Abstract
Viral metagenomic analysis of the liver of a black headed python (Aspidites melanocephalus) euthanized for a proliferative spinal lesion of unknown etiology yielded the first characterized genome of a reptile-infecting circovirus (black-headed python circovirus or BhPyCV). BhPyCV-specific in situ hybridization (ISH) showed that viral nucleic acids were strongly expressed in the intestinal lining and mucosa and multifocally in the liver. To investigate the presence of this virus in other snakes and its possible pathogenicity, 17 snakes in the python family with spinal disease were screened with ISH yielding a second BhP positive in intestinal tissue, and a Boelen’s python (Morelia boeleni) positive in the liver. BhPyCV specific PCR was used to screen available frozen tissues from 13 of these pythons, four additional deceased pythons with and without spinal disease, and fecal samples from 37 live snakes of multiple species with unknown disease status. PCR detected multiple positive tissues in both of the ISH positive BhP and in the feces of another two live BhP and two live annulated tree boas (Corallus annulatus). Preliminary analysis indicates this circovirus can infect BhPs where it was found in 4/5 BhPs tested (2/2 with spinal disease, 2/3 live with unknown status), Boelen’s python (1/2 with spinal disease), and annulated tree boa (2/6 live with unknown status) but was not detected in other python species with the same spinal lesions. This circovirus’ causal or contributory role in spinal disease remains speculative and not well supported by these initial data.
Electronic supplementary material
The online version of this article (10.1186/s13567-019-0653-z) contains supplementary material, which is available to authorized users.
Introduction
Circoviruses have some of the smallest viral genomes ranging in size from 1.7 to 2.1 kb of circular ssDNA encoding two major proteins, the replication-associated protein (Rep) involved in genome replication and the nucleocapsid (Cap) [1]. Most notorious for causing diseases of birds and swine, circovirus infections have also been documented in other species of mammals, and more recently in amphibians and fish [2–5]. Endogenized sections of circovirus genomes have also been detected in the germ line of mammals, birds, amphibian, reptiles, hagfish, bony fish, and in seven species of snakes in the Viperidae family and one member of the Pythonidae family (Burmese python or Python bivittatus) [6, 7]. The current and past host range of circoviruses therefore appears to include a wide range of vertebrates. A sister clade in the Circoviridae family named cyclovirus, whose complementary strand is packaged into virions, has been reported in insects and feces from different vertebrates [1]. Endogenized cyclovirus genomes have so far been detected only in the genome of one insect species (elongate twig ant or Pseudomyrmex gracilis) indicating that their tropism may be restricted to invertebrates [8].
In general, circoviruses cause diseases of the immune system, with lymphoid necrosis and depletion in primary or secondary lymphoid tissues linked with immunosuppression [9, 10]. Circovirus pathogenicity is also linked to opportunistic secondary infections and has been studied most extensively in pigs infected with porcine circovirus type 2 (PCV2) [11] and in birds infected with Psittacine beak and feather disease virus (BFDV) [12]. In swine, a number of factors including co-infections, viral load and genotype, management conditions, and serostatus have been shown to influence the outcome of circovirus infections [13–15]. Other circoviruses with described disease associations include canine circovirus [16–18], mink circovirus and other avian circoviruses [12, 19, 20].
So called “spinal osteopathy” is an umbrella term used to describe a well-recognized problem in captive reptiles that present with proliferative and degenerative osteochondral lesions in the vertebrae. In some cases, the characteristic lesions can be clearly associated with previous trauma or infections, while others have no clear etiology leaving metabolic, toxic, or nutritional etiologies as additional considerations [21, 22]. In the latter cases, lesions have been likened to osteitis deformans, or Paget’s Disease, of humans. In addition to similar pathologic findings seen in spinal osteopathy in reptiles, Paget’s Disease has no clear etiology, with genetic and viral origins considered possibilities [23, 24]. For both diseases, the development of lesions is likely complicated and multifactorial.
Here we used available tissues from a black-headed python (BhP) that was euthanized with severe spinal osteopathy of unknown etiology and characterize a novel circovirus genome named Black-headed python circovirus 1 strain SDZ9/2017/USA (BhPyCV-1). We then developed specific PCR and in situ hybridization probes to screen additional snakes in the Pythonidae and other families with spinal disease and fecal samples from live snakes with unknown disease status.
Materials and methods
Case material
Liver collected at post-mortem exam from a 3-year old male BhP euthanized for severe, progressive spinal disease was frozen at −70 and used for viral metagenomics. Archived formalin-fixed, paraffin-embedded sections of GI tract and liver from 17 similarly affected pythons were screened by in situ hybridization (ISH) for the presence of BhPyCV-1 RNA. Archived, frozen tissues from 13 of those snakes, four additional pythons, and prospectively collected fecal samples from a total of 37 additional live snakes of unknown disease status from various species were also used for PCR. All snakes were from the San Diego zoo.
Viral discovery
Liver tissue from the index BhP case was homogenized with a hand-held rotor in approximately 10× volume of Phosphate-buffered saline and rapidly frozen and thawed on dry ice 5 times. After centrifugation for 10 min in a table-top microfuge (15 000 × g), supernatant (400 mL) was collected and filtered through a 0.45 mm filter (Millipore). The filtrates were treated with a mixture of DNases (Turbo DNase [Ambion], Baseline-ZERO [Epicentre], benzonase [Novagen]) and RNase (Fermentas) at 37 °C for 90 min to enrich for viral capsid-protected nucleic acids [25]. Nucleic acids were then extracted using magnetic beads of the MagMAX Viral RNA Isolation kit (Ambion) according to the manufacturer’s instructions. Random RT-PCR followed by Nextera™ XT Sample Preparation Kit (Illumina) was used to generate a library for Illumina MiSeq (2 × 250 bases) with dual barcoding as previously described [26]. An in-house analysis pipeline was used to analyze sequence data. Paired-end reads of 250 bp generated by MiSeq were debarcoded using vendor software from Illumina. Adaptor and primer sequences are trimmed using the default parameters of VecScreen [27]. We developed a strategy that integrates the sequential use of de Bruijn graph (DBG) and overlap-layout-consensus assemblers (OLC) with a novel partitioned sub-assembly approach called ENSEMBLE [28]. Sequence reads were first analyzed using BLASTx (version 2.2.7) for translated protein sequence similarity to all viral protein sequences in GenBank’s virus RefSeq database plus protein sequences taxonomically annotated as viral in GenBank’s non-redundant database using E-value cutoff of 0.001. To remove background due to sequence misclassification these initial viral hits were then compared to all protein sequences in NR using the program DIAMOND (version 0.9.6) and retained only when the top hit was to a sequence annotated as viral. To align reads and contigs to reference viral genomes from GenBank and generate complete genome sequences the Geneious R10 program was used.
The amino acid (aa) pairwise alignments of Rep and Cap protein were performed by the Genious using the in-built MUSCLE algorithm. The amino acid phylogenetic trees were constructed using the Maximum likelihood method with two substitution models: Le Gascuel 2008 model based model with gamma distributed (G+) for Rep and Cap in MEGA software version 6 [29, 30].
PCR to confirm long intergenic region
After generation of a complete genome, the repeated region was shown in the long intergenic region. To confirming that region, we designed nested PCR primers as follows: for black-headed python circovirus, primers BhPyCV-817F1 and BhPyCV-277R1 were used for the first round of PCR, and primers BhPyCV-1607F2 and BhPyCV-147R2 for the second round of PCR (Table 1). The PCR conditions in these two assays were 95 °C for 5 min, 35 cycles 95 °C for 30 s, 58 °C (for the first or second round) for 30 s, and 72 °C for 1 min, a final extension at 72 °C for 10 min, resulting in an expected amplicon of 649 bp for BhPyCV long intergenic region.
Table 1.
PCR primers used in this study
| Primer ID | Sequence (5′ to 3′) | Size | Target |
|---|---|---|---|
| BhPyCV-817F1 | ATAAGGGTGGCACGGTTGAG | 1 round 1571 bp |
long intergenic region |
| BhPyCV-277R1 | ACAAACCCCTGGATATGCGG | ||
| BhPyCV-1607F2 | CAAAGTCCAGTACCGGTCCC | 2 round 649 bp |
|
| BhPyCV-147R2 | AACACCATCTCTTGCTCGGG | ||
| BhPyCV-642F | TGTTATATGGGCCCCCTGGA | 1 round 752 bp |
rep gene |
| BhPyCV-1393R1 | ACTTCAGACCACGTCCGAAC | ||
| BhPyCV-874R2 | CTCAACCGTGCCACCCTTAT | 2 round 233 bp |
In situ RNA hybridization
RNA in situ hybridization was performed using the RNAscope® 2.5 HD Red Chromogenic Reagent Kit according to the manufacturer’s instructions (Advanced Cell Diagnostics, Newark, CA, USA). Target probes were designed using the custom software as described previously [31], based on BhPyCV GenBank accession number MH368042. 5 μm sections of formalin fixed, paraffin embedded (FFPE) tissue were mounted on AutoFrost® charged adhesion slides (Cancer Diagnostics, Inc, Durham, NC, USA), baked at 60 °C in a dry oven, and deparaffinized. The sections were treated with an endogenous peroxidase blocker for 10 min at room temperature before boiling in a target retrieval solution for 15 min. Protease plus was then applied for 30 min at 40 °C. Target probes were hybridized for 2 h at 40 °C, followed by a series of signal amplification and washing steps. Hybridization signals were detected by chromogenic reactions using Fast Red. Slides were counterstained in 50% hematoxylin for 2 min and decolorized with 0.2% ammonium hydroxide. After rinsing in deionized water, they were dried in a 60 °C oven, dipped in xylene, and cover-slipped using xylene based SHUR/Mount medium (Triangle Biomedical Sciences, Durham, NC, USA). RNA staining signal was identified as red punctate dots. Negative control background staining was evaluated using a probe specific to a bacterial DapB gene.
Rep gene polymerase chain reaction assays (PCR)
DNA was extracted from available, archived tissues belonging to python species that had been frozen at −70 °C, and from prospectively collected fecal samples from multiple species. A total 67 tissues or fecal samples were tested representing 53 snakes and 18 different species. The tissue extraction was performed using the Qiagen Dneasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) was utilized following the manufacturer’s tissue extraction protocol. The sample DNA was eluted at 100 µL. DNA concentration and purity was assessed utilizing the Biotek Synergy Take3 plate system (Biotek, Winooski, VT, USA). The DNA was stored at 4 °C prior to use and stored long-term at −80 °C. Two primer sets designed in house utilizing the Geneious software v10.2.5 were used independently or as a semi-nested PCR reaction to screen frozen tissues for BhPyCV. The first set included forward primer sequence BhPyCV_642F and reverse primer BhPyCV_1393R1 targeting a ~752 bp fragment of the rep gene. The second set consisted of forward primer BhPyCV_642F and reverse primer BhPyCV_874R2 targeting a ~233 bp of the same region (Table 1). PCR assays were performed using the Eppendorf Mastercycler Pro S thermocycling system (Eppendorf, Hauppauge, NY, USA). Each 25 µL reaction contained the following reagents; 12.5 µL of 2× My Taq HS Mix (Bioline), 400 nM of each primer (Eton Bioscience, San Diego, CA, USA). 2 µL of the tissue extracted DNA was added to each reaction. Each sample was run with a known positive control, negative extraction control, and PCR negative control. The first PCR round had the following reaction parameters: 95 °C for 5 min, [95 °C for 30 s, 53.5 °C for 30 s, 72 °C for 90 s] × 40 cycles, 72 °C for 10 min, and held at 4 °C. The second round PCR was identical except that the annealing temperature was at 53.7 °C. Bands of the expected size were identified using a 1.8% agarose gel stained with ethidium bromide, gel extracted using the EMD Millipore Ultrafree™-DA (Millipore, Billerica, MA, USA), and Sanger sequenced.
Results
Genome characterization
The liver of a BhP with spinal cord defect was processed for viral metagenomics. Viral like particle (VLP)-associated nucleic acids were enriched, randomly amplified and processed for sequencing using the Illumina MiSeq platform (Materials and methods). Nearly 350,000 reads (250 bases long) were generated. The raw sequence data is available at NCBI’s Short Reads Archive under GenBank accession number SRR7982872. Following BLASTx against all known viral protein sequences available in GenBank, a total of 817 reads yielded matches to different circovirus proteins with E score ranging from 1.18e−67 to 5.6e−4 including a 1527 bases contigs. No other viral contigs nor credible matches to other viral proteins were identified. The contigs were used to design outward-facing PCR primers which were used to fill the gap of the circular DNA genomes by PCR and Sanger sequencing resulting in a 2110 bases genome. Typical of circoviruses, an origin of replication consisting of a 9 base-pair stem-loop topped by unpaired canonical TAGTATTAC sequence was flanked by a Rep protein open reading frame (ORF) encoded on the same strand and a Cap protein ORF encoded on the complementary strand (Figure 1A). The expected Rep rolling circle replication and SF3 helicase motifs were all detected (Figure 1A, Additional file 1) [1]. As typical of circovirus nucleocapsid sequences, its amino acid terminus was rich in basic amino acids (MYRRHFRRRFHHKRHTHFRRYKRHFHHKKR).
Figure 1.
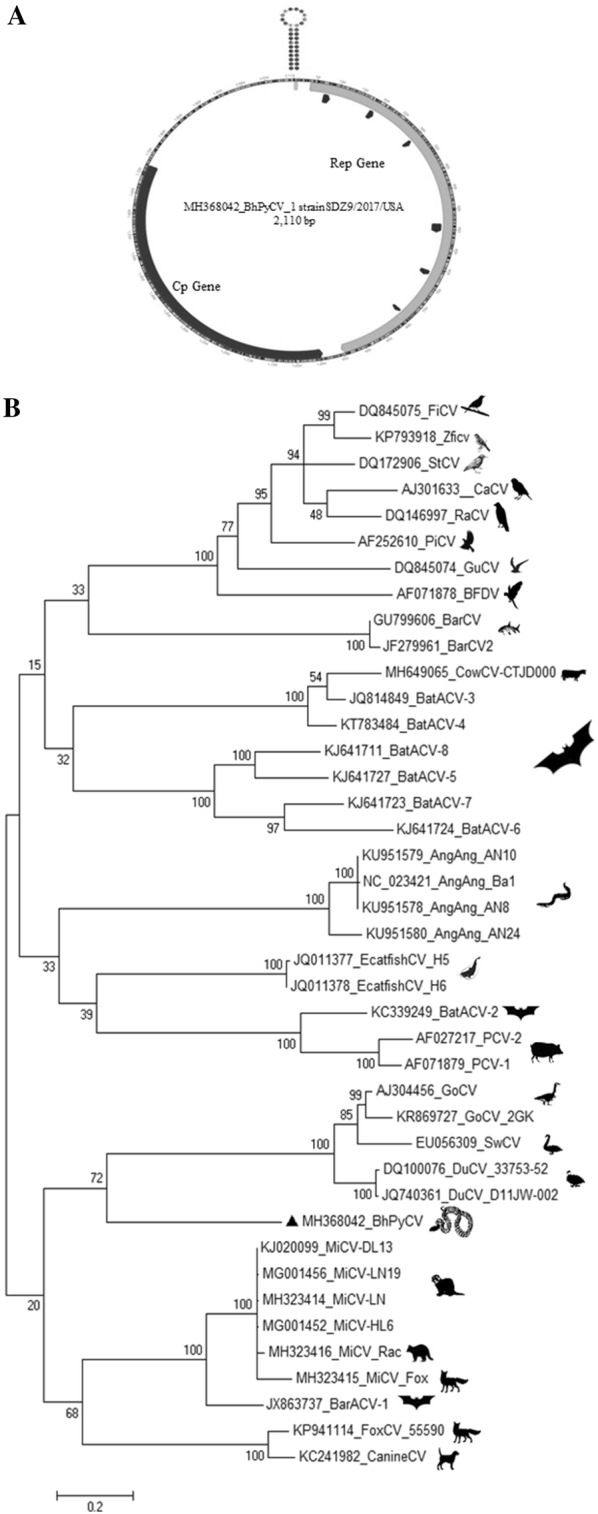
Characteristics of BhPyCV genome. A Open reading frames and stem-loop origin of replication of BhPyCV genome. Conserved domains in Rep protein are highlighted. B Phylogenetic analysis of circovirus Rep proteins using Maximum Likelihood.
An unusually large untranslated region (UTR) of 508 bases was found between the start codon of the Rep and Cap genes (Figure 1A). The length of this UTR was confirmed by nested PCR using PCR primers located within the Rep and Cap genes and Sanger sequencing. The Rep protein showed closest, yet distant, similarity with the Rep of mink circovirus (GB MG001456) with 51% identity (over 92% protein) followed by goose circovirus (GB KR869727) with 50% identity (over 93% protein) (Additional file 2). Sequence alignments showed that the Cap protein of the BhPyCV showed closest (although distant) similarity to a circovirus (BtCV LJR22) of a Least horseshoe bat (Rhinolophus pusillus) from China (GB MF278661) with 26% identity (over 81% of protein) (Additional file 2). That bat circovirus genome also showed an unusually long UTR of 450 bases while other circovirus UTR have length ranging from ~70 to 240 bases. The next best capsid protein alignment was with PCV2 with 21% identity over 78% of protein followed by capsids from bird circoviruses (Additional file 2). Phylogenetic analysis of the python circovirus Rep is shown showing its deep branch to other circovirus Rep with a weakly supported branch (72/100) with water fowl-infecting circoviruses (Figure 1B).
RNA in situ hybridization
RNA ISH was used to confirm infection of the original snake as well as screen available tissues from additional Pythonidae species. In the index BhP (#60415), positive ISH signal was intense throughout the cell lining and mucosa of the small intestine, extending from the superficial submucosa to the tips of villi (Figure 2). Signal was strongest along the epithelial lining with numerous positive cells in the lamina propria which was mildly edematous with some loosely scattered lymphocytes identified on routine staining. Individualized to coalescent signal was seen in submucosal lymphoid follicles and more loosely dispersed in the submucosal stroma. In the liver, positive signal was interpreted as similar bright red and punctate dots within sinusoidal melanomacrophage centers.
Figure 2.
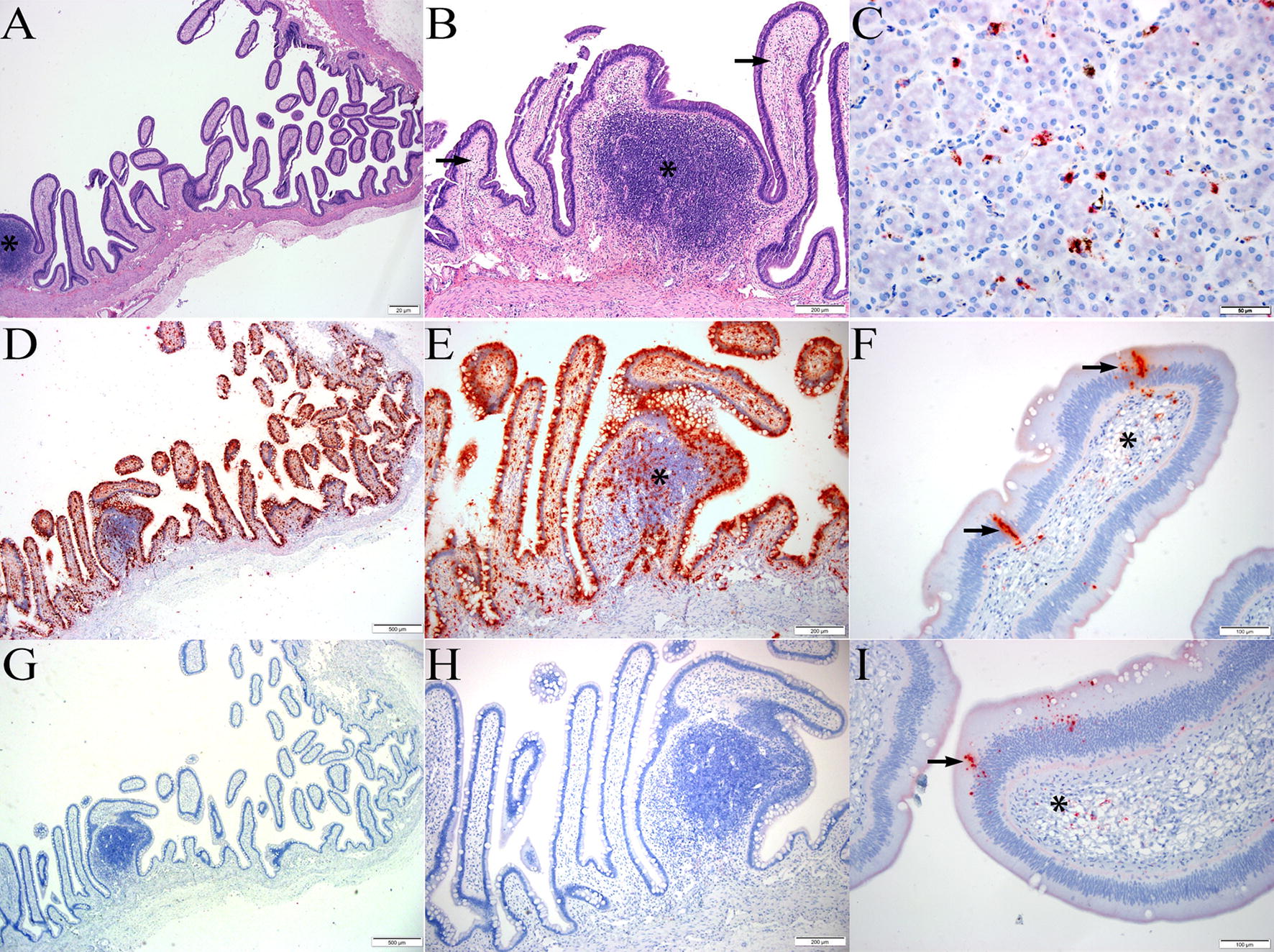
BhPyCV infection. A, B Longitudinal section of small intestine from index BhP at low (×4, A) and higher magnification (×10, B) showing a mucosal lymphoid aggregate (asterisk) and villi mildly expanded by edema and lymphocytes (arrows), H&E. D, E Serial sections from A–B showing abundant, deep red, punctate to coalescing ISH signal lining the mucosal epithelium, filling intestinal villi, and scattered in lymphoid follicle (asterisk). Hematoxylin counterstain, ×4 (D); ×10 (E). G, H Serial histologic sections from A–B, D–E. Negative control using bacterial DapB gene probe with no positive ISH signal, hematoxylin counterstain, ×4 (G); ×10 (H). C BhPyCV ISH probe on liver from index BhP showing discrete, punctate red staining within sinusoidal macrophages. Hematoxylin counterstain, ×40. F, I Medium–high magnification of intestinal villi from second BhP showing similar yet less dense and frequent staining within the epithelium (arrows) and scattered in the lamina propria (asterisk). Hematoxylin counterstain, ×20.
An additional 17 snakes in the family Pythonidae, all of which have been diagnosed with spinal osteoarthropathy, had both liver and GI tissues screened with ISH. One other BhP showed strong but much less frequent staining in the intestinal mucosa only, and a Boelen’s python had similar staining in the liver (Figure 2). The remainder of pythons had what was considered negative or equivocal staining characterized by rare, single, punctate dots at the basolateral aspect of intestinal epithelium or localized to the nuclear membrane (not shown). No other staining was seen in the liver or intestine of the snakes screened and no similar staining appeared in any of the negative control slides stained with unrelated probes. Negative by ISH were gut and liver tissues from 10 Green tree pythons, 2 carpet pythons, 2 diamond python, and another Boelen’s pythons all exhibiting the spinal defect (Table 2).
Table 2.
Deceased snakes tissues with spinal defect screened for BhPyCV with ISH and PCR
| ISH liver and gut | PCR liver | |
|---|---|---|
| Species | Animal tested/positive | Animal tested/positive |
| Black-headed python | 2/2a | 2/2a |
| Ball python | 0/0 | 1/0 |
| Boelen’s python | 2/1b | 1/0b |
| Burmese python | 0/0 | 2/0 |
| Carpet python | 2/0 | 0/0 |
| Diamond python | 2/0 | 0/0 |
| Green tree python | 10/0c | 10/0c |
| Scrub python | 0/0 | 1/0 |
aSame two animals tested by ISH and PCR. Tissues tested/PCR positive: liver (2/2), spleen (2/2), bone (2/2), lung (1/2), kidney (1/2).
bOnly the ISH negative Boelen’s python tested by PCR.
cSame ten snakes tested by ISH and PCR.
PCR
Archived tissues from python tissues frozen at −70 were screened by hemi-nested PCR using BhPyCV-specific primers. Additionally, DNA from fecal samples collected prospectively to screen for Cryptosporidium infection were opportunistically tested for BhPyCV DNA. A summary of PCR results is presented in Table 2. In total six snakes tested positive, the two ISH positive BhP which died with spinal disease (including the index BhP) from whom multiple tissues including liver, spleen, lung, bone, and spleen were PCR positive (Table 2). Feces from another four live snakes were PCR positive, 2/3 BhP and 2/6 annulated boas with unknown disease status (Table 3). Feces (n = 28) from another 10 snake species and tissues (n = 4) from another 3 snakes species also with unknown status were also PCR negative. PCR products were Sanger sequenced showing nucleotide identity of 99.5 to 100% with the sequence from the index BhP.
Table 3.
Feces from live snakes with unknown diseases status screened for BhPyCV by PCR
| Species | Animal tested | Animal positive |
|---|---|---|
| African bush viper | 2 | 0 |
| Angolan python | 8 | 0 |
| Annulated tree boa | 6 | 2 |
| Banded water cobra | 1 | 0 |
| Black-headed python | 3 | 2 |
| Eastern indigo snake | 2 | 0 |
| Eyelash viper | 2 | 0 |
| Green anaconda | 2 | 0 |
| King cobra | 1 | 0 |
| Madagascar tree boa | 1 | 0 |
| Mangshan pitviper | 2 | 0 |
| Rattleless rattlesnake | 7 | 0 |
Discussion
We describe here the first circovirus genome replicating in a reptile, the Black-headed Python (Aspidites melanocephalus). The detection of an exogenous snake circovirus was anticipated by the report of endogenized circovirus genome remnant in the chromosomes of species in the Pythonidae and Viperidae families [6, 7]. The exogenous circovirus genome described here was not the closest extant relative of the endogenized rep sequences (over an only partially retained Rep encoding region) found integrated in genomes of the family Viperidae (the rattlesnakes Crotalus horridus and Crotalus mitchellii) and family Pythonidae (the Burmese Python, Python bivittatus) (Additional file 3). These results indicate that further exogenous snake circoviruses likely remain to be characterized including lineages more closely related to endogenized circoviral elements [6, 7].
BhPyCV showed typical bidirectional rep and cap ORFs genome architecture with low sequence identity with other circoviruses and an unusually long UTR over the stem loop origin of replication.
The virus was detected by ISH in liver and/or gut of three deceased snakes with severe, non-inflammatory lesions of bone (two BhP and one Boelen’s python). In both BhP cases, DNA extracted from bone was also PCR positive. Feces from another 2/3 BhP and from 2/5 annulated boas of unknown disease status were also PCR positive indicating that enteric replication likely also occurs in both species. However, a number of other pythonidae snakes with the same syndrome were PCR negative in liver and by ISH in liver and GI tract, making the correlation between circoviral infection and spinal disease speculative and currently unsupported by this preliminary screening.
While spinal osteopathy has been linked to infections [21], our cases were considered less likely to be infectious given the lack of an inflammatory response and the dramatic nature to the lesions. Metabolic, toxic or nutritional etiologies remain under strong consideration. Nevertheless, the discovery of a novel virus in this population warrants additional investigation. Not all infections, particularly with viruses, cause significant inflammatory reactions. Many viruses are capable of replicating in tissue and employing mechanisms to evade surveillance and removal by the immune system, including circoviruses [32]. Tissue inflammatory mediators in the absence of a cellular response could also play a role in the development of spinal osteopathy. While numerous species in this collection are affected, often the affected tissues (vertebral bone) are unavailable for screening by ISH and PCR. Routinely collected samples at post-mortem do not typically include bone, and traditional decalcification in hydrochloric acid precludes the ability to run RNA ISH. Therefore, prospective collection of target tissues including affected and unaffected bone as well as optimization of the ISH on decalcified bone is warranted.
Lastly, a previous inflammatory process seeding the vertebral vessels and affecting the bones and joints of the vertebrae cannot be ruled out. Veterinary pathologists with exotic experience consider reptilian bone a dynamic tissue with the propensity to react to different injuries in an exuberate fashion. Therefore prior bone and joint damage could lead to instability and subsequent, ongoing osteochondral proliferation. There is evidence that once infected, circoviruses persist in tissue [33–35]. Stress, co-infections, and other factors could be associated with recurrent bouts of virus replication, compounding inflammatory tissue damage over time. This emphasizes the importance of prospective screening of bone. With more target tissue to evaluate, case–control studies should yield stronger conclusions about the association between circovirus and spinal osteopathy.
Regardless of the relationship of circovirus to spinal osteopathy in snakes, its role in other diseases, particularly as a co-factor in infectious/inflammatory diseases, should also be considered. It is not uncommon for snakes to have inflammatory diseases of the GI, respiratory and reproductive tracts, often with either commensal or opportunistic organisms detected via culture. The possibility of a predisposing circovirus infection can therefore also be considered.
Additional files
Additional file 1. Conserved motifs of Rep detected in Black headed python circovirus 1 based on [ 1 ].
Additional file 2. Pairwise amino acid sequence identity values between the Rep and Cap of BhPyCV and those of the most closely related circoviruses.
Additional file 3. Alignment, protein distances, and phylogenetic analysis of partial Rep of BhPyCV, endogenized snake circoviruses (Serpentes taxa) and exogeneous circoviruses.
Acknowledgements
We acknowledge the support from Vitalant Research Institute (formerly Blood Systems Research Institute), the Virginia Friedhofer Charitable Trust, and the Institute for Conservation Research, San Diego Zoo Global.
Abbreviations
- Rep
replication-associated protein
- Cap
nucleocapsid
- PCV2
porcine circovirus type 2
- BFDV
Psittacine beak and feather disease virus
- BhP
black-headed python
- BhPyCV
black-headed python circovirus
- ISH
in situ hybridization
- DBG
de Bruijn graph
- OLC
overlap-layout-consensus assemblers
- aa
amino acid
- G+
gamma distributed
- FFPE
formalin fixed, paraffin embedded
- PCR
polymerase chain reaction assays
- VLP
viral like particle
- ORF
open reading frame
- UTR
untranslated region
- GB
GenBank
Authors’ contributions
ED, EA and SVK, designed the study. SVK and EB provided samples. EA, SVK and JB performed the experiments. ED, EA, SVK and XD, analyzed the data. ED, SVK and EA wrote the manuscript. ED, SVK and EB edited the manuscript and provided funding. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Samples from live snakes tested via PCR were utilized under San Diego Zoo Global’s Institutional Animal Care and Use Committee (#15-017 and #18-024). Diseased snakes that contributed tissues for metagenomics, PCR, and ISH were humanely euthanized due to poor or grave prognosis for recovery in accordance with the American Association of Zoo Veterinarians Guidelines for Euthanasia of Non Domestic Animals (2006) and the American Veterinary Medical Association Guidelines for the Euthanasia of Animals (2013).
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Eda Altan, Email: edaltan@hotmail.com.
Steven V. Kubiski, Email: skubiski@gmail.com
Jennifer Burchell, Email: JBurchell@sandiegozoo.org.
Elizabeth Bicknese, Email: BBicknese@sandiegozoo.org.
Xutao Deng, Email: XDeng@vitalant.org.
Eric Delwart, Email: EDelwart@vitalant.org.
References
- 1.Rosario K, Breitbart M, Harrach B, Segales J, Delwart E, Biagini P, Varsani A. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol. 2017;162:1447–1463. doi: 10.1007/s00705-017-3247-y. [DOI] [PubMed] [Google Scholar]
- 2.Delwart E, Li L. Rapidly expanding genetic diversity and host range of the Circoviridae viral family and other Rep encoding small circular ssDNA genomes. Virus Res. 2012;164:114–121. doi: 10.1016/j.virusres.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarjan ZL, Penzes JJ, Toth RP, Benko M. First detection of circovirus-like sequences in amphibians and novel putative circoviruses in fishes. Acta Vet Hung. 2014;62:134–144. doi: 10.1556/AVet.2013.061. [DOI] [PubMed] [Google Scholar]
- 4.Lorincz M, Csagola A, Farkas SL, Szekely C, Tuboly T. First detection and analysis of a fish circovirus. J Gen Virol. 2011;92:1817–1821. doi: 10.1099/vir.0.031344-0. [DOI] [PubMed] [Google Scholar]
- 5.Lorincz M, Dan A, Lang M, Csaba G, Toth AG, Szekely C, Csagola A, Tuboly T. Novel circovirus in European catfish (Silurus glanis) Arch Virol. 2012;157:1173–1176. doi: 10.1007/s00705-012-1291-1. [DOI] [PubMed] [Google Scholar]
- 6.Dennis TPW, de Souza WM, Marsile-Medun S, Singer JB, Wilson SJ, Gifford RJ. The evolution, distribution and diversity of endogenous circoviral elements in vertebrate genomes. Virus Res. 2019;262:15–23. doi: 10.1016/j.virusres.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert C, Meik JM, Dashevsky D, Card DC, Castoe TA, Schaack S. Endogenous hepadnaviruses, bornaviruses and circoviruses in snakes. Proc Biol Sci. 2014;281:20141122. doi: 10.1098/rspb.2014.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dennis TPW, Flynn PJ, de Souza WM, Singer JB, Moreau CS, Wilson SJ, Gifford RJ. Insights into circovirus host range from the genomic fossil record. J Virol. 2018;92:e00145-18. doi: 10.1128/JVI.00145-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mankertz A, Caliskan R, Hattermann K, Hillenbrand B, Kurzendoerfer P, Mueller B, Schmitt C, Steinfeldt T, Finsterbusch T. Molecular biology of Porcine circovirus: analyses of gene expression and viral replication. Vet Microbiol. 2004;98:81–88. doi: 10.1016/j.vetmic.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Parrish C. Circoviridae. Fenner’s veterinary virology. 4. London: Elsevier; 2011. [Google Scholar]
- 11.Allan G, Krakowka S, Ellis J, Charreyre C. Discovery and evolving history of two genetically related but phenotypically different viruses, porcine circoviruses 1 and 2. Virus Res. 2012;164:4–9. doi: 10.1016/j.virusres.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 12.Todd D. Circoviruses: immunosuppressive threats to avian species. Avian Pathol. 2000;29:373–394. doi: 10.1080/030794500750047126. [DOI] [PubMed] [Google Scholar]
- 13.Allan GM, McNeilly F, Cassidy JP, Reilly GA, Adair B, Ellis WA, McNulty MS. Pathogenesis of porcine circovirus; experimental infections of colostrum deprived piglets and examination of pig foetal material. Vet Microbiol. 1995;44:49–64. doi: 10.1016/0378-1135(94)00136-K. [DOI] [PubMed] [Google Scholar]
- 14.Ellis JA, Bratanich A, Clark EG, Allan G, Meehan B, Haines DM, Harding J, West KH, Krakowka S, Konoby C, Hassard L, Martin K, McNeilly F. Coinfection by porcine circoviruses and porcine parvovirus in pigs with naturally acquired postweaning multisystemic wasting syndrome. J Vet Diagn Investig. 2000;12:21–27. doi: 10.1177/104063870001200104. [DOI] [PubMed] [Google Scholar]
- 15.Opriessnig T, Halbur PG. Concurrent infections are important for expression of porcine circovirus associated disease. Virus Res. 2012;164:20–32. doi: 10.1016/j.virusres.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson A, Hartmann K, Leutenegger CM, Proksch AL, Mueller RS, Unterer S. Role of canine circovirus in dogs with acute haemorrhagic diarrhoea. Vet Rec. 2017;180:542. doi: 10.1136/vr.103926. [DOI] [PubMed] [Google Scholar]
- 17.Decaro N, Martella V, Desario C, Lanave G, Circella E, Cavalli A, Elia G, Camero M, Buonavoglia C. Genomic characterization of a circovirus associated with fatal hemorrhagic enteritis in dog, Italy. PLoS One. 2014;9:e105909. doi: 10.1371/journal.pone.0105909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, McGraw S, Zhu K, Leutenegger CM, Marks SL, Kubiski S, Gaffney P, Jr Dela Cruz FN, Wang C, Delwart E, Pesavento PA. Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerg Infect Dis. 2013;19:534–541. doi: 10.3201/eid1904.121390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kundu S, Faulkes CG, Greenwood AG, Jones CG, Kaiser P, Lyne OD, Black SA, Chowrimootoo A, Groombridge JJ. Tracking viral evolution during a disease outbreak: the rapid and complete selective sweep of a circovirus in the endangered Echo parakeet. J Virol. 2012;86:5221–5229. doi: 10.1128/JVI.06504-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lian H, Liu Y, Li N, Wang Y, Zhang S, Hu R. Novel circovirus from mink, China. Emerg Infect Dis. 2014;20:1548–1550. doi: 10.3201/eid2009.140015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isaza R, Garner M, Jacobson E. Proliferative osteoarthritis and osteoarthrosis in 15 snakes. J Zoo Wildl Med. 2000;31:20–27. doi: 10.1638/1042-7260(2000)031[0020:POAOIS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 22.Kiel JL. Spinal osteoarthropathy in two southern copperheads. J Zoo Anim Med. 1977;8:21–24. doi: 10.2307/20460146. [DOI] [Google Scholar]
- 23.Mills BG, Singer FR. Nuclear inclusions in Paget’s disease of bone. Science. 1976;194:201–202. doi: 10.1126/science.959849. [DOI] [PubMed] [Google Scholar]
- 24.Ralston SH, Langston AL, Reid IR. Pathogenesis and management of Paget’s disease of bone. Lancet. 2008;372:155–163. doi: 10.1016/S0140-6736(08)61035-1. [DOI] [PubMed] [Google Scholar]
- 25.Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, Naeem A, Zaidi S, Delwart E. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Deng X, Mee ET, Collot-Teixeira S, Anderson R, Schepelmann S, Minor PD, Delwart E. Comparing viral metagenomics methods using a highly multiplexed human viral pathogens reagent. J Virol Methods. 2015;213:139–146. doi: 10.1016/j.jviromet.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye J, McGinnis S, Madden TL. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006;34:W6–W9. doi: 10.1093/nar/gkl164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng X, Naccache SN, Ng T, Federman S, Li L, Chiu CY, Delwart EL. An ensemble strategy that significantly improves de novo assembly of microbial genomes from metagenomic next-generation sequencing data. Nucleic Acids Res. 2015;43:e46. doi: 10.1093/nar/gkv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNeilly F, Allan GM, Foster JC, Adair BM, McNulty MS, Pollock J. Effect of porcine circovirus infection on porcine alveolar macrophage function. Vet Immunol Immunopathol. 1996;49:295–306. doi: 10.1016/0165-2427(95)05476-6. [DOI] [PubMed] [Google Scholar]
- 33.Bolin SR, Stoffregen WC, Nayar GP, Hamel AL. Postweaning multisystemic wasting syndrome induced after experimental inoculation of cesarean-derived, colostrum-deprived piglets with type 2 porcine circovirus. J Vet Diagn Investig. 2001;13:185–194. doi: 10.1177/104063870101300301. [DOI] [PubMed] [Google Scholar]
- 34.Duchatel JP, Todd D, Smyth JA, Bustin JC, Vindevogel H. Observations on detection, excretion and transmission of pigeon circovirus in adult, young and embryonic pigeons. Avian Pathol. 2006;35:30–34. doi: 10.1080/03079450500465692. [DOI] [PubMed] [Google Scholar]
- 35.Madson DM, Patterson AR, Ramamoorthy S, Pal N, Meng XJ, Opriessnig T. Reproductive failure experimentally induced in sows via artificial insemination with semen spiked with porcine circovirus type 2. Vet Pathol. 2009;46:707–716. doi: 10.1354/vp.08-VP-0234-O-FL. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Conserved motifs of Rep detected in Black headed python circovirus 1 based on [ 1 ].
Additional file 2. Pairwise amino acid sequence identity values between the Rep and Cap of BhPyCV and those of the most closely related circoviruses.
Additional file 3. Alignment, protein distances, and phylogenetic analysis of partial Rep of BhPyCV, endogenized snake circoviruses (Serpentes taxa) and exogeneous circoviruses.


