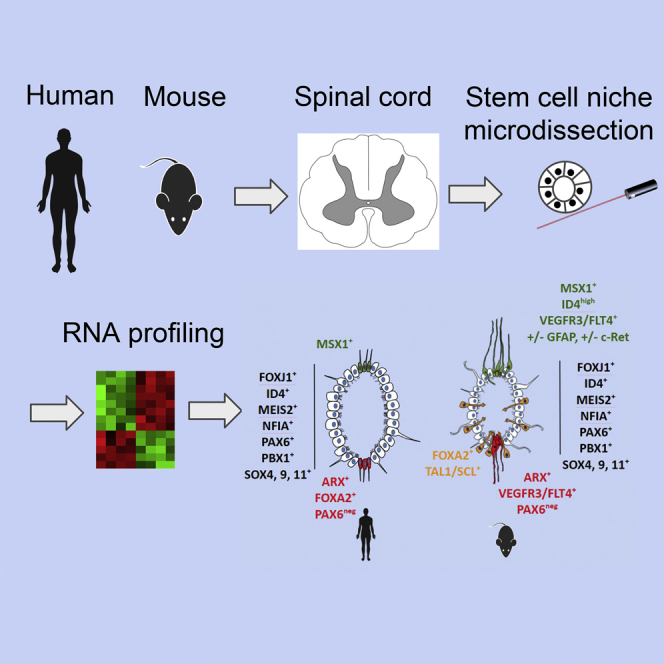
Summary
Anamniotes, rodents, and young humans maintain neural stem cells in the ependymal zone (EZ) around the central canal of the spinal cord, representing a possible endogenous source for repair in mammalian lesions. Cell diversity and genes specific for this region are ill defined. A cellular and molecular resource is provided here for the mouse and human EZ based on RNA profiling, immunostaining, and fluorescent transgenic mice. This uncovered the conserved expression of 1,200 genes including 120 transcription factors. Unexpectedly the EZ maintains an embryonic-like dorsal-ventral pattern of expression of spinal cord developmental transcription factors (ARX, FOXA2, MSX1, and PAX6). In mice, dorsal and ventral EZ cells express Vegfr3 and are derived from the embryonic roof and floor plates. The dorsal EZ expresses a high level of Bmp6 and Gdf10 genes and harbors a subpopulation of radial quiescent cells expressing MSX1 and ID4 transcription factors.
Keywords: spinal cord, niche, neural stem cells, regionalization, ependyma, ependymal cells, radial glial cells, transcription factors, Msx1, roof plate, floor plate
Graphical Abstract
Highlights
-
•
A molecular resource for the human and mouse spinal cord ependymal zone
-
•
Identification of 120 transcription factors in the human and mouse ependymal zone
-
•
Embryonic-like organization of the adult spinal cord ependymal zone
-
•
Dorsal ependymal cells expressing Msx1 are derived from the embryonic roof plate
A niche of stem cells is present around the central canal of the adult spinal cord. A better description of cell diversity and genes expressed in this niche may help to use it to promote spinal cord regeneration after lesions. In this article, based on several techniques, Ghazale and colleagues provide a cellular and molecular resource for the adult human and mouse stem cell niches.
Introduction
The adult central nervous system maintains neural stem cells in specific areas called niches (Gage and Temple, 2013). The main stem cell pools are in the subventricular zone (SVZ) and in the hippocampus. A third stem cell niche is found in the adult spinal cord around the central canal in anamniotes, rodents, and humans (Becker et al., 2018, Marichal et al., 2017, Stenudd et al., 2015). This niche originates from the embryonic neuroepithelium and forms the spinal cord ependymal zone (EZ) organized as a pseudo-epithelium. As in the brain, this stem cell niche is highly organized and contains stem and non-stem cells. Four different cell types, namely ependymocytes, cerebrospinal-fluid-contacting neurons (CSF-N), vessels, and long radial cells have been described in mice. In particular, the EZ presents a dorsal-ventral regionalization with long radial glial fibrillary acidic protein (GFAP)+ cells in the dorsal part (Sabourin et al., 2009). In contrast to brain, spinal cord ependymal cells slowly proliferate to self-renew (Alfaro-Cervello et al., 2012, Pfenninger et al., 2011). In cultures, a fraction of these cells can also generate passageable neurospheres (i.e. clonal expansion of neural precursor cells) which can generate astrocytes, oligodendrocytes, and neurons after differentiation (Weiss et al., 1996). The identity of these neurosphere-forming cells in the EZ is still not completely clear as both GFAP+ and GFAP− ependymal cells can behave as neural stem cells in vitro (Barnabé-Heider et al., 2010, Fiorelli et al., 2013, Sabourin et al., 2009, Xu et al., 2017). Recent single cell analysis has identified neurogenesis in the adult spinal cord (Habib et al., 2016); however, whether these new neurons are derived from the EZ is not yet established. It has been known since 1962 (Adrian and Walker, 1962) that the EZ can readily activate and produce new cells upon injury (Becker et al., 2018). Depending on the lesion type and severity, EZ-derived cells can significantly contribute to the glial scar formation (Ren et al., 2017, Stenudd et al., 2015).
In comparison with the brain niches, less is known about the adult spinal cord EZ. Reminiscent of the mouse niche, in human, ependymal cells around the central canal display immature features such as expression of NES (nestin), VIM (vimentin), and SOX2 (Becker et al., 2018). However, with aging the central lumen can disappear and the EZ is disorganized (Garcia-Ovejero et al., 2015). Multipotent neurospheres with a limited proliferation ability have been derived from the human spinal cord (Dromard et al., 2008) and using alternative culture conditions, Mothe et al. (2011) were able to maintain a sustained proliferation of multipotent human-derived neural stem cells.
A detailed transcriptomic profiling of the human and mouse EZ is currently lacking. This would help us to understand the specificity and diversity of these cells as well as identify gene expressions and molecular pathways conserved between primates and rodents. It would also provide important insights into why, in contrast to anamniotes, mammalian ependymal cells cannot regenerate neurons after spinal cord injury (Becker et al., 2018).
Here we provide a cellular and molecular resource for the mouse and human EZ based on RNA profiling, immunostaining, and fluorescent transgenic mice. This uncovered the conserved expression of 1,200 genes specifically expressed in the EZ, including 120 transcription factors (TFs). Unexpectedly, the EZ maintains an embryonic-like dorsal-ventral pattern of expression of spinal cord developmental TFs. New subpopulations of cells expressing specific genes were identified in the dorsal and ventral part of the EZ. In mice, dorsal ependymal cells were found to be derived from the embryonic spinal cord roof plate.
Results
Genes Enriched in the Adult Human and Mouse EZ
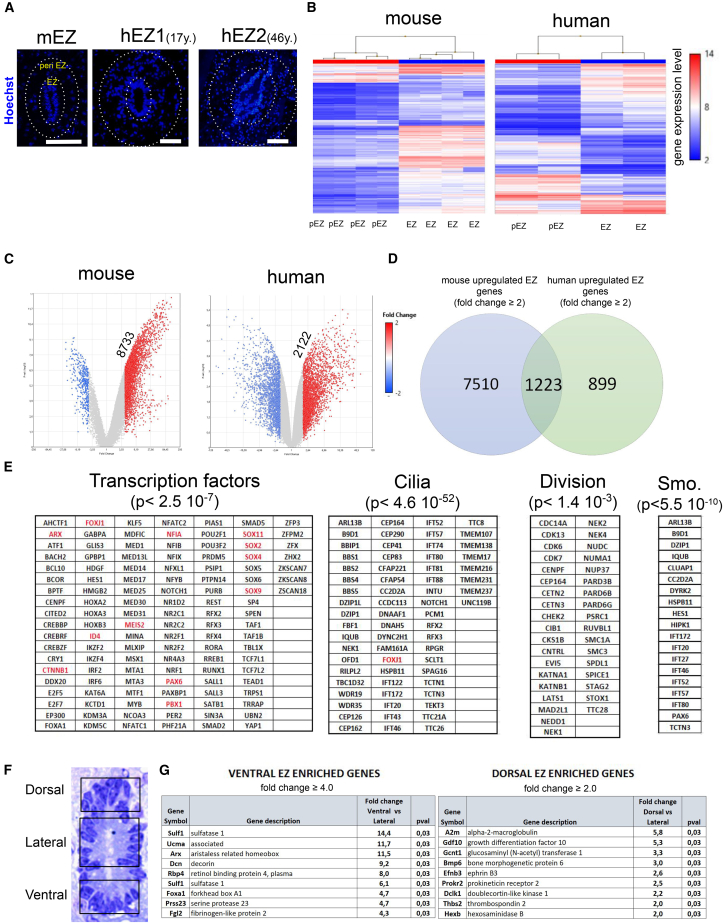
An epithelial organization of the EZ is observed both in human and mouse as evidenced by CTNNB1 (β-catenin) and CD24 stainings (Figures 2 and 3). To identify gene expression enriched in the EZ, we microdissected this region and adjacent tissue in two human samples and four mice (Figure 1A). For human, we selected two samples with a lumen from patients aged 17 and 46 years. Microarrays were used for RNA profiling and heatmaps indicated adequate clustering of EZ samples (Figure 1B). Volcano plots showed 8,733 and 2,122 genes enriched (fold change ≥2) in the mouse and human EZ, respectively, and 1,223 genes commonly enriched in both species (Figures 1C and 1D). Table 1 shows the top 15 genes commonly enriched in the human and mouse EZ and genes that are more specifically enriched in human or mice. Figure S1A shows examples of identified genes whose specific expression in the mouse EZ is confirmed in the Allen brain atlas (Lein et al., 2007). In mouse and human, GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analyses revealed an enrichment for genes involved in cilia formation, smoothened and hippo pathways, cell division, and transcription (Figure 1E and Tables S1, S2, and S3). In addition, consistent with their position at the interface between CSF and the nervous parenchyma, ependymal cells also expressed 129 and 34 members of the solute carrier family in mice and human, respectively (Tables S1, S2, and S3). Some of them are very specifically (fold change >30) expressed in the mouse EZ such as Slc26A3, Slc14a1, and Slc16a12, which transport chloride, urea, and monocarboxylic acid, respectively. Unexpectedly, Cftr, a chloride transporter responsible for cystic fibrosis, was found to be specifically expressed in the EZ in both species (Table 1). Using a single cell transcriptome approach in the mouse spinal cord, two recent studies (Rosenberg et al., 2018, Zeisel et al., 2018) provided a limited number of genes (<50) enriched in spinal cord ependymal cells and CSF-N, and most of them were identified in our study (Table S4). Interestingly, only Tmem212, a gene coding for a transmembrane protein with few annotations, was identified in the three studies (including ours) as being enriched in the ependymal cells. With regard to CSF-N, Espn (Espin) and Pkd2l1 were identified in the three studies (Table S4).
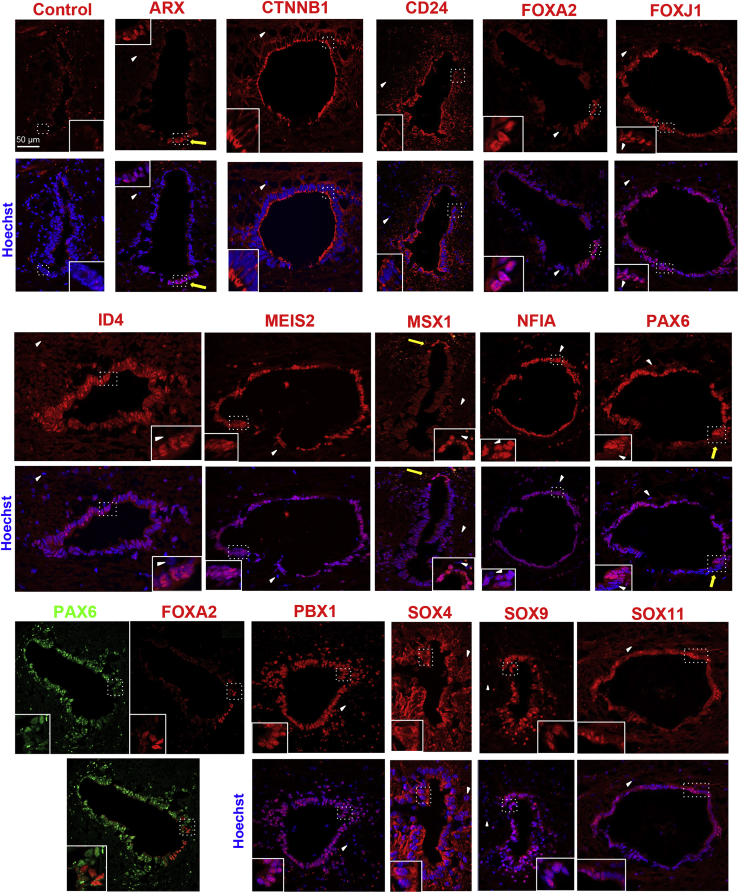
Figure 2.
Human EZ Characterization
IF of the indicated proteins in the human EZ (patient aged 17 years). White arrowheads show negative cells indicative of the staining specificity. Yellow arrows show ventral ARX+ cells, dorsal MSX1+ cells, and ventral PAX6− cells. Note that SOX4 is mostly cytoplasmic. Images are oriented with ventral part at bottom. Labelings were performed at the thoracic (control, ARX, CD24, FOXA2, MSX1, SOX4, SOX9) or lumbar level (CTNNB1, FOXJ1, ID4, MEIS2, NFIA, PAX6, PBX1, SOX11). These images are representative of at least eight sections. Scale bar, 50 μm (applies to all images).
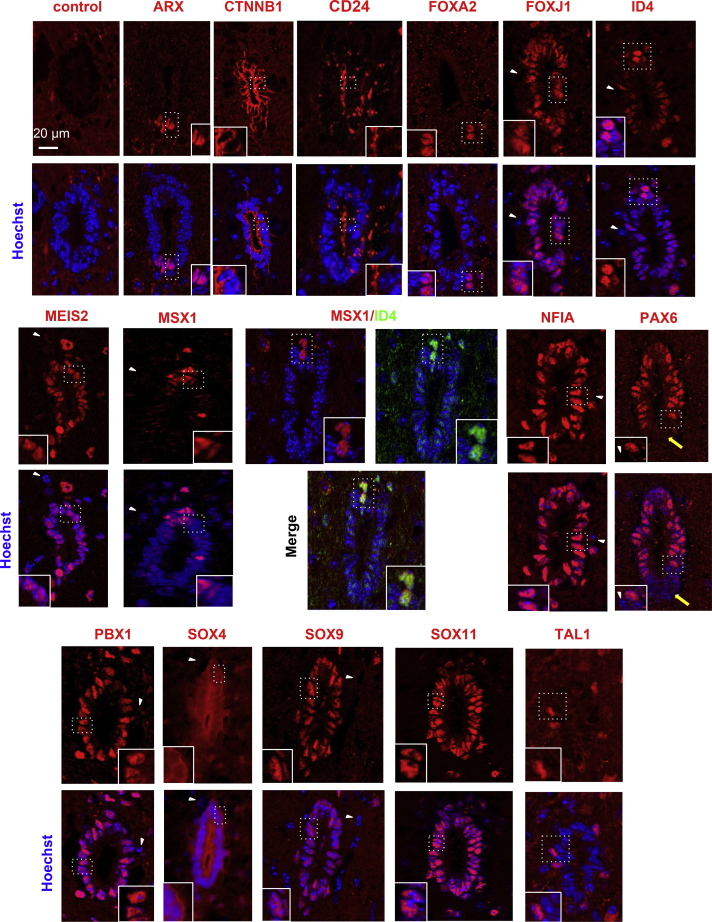
Figure 3.
Mouse EZ Characterization
IF for the indicated proteins in the adult mouse EZ (lumbar level). Images are oriented with ventral part at bottom. The yellow arrows on PAX6 staining show negative cells in the ventral part. Note the higher expression of ID4 in cells localized in the dorsal part, which also express MSX1. FOXA2+ and TAL1+ cells are localized in a subependymal position and express PKD2L1 a marker specific for CSF-N (Figure S2). Note that SOX4 is mostly cytoplasmic. These images are representative of ten sections per animal, n = 4 mice analyzed. Scale bar, 20 μm (applies to all images).
Figure 1.
RNA Profiles of the Mouse and Human EZ
(A) Aspect of EZ in the mouse (mEZ, thoraco level) and human samples (hEZ1 and hEZ2, thoracolumbar level). A lumen was present in mice and in the two human (aged 17 and 46 years) samples. Microdissected EZ and peri EZ regions are delimited with dotted circles. Scale bars, 100 μm.
(B) Heatmap of hierarchical clustering of genes expressed in EZ and peri EZ (pEZ) regions in the four mouse and two human samples.
(C) Volcano plots of genes whose expression is enriched in the mouse and human EZ (fold change ≥2).
(D) Venn diagram of genes enriched in the mouse and human EZ.
(E) Genes enriched for ciliogenesis, smoothened pathway (Smo.), division, and transcription factors in the mouse and human EZ. Expression of genes in red was subsequently confirmed at the protein level (Figures 2 and 3).
(F) Microdissected subregions of the mouse EZ.
(G) Genes enriched (top 9) in the ventral and dorsal EZ (full lists are in Table S5).
Table 1.
Three Lists of Genes (Top 15) Enriched in Mouse, Human, and Mouse and Human EZ
| Top 15 Human EZ-Specific Genes |
Top 15 Mouse EZ-Specific Genes |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Description | hEZ (log2) | Peri hEZ (log2) | Fold Change | p Value | Gene | Description | mEZ (log2) | Peri mEZ (log2) | Fold Change | p Value |
| C7orf57 | chromosome 7 open reading frame 57 | 10.9 | 4.9 | 61.8 | 0.0013 | Rsph4a | radial spoke head 4 homolog A | 10.9 | 3.8 | 133.1 | 2.32 × 10−13 |
| VWA3B | von Willebrand factor A domain-containing 3B | 11.4 | 5.7 | 52.8 | 7.43 × 10−5 | 1500015O10Rik | RIKEN cDNA 1500015O10 gene | 11.1 | 4.0 | 132.7 | 1.38 × 10−12 |
| NEK5 | NIMA-related kinase 5 | 11.1 | 5.5 | 47.9 | 6.43 × 10−5 | Stoml3 | stomatin (Epb7.2)-like 3 | 10.4 | 3.5 | 121.7 | 8.47 × 10−13 |
| CD36 | CD36 molecule | 10.5 | 5.0 | 45.8 | 3.50 × 10−5 | 1700007K13Rik | RIKEN cDNA 1700007K13 gene | 11.1 | 4.2 | 120.3 | 2.66 × 10−11 |
| EFHB | EF-hand domain family, member B | 11.2 | 5.7 | 44.0 | 0.0004 | Slc26a3 | solute carrier family 26, member 3 | 11.2 | 4.4 | 110.5 | 2.05 × 10−12 |
| MYLK3 | myosin light-chain kinase 3 | 11.4 | 6.0 | 42.6 | 0.0044 | Tnnc2 | troponin C2, fast | 10.1 | 3.4 | 105.2 | 8.39 × 10−5 |
| CCDC39 | coiled-coil domain-containing 39 | 10.0 | 4.6 | 42.4 | 0.0001 | 2810047C21Rik1 | RIKEN cDNA 2810047C21 | 9.8 | 3.2 | 103.2 | 4.85 × 10−11 |
| ODF3B | outer dense fiber of sperm tails 3B | 9.9 | 4.5 | 41.5 | 0.0008 | Chil3; Chil4 | chitinase-like 3; chitinase-like 4 | 11.5 | 4.8 | 102.2 | 4.87 × 10−5 |
| ZBBX | zinc finger, B-box domain-containing | 10.6 | 5.3 | 41.3 | 2.38 × 10−5 | C1qtnf3 | C1q and tumor necrosis factor related protein 3 | 10.7 | 4.1 | 97.9 | 1.63 × 10−11 |
| CCDC114 | coiled-coil domain-containing 114 | 11.3 | 6.0 | 40.3 | 0.0005 | Cfap161 | cilia- and flagella-associated protein 161 | 10.5 | 3.9 | 95.3 | 5.65 × 10−13 |
| FAM216B | family with sequence similarity 216, member B | 9.9 | 4.5 | 39.8 | 0.0012 | Fam183b | family with sequence similarity 183, member B | 11.5 | 4.9 | 91.8 | 1.06 × 10−11 |
| CFAP43 | cilia- and flagella-associated protein 43 | 11.3 | 6.0 | 39.1 | 0.0003 | Gm11992 | predicted gene 11.992 | 9.8 | 3.3 | 91.6 | 7.02 × 10−13 |
| FMO3 | flavin-containing monooxygenase 3 | 9.9 | 4.6 | 38.9 | 0.0014 | Iqca | IQ motif containing with AAA domain | 11.0 | 4.5 | 90.4 | 1.12 × 10−10 |
| CFAP70 | cilia- and flagella- associated protein 70 | 11.3 | 6.0 | 38.5 | 0.0023 | Capsl | calcyphosin-like | 9.6 | 3.1 | 87.3 | 5.95 × 10−12 |
| SPATA17 | spermatogenesis-associated 17 | 10.3 | 5.1 | 37.1 | 0.0003 | Acta1 | actin, alpha 1, skeletal muscle | 10.7 | 4.2 | 85.5 | 0.0002 |
| TOP15 Human and mouse EZ-specific genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Description | hEZ (log2) | peri hEZ (log2) | Fold Change | p Value | mEZ (log2) | peri mEZ (log2) | Fold Change | p Value |
| CCDC39 | coiled-coil domain containing 39 | 10.0 | 4.6 | 42.4 | 0.0001 | 9.6 | 3.5 | 68.5 | 9.13 × 10−12 |
| ODF3B | outer dense fiber of sperm tails 3B | 9.9 | 4.5 | 41.5 | 0.0008 | 10.5 | 4.4 | 68.7 | 1.01 × 10−10 |
| RSPH4A | radial spoke head 4 homolog A (Chlamydomonas) | 9.8 | 4.9 | 30.0 | 0.0004 | 10.9 | 3.8 | 133.1 | 2.32 × 10−13 |
| ARMC4 | armadillo repeat containing 4 | 10.3 | 5.1 | 36.7 | 3.27 × 10−5 | 9.9 | 4.1 | 58.9 | 9.43 × 10−12 |
| CAPSL | calcyphosine-like | 11.3 | 6.4 | 31.0 | 0.0001 | 9.6 | 3.1 | 87.3 | 5.95 × 10−12 |
| MNS1 | meiosis-specific nuclear structural 1 | 11.0 | 6.1 | 30.7 | 0.0014 | 10.4 | 4.2 | 72.3 | 3.96 × 10−13 |
| DYNLRB2 | dynein, light chain, roadblock-type 2 | 11.2 | 6.2 | 30.6 | 0.0007 | 11.3 | 5.1 | 72.4 | 1.63 × 10−12 |
| TTC29 | tetratricopeptide repeat domain 29 | 9.7 | 4.9 | 28.4 | 6.06 × 10−5 | 9.5 | 3.4 | 67.9 | 8.98 × 10−13 |
| TEKT1 | tektin 1 | 9.8 | 5.0 | 27.3 | 0.0004 | 10.1 | 4.1 | 64.7 | 2.59 × 10−11 |
| WDR63 | WD repeat domain 63 | 10.0 | 5.3 | 26.3 | 0.0005 | 9.6 | 3.5 | 66.4 | 2.21 × 10−12 |
| ARMC3 | armadillo repeat containing 3 | 9.7 | 5.0 | 26.0 | 0.0002 | 9.1 | 2.9 | 73.2 | 4.72 × 10−11 |
| ZMYND10 | zinc finger, MYND-type containing 10 | 9.0 | 4.5 | 23.5 | 0.0002 | 9.8 | 3.8 | 62.8 | 5.05 × 10−12 |
| STOML3 | stomatin (EPB72)-like 3 | 9.3 | 4.8 | 23.4 | 7.92 × 10−5 | 10.4 | 3.5 | 121.7 | 8.47 × 10−13 |
| SPEF2 | sperm flagellar 2 | 9.3 | 4.8 | 22.0 | 1.46 × 10−5 | 7.2 | 3.2 | 15.6 | 2.52 × 10−9 |
| CFAP52 | cilia and flagella associated protein 52 | 12.0 | 7.6 | 21.3 | 0.0013 | 10.6 | 4.3 | 81.3 | 1.07 × 10−11 |
| Top 15 Human and Mouse EZ Transcription Factors | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Description | hEZ (log2) | Peri hEZ (log2) | Fold Change | p Value | mEZ (log2) | Peri mEZ (log2) | Fold Change | p Value |
| ARX | aristaless-related homeobox | 8.6 | 4.6 | 16.4 | 0.0012 | 6.7 | 3.3 | 10.6 | 2.15 × 10−9 |
| RFX2 | regulatory factor X, 2 | 9.6 | 5.6 | 16.2 | 0.0018 | 7.6 | 3.5 | 17.3 | 5.10 × 10−8 |
| SOX6 | SRY box 6 | 10.4 | 8.3 | 4.5 | 0.0016 | 9 | 5.3 | 13.1 | 1.84 × 10−8 |
| PAX6 | paired box 6 | 9.7 | 7.5 | 4.5 | 0.0385 | 7.6 | 3.8 | 13.6 | 4.18 × 10−10 |
| NFIB | nuclear factor I/B | 9.4 | 7.2 | 4.4 | 0.0582 | 9.3 | 5.7 | 12.8 | 1.18 × 10−8 |
| ID4 | inhibitor of DNA binding 4 | 11.6 | 9.5 | 4.4 | 0.0724 | 11.8 | 8.5 | 10.1 | 2.79 × 10−8 |
| MYB | v-myb avian myeloblastosis viral oncogene homolog | 6.4 | 4.3 | 4.3 | 0.0001 | 10.2 | 4.4 | 59 | 1.48 × 10−11 |
| NR4A3 | nuclear receptor subfamily 4, member 3 | 6.3 | 4.3 | 4 | 0.0037 | 11.2 | 5.7 | 45.6 | 1.02 × 10−11 |
| REST | RE1-silencing transcription factor | 9.1 | 7.2 | 3.8 | 0.0032 | 7.1 | 4.1 | 8 | 2.53 × 10−9 |
| KDM3A | lysine (K)-specific demethylase 3A | 8.3 | 6.5 | 3.6 | 0.0262 | 8.9 | 5.8 | 8.9 | 1.86 × 10−9 |
| SOX2 | SRY box 2 | 12.5 | 10.7 | 3.5 | 0.0301 | 12.1 | 9.2 | 7.3 | 3.92 × 10−8 |
| SOX9 | SRY box 9 | 11.5 | 9.8 | 3.4 | 0.0255 | 9 | 4.4 | 24.2 | 2.16 × 10−10 |
| NR2F1 | nuclear receptor subfamily 2, member 1 | 11.4 | 9.7 | 3.3 | 0.0165 | 11 | 7.8 | 8.9 | 1.46 × 10−9 |
| SALL1 | spalt-like transcription factor 1 | 11.5 | 9.9 | 3.1 | 0.0278 | 9.9 | 4.6 | 38 | 2.76 × 10−10 |
| RFX3 | regulatory factor X, 3 | 5.4 | 3.8 | 2.9 | 0.0031 | 9.2 | 5.2 | 16.5 | 1.01 × 10−9 |
| Top 15 Human and Mouse EZ Transport-related genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Description | hEZ (log2) | Peri hEZ (log2) | Fold Change | p Value | mEZ (log2) | Peri mEZ (log2) | Fold Change | p Value |
| CFTR | cystic fibrosis transmembrane conductance regulator | 9.5 | 5.1 | 21.2 | 3.86 × 10−6 | 8.5 | 3.4 | 34.8 | 6.82 × 10−11 |
| SLC44A1 | solute carrier family 44, member 1 | 9.5 | 7.3 | 4.5 | 0.0082 | 8.9 | 6.5 | 5.3 | 2.58 × 10−8 |
| OCA2 | oculocutaneous albinism II | 8.2 | 6.2 | 4.1 | 0.0026 | 6.2 | 4.0 | 4.6 | 6.24 × 10−6 |
| SLC15A2 | solute carrier family 15, member 2 | 8.2 | 6.3 | 3.8 | 0.0069 | 7.5 | 4.7 | 7.1 | 0.0003 |
| SLC38A6 | solute carrier family 38, member 6 | 7.5 | 5.6 | 3.6 | 0.0842 | 7.0 | 5.5 | 3.0 | 5.09 × 10−5 |
| SLC40A1 | solute carrier family 40, member 1 | 10.2 | 8.6 | 3.1 | 0.0473 | 7.2 | 5.8 | 2.7 | 0.0083 |
| LRP6 | low-density lipoprotein receptor-related protein 6 | 10 | 8.4 | 3 | 0.0099 | 7.7 | 6.4 | 2.4 | 0.0016 |
| FOLR1 | folate receptor 1 (adult) | 6.6 | 5.1 | 2.9 | 0.0164 | 9.0 | 4.3 | 25.0 | 4.04 × 10−11 |
| SLC16A9 | solute carrier family 16, member 9 | 8.7 | 7.2 | 2.9 | 0.006 | 8.1 | 5.6 | 5.4 | 8.88 × 10−7 |
| SLC22A4 | solute carrier family 22, member 4 | 9.3 | 7.8 | 2.9 | 0.0151 | 7.0 | 4.0 | 7.7 | 3.30 × 10−7 |
| SLC35D1 | solute carrier family 35 D1 | 8.5 | 7.1 | 2.6 | 0.0042 | 7.7 | 4.6 | 8.4 | 6.28 × 10−9 |
| SLC12A7 | solute carrier family 12, member 7 | 8.6 | 7.4 | 2.4 | 0.0143 | 8.1 | 5.3 | 7.0 | 1.03 × 10−7 |
| SLC2A12 | solute carrier family 2, member 12 | 6.8 | 5.6 | 2.4 | 0.0335 | 8.4 | 4.8 | 12.0 | 1.18 × 10−9 |
| SLC30A5 | solute carrier family 30, member 5 | 5.8 | 4.6 | 2.3 | 0.0341 | 8.5 | 7.0 | 2.7 | 0.0018 |
| ABCC1 | ATP-binding cassette, member 1 | 9.3 | 8.2 | 2.2 | 0.0194 | 8.2 | 5.2 | 7.9 | 5.16 × 10−9 |
The table also shows lists of transporters and transcription factors (top 15) enriched in the mouse and human EZ. mEZ, mouse EZ; hEZ, human EZ.
We and others have reported that the dorsal and ventral parts of the mouse EZ have distinctive features such as the presence of radial cells expressing NES protein (Alfaro-Cervello et al., 2012, Becker et al., 2018, Hamilton et al., 2009) and the preferential expression of the TF ZEB1 in the dorsal part (Sabourin et al., 2009). Further exploration of regional gene expression was done by microdissecting the dorsal, lateral, and ventral parts of the mouse EZ (Figure 1F, n = 4 mice). Only few regionally-expressed genes were identified (Figure 1G and Table S5). In the ventral part, we found a strong expression of three genes (Arx, Foxa1, Sulf1) involved in SHH signaling, a pathway known to be crucial for ventral specification of spinal cord during development. The proteoglycan Decorin (Dcn) was also expressed in the ventral part, a gene positively regulated by SHH signaling (Ingram et al., 2008). To identify the cellular origin of Dcn, we performed double immunofluorescence (IF) for DCN and PKD2L1, a specific marker for CSF-N (Becker et al., 2018) (Figure S2). Indeed, co-localization indicated that these neurons express a high level of DCN, a result supported by recent single cell RNA-sequencing (RNA-seq) databases (Zeisel et al., 2018) (Figure S5A). Contrasting with ventral SHH signaling, a robust expression of two bone morphogenetic protein (BMP) morphogens (namely Bmp6 and Gdf10/Bmp3b) involved in the dorsal patterning of the developing spinal cord (Wilson and Maden, 2005) were detected in the dorsal EZ (Figure 1G and Table S5). Two genes, Prokr2 and Thbs2, modulating neural stem cell fate in the SVZ (Benner et al., 2013, Prosser et al., 2007), were also enriched in the dorsal part. The dorsal or ventral expression of Arx, Bmp6, Dcn, Foxa1, Gdf10, Prokr2, Sulf1, and Thbs2 in the EZ was confirmed in the Allen brain atlas (Figure S3A).
The Mouse and Human EZ Show a Conserved Dorsal-Ventral Regionalization of Transcription Factor Expression
One hundred twenty-one TFs were found more specifically-expressed (fold change EZ/periEZ ≥ 2) in the human and mouse EZ (Figure 1E). Examples of expression in the Allen brain atlas are illustrated in Figure S1B. IF confirmed the presence of corresponding proteins for some of them (ARX, FOXJ1, ID4, MEIS2, MSX1, NFIA, PAX6, PBX1, SOX4, SOX9, and SOX11) both in human (17-year-old) and mice (Figures 2 and 3). The histological quality of the second human spinal cord (46-year-old) was reduced compared with that of the first patient, although we could confirm protein expression for FOXJ1, ID4, MEIS2, NFIA, and PAX6 in this sample (Figure S4A). Unexpectedly, some of these TFs showed a dorsal-ventral asymmetric expression. Both in human (17-year-old) and in mouse, PAX6 protein stained dorsal and lateral cells while ventral cells were negative (Figures 2 and 3), which was also observed in the GENSAT gene expression atlas (Figure S1B). Another clear regionalized-expression was found for MSX1 protein in mouse and in human (17-year-old). In the human sample, MSX1 was weakly expressed by ependymal cells but cells in the dorsal part had a much stronger staining (Figure 2). In mice, MSX1 protein was confined to a few cells constituting the roof of the EZ (Figure 3), which was confirmed in gene expression atlases (Figure S3B). These MSX1+ cells typically exhibit a higher level of ID4 staining in mice (Figure 3). Contrasting with the dorsally-expressed proteins, we detected a ventral expression of ARX, a TF expressed by the floor-plate during spinal cord development. ARX protein was confined to a group of ventral cells both in human and mouse EZ (Figures 2 and 3). This led us to investigate by IF the expression of FOXA2, another TF involved in floor plate formation (Cho et al., 2014). Foxa2 specificity for the EZ was under our selected threshold in mice and in human (fold change EZ/periEZ = 1.7 and 1.3, respectively) however the FOXA2 protein was detected in a group of ventral and ventral/ventro-lateral cells in human and mice respectively (Figures 2 and 3). In human, the same population of ventral cells expressed ARX and FOXA2 (Figure S4B) whereas in mice, FOXA2 was expressed by ventral CSF-N expressing DCN and PKD2L1 but did not express ARX (Figure S2). In addition to these evolutionarily-conserved TFs, we also detected genes whose expression appears to be specifically expressed in the mouse or the human EZ (Table S3). This was checked at the protein level for Tal1 (also known as Scl), a well-known TF involved in hematopoiesis but also expressed during spinal cord development (Smith et al., 2002). IF for TAL1 revealed the presence of positive subpopulations in the mouse EZ (Figure S2) which co-labeled with PDK2L1, thus identifying these cells as CSF-N. This was confirmed by single cell CNS RNA-seq database (Zeisel et al., 2018) (Figure S5A). In contrast, no convincing staining for TAL1 and PDK2L1 proteins was detected in the two human spinal cord EZs (not shown), and DCN labeling was restricted to vessels in the parenchyma (Figure S5B) as expected from the literature (Jarvelainen et al., 2015). A recent single cell CNS RNA-seq database (Zeisel et al., 2018) identified six markers for mouse CSF-N (Crct1, Dcn, Espn, Pkd1l2, Pkd2l1, and Pdzk1ip1), which we found enriched at the transcriptional level in the mouse EZ but not in the human EZ (Figure S5C).
Collectively, these results indicate a conserved dorsal-ventral regionalization of TF expression in the mouse and human EZ; however, no evidence could be found for the presence of CSF-N in human.
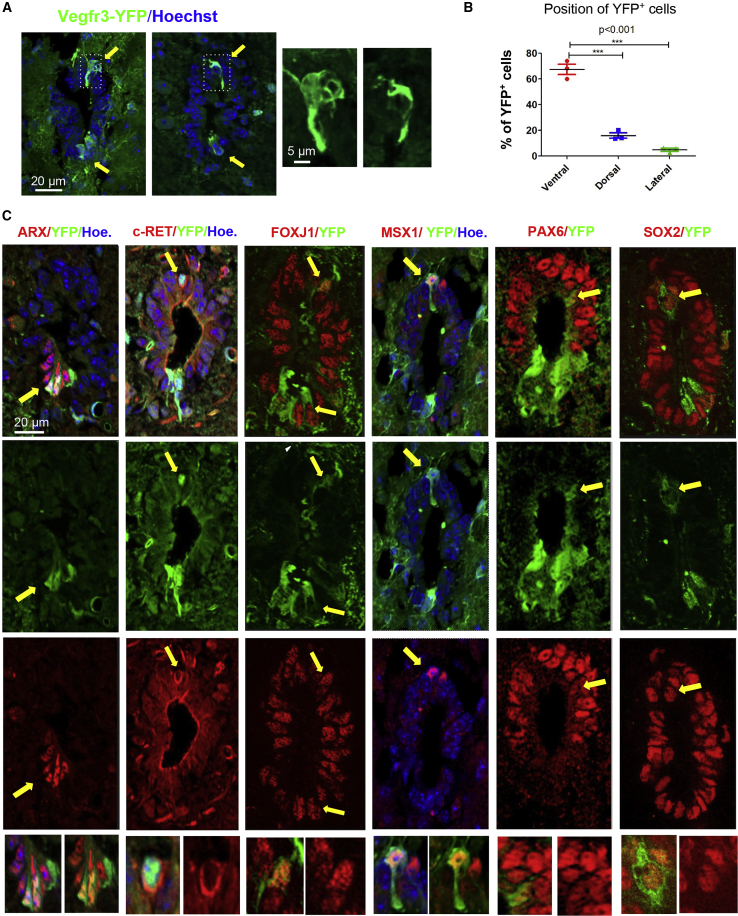
Vegfr3-YFP Transgenic Mice Reveal Distinct MSX1+ and ARX+ Cells in the Roof and Floor of EZ
The regionalized expression of TFs in the EZ was indicative of specific cells located in the roof and floor of the EZ. This was also suggested by the specific expression of the immature neural marker NES in these regions (Becker et al., 2018). The Vegfr3-YFP transgenic mouse has been used to detect subpopulations of neural precursor cells in the neurogenic SVZ (Calvo et al., 2011), which prompted us to use it in the spinal cord EZ context. Indeed, subpopulations of EZ cells expressing YFP were observed in all sections and were specifically enriched at the roof and floor portions (Figures 4A and 4B). These cells were positive for TFs strongly expressed by ependymal cells such as FOXJ1 and SOX2 while only the dorsal YFP+ cells express PAX6 (Figure 4C). In addition, in the roof part, 67% (41 among 61 cells, two mice) of YFP+ cells expressed the MSX1 TF (Figure 4C), which was further confirmed by generating a double transgenic mouse (Vegfr3-YFP × Msx1-Tomato, see below, Figure 6F) in which approximately 77% of the dorsal YFP+ cells were also Tomato+ (37 among 48 cells, two mice). It was previously reported that the c-RET receptor is present in a subpopulation of radial dorsal cells in the EZ (Pfenninger et al., 2011), and we observed that 71% of YFP+ (20 among 28 cells, two mice) were positive for this receptor (Figure 4C). In the EZ floor, 90% of YFP+ cells (60 among 66 cells, three mice) expressed the ARX TF (Figure 4C). Finally, some dorsal Vegfr3-YFP+ cells were clearly positive for GFAP and NES intermediate filaments (Figure S6A), whereas we could not establish this with confidence for ventral cells. These results support the existence of distinct EZ roof and floor cells expressing Vegfr3 and specific TFs.
Figure 4.
EZ in Vegfr3-YFP Mice
(A) Representative images of the spinal cord EZ (lumbar level) in Vegfr3-YFP mice (IF anti-GFP). Images are oriented with ventral part at bottom. YFP+ cells are mainly present in the dorsal and ventral regions (yellow arrows). Boxed areas show dorsal cells sending a process toward the lumen. These images are representative of at least 20 sections per animal (n = 3 mice analyzed). Ventral YFP+ cells are present in >95% of sections whereas dorsal YFP+ cells are present in approximately 25% of examined sections.
(B) Quantification of YFP+ cells in the lumbar EZ (187 YFP+ cells counted) indicated a preferential ventral and dorsal localization. One-way ANOVA + Tukey’s post test (n = 3 mice).
(C) Phenotypic characterization of YFP+ cells with indicated protein (images are representative of 20 sections, n = 3 mice). Yellow arrows show double-positive cells. Images at the bottom are high magnification of arrow-pointed areas. Hoe., Hoechst. Scale bar, 20 μm (applies to all images).
Figure 6.
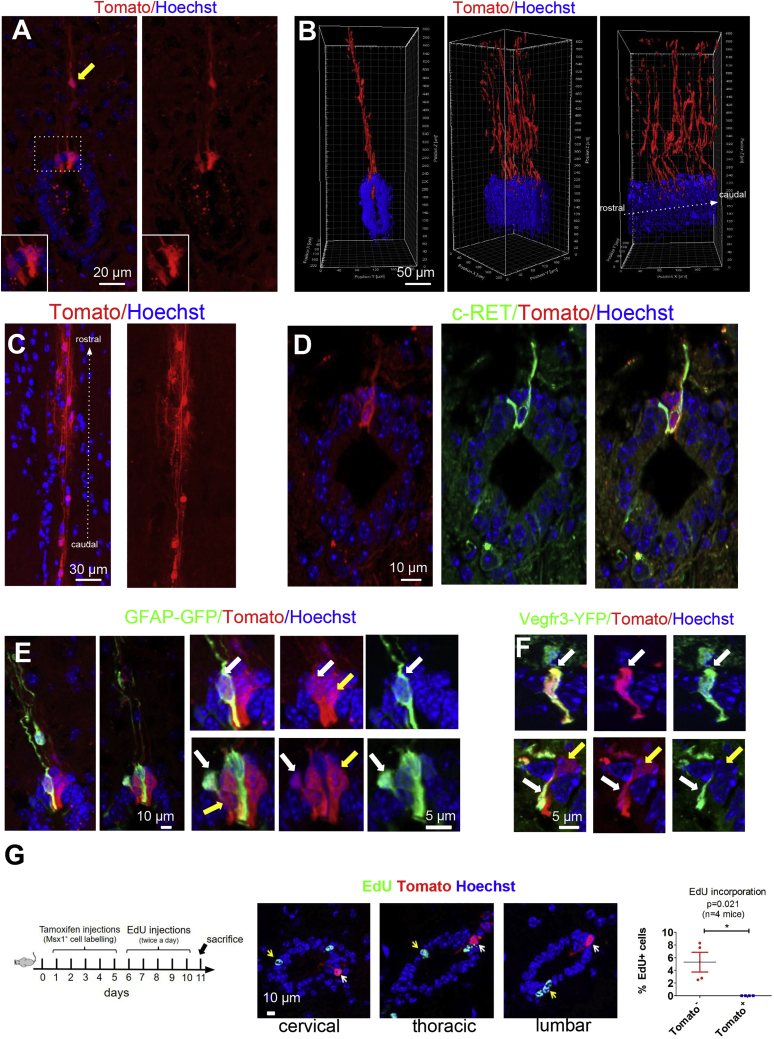
Characterization of Dorsal MSX1+ Cells
(A) Radial morphology of MSX1+ cells observed in Msx1-CreERT2/Rosa-Tomato mice. Yellow arrow shows rare radial Tomato+ cells outside EZ. Images are oriented with ventral part at bottom.
(B) 3D reconstruction of Tomato+ cells (coronal, intermediate, and lateral views).
(C) Dorsal view of the dorsal EZ region showing rostral-caudal oriented Tomato+ processes.
(D) Expression of c-RET receptor in Tomato+ cells (n = 10 sections).
(E and F) Representative images (n = 20 sections, two mice) of the EZ in double transgenic hGFAP-GFP/Msx1-CreERT2/Rosa-Tomato (E) and Vegfr3-YFP/Msx1-CreERT2/Rosa-Tomato (F) mice. White arrows indicate double-positive cells and yellow arrows Tomato+-only cells.
(G) EdU incorporation (5 days) in the EZ. Images show EdU+ cells (yellow arrows) not positive for Tomato (white arrows) in Msx1-CreERT2/Rosa-Tomato mouse sections. Quantifications are provided on right-hand graph (112 spinal cord sections, 5,120 cells examined, unpaired t test, n = 4 mice).
Roof MSX1+ and Floor ARX+ Ependymal Cells are Born Early during Development
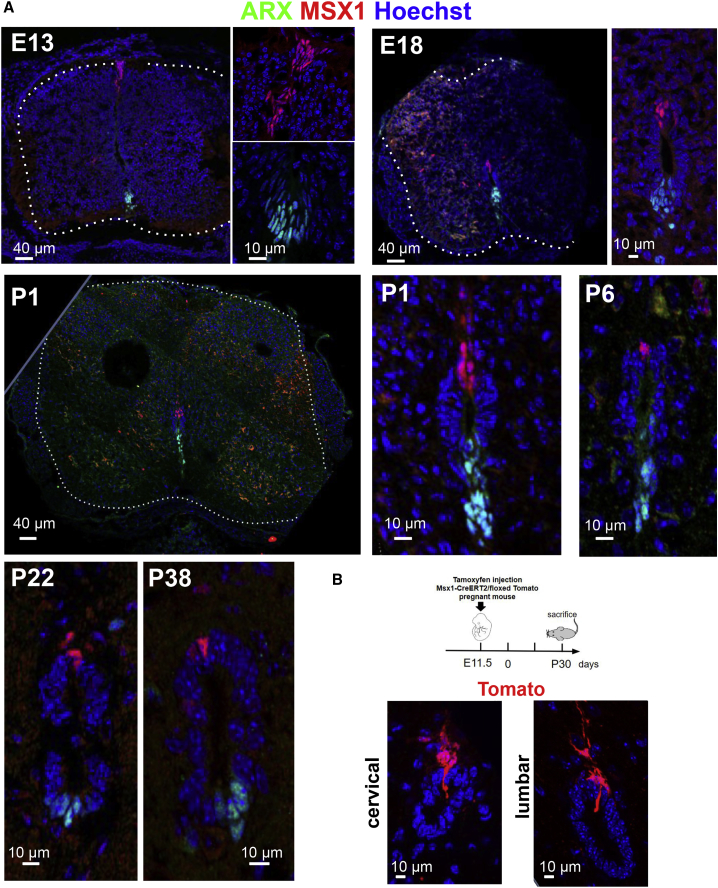
We next questioned the developmental origin of MSX1+ and ARX+ cells by performing IF at multiple developmental stages (embryonic day 13 [E13], E18, postnatal day 1 [P1], P6, P22, and P38) (Figure 5A). At E13, a group of MSX1+ cells was detected at the dorsalmost part of the developing spinal cord while a group of ARX+ cells were present in the ventralmost region. These cells, situated at positions corresponding respectively to the embryonic floor and roof plates, had elongated nuclei oriented along the dorsal-ventral axis, evocating migration. At E18, ARX+ and MSX1+ cells appeared to have migrated centrally to delimit a small group of cells presumptive of the adult EZ. At P1 and P6, the number of ARX+ and MSX1+ cells was reduced and at P22 and P38 only a few cells remained in the roof and floor of the EZ (Figure 5A). To analyze further the developmental origin of adult EZ MSX1+ cells, we used a genetic tracing approach based on a Msx1-CreERT2/Rosa-Loxed Tomato transgenic line (hereafter referred to as Msx1-Tomato [Lallemand et al., 2013]). Tamoxifen injection in adult mice revealed the presence of Tomato+ cells residing in the EZ roof (Figures 6A and S6B), which validated this tool. Tomato+ cells were very rarely observed in the ventral part of the EZ or in the parenchyma (not shown). Tamoxifen was then injected in pregnant transgenic mice at E11.5 to permanently label embryonic MSX1+ cells and explore their fate (Figure 5B). Analysis of sections taken from cervical and lumbar spinal cord levels of P30 mice (n = 2 mice) revealed the constant presence of Tomato+ cells situated in the EZ roof (Figure 5B). Altogether, these data demonstrate an early embryonic origin of ARX+ and MSX1+ spinal cord EZ cells.
Figure 5.
MSX1 and ARX Expression in the Developing Spinal Cord
(A) IF for MSX1 and ARX during spinal cord embryonic development (E13 and E18) and postnatal stages (P1, P6, P22, P38). All images are oriented with ventral part at bottom. Images are representative of ten sections (n = 2 embryos and pups analyzed per stage).
(B) Representative images (n = 20 sections each levels, two mice) of the EZ of a P30 Msx1-CreERT2/Rosa-Tomato mouse derived from an embryo subjected to tamoxifen at E11.5.
Roof MSX1+ Cells Are Radial Quiescent Cells
We previously reported an enrichment of neurosphere-forming cells in the dorsal half of the EZ (Sabourin et al., 2009). This was associated with a high content of GFAP+ radial glial-like cells. As MSX1+ cells were found almost exclusively in the roof of the EZ, further characterization of these cells was done. This was also motivated by the reported expression of Msx1/2 in some stem cells during eye development (Bélanger et al., 2017) and its role in promoting regeneration of amputated tail including spinal cord (Beck et al., 2003) in Xenopus. The intense fluorescence observed in Msx1-Tomato mice enabled morphological characterization of these cells, notably using clarification of the whole spinal cord and 3D reconstruction. Tomato+ cells have a long radial morphology and send their process toward the pial surface (Figures 6A, 6B, and S6B). Their soma make contact with the lumen, but radial Tomato+ cells can also be observed more dorsally and at distance from the EZ (Figure 6A, yellow arrow). Horizontal sections also revealed the existence of Tomato+ processes running longitudinally along the EZ roof (Figure 6C).
By performing IF for c-RET, FOXJ1, GFAP, NES, and SOX2, we observed that most Tomato+ cells also express these markers (Figures 6D and S6C). We next crossed Msx1-Tomato mice with hGFAP-GFP transgenic mice, which have been widely used to purify adult neural stem cells (Nolte et al., 2001). Contrasting with GFAP immunostaining (Figure S6C), only a small fraction of Tomato+ cells (15%, 8 double-positive cells among 54 Tomato+ cells; n = 2 mice) were GFP+ (Figure 6E). Often GFP+ and Tomato+ cells were found to be associated, suggesting close interactions between them. We then explored the proliferation rate of Tomato+ cells by injecting tamoxifen for 5 days in Msx1-Tomato mice to label MSX1+ cells, then 5-ethynyl-2′-deoxyuridine (EdU) injections were performed for 5 additional days (twice a day) before sacrificing on day 11 (Figure 6G). As previously reported, few EdU+ cells were found in the EZ (approximately one cell per section), however no Tomato+ EdU+ could be observed (Figure 6G). This indicates that MSX1+ cells are quiescent or proliferate at a much slower rate compared with the other ependymal cells.
Discussion
In this article, RNA profiling was used to generate a cellular and molecular resource for the adult human and mouse spinal cord EZ. Our findings reveal important and new characteristics of this poorly defined region.
In accordance with the presence of neural stem cells in the EZ, this region shows enrichment for smoothened/SHH and Hippo/YAP signaling genes, which are involved in stem cell maintenance (Alvarez-Buylla and Ihrie, 2014, Mo et al., 2014). Echoing their localization at the interface between CSF and the spinal cord parenchyma, ependymal cells also highly express genes involved in transport and ciliogenesis including three TFs of the RFX family and FOXJ1 (Figure 1E). Compared with brain, spinal cord ependymal cells proliferate (Alfaro-Cervello et al., 2012, Pfenninger et al., 2011), which is also reflected at the RNA level by enrichment for genes involved in cell division (Figure 1E; Tables S1 and S2). A set of 120 TFs conserved between mice and humans was identified and the expression at the protein level was confirmed for 12 of them. MEIS2 and PBX1 expressions are particularly interesting, as these TFs dimerize and are essential regulators of adult SVZ neurogenesis (Grebbin et al., 2016). Ependymal cells also express high levels of NFIA and SOX9, two TFs involved in gliogenesis during development (Kang et al., 2012). Post lesion, these cells mostly generate astrocytes (Barnabé-Heider et al., 2010) and the expression of NFIA and SOX9 may be responsible for their glial fate restriction.
One unexpected observation was the conserved and regional expression of four homeodomain-containing TFs in the niche, namely ARX, FOXA2, MSX1, and PAX6. This situation is reminiscent of the developing spinal cord neuroepithelium where cells along the dorsal-ventral axis express different combinations of homeogenes (Wilson and Maden, 2005). Both in human and mouse, ARX and FOXA2 are expressed by ventralmost cells that do not co-express PAX6. Of note, whereas FOXA2 was expressed by ventral CSF-N in mice (Figure S2 and Petracca et al., 2016), no such cells have been identified in human so far. The two other markers we identified in mice for these cells, DCN, an extracellular matrix protein (Jarvelainen et al., 2015), and TAL1, a hematopoietic transcription factor, were not observed in the human EZ. This indicates a divergent organization of the ventral EZ between rodents and humans. In the dorsal portion of the EZ, high MSX1 expression was restricted to few cells located in the EZ roof both in mouse and human. By tracing the origin of ARX+ and MSX1+ cells during spinal cord development in mice, we observed that they are already present at E13.5 in the spinal cord roof and floor plates. Their number is then reduced and they appear to migrate centrally to generate the roof and floor of the adult EZ. The early developmental origin (at least E11.5) of MSX1+ cells was further demonstrated using genetic tracing. Thus, in contradiction to what was previously established (Fu et al., 2003), ependymal cells are not entirely derived from the ventral neuroepithelium but also incorporate cells from the dorsal part of the developing spinal cord. Another distinguishing feature of the dorsal and ventral EZ cells is the expression of Vegfr3 (also known as Flt4, the receptor for VEGFC), which was observed using Vegfr3-YFP transgenic mice. Part of these Vegfr3-YFP+ cells express MSX1 and ARX. During brain development Vegfr3 is expressed in the ventricular zone and radial glial cells (Ward and Cunningham, 2015). It is also expressed in brain adult neural stem cells where it regulates their activation and proliferation (Calvo et al., 2011). A similar role for Vegfr3 may apply for adult spinal cord ependymal cells.
These results provide evidence for the persistence of embryonic floor and roof plate cells in the adult spinal cord niche. Recent studies, based on a Wnt-reporter mouse labeling dorsal neural tube cells (Xing et al., 2018) and on a Nato3-reporter mouse labeling floor plate cells (Khazanov et al., 2017), reached similar conclusions. During development, these floor and roof plate cells secrete morphogens such as SHH and BMP6, acting as growth factors and patterning signals (Wilson and Maden, 2005). Our results (Figure 1G) and expression atlas (Figure S3A) indicate that dorsal EZ cells express morphogen genes (Bmp6 and Gdf10). This suggests a specific role for these cells in the spinal cord niche, which warrants further exploration.
As the presence of MSX1+ cells has not been reported in the spinal cord EZ, we characterized these cells further and observed that they have distinct features compared with the other cells of the niche. They have a long radial morphology and express FOXJ1, GFAP, NES, and SOX2 proteins. They also express c-RET, the receptor for GDNF and NTN growth factors, which is important for hematopoietic stem cells and neural crest cells (Kubota et al., 2004). By crossing Msx1-CreERT2/Tomato and hGFAP-GFP transgenic mice, only a minority of Tomato+ cells were double-positive, thus revealing the presence of a cellular heterogeneity in the dorsal part of the niche and of MSX1+ cells.
The existence of subpopulations of quiescent neural stem cells has been reported in the hippocampus and the SVZ niches (Codega et al., 2014, Llorens-Bobadilla and Martin-Villalba, 2017, Llorens-Bobadilla et al., 2015). Importantly, data mining of single cell RNA-seq analysis performed in the SVZ revealed that Msx1 and Id4 are highly enriched in quiescent neural stem cells (Llorens-Bobadilla et al., 2015) (Figure S5D). These two TFs are regulated by the BMP signaling (Ramos and Robert, 2005, Ruzinova and Benezra, 2003) and, indeed, expression of Bmp6 and Bmp3b/Gdf10 is also restricted to SVZ quiescent stem cells (Figure S5D). A similar situation may also be present in the dorsal part of the spinal cord EZ. Indeed, we found that Bmp6, Bmp3b/Gdf10, and Msx1 are highly expressed in the dorsal part of the spinal cord EZ and that MSX1+ cells express a higher level of ID4 transcription factor (Figure 3). EdU incorporation also revealed that MSX1+ cells are less proliferative than the other cells of the niche (Figure 6G). This suggests that dorsal radial MSX1+ cells may behave as quiescent neural stem cells in the adult spinal cord. Further work is needed to support this hypothesis.
In summary, our results uncovered that the adult spinal cord EZ region is conserved, regionalized, and composed of a mosaic of cells with different embryonic origin and expressing different types of TFs. This corpus of knowledge on the organization and genes expressed in the EZ will help to explore this adult stem cell niche further and will also be useful to shed light on ependymoma, a rare type of tumor that can arise in the human spinal cord EZ.
Experimental Procedures
Human Samples
Human spinal cords were collected at the Montpellier Hospital from two organ-donor patients (17 [male] and 46 [female] years old, accidental death) in strict agreement with the French bioethics laws (articles L1232-1 and -6) and after approval by the French institution for organ transplantation. An informed consent from the families was obtained by the organ procurement organization for this study. Surgery was performed as described previously (Bauchet et al., 2013), and the thoracolumbar segments were immediately placed in liquid nitrogen before processing for microdissection and immunofluorescence.
Animals
Mice were handled following the guidelines of the Animal Care and Use Committee of the National Institute of Health and Medical Research (INSERM) who approved this study in accordance with the European Council directive (2010/63/UE) for the protection and use of vertebrate animals. Adult CD1 mice (3 months, Charles River, France) were used for microdissection, RNA profiling, and histology. Msx1-CreERT2/Rosa-Loxed Tomato transgenic line (Lallemand et al., 2013) was obtained from Y. Lallemand (Pasteur Institute, Paris). To induce recombination in Msx1-CreERT2 animals, we injected 100 μL of tamoxifen (Sigma, T5648, 20 mg/mL dissolved in corn oil) intraperitoneally for 4–5 days. hGFAP-GFP (Nolte et al., 2001) and Vegfr3-YFP (Calvo et al., 2011) transgenic mice were obtained from Prof. H. Kettenmann (MDC, Berlin) and Dr. J.L. Thomas (ICM, Paris), respectively.
Tissue Microdissection
For human and mouse, after collection the spinal cords were flash frozen in N2 without chemical fixation. Frozen sections (30 μm thick, T9-T10 thoracic part for mouse and thoracolumbar for human) were obtained at −23°C to prevent RNA degradation using a CM3050S microtome (Leica Microsystems, Wetzlar, Germany) and were mounted on PEN-membrane 1-mm glass slides (P.A.L.M. Microlaser Technologies, Bernried, Germany) that had been pretreated to inactivate RNase. Sections were then fixed in a series of pre-cooled ethanol baths (40 s in 95%, 75% and 30 s in 50%), stained with cresyl violet 1% for 30 s, and dehydrated in a series of pre-cooled ethanol baths (30 s in 50%, 75%, and 40 s in 95% and 100%). Immediately after dehydration laser microdissection was performed using a PALM MicroBeam microdissection system version 4.6 equipped with PALM RoboSoftware (P.A.L.M. Microlaser Technologies). Laser power and duration were adjusted to optimize capture efficiency, and microdissection was performed at 63× magnification. Samples were collected in adhesive caps (P.A.L.M. Microlaser Technologies). To limit RNA degradation, we collected samples for up to 15 min per slide and lysed microdissected tissue with 250 μL of lysis buffer (Promega, Madison, WI, USA). The samples were stored at −80°C until extraction was performed using the ReliaPrep RNA cell Miniprep System (Promega) according to the manufacturer's protocol and eluted with 14 μL of RNase-free water. The concentration of RNA was determined using Nanodrop 1000 and the integrity of RNA was determined using the RNA 6000 Pico Kit and Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The RNA integrity number was above 7/8.
RNA Profiling and Bioinformatics Analysis
RNA profiling was performed using Affymetrix microarray technology. Hybridization targets were obtained following a double-amplification procedure according to the protocol developed by Affymetrix (GeneChipTwo-Cycle Eukaryotic Target Labeling Assay; Affymetrix, USA). A hybridization mixture containing 10 μg of biotinylated cRNA was generated. The biotinylated cRNA was hybridized to HT_HG-U133_Plus_PM (human) and HT_MG-430_PM (mouse) Affymetrix microarrays. Four mouse and two human samples (EZ and peri EZ regions each, Figure 1A) were analyzed in total. The microarrays were scanned using the Affymetrix Gene Atlas scanner. The data files were generated with Affymetrix Expression Console v1.2.1 and gene expression data were normalized with the GC-RMA algorithm. Gene expression profiles were analyzed using the Affymetrix TAC 4.0 software (Transcriptome Analysis Console). The filter criteria were set to a linear fold change ≥2 between EZ and peri EZ regions. Gene lists were analyzed with DAVID Bioinformatics Resources 6.83 for gene enrichment analysis (Huang et al., 2008).
Human and Mouse Histology
Mice were anesthetized by intraperitoneal injection of sodium pentobarbital (50 mg/kg) and perfused intracardially with 10 mL of PBS followed by 50 mL of 4% formaldehyde-PBS solution (pH 7.0). After dissection, spinal cords were post-fixed in the same solution for 1 h at 4°C and cryopreserved by successive immersion in 10%, 20%, and 30% sucrose solutions in PBS for at least 6 h. Cervical, thoracic and lumbar parts of the spinal cord were cut, embedded in OCT medium, rapidly frozen in liquid N2-cooled isopentane, and cryosectioned (14 μm) (Leica apparatus). For developmental studies using time-mated embryos (Figure 5A), the day the plug was found was considered as E0.5. The embryos and pups (entire animal for P1, P6, and dissected spinal cord for P22) were fixed by direct immersion in 4% formaldehyde-PBS solution for 1 h then processed as for the adult spinal cord. P38 animals were perfused intracardially and post-fixed as for adults. To access the proliferation rate of Tomato+ cells (Figure 6G), we injected adult mice with EdU twice a day for 5 days (50 mg/kg), perfused them intracardially, and processed them for EdU staining (Baseclick kit).
For human, unfixed spinal cords at thoraco or thoracolumbar levels were directly cryosectioned (20 μm) and sections were immediately fixed by direct immersion in ice-cooled 4% formaldehyde-PBS solution for 20 min, followed by three washes with PBS.
Immunofluorescences were performed with primary antibodies (listed in Table S6) on sections permeabilized for 1 h with 0.1% Triton X-100 and 5% donkey serum. Secondary antibodies (Alexa 488- or Alexa 594-conjugated species-specific anti-mouse, -rabbit, or -goat) were purchased from The Jackson Laboratories. Incubations without primary antibody or with antibody recognizing antigens not present in the sections (monoclonal anti-DYKDDDDK tag or polyclonal antibodies against GFP) were used as negative controls. Nuclei (blue in all images) were stained with 1 μg/mL Hoechst for 10 min. The quality of staining was evaluated by two independent investigators (J.-P.H. and H.G. or C.R.). Images were taken using optical sectioning with structured illumination (Zeiss apotome microscope) or using a multi-photonic microscope (Zeiss LSM 7MP OPO) (Figure 6B). All presented images for mice are representative images, and the number of examined sections and animals are indicated in the figure legends. For human labelings, images are representative of at least eight sections for the first (17-year-old) patient and four sections for the second (46-year-old) patient. Clarification of mouse spinal cord was performed with protocol described in Tomer et al. (2014), and Imaris software was used for image treatments and 3D reconstruction.
Statistical Analysis
All experiments and stainings were performed at least twice, most of them three times. Data are represented as means ± standard error of mean. Statistical differences in experiments were analyzed with tests indicated in the figure legends (GraphPad Prism software). Significance is denoted by ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p ≤ 0.05.
Author Contributions
C.R., H.G., N.L., D.M., and J.-P.H. performed and analyzed most of the experiments. J.-P.H. wrote the article. F.E.P., H.N.N., S.A., S.M., L.J., and B.R. contributed to spinal cord histology. Y.G. performed clarification and 3D reconstruction of spinal cord. L.B. performed surgery for human spinal cord and V.R. provided human spinal cord tissues. C.-F.C, J.-L.T., Y.L., and E.H. provided reagents and mouse models as well as intellectual input.
Acknowledgments
This work was supported by grants from IRP (Switzerland), IRME (France), AFM (France), ANR EU ERANET Neuroniche (J.-P.H.), and ANR Brainwash (J.-L.T., L.J.). H.G. was supported by an AFM PhD fellowship. We thank all Montpellier biocampus facilities for help (RHEM, MRI RIO, RAM) and excellent technical work. We are very grateful to Dr. H Boukhaddaoui (imaging), Dr. C Duperray (cytometry), M. Maistre (laser microdissection facilities, Bordeaux), V. Pantesco (Affymetrix facilities), and M. Goussard/P. Guigue (animals) for providing technical expertise in this work. We warmly thank Prof. Morohashi (Kyushu University, Japan) for anti-Arx antibody. The authors declare that there is no conflict of interest regarding the publication of this article.
Published: April 25, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.04.001.
Accession Numbers
Data are available at the functional genomics data Gene Expression Omnibus (GEO: GSE118445).
Supplemental Information
References
- Adrian E.K., Jr., Walker B.E. Incorporation of thymidine-H3 by cells in normal and injured mouse spinal cord. J. Neuropathol. Exp. Neurol. 1962;21:597–609. doi: 10.1097/00005072-196210000-00007. [DOI] [PubMed] [Google Scholar]
- Alfaro-Cervello C., Soriano-Navarro M., Mirzadeh Z., Alvarez-Buylla A., Garcia-Verdugo J.M. Biciliated ependymal cell proliferation contributes to spinal cord growth. J. Comp. Neurol. 2012;520:3528–3552. doi: 10.1002/cne.23104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla A., Ihrie R.A. Sonic hedgehog signaling in the postnatal brain. Semin. Cell Dev. Biol. 2014;33:105–111. doi: 10.1016/j.semcdb.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnabé-Heider F., Göritz C., Sabelström H., Takebayashi H., Pfrieger F.W., Meletis K., Frisén J. Origin of new glial cells in intact and injured adult spinal cord. Cell Stem Cell. 2010;7:470–482. doi: 10.1016/j.stem.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Bauchet L., Lonjon N., Vachiery-Lahaye F., Boularan A., Privat A., Hugnot J.-P. Isolation and culture of precursor cells from the adult human spinal cord. Methods Mol. Biol. 2013;1059:87–93. doi: 10.1007/978-1-62703-574-3_8. [DOI] [PubMed] [Google Scholar]
- Beck C.W., Christen B., Slack J.M. Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev. Cell. 2003;5:429–439. doi: 10.1016/s1534-5807(03)00233-8. [DOI] [PubMed] [Google Scholar]
- Becker C.G., Becker T., Hugnot J.-P. The spinal ependymal zone as a source of endogenous repair cells across vertebrates. Prog. Neurobiol. 2018;170:67–80. doi: 10.1016/j.pneurobio.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Bélanger M.-C., Robert B., Cayouette M. Msx1-positive progenitors in the retinal ciliary margin give rise to both neural and non-neural progenies in mammals. Dev. Cell. 2017;40:137–150. doi: 10.1016/j.devcel.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Benner E.J., Luciano D., Jo R., Abdi K., Paez-Gonzalez P., Sheng H., Warner D.S., Liu C., Eroglu C., Kuo C.T. Protective astrogenesis from the SVZ niche after injury is controlled by Notch modulator Thbs4. Nature. 2013;497:369. doi: 10.1038/nature12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo C.-F., Fontaine R.H., Soueid J., Tammela T., Makinen T., Alfaro-Cervello C., Bonnaud F., Miguez A., Benhaim L., Xu Y. Vascular endothelial growth factor receptor 3 directly regulates murine neurogenesis. Genes Dev. 2011;25:831–844. doi: 10.1101/gad.615311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho G., Lim Y., Cho I.T., Simonet J.C., Golden J.A. Arx together with FoxA2, regulates Shh floor plate expression. Dev. Biol. 2014;393:137–148. doi: 10.1016/j.ydbio.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codega P., Silva-Vargas V., Paul A., Maldonado-Soto A.R., DeLeo A.M., Pastrana E., Doetsch F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82:545–559. doi: 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dromard C., Guillon H., Rigau V., Ripoll C., Sabourin J.-C., Perrin F., Scamps F., Bozza S., Sabatier P., Lonjon N. Adult human spinal cord harbors neural precursor cells that generate neurons and glial cells in vitro. J. Neurosci. Res. 2008;86:1916–1926. doi: 10.1002/jnr.21646. [DOI] [PubMed] [Google Scholar]
- Fiorelli R., Cebrian-Silla A., Garcia-Verdugo J.M., Raineteau O. The adult spinal cord harbors a population of GFAP-positive progenitors with limited self-renewal potential. Glia. 2013;61:2100–2113. doi: 10.1002/glia.22579. [DOI] [PubMed] [Google Scholar]
- Fu H., Qi Y., Tan M., Cai J., Hu X., Liu Z., Jensen J., Qiu M. Molecular mapping of the origin of postnatal spinal cord ependymal cells: evidence that adult ependymal cells are derived from Nkx6. 1+ ventral neural progenitor cells. J. Comp. Neurol. 2003;456:237–244. doi: 10.1002/cne.10481. [DOI] [PubMed] [Google Scholar]
- Gage F.H., Temple S. Neural stem cells: generating and regenerating the brain. Neuron. 2013;80:588–601. doi: 10.1016/j.neuron.2013.10.037. [DOI] [PubMed] [Google Scholar]
- Garcia-Ovejero D., Arevalo-Martin A., Paniagua-Torija B., Florensa-Vila J., Ferrer I., Grassner L., Molina-Holgado E. The ependymal region of the adult human spinal cord differs from other species and shows ependymoma-like features. Brain. 2015;138:1583–1597. doi: 10.1093/brain/awv089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebbin B.M., Hau A.-C., Groß A., Anders-Maurer M., Schramm J., Koss M., Wille C., Mittelbronn M., Selleri L., Schulte D. Pbx1 is required for adult SVZ neurogenesis. Development. 2016;143:2281–2291. doi: 10.1242/dev.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N., Li Y., Heidenreich M., Swiech L., Avraham-Davidi I., Trombetta J.J., Hession C., Zhang F., Regev A. Div-Seq: single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton L., Truong M., Bednarczyk M., Aumont A., Fernandes K. Cellular organization of the central canal ependymal zone, a niche of latent neural stem cells in the adult mammalian spinal cord. Neuroscience. 2009;164:1044–1056. doi: 10.1016/j.neuroscience.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ingram W., McCue K., Tran T., Hallahan A., Wainwright B. Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene. 2008;27:1489–1500. doi: 10.1038/sj.onc.1210767. [DOI] [PubMed] [Google Scholar]
- Jarvelainen H., Sainio A., Wight T.N. Pivotal role for decorin in angiogenesis. Matrix Biol. 2015;43:15–26. doi: 10.1016/j.matbio.2015.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P., Lee H.K., Glasgow S.M., Finley M., Donti T., Gaber Z.B., Graham B.H., Foster A.E., Novitch B.G., Gronostajski R.M. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74:79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazanov S., Paz Y., Hefetz A., Gonzales B.J., Netser Y., Mansour A.A., Ben-Arie N. Floor plate descendants in the ependyma of the adult mouse central nervous system. Int. J. Dev. Biol. 2017;61:257–265. doi: 10.1387/ijdb.160232nb. [DOI] [PubMed] [Google Scholar]
- Kubota H., Avarbock M.R., Brinster R.L. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. U S A. 2004;101:16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand Y., Moreau J., Saint Cloment C., Vives F.L., Robert B. Generation and characterization of a tamoxifen inducible Msx1CreERT2 knock-in allele. Genesis. 2013;51:110–119. doi: 10.1002/dvg.22350. [DOI] [PubMed] [Google Scholar]
- Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Martin-Villalba A. Adult NSC diversity and plasticity: the role of the niche. Curr. Opin. Neurobiol. 2017;42:68–74. doi: 10.1016/j.conb.2016.11.008. [DOI] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K., Martin-Villalba A. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Marichal N., Reali C., Trujillo-Cenóz O., Russo R.E. Spinal cord stem cells in their microenvironment: the ependyma as a stem cell niche. Adv. Med. Exp. Biol. 2017;1041:55–79. doi: 10.1007/978-3-319-69194-7_5. [DOI] [PubMed] [Google Scholar]
- Mo J.S., Park H.W., Guan K.L. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15:642–656. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothe A.J., Zahir T., Santaguida C., Cook D., Tator C.H. Neural stem/progenitor cells from the adult human spinal cord are multipotent and self-renewing and differentiate after transplantation. PLoS One. 2011;6:e27079. doi: 10.1371/journal.pone.0027079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte C., Matyash M., Pivneva T., Schipke C.G., Ohlemeyer C., Hanisch U.K., Kirchhoff F., Kettenmann H. GFAP promoter-controlled EGFP-expressing transgenic mice: a tool to visualize astrocytes and astrogliosis in living brain tissue. Glia. 2001;33:72–86. [PubMed] [Google Scholar]
- Petracca Y.L., Sartoretti M.M., Di Bella D.J., Marin-Burgin A., Carcagno A.L., Schinder A.F., Lanuza G.M. The late and dual origin of cerebrospinal fluid-contacting neurons in the mouse spinal cord. Development. 2016;143:880–891. doi: 10.1242/dev.129254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenninger C.V., Steinhoff C., Hertwig F., Nuber U.A. Prospectively isolated CD133/CD24-positive ependymal cells from the adult spinal cord and lateral ventricle wall differ in their long-term in vitro self-renewal and in vivo gene expression. Glia. 2011;59:68–81. doi: 10.1002/glia.21077. [DOI] [PubMed] [Google Scholar]
- Prosser H.M., Bradley A., Caldwell M.A. Olfactory bulb hypoplasia in Prokr2 null mice stems from defective neuronal progenitor migration and differentiation. Eur. J. Neurosci. 2007;26:3339–3344. doi: 10.1111/j.1460-9568.2007.05958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos C., Robert B. msh/Msx gene family in neural development. Trends Genet. 2005;21:624–632. doi: 10.1016/j.tig.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Ren Y., Ao Y., O’Shea T.M., Burda J.E., Bernstein A.M., Brumm A.J., Muthusamy N., Ghashghaei H.T., Carmichael S.T., Cheng L. Ependymal cell contribution to scar formation after spinal cord injury is minimal, local and dependent on direct ependymal injury. Sci. Rep. 2017;7:41122. doi: 10.1038/srep41122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg A.B., Roco C.M., Muscat R.A., Kuchina A., Sample P., Yao Z., Graybuck L.T., Peeler D.J., Mukherjee S., Chen W. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360:176–182. doi: 10.1126/science.aam8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzinova M.B., Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Sabourin J.C., Ackema K.B., Ohayon D., Guichet P.O., Perrin F.E., Garces A., Ripoll C., Charité J., Simonneau L., Kettenmann H. A mesenchymal-like ZEB1+ niche harbors dorsal radial glial fibrillary acidic protein-positive stem cells in the spinal cord. Stem Cells. 2009;27:2722–2733. doi: 10.1002/stem.226. [DOI] [PubMed] [Google Scholar]
- Smith E., Hargrave M., Yamada T., Begley C.G., Little M.H. Coexpression of SCL and GATA3 in the V2 interneurons of the developing mouse spinal cord. Dev. Dyn. 2002;224:231–237. doi: 10.1002/dvdy.10093. [DOI] [PubMed] [Google Scholar]
- Stenudd M., Sabelström H., Frisén J. Role of endogenous neural stem cells in spinal cord injury and repair. JAMA Neurol. 2015;72:235–237. doi: 10.1001/jamaneurol.2014.2927. [DOI] [PubMed] [Google Scholar]
- Tomer R., Ye L., Hsueh B., Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 2014;9:1682. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M., Cunningham A. Developmental expression of vascular endothelial growth factor receptor 3 and vascular endothelial growth factor C in forebrain. Neuroscience. 2015;303:544–557. doi: 10.1016/j.neuroscience.2015.04.063. [DOI] [PubMed] [Google Scholar]
- Weiss S., Dunne C., Hewson J., Wohl C., Wheatley M., Peterson A.C., Reynolds B.A. Multipotent CNS stem cells are present in the adult mammalian spinal cord and ventricular neuroaxis. J. Neurosci. 1996;16:7599–7609. doi: 10.1523/JNEUROSCI.16-23-07599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., Maden M. The mechanisms of dorsoventral patterning in the vertebrate neural tube. Dev. Biol. 2005;282:1–13. doi: 10.1016/j.ydbio.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Xing L., Anbarchian T., Tsai J.M., Plant G.W., Nusse R. Wnt/β-catenin signaling regulates ependymal cell development and adult homeostasis. Proc. Natl. Acad. Sci. U S A. 2018;115:E5954–E5962. doi: 10.1073/pnas.1803297115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Sachewsky N., Azimi A., Hung M., Gappasov A., Morshead C.M. Myelin basic protein regulates primitive and definitive neural stem cell proliferation from the adult spinal cord. Stem Cells. 2017;35:485–496. doi: 10.1002/stem.2488. [DOI] [PubMed] [Google Scholar]
- Zeisel A., Hochgerner H., Lonnerberg P., Johnsson A., Memic F., van der Zwan J., Haring M., Braun E., Borm L.E., La Manno G. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.