Abstract
Introduction
Cervical cancer is the second most common type of cancer and the third leading cause of cancer deaths in females in developing countries. Recent studies showed that long non-coding RNAs play a key role in human cancers. However, the molecular mechanisms underlying the initiation and progression of cervical cancer remained to be further explored.
Material and methods
In this study, we explored the differential expression of lncRNAs and mRNAs in cervical cancer progression by analyzing the public dataset GSE63514. Next, PPI and co-expression networks were constructed to reveal the potential roles of cervical cancer related mRNAs and lncRNAs. Furthermore, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed to explore functions of differentially expressed genes (DEGs) in cervical cancer.
Results
In the present study, we observed that 3021 mRNAs were up-regulated and 1605 mRNAs were down-regulated in cervical cancer progression. Meanwhile, we for the first time found that 172 lncRNAs were up-regulated and 106 lncRNAs were down-regulated in cervical cancer progression. Co-expression network analysis showed that lncRNAs were widely co-expressed with cell cycle related genes in cervical cancer, implicating the important roles of these lncRNAs in cell proliferation regulation. Of note, we identified two hub lncRNA-mRNA networks involved in regulating various biological processes in cervical cancer progression.
Conclusions
Our results identified key mRNAs and lncRNAs in cervical cancer progression. This study will provide novel insights to explore the potential mechanisms underlying cervical cancer progression.
Keywords: long non-coding RNAs, cervical cancer, protein-protein interaction analysis, expression profiling
Introduction
Cervical cancer is the second most common type of cancer and the third leading cause of cancer deaths in women in developing countries [1–3]. Previous studies had revealed that transcription factors, miRNAs, and RNA binding proteins were involved in cervical cancer progression. For example, AP-2β suppressed cervical cancer cell proliferation by inducing the degradation of β-catenin [4], and microRNA-221-3p promoted cervical cancer metastasis by targeting THBS2 [5]. However, the molecular mechanisms underlying the initiation and progression of cervical cancer remained to be further explored.
Long non-coding RNAs (lncRNAs) were a type of non-coding RNAs more than 200 nucleotides in length [6, 7]. Interestingly, several studies showed that a few lncRNAs had protein coding potential. Recent reports demonstrated the important roles of lncRNAs in cancer progression [8]. LncRNAs were differently expressed in various human cancers, such as prostate cancer, lung cancer and breast cancer, and were involved in regulating widespread biological processes, including cell proliferation, apoptosis, metastasis and autophagy [9–11]. In cervical cancer, lncRNAs had been revealed to be involved in tumor proliferation, radio-resistance and metastasis regulation. For example, HOTAIR promoted HeLa cell migration and invasion by inhibiting miR206 [12], and HOXD-AS1 regulated cervical cancer proliferation by activating Ras/ERK signaling [13]. However, the roles of lncRNAs in cervical cancer progression remained to be further explored.
In the present study, we aimed to identify differentially expressed lncRNAs and mRNAs in cervical cancer progression by analyzing the public dataset GSE63514, including 24 normal, 14 CIN1 lesions, 22 CIN2 lesions, 40 CIN3 lesions, and 28 cervical cancer samples. We constructed a protein-protein interaction network and lncRNA co-expression network to reveal the potential roles of cervical cancer related mRNAs and lncRNAs. This study will provide useful information to explore the potential candidate biomarkers for diagnosis, prognosis, and drug targets for cervical cancer.
Material and methods
Microarray data and data preprocessing
In this study, we downloaded a public dataset, GSE63514 [14], from NCBI GEO datasets (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63514). GSE63514 dataset including 24 normal, 14 CIN1 lesions, 22 CIN2 lesions, 40 CIN3 lesions, and 28 cervical cancer samples. All samples were cryosectioned and used for laser-capture, RNA extraction, two rounds of T7-mediated amplification, and cRNA biotinylation. Then, bio-cRNA was hybridized to Affymetrix U133-Plus 2.0 arrays, and scanned signals were processed through GC-RMA. We used the limma package in R software to normalize the raw data (https://www.r-project.org/). The differentially expressed mRNA and lncRNAs were identified by the linear models for microarray analysis (Limma) method [15]. The gene expression among 24 normal, 14 CIN1 lesions, 22 CIN2 lesions, 40 CIN3 lesions, and 28 cervical cancer samples with thresholds of |log FC| > 1.5 and p < 0.001 were identified to be differentially expressed. Hierarchical cluster analysis of differentially expressed mRNAs and lncRNAs was performed using CLUSTER 3.0, and the hierarchical clustering heat map was visualized by Tree View [16].
LncRNA classification pipeline
Previous studies indicated many mRNA microarray datasets including many long non-coding RNAs probes. In this study, we screened differently expressed lncRNAs in cervical cancer progression by applying a pipeline reported by Zhang et al. [17]. Briefly, first, the GPL570 platform of Affymetrix Human Genome U133 Plus 2.0 Array (Affymetrix Inc., Santa Clara, California, USA) probe set ID was mapped to the NetAffx Annotation Files (HG-U133 Plus 2.0 Annotations, CSV format, release 31, 08/23/10). The annotations included the probe set ID, gene symbol, and Refseq transcript ID. Second, the probe sets that were assigned with a Refseq transcript ID in the NetAffx annotations were extracted. In this study, we only retained those labeled as “NR_” (NR indicates non-coding RNA in the Refseq database). Finally, 2448 annotated lncRNA transcripts with corresponding Affymetrix probe IDs were generated. LncRNAs having fold changes ≥ 2 and p < 0.05 were selected as of significantly differential expression.
GO and KEGG pathway analysis
To identify functions of DEGs in cervical cancer, we performed GO function enrichment analysis in 3 functional ontologies: biological process (BP), cellular component (CC) and molecular function (MF). KEGG pathway enrichment analysis was also performed to identify pathways enriched in smoking related lung cancer using the DAVID system (https://david.ncifcrf.gov/). The p-value less than 0.05 was considered as significant.
Construction of PPI network and module analysis
In order to predict protein interactions, which included physical and functional associations, the present study used the Search Tool for the Retrieval of Interacting Genes (STRING) to construct the PPI network for DEGs (minimum required interaction score > 0.4) [18]. The interaction relationships of the proteins encoded by DEGs were searched by STRING online software, and the combined score > 0.4 was used as the cut-off criterion. In addition, Cytoscape software version 3.4.0 (http://cytoscape.org/download_old_versions.html) was used for visualization of the PPI networks [19]. Following the construction of the PPI network, a module analysis of the network was performed using the MCODE plug-in (degree cut-off ≥ 2 and the nodes with edges ≥ 2-core) [20]. Additionally, the Network Analyzer was used to compute the basic properties of the PPI network, including average clustering co-efficient distribution, closeness centrality, average neighborhood connectivity, node degree distribution, shortest path length distribution, and topological coefficients [21].
Co-expression network construction and analysis
In this study, the Pearson correlation coefficient of DEG-lncRNA pairs was calculated according to their expression value. The co-expressed DEG-lncRNA pairs with the absolute value of Pearson correlation coefficient ≥ 0.75 were selected and the co-expression network was established using Cytoscape software. Cytoscape MCODE plug-in (Version 3.4.0, available online: http://www.cytoscape.org/) was applied for visualization of the co-expression networks.
Statistical analysis
All statistical analysis of this research was carried out using SPSS 18.0 statistical software. Statistical comparisons between groups of normalized data were performed using the t-test or Mann-Whitney U-test according to the test condition. A p < 0.05 was considered statistically significant with a 95% confidence level.
Results
Identification of differentially expressed mRNAs and lncRNAs in cervical cancer
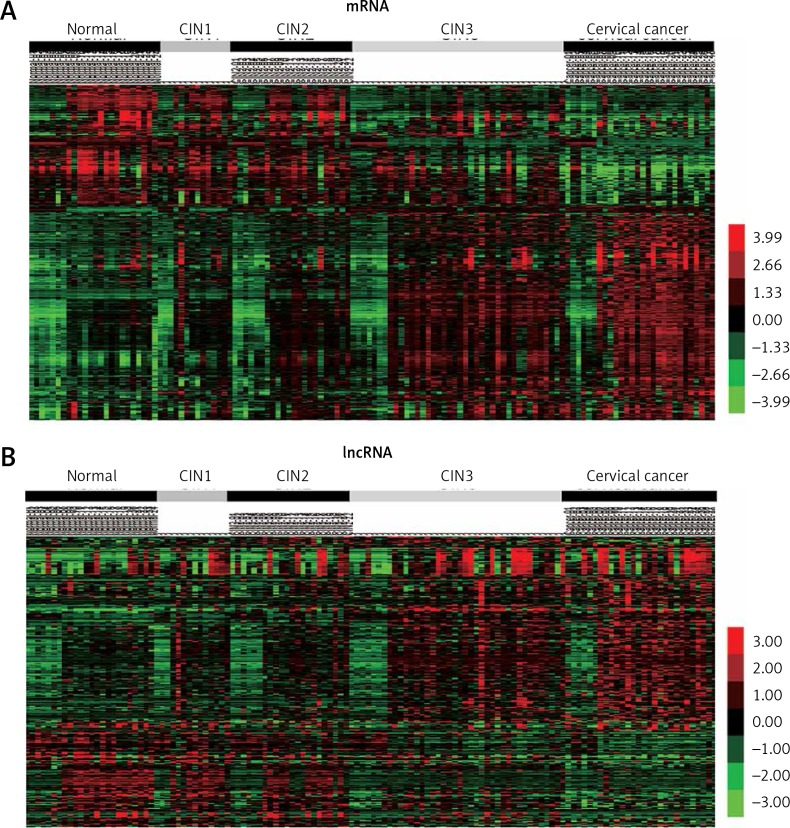
In the present study, we analyzed the GSE63514 dataset to identify differentially expressed genes in cervical cancer progression. The gene expression with |logFC| > 1 and p < 0.05 among 24 normal samples, 14 CIN1 lesions, 22 CIN2 lesions, 40 CIN3 lesions, and 28 cervical cancer samples was identified to be differently expressed. A total of 4799 mRNAs were identified as cervical cancer progression related genes. Among these mRNAs, 3021 mRNAs were up-regulated and 1605 mRNAs were down-regulated in cervical cancer progression. The top up-regulated and down-regulated 10 mRNAs involved in cervical cancer progression are listed in Tables I and II. As shown in Figure 1 A, hierarchical clustering analysis was performed to show the DEGs in the progression of cervical cancer from normal to cancer.
Table I.
The top 10 up-regulated mRNAs in cervical cancer progression
| Gene name | Ave (normal) | Ave (CIN1) | Ave (CIN2) | Ave (CIN3) | Ave (tumor) | FC (CIN1 vs. normal) | FC (CIN2 vs. normal) | FC (CIN3 vs. normal) | FC (tumor vs. normal) | P (CIN1 vs. normal) | P (CIN2 vs. normal) | P (CIN3 vs. normal) | P (tumor vs. normal) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NUP210 | 2.97 | 4.34 | 4.59 | 7.33 | 7.44 | 2.58 | 3.06 | 20.51 | 22.22 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| MMP12 | 3.83 | 5.28 | 5.70 | 7.25 | 8.88 | 2.73 | 3.65 | 10.65 | 33.06 | 0.10 | 0.02 | < 0.001 | < 0.001 |
| SYNGR3 | 3.50 | 5.36 | 5.67 | 8.20 | 7.51 | 3.62 | 4.49 | 25.95 | 16.04 | 0.01 | < 0.001 | < 0.001 | < 0.001 |
| HS6ST2 | 4.23 | 6.63 | 6.88 | 8.58 | 8.61 | 5.30 | 6.29 | 20.45 | 20.86 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| HS6ST2 | 5.93 | 8.61 | 8.58 | 10.49 | 10.02 | 6.43 | 6.31 | 23.65 | 17.02 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| ZIC2 | 2.79 | 3.22 | 4.20 | 5.82 | 8.31 | 1.35 | 2.66 | 8.19 | 46.13 | 0.12 | 0.02 | < 0.001 | < 0.001 |
| CTHRC1 | 5.95 | 6.34 | 7.60 | 9.07 | 11.68 | 1.31 | 3.14 | 8.73 | 53.10 | 0.69 | 0.05 | < 0.001 | < 0.001 |
| ELAVL2 | 2.31 | 4.32 | 5.32 | 6.82 | 7.69 | 4.02 | 8.06 | 22.73 | 41.52 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| SYCP2 | 4.11 | 5.56 | 7.57 | 9.23 | 9.26 | 2.72 | 11.02 | 34.73 | 35.41 | 0.05 | < 0.001 | < 0.001 | < 0.001 |
| CDKN2A | 4.07 | 6.90 | 9.37 | 10.67 | 10.47 | 7.15 | 39.46 | 96.88 | 84.76 | 0.02 | < 0.001 | < 0.001 | < 0.001 |
Table II.
The top 10 down-regulated mRNAs in cervical cancer progression
| Gene name | Ave (normal) | Ave (CIN1) | Ave (CIN2) | Ave (CIN3) | Ave (tumor) | FC (CIN1 vs. normal) | FC (CIN2 vs. normal) | FC (CIN3 vs. normal) | FC (tumor vs. normal) | P (CIN1 vs. normal) | P (CIN2 vs. normal) | P (CIN3 vs. normal) | P (tumor vs. normal) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRISP3 | 13.32 | 9.88 | 10.49 | 7.67 | 4.41 | 0.09 | 0.14 | 0.02 | 0.00 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| CRISP2 | 8.02 | 4.72 | 5.37 | 3.59 | 2.91 | 0.10 | 0.16 | 0.05 | 0.03 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| RPTN | 8.05 | 5.01 | 5.96 | 4.33 | 3.45 | 0.12 | 0.24 | 0.08 | 0.04 | 0.02 | 0.06 | < 0.001 | < 0.001 |
| UPK1A | 8.11 | 6.41 | 5.78 | 3.88 | 2.43 | 0.31 | 0.20 | 0.05 | 0.02 | 0.22 | 0.05 | < 0.001 | < 0.001 |
| ZBED2 | 8.14 | 6.60 | 5.60 | 4.44 | 3.53 | 0.34 | 0.17 | 0.08 | 0.04 | 0.15 | 0.01 | < 0.001 | < 0.001 |
| LCE3D | 10.36 | 8.04 | 8.73 | 7.30 | 5.00 | 0.20 | 0.32 | 0.12 | 0.02 | 0.13 | 0.22 | 0.01 | < 0.001 |
| CRCT1 | 8.67 | 7.13 | 6.64 | 5.25 | 3.33 | 0.34 | 0.25 | 0.09 | 0.02 | 0.30 | 0.12 | < 0.001 | < 0.001 |
| KRT2 | 5.37 | 3.80 | 2.85 | 3.08 | 1.83 | 0.34 | 0.17 | 0.20 | 0.09 | 0.22 | 0.02 | 0.01 | < 0.001 |
| EPB41L3 | 8.22 | 6.83 | 5.58 | 5.20 | 5.79 | 0.38 | 0.16 | 0.12 | 0.19 | 0.09 | < 0.001 | < 0.001 | < 0.001 |
| GYS2 | 6.09 | 4.65 | 4.29 | 3.17 | 2.35 | 0.37 | 0.29 | 0.13 | 0.07 | 0.21 | 0.07 | < 0.001 | < 0.001 |
Figure 1.
Identification of differentially expressed mRNAs and lncRNAs in cervical cancer. A – Hierarchical clustering analysis shows differential mRNAs expression in cervical cancer by using GSE63514. B – Hierarchical clustering analysis shows differential lncRNAs expression in cervical cancer by using GSE63514
Here, we found that GSE63514 provided a good platform to evaluate the functional roles of lncRNAs in cervical cancer tumorigenesis. After applying the lncRNA classification pipeline reported by Zhang et al., 3687 lncRNA probes were found in the GSE63514 dataset. We then compared the lncRNA expression patterns between normal and all grades of lesions. We for the first time found that 278 lncRNAs were associated with cervical cancer progression. Among these lncRNAs, 172 lncRNAs were up-regulated and 106 lncRNAs were down-regulated in the cervical cancer progression. Of note, several lncRNAs, including TUG1, MEG3 and XIST, had been reported to be associated with cervical cancer proliferation and metastasis. The top up-regulated and down-regulated 10 lncRNAs involved in cervical cancer progression are listed in Tables III and IV. Hierarchical clustering analysis of the lncRNAs in cervical cancer is shown in Figure 1 B.
Table III.
The top 10 up-regulated lncRNAs in cervical cancer progression
| Gene name | Ave (normal) | Ave (CIN1) | Ave (CIN2) | Ave (CIN3) | Ave (tumor) | FC (CIN1 vs. normal) | FC (CIN2 vs. normal) | FC (CIN3 vs. normal) | FC (tumor vs. normal) | P (CIN1 vs. normal) | P (CIN2 vs. normal) | P (CIN3 vs. normal) | P (tumor vs. normal) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RP11-1L12.3 | 2.78 | 3.90 | 4.28 | 5.58 | 5.23 | 2.16 | 2.82 | 6.96 | 5.46 | 0.07 | 0.02 | < 0.001 | < 0.001 |
| LINC01305 | 2.59 | 3.24 | 4.01 | 5.41 | 5.46 | 1.57 | 2.67 | 7.06 | 7.31 | 0.09 | 0.03 | < 0.001 | < 0.001 |
| LOC375196 | 2.87 | 3.44 | 4.47 | 6.16 | 5.69 | 1.49 | 3.04 | 9.83 | 7.08 | 0.19 | < 0.001 | < 0.001 | < 0.001 |
| ANKRD36BP2 | 5.03 | 7.32 | 6.26 | 7.00 | 8.65 | 4.90 | 2.35 | 3.93 | 12.34 | 0.01 | 0.07 | 0.01 | < 0.001 |
| LINC00925 | 3.07 | 3.72 | 4.91 | 7.00 | 5.82 | 1.57 | 3.57 | 15.19 | 6.71 | 0.16 | < 0.001 | < 0.001 | < 0.001 |
| CTA-384D8.35 | 2.59 | 4.04 | 3.93 | 6.29 | 6.50 | 2.73 | 2.52 | 12.96 | 14.96 | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| ABCA17P | 2.92 | 4.58 | 5.28 | 7.56 | 6.72 | 3.17 | 5.15 | 24.98 | 13.97 | 0.01 | < 0.001 | < 0.001 | < 0.001 |
| TCAM1P | 3.30 | 4.71 | 6.62 | 8.09 | 7.94 | 2.66 | 9.97 | 27.68 | 24.85 | 0.01 | < 0.001 | < 0.001 | < 0.001 |
| RP1-93H18.6 | 4.64 | 5.97 | 5.64 | 6.94 | 6.97 | 2.51 | 2.01 | 4.94 | 5.04 | 0.01 | 0.02 | < 0.001 | < 0.001 |
| AC128677.4 | 2.78 | 3.43 | 3.61 | 4.83 | 5.54 | 1.57 | 1.77 | 4.15 | 6.77 | 0.22 | 0.12 | < 0.001 | < 0.001 |
Table IV.
The top 10 down-regulated lncRNAs in cervical cancer progression
| Gene name | Ave (normal) | Ave (CIN1) | Ave (CIN2) | Ave (CIN3) | Ave (tumor) | FC (CIN1 vs. normal) | FC (CIN2 vs. normal) | FC (CIN3 vs. normal) | FC (tumor vs. normal) | P (CIN1 vs. normal) | P (CIN2 vs. normal) | P (CIN3 vs. normal) | P (tumor vs. normal) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP2B7P | 7.43 | 6.38 | 5.77 | 5.19 | 4.23 | 0.48 | 0.32 | 0.21 | 0.11 | 0.30 | 0.06 | < 0.001 | < 0.001 |
| PRSS3P2 | 7.03 | 5.98 | 5.85 | 4.25 | 4.01 | 0.48 | 0.44 | 0.15 | 0.12 | 0.03 | < 0.001 | < 0.001 | < 0.001 |
| RAB11B-AS1 | 7.15 | 6.14 | 6.03 | 4.75 | 3.99 | 0.50 | 0.46 | 0.19 | 0.11 | 0.07 | 0.02 | < 0.001 | < 0.001 |
| LINC01214 | 4.51 | 3.38 | 3.15 | 2.65 | 2.44 | 0.46 | 0.39 | 0.28 | 0.24 | 0.16 | 0.02 | < 0.001 | < 0.001 |
| RP11-774O3.3 | 5.60 | 4.84 | 4.43 | 3.16 | 3.06 | 0.59 | 0.44 | 0.18 | 0.17 | 0.17 | 0.02 | < 0.001 | < 0.001 |
| HCG22 | 5.20 | 3.73 | 4.31 | 3.40 | 3.15 | 0.36 | 0.54 | 0.29 | 0.24 | 0.05 | 0.17 | < 0.001 | < 0.001 |
| ZNF300P1 | 7.06 | 6.53 | 5.79 | 4.90 | 3.90 | 0.69 | 0.41 | 0.22 | 0.11 | 0.47 | 0.03 | < 0.001 | < 0.001 |
| FLG-AS1 | 7.34 | 6.64 | 6.48 | 4.81 | 4.00 | 0.62 | 0.55 | 0.17 | 0.10 | 0.33 | 0.20 | < 0.001 | < 0.001 |
| LOC100507487 | 6.58 | 6.24 | 5.39 | 3.62 | 3.50 | 0.79 | 0.44 | 0.13 | 0.12 | 0.62 | 0.02 | < 0.001 | < 0.001 |
| LINC01094 | 5.37 | 4.78 | 4.13 | 3.55 | 3.60 | 0.66 | 0.42 | 0.28 | 0.29 | 0.48 | 0.07 | < 0.001 | < 0.001 |
Construction of PPI networks for differentially expressed mRNAs in cervical cancer
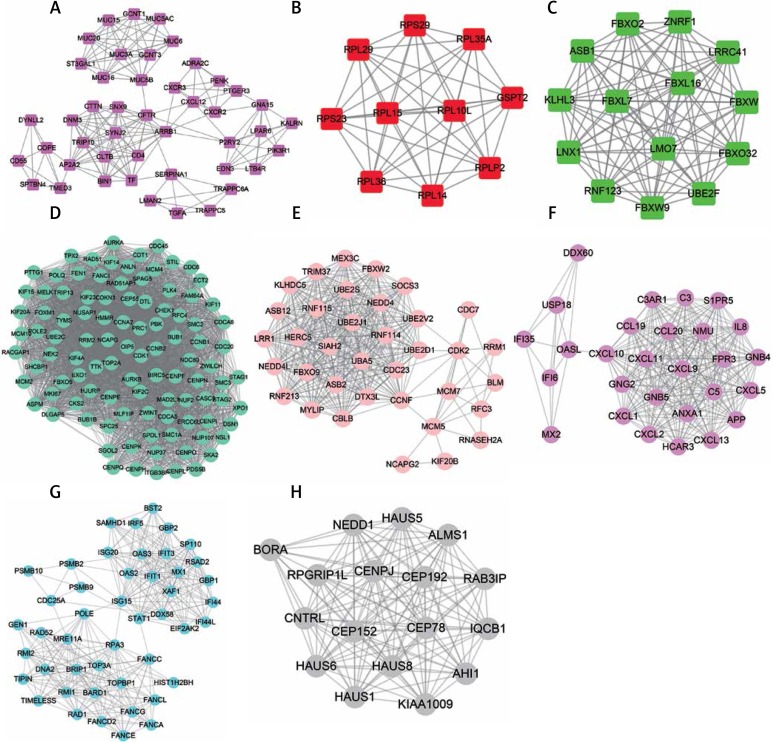
We constructed the protein-protein interaction network analysis for 4799 differently expressed mRNAs by using the STRING database. We then used the Mcode plug-in to identify key networks with degree cut-off ≥ 2 and the nodes with edges ≥ 2-core in the total PPI network. The top 5 up-regulated hub-networks and top 3 down-regulated hub-networks are shown in Figure 2.
Figure 2.
Construction of PPI networks for differentially expressed mRNAs in cervical cancer. The top 5 up-regulated hub-networks and top 3 down-regulated hub-networks
Co-expression network analysis of differently expressed lncRNAs in cervical cancer progression
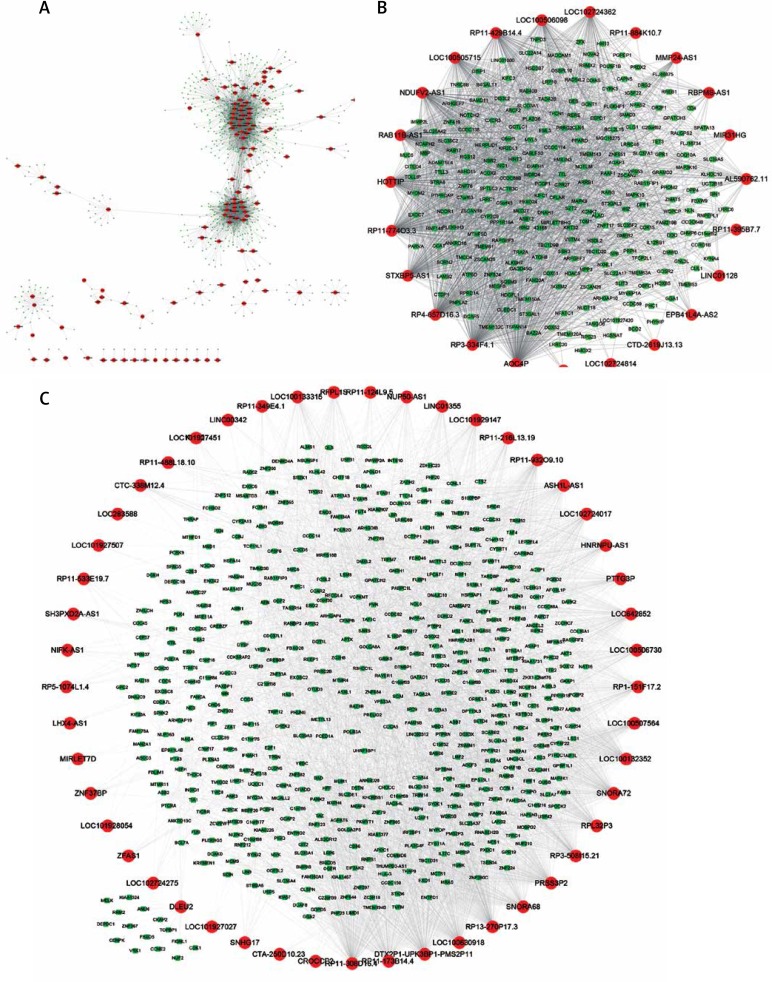
We next constructed the gene co-expression network between lncRNAs and mRNAs in cervical cancer progression. We performed the correlation analysis for mRNAs and lncRNAs by calculating the Pearson correlation coefficient in all samples and selected lncRNA-mRNA pairs with |R| > 0.65 for co-expression network construction. As shown in Figure 3, a total of 278 lncRNAs and 786 mRNAs were included in this network. Interestingly, we observed many antisense lncRNAs co-expressed with sense mRNAs, including the MFI2-AS1-MFI2 pair, PSMB8-AS1-PSMB9 pair, and HOXB-AS3-HOXB6 pair (Figure 3 A). Several well-known lncRNAs, such as MALAT1 and DLEU2, were also revealed to play important roles in cervical cancer progression (Figure 3 B). Among these lncRNAs, lncRNA RP11-173B14.4, RP11-308D16.4, LOC100630918, and RP13-270P17.3 were identified as key lncRNAs in this network by regulating about 190 mRNAs (Figure 3 C).
Figure 3.
Co-expression network analysis of differentially expressed lncRNAs in cervical cancer progression. The gene co-expression network between lncRNAs and mRNAs in cervical cancer progression. 278 lncRNAs and 786 mRNAs were included in this network
GO and KEGG analysis of differentially expressed lncRNAs in cervical cancer progression
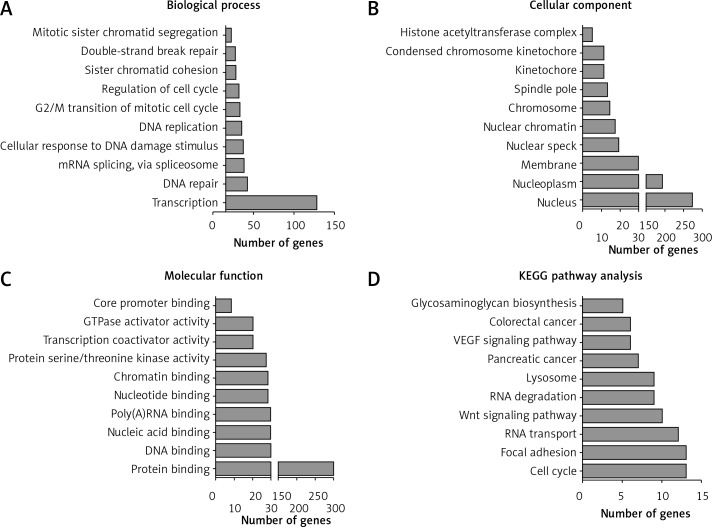
Furthermore, we performed GO and KEGG analysis for differentially expressed lncRNAs (Figures 4 A–C). GO analysis showed that differentially expressed lncRNAs were mainly involved in regulating transcription, cell division, DNA repair, mRNA splicing, via spliceosome, cellular response to DNA damage stimulus, DNA replication, G2/M transition of mitotic cell cycle, regulation of the cell cycle, sister chromatid cohesion, and double-strand break repair.
Figure 4.
GO and KEGG analysis of differentially expressed lncRNAs in cervical cancer progression. A–C – GO analysis shows differentially expressed lncRNA associated biological processes (A), cellular component (B) and molecular function (C). D – KEGG pathway analysis shows differentially expressed lncRNAs associated pathways
KEGG pathway analysis revealed that cervical cancer progression related lncRNAs were primarily enriched in pathways associated with the cell cycle, focal adhesion, RNA transport, Wnt signaling pathway, RNA degradation, lysosome, pancreatic cancer, VEGF signaling pathway, colorectal cancer, and glycosaminoglycan biosynthesis (Figure 4 D).
Functional prediction of key lncRNA-mRNAs network
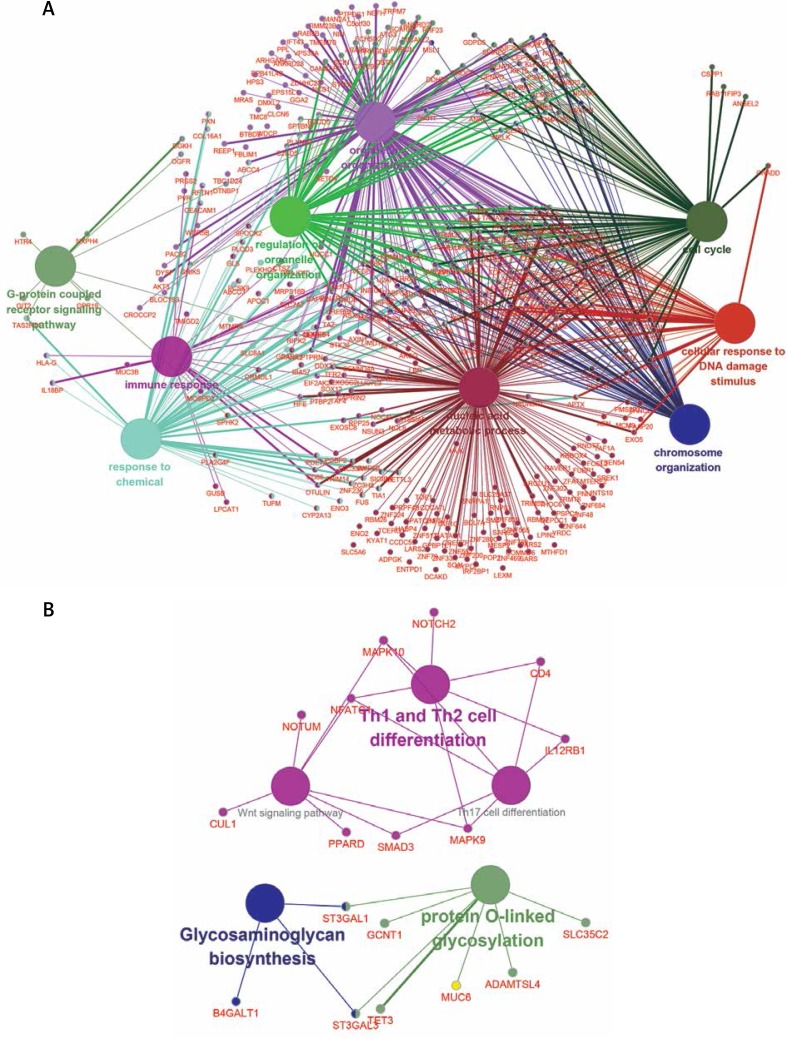
In this study, we observed 2 key lncRNA-mRNAs’ co-expression network. In order to predict the function of three key lncRNA-mRNA networks, the intersection mRNA was analyzed by the Cytoscape plug-in ClueGo and DAVID. Our results showed that hub lncRNA-mRNAs network 1 was involved in regulating the cell cycle, cellular response to DNA damage stimulus, chromosome organization, nucleic acid metabolic process, immune response and the G-protein coupled receptor signaling pathway (Figure 5 A). Hub lncRNA-mRNAs network 2 was involved in regulating Th1 and Th2 cell differentiation, the Wnt signaling pathway, glycosaminoglycan biosynthesis and protein O-linked glycosylation (Figure 5 B).
Figure 5.
Functional prediction of key lncRNA-mRNAs network. A – Hub lncRNA-mRNAs network 1 was involved in regulating Th1 and Th2 cell differentiation, Wnt signaling pathway, glycosaminoglycan biosynthesis and protein O-linked glycosylation. B – Hub lncRNA-mRNAs network 2 was involved in regulating cell cycle, cellular response to DNA damage stimulus, chromosome organization, and immune response
Discussion
Cervical cancer is one of the most common types of cancer in females [1]. However, the molecular mechanisms underlying the initiation and progression of cervical cancer remained to be further explored. In this study, we analyzed the GSE63514 dataset to identify differentially expressed genes in cervical cancer progression. We observed that 3021 mRNAs were up-regulated and 1605 mRNAs were down-regulated in cervical cancer progression. Meanwhile, we for the first time found that 172 lncRNAs were up-regulated and 106 lncRNAs were down-regulated in cervical cancer progression.
Previous studies had revealed that several proteins were involved in regulating cervical cancer progression. For example, GADD45α increased cytoplasmic APE1 levels to sensitize cervical cancer cells to radiotherapy [22]. PD-L1 was found to promote glucose metabolism in cervical cancer [23]. However, there was still no system-wide identification of cervical cancer progression related genes. Here, we analyzed the GSE63514 dataset, which included 24 normal samples, 14 CIN1 lesions, 22 CIN2 lesions, 40 CIN3 lesions, and 28 cervical cancer samples. A total of 3021 mRNAs were up-regulated and 1605 mRNAs were down-regulated in cervical cancer progression. In order to explore the potential roles of these genes, we constructed PPI networks. Our results showed that several genes, including CDK1, OIP5, NCAPG, CCNA2, AURKB and HMMR, played key roles in cervical cancer progression.
Recent studies showed lncRNAs played crucial roles in tumor proliferation, apoptosis, metastasis, and invasion. In cervical cancer, several lncRNAs were found to be dysregulated in tumor samples. For instance, lncRNA PCAT-1 regulated cervical cancer cell proliferation, metastasis and invasion [24]. Silencing HOXD-AS1 remarkably suppressed proliferation of cervical cancer cells by regulating Ras/ERK signaling [13]. LncRNA TUG1 promoted cervical cancer progression by affecting the miR-138-5p-SIRT1 axis [25]. Meanwhile, HOTAIR was reported to be an oncogene to promote cervical cancer growth and migration [26]. However, the roles of lncRNAs in cervical cancer progression remained unclear. In this study, we identified 172 lncRNAs that were up-regulated and 106 lncRNAs that were down-regulated in cervical cancer progression. Of note, several lncRNAs, including TUG1, MEG3 and XIST, had been reported to be associated with cervical cancer proliferation and metastasis [27, 28]. However, most of these lncRNAs were never reported in human cancers. Our study for the first time comprehensively revealed cervical cancer progression related lncRNAs.
The function roles of most lncRNAs remained largely unclear. Recently, gene co-expression network analysis was widely used to evaluate potential roles of lncRNAs. For example, Du et al. [29] performed lncRNA-mRNA co-expression analysis in thyroid cancer and Li et al. [30] constructed lncRNAs and circRNAs mediated co-expression and ceRNA networks in mouse germline stem cells. Here, we also performed co-expression network analysis of differentially expressed lncRNAs in cervical cancer progression. Interestingly, we observed many antisense lncRNAs co-expressed with sense mRNAs, including the MFI2-AS1-MFI2 pair, PSMB8-AS1-PSMB9 pair, and HOXB-AS3-HOXB6 pair. Bioinformatics analysis showed that these lncRNAs played important roles in regulating the cell cycle, cell division, DNA repair, and mRNA splicing. Moreover, we identified two hub lncRNA-mRNA networks. Hub network 1 was involved in regulating the cell cycle, cellular response to DNA damage stimulus, chromosome organization, and the immune response, and Hub l network 2 was involved in regulating Th1 and Th2 cell differentiation and the Wnt signaling pathway.
In conclusion, we identified that 4799 mRNAs and 278 lncRNAs were differentially expressed in cervical cancer progression. Co-expression network analysis showed that lncRNAs were widely co-expressed with cell cycle related genes in cervical cancer, implicating the important roles of these lncRNAs in cell proliferation regulation. Of note, we identified two hub lncRNA-mRNA networks. We believe that our analysis could provide novel insights to explore the potential mechanisms underlying cervical cancer progression.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Baoning Q, Chuandao S, Na S, Xinan W, Yuanping H. Efficacy and safety of radio-chemotherapy combined with thermotherapy for advanced cervical cancer in Chinese women: a meta-analysis. Arch Med Sci Civil Dis. 2017;2:182–90. [Google Scholar]
- 4.Wang F, Huang W, Hu X, et al. Transcription factor AP – 2beta suppresses cervical cancer cell proliferation by promoting the degradation of its interaction partner beta-catenin. Mol Carcinog. 2017;56:1909–23. doi: 10.1002/mc.22646. [DOI] [PubMed] [Google Scholar]
- 5.Wei WF, Zhou CF, Wu XG, et al. MicroRNA-221-3p, a TWIST2 target, promotes cervical cancer metastasis by directly targeting THBS2. Cell Death Dis. 2017;8:3220.. doi: 10.1038/s41419-017-0077-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–24. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 7.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan X, Huang W, Yang S, et al. Identification of androgen-responsive lncRNAs as diagnostic and prognostic markers for prostate cancer. Oncotarget. 2016;7:60503. doi: 10.18632/oncotarget.11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–66. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655–69. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: a new player in cancer. J Hematol Oncol. 2013;6:37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng P, Yin Z, Wu Y, Xu Y, Luo Y, Zhang TC. LncRNA HOTAIR promotes cell migration and invasion by regulating MKL1 via inhibition miR206 expression in HeLa cells. Cell Commun Signal. 2018;16:5. doi: 10.1186/s12964-018-0216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Wang A, Lu J, Cen R, Liu L. Long noncoding RNA HOXD-AS1 regulates proliferation of cervical cancer cells by activating Ras/ERK signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:5049–55. doi: 10.26355/eurrev_201711_13817. [DOI] [PubMed] [Google Scholar]
- 14.Manawapat-Klopfer A, Thomsen LT, Martus P, et al. TMEM45A, SERPINB5 and p16INK4A transcript levels are predictive for development of high-grade cervical lesions. Am J Cancer Res. 2016;6:1524–36. [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth GK. Bioinformatics and Computational Biology Solutions using R and Bioconductor. 2005. Springer. Limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- 16.Page RD. TreeView. Glasgow, UK: Glasgow University; 2001. [Google Scholar]
- 17.Zhang X, Sun S, Pu JKS, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–8. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. Bmc Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Assenov Y, Ramirez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–4. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 22.Li Q, Wei X, Zhou ZW, et al. GADD45α sensitizes cervical cancer cells to radiotherapy via increasing cytoplasmic APE1 level. Cell Death Disease. 2018;9:524. doi: 10.1038/s41419-018-0452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang W, Song Y, Lu YL, Sun JZ, Wang HW. Increased expression of programmed death (PD)-1 and its ligand PD-L1 correlates with impaired cell-mediated immunity in high-risk human papillomavirus-related cervical intraepithelial neoplasia. Immunology. 2013;139:513–22. doi: 10.1111/imm.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L, Liao LM, Liu AW, et al. Overexpression of long noncoding RNA HOTAIR predicts a poor prognosis in patients with cervical cancer. Arch Gynecol Obstet. 2014;290:717–23. doi: 10.1007/s00404-014-3236-2. [DOI] [PubMed] [Google Scholar]
- 25.Zhu J, Shi H, Liu H, Wang X, Li F. Long non-coding RNA TUG1 promotes cervical cancer progression by regulating the miR-138-5p-SIRT1 axis. Oncotarget. 2017;8:65253–64. doi: 10.18632/oncotarget.18224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.KIm HJ, Lee DW, Yim GW, et al. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521–30. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Yao T, Wang Y, Yu J, Liu Y, Lin Z. Long noncoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR-21. Cancer Biol Ther. 2016;17:104–13. doi: 10.1080/15384047.2015.1108496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng L, Yuan X, Jiang B, Tang Z, Li GC. LncRNAs: key players and novel insights into cervical cancer. Tumor Biol. 2016;37:2779–88. doi: 10.1007/s13277-015-4663-9. [DOI] [PubMed] [Google Scholar]
- 29.Du Y, Xia W, Zhang J, Wan D, Yang Z, Li X. Comprehensive analysis of long noncoding RNA-mRNA co-expression patterns in thyroid cancer. Mol BioSystems. 2017;13:2107115. doi: 10.1039/c7mb00375g. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Ao J, Wu J. Systematic identification and comparison of expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in mouse germline stem cells. Oncotarget. 2017;8:26573–90. doi: 10.18632/oncotarget.15719. [DOI] [PMC free article] [PubMed] [Google Scholar]