lett. 2002, 12 (22), 3345–3347. 10.1016/S0960-894X(02)00744-8. [DOI] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66 (2), 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40 (4), 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66 (4), 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren A.; Vusurovic N.; Gebetsberger J.; Gao P.; Juen M.; Kreutz C.; Micura R.; Patel D. J. Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage. Nat. Chem. Biol. 2016, 12 (9), 702–708. 10.1038/nchembio.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. A.; Wang J.; Steitz T. A. Crystal structure of Pistol, a class of self-cleaving ribozyme. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (5), 1021–1026. 10.1073/pnas.1611191114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. S.; Silva Lopez C.; Giambasu G. M.; Martick M.; Scott W. G.; York D. M. Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation. J. Am. Chem. Soc. 2008, 130 (10), 3053–3064. 10.1021/ja076529e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuner S.; Falschlunger C.; Fuchs E.; Himmelstoss M.; Ren A.; Patel D. J.; Micura R. Atom-specific mutagenesis reveals structural and catalytic roles for an active-site adenosine and hydrated Mg2+ in pistol ribozymes. Angew. Chem., Int. Ed. 2017, 56 (50), 15954–15958. 10.1002/anie.201708679. [DOI] [PubMed] [Google Scholar]
- Aurup H.; Tuschl T.; Benseler F.; Ludwig J.; Eckstein F. Oligonucleotide duplexes containing 2′-amino-2′-deoxycytidines: thermal stability and chemical reactivity. Nucleic Acids Res. 1994, 22 (1), 20–24. 10.1093/nar/22.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonarski F.; D’Ascenzo L.; Auffinger P. Mg2+ ions: do they bind to nucleobase nitrogens?. Nucleic Acids Res. 2017, 45 (2), 987–1004. 10.1093/nar/gkw1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boots J. L.; Canny M. D.; Azimi E.; Pardi A. Metal ion specificities for folding and cleavage activity in the Schistosoma hammerhead ribozyme. RNA 2008, 14 (10), 2212–2222. 10.1261/rna.1010808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M.; Schultz E. P.; Martick M.; Scott W. G. Active-site monovalent cations revealed in a 1.55-A-resolution hammerhead ribozyme structure. J. Mol. Biol. 2013, 425 (20), 3790–3798. 10.1016/j.jmb.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.