Abstract
Background
Environmental risk factors have been shown to alter DNA copy number variations (CNVs). Recently, CNVs have been described to arise after low-dose ionizing radiation in vitro and in vivo. Development of cost- and size-effective laser-driven electron accelerators (LDEAs), capable to deliver high energy beams in pico- or femtosecond durations requires examination of their biological effects. Here we studied in vitro impact of LDEAs radiation on known CNV hotspots in human peripheral blood lymphocytes on single cell level.
Results
Here CNVs in chromosomal regions 1p31.1, 7q11.22, 9q21.3, 10q21.1 and 16q23.1 earlier reported to be sensitive to ionizing radiation were analyzed using molecular cytogenetics. Irradiation of cells with 0.5, 1.5 and 3.0 Gy significantly increased signal intensities in all analyzed chromosomal regions compared to controls. The latter is suggested to be due to radiation-induced duplication or amplification of CNV stretches. As significantly lower gains in mean fluorescence intensities were observed only for chromosomal locus 1p31.1 (after irradiation with 3.0 Gy variant sensitivites of different loci to LDEA is suggested. Negative correlation was found between fluorescence intensities and chromosome size (r = − 0.783, p < 0.001) in cells exposed to 3.0 Gy irradiation and between fluorescence intensities and gene density (r = − 0.475, p < 0.05) in cells exposed to 0.5 Gy irradiation.
Conclusions
In this study we demonstrated that irradiation with laser-driven electron bunches can induce molecular-cytogenetically visible CNVs in human blood leukocytes in vitro. These CNVs occur most likely due to duplications or amplification and tend to inversely correlate with chromosome size and gene density. CNVs can last in cell population as stable chromosomal changes for several days after radiation exposure; therefore this endpoint can be used for characterization of genetic effects of accelerated electrons. These findings should be complemented with other studies and implementation of more sophisticated approaches for CNVs analysis.
Keywords: AREAL, Accelerated electrons, Copy number variations (CNVs), Chromosome size, Duplication, Gene density
Background
Copy number variations (CNVs) that arise due to deletions and duplications in the genome are major contributors to genetic diversity in human population [1]. These changes may lead to phenotypic expression and/or various diseases (cancer, infertility, neurodevelopmental disorders etc.) [2–4], have adaptive effects [5], or can be neutral without significant consequences [1]. Recent studies demonstrated that DNA replication inhibitors have potential to induce CNVs in vitro and in vivo [6–10]. However, natural and artificial environmental factors that may induce CNVs are still poorly studied.
Recent achievements in the field of particle acceleration technologies has led to development of cost- and size-effective laser-driven electron accelerators (LDEAs) [11, 12]. This technology permits to deliver high energy beams (from few MeV up to several hundred MeV) into deep layers of tissue in pico- or femtosecond durations with little lateral spread [13–15]. Regarding the long-term goal to develop and establish laser-based particle accelerators for a future radiotherapeutic treatment of cancer, the radiobiological consequences of laser-driven beams have to be investigated [16].
Different types of DNA damage-associated biomarkers have shown potential as predictors of radiation effects, including cytogenetics (e.g. micronuclei, translocations, dicentrics), proteomics (e.g. g-H2AX, pATM, pP53), genomics (e.g. mRNA, SNPs), or epigenomics (e.g. miRNA, lncRNA) [17]. One of the widely used methods in radiation biology is the comet assay which enables detection of initial radiation-induced DNA breaks [18–20] and analysis of the inter-individual [21] and inter-cellular [22] differences in response to radiation. DNA double-strand breaks (DSBs) represent an important radiation-induced lesion that can be monitored by the gammaH2AX foci analysis [23]. Misrepair of DSBs can produce many types of chromosomal aberrations, in particular DNA rearrangements, including CNVs [9]. While majority of DNA damage can be repaired in several hours or days, chromosomal aberrations are more persistent and even can last up to several years in human. Therefore, evaluation of chromosome damage is a crucial predictor for the degree of radiation induced damage [24–28]. In several studies effects of radiation have been studied focusing on CNVs. In particular, low-dose ionizing radiation was shown to induce de novo CNVs in in vitro studies, with slight prevalence of duplications over deletions. Moreover, hotspots of radiation-induced CNVs have been identified in chromosomal regions 1q44, 3q13.31, 7q11.22, 9p21.3, 10q11.23-q21.1 and 16q23.1 [7]. Furthermore, the frequency of de novo CNVs was significantly elevated in offspring of laboratory mice, exposed to ionizing radiation [9], as well as in the progeny of a human subpopulation accidentally exposed during a radiological accident [10].
Studies of the genetic effects of accelerated particles are still limited. Recently genetic effects of irradiation with LDEA were estimated using comet assay [29, 30], micronucleus test [12] and gammaH2AX foci, reflecting level of DSBs [16, 31, 32]. But for all we know, the effect of accelerated particles on CNVs has not yet been studied. The introduction of additional radiobiological endpoints would greatly expand our understanding of biological effectiveness of laser-driven electron beams.
The efficiency of CNVs as endpoint of genetic effects of ionizing radiation supports the possibility of their application in studies of accelerated electrons. Here we examine in vitro laser-generated ultrashort electron beam irradiation effect on CNVs hotspots in blood lymphocytes of healthy individuals, using parental origin determination fluorescence in situ hybridization (POD-FISH) technique.
Methods
Blood cultivation and irradiation
Blood samples were collected by venipuncture from four healthy nonsmoking donors (two female and two male) aged 27–29 years, with normal 46,XX and 46,XY karyotypes, respectively. This study was approved by the Ethic Committee of the National Center of Bioethics (Yerevan State University, Faculty of Biology), and informed consent was obtained from all study donors. The venous blood (2 ml from each donor) was collected into vacutainers with heparin and irradiated by a laser-driven radiofrequency gun-based linear AREAL accelerator (CANDLE, Synchrotron Research Institute, Armenia). Vacutainers with blood samples were placed in the sample holder facing vertically towards the beam coming from the direction of the vacuum window. Earlier the levels of DNA damage in human K-562 cells were studied after irradiation with LDEAs at 0, 2, 4 and 8 Gy at 3.6 and 36 Gy/min dose rates [30]. In current study, relatively low radiation doses and dose rate were selected to avoid pronounced increase of DNA damage. Samples were irradiated with doses of 0.5, 1.5 and 3.0 Gy with a dose rate of 2 Gy/min, the beam charge was 10 pC with the energy of electrons 3 MeV, pulse duration 0.42 fs and pulse repetition rate 2 Hz. After irradiation blood samples were cultivated in RPMI-1640 medium, containing 10% fetal bovine serum, 1% penicillin/streptomycin, and 10 μg/ml phytohemagglutinin-L at 37 °C for 72 h.
Metaphase chromosome preparation
Metaphase chromosomes were prepared as previously described [33]. Colcemid (0.1 μg/ml final concentration) was added to the culture 1.5 h before harvesting and incubated at 37 °C to achieve metaphase block. At the end of cultivation cells were harvested and centrifuged at 1500 rpm (7 min). The medium was removed completely except for about 0.5 ml of supernatant remaining above the cell pellet. 10 ml of pre-warmed (37 °C) hypotonic solution (0.075 M KCl) was added to the tubes and the contents were mixed gently and incubated for 15 min at 37 °C. After centrifugation and discarding supernatant, cells were fixed in 10 ml of ice-cold fixative (methanol/glacial acetic acid, 3:1 v/v). After incubation 10–15 min at room temperature the cells were centrifuged, supernatant was discarded and 10 ml of fixative was added. After the last centrifugation, cells were resuspended in a small amount of fixative and the suspension was dropped onto a microscope slide, prewashed by fixative. Then the slide was placed on hotplate (51 °C) covered by wet tissue paper and kept until the surface of the slide was dried.
FISH analysis
POD-FISH has already been successfully used to identify CNVs in human cells [8, 34, 35]. BAC clones for CNV regions were purchased from the Children’s Hospital Oakland Research Institute, Oakland, CA, USA, or kindly provided by the Sanger Centre, UK. BAC DNA was isolated, PCR amplified, and labeled by Nick translation (Roche, Karlsruhe, Germany) [36]. The following BACs were used: RP11-393 N21 for 1p31.1 (TexasRed), RP11-1129E22 for 7q11.22 (SpectrumGreen), RP11-174 K23 for 9q21.3 (TexasRed), RP11-123 L21 for 10q21.1 (SpectrumOrange) and RP11-264 M12 for 16q23.1 (SpectrumGreen). The set of CNV regions was selected on the base of results of genomic distribution of low-dose ionizing radiation-induced CNVs [7]. Image capturing and acquisition were processed with the Isis imaging system (MetaSystems, GmbH, Altlussheim, Germany). For analysis of POD-FISH signals the ImageJ freeware was applied (https://imagej.nih.gov/ij/) [37]. For that purpose images were imported into ImageJ program and size of CNVs was measured on the base of fluorescence intensities of signals from 50 to 60 metaphases for each chromosome region and expressed in arbitrary units (a. u.).
Statistical analysis
The normality of distribution of FISH signals intensity was analyzed by the Kolmogorov-Smirnov test. Differences between fluorescence intensities of studied chromosome loci of male and female donors were analyzed by Multiple Range test. Significance of difference between untreated and irradiated cells was tested by Student’s t-test. Pearson’s correlation was applied for analysis of relation between fluorescence intensities of chromosome loci after irradiation and chromosome size, gene density and interphase position. Statistical analysis was performed using the statistical package Statgraphics Centurion 16.2 and a p value < 0.05 was considered statistically significant.
Results
Comparison of CNVs in control and irradiated cells
CNVs of 5 chromosomal regions were analyzed by POD-FISH [8]. Fluorescence intensities of signals reflecting the sizes of the CNVs were compared between treated and untreated samples (Fig. 1). No significant difference in the fluorescence intensities of CNVs was found between males and females; so the pooled data from the four donors are presented in Table 1. Irradiation of cells with 0.5, 1.5 and 3.0 Gy significantly increased signal intensities in all analyzed chromosome regions compared with control due to induced duplications or amplifications. Non-significant increase was shown only in 7q11.22 after irradiation with 1.5 Gy. Studied chromosomal loci demonstrated minor differences in sensitivity to irradiation with LDEA (Table 1). Multiple Range test revealed significantly lower gains in fluorescence intensities in chromosome locus 1p31.1 after irradiation with 3.0 Gy compared to 7q11.22, 9q21.3, 10q21.1 and 16q23.1 loci indicating less LDEA sensitivity of this locus. Significant differences were not observed between loci 7q11.22, 9q21.3, 10q21.1 and 16q23.1 after irradiation with 3.0 Gy, as well as between all studied loci after irradiation with 0.5 and 1.5 Gy.
Fig. 1.
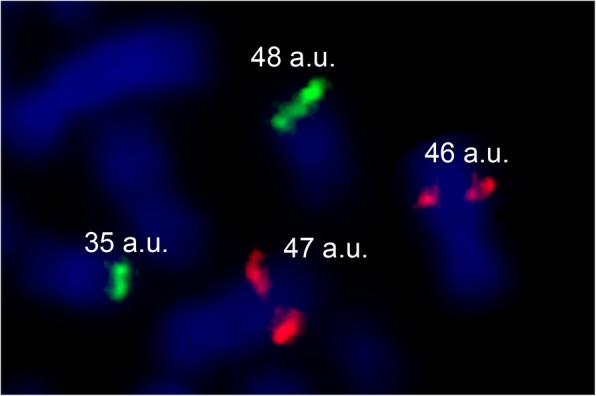
Sample of evaluation of signal intensities by ImageJ program. Signal intensities measurements in arbitrary units (a.u.) were done on homologous chromosomes of 9q21.3 (TexasRed) and 16q23.1 (SpectrumGreen) before and after irradiation with accelerated electrons by ImageJ program. Duplication (48 a.u.) was detected as increase of fluorescence intensity of BAC probe for 16q23.1 locus
Table 1.
Fluorescence intensity of BAC signals (mean ± SD of 50–60 measurements) in different chromosome loci after irradiation
| Dose (Gy) | 1p31.1 | 7q11.22 | 9q21.3 | 10q21.1 | 16q23.1 |
|---|---|---|---|---|---|
| 0 | 57.59 ± 1.75 | 56.06 ± 4.53 | 55.13 ± 1.09 | 56.22 ± 1.71 | 57.03 ± 1.81 |
| 0.5 | 67.20 ± 1.37* | 66.69 ± 2.80* | 64.02 ± 2.18* | 67.88 ± 3.15* | 69.81 ± 0.86* |
| 1.5 | 64.43 ± 1.79* | 63.64 ± 4.40 | 66.14 ± 2.07* | 64.51 ± 4.80* | 64.80 ± 0.60* |
| 3.0 | 64.83 ± 0.94* | 69.75 ± 1.37*a | 69.56 ± 0.91* a | 68.86 ± 1.01* a | 70.02 ± 2.01*a |
*p < 0.05—significant difference compared to non-irradiated cells
ap < 0.05—significantly higher gain in fluorescence intensity compared to 1p31.1 after irradiation with 3.0 Gy
Correlation of CNVs with chromosome size, gene density and interphase position
To study the involvement of different chromosomes in CNVs instability the Pearson (r) correlations between fluorescence intensities of studied chromosome loci after irradiation with doses 0.5, 1.5 and 3.0 Gy and chromosomes size (bp), gene density (gene/Mb) and interphase position [38, 39] were analyzed (Table 2). Negative correlation was found between fluorescence intensity and chromosome size (r = − 0.783, p < 0.001) in cells exposed to 3.0 Gy irradiation and between gene density (r = − 0.475, p < 0.05) in cells exposed to 0.5 Gy irradiation. Statistically significant correlation between fluorescence intensity in irradiated cells and 3D localization of chromosomes in the nucleus was not revealed (p > 0.05).
Table 2.
Correlations of fluorescence intensities in CNVs loci with chromosome size, gene density and interphase position
| 0.5 Gy | 1.5 Gy | 3.0 Gy | |
|---|---|---|---|
| Chromosome size | r = − 0.186 | r = −0.072 | r = − 0.783** |
| Gene density | r = −0.475* | r = −0.001 | r = 0.268 |
| Interphase position | r = −0.395 | r = −0.112 | r = − 0.170 |
Statistically significant negative correlations are indicated at *p < 0.05 and **p < 0.001
Discussion
Spontaneously arising CNVs as a source of genetic diversity in human population have been studied extensively [35, 40, 41] and their clinical impact was also demonstrated [42, 43]. Nevertheless, little is known about environmental factors that can induce de novo CNVs. It was shown that de novo CNVs may occur due to influence of replication inhibitors (aphidicolin, hydroxyurea) in vitro in normal human fibroblasts [6, 44]. Earlier we have confirmed these results using mycotoxin aflatoxin B1 as replication inhibitor in cultured human normal leukocytes [8].
Here we demonstrated that laser-driven electron bunches, a direct DNA damaging agent, may induce CNVs in chromosome loci 1p31.1, 7q11.22, 9q21.3, 10q21.1 and 16q23.1 in cultured normal human blood leukocytes. Our data confirmed that hotspots of de novo CNVs mutations defined in normal human fibroblast cell line after ionizing radiation [7] represent also targets for accelerated electrons. Flunkert et al. [45] showed that clones of primary human fibroblasts irradiated with X-ray displayed an increased rate of CNVs in 3p14.2 and 7q11.21. Consistent with this study, our results suggest that locus 7q11.2 is one of the most radiation sensitive sites. We showed that CNVs occurred as duplications or amplifications in all studied chromosome loci which is consistent with results of Arlt et al. [7] where excess of copy number gains over losses was detected. We found only minor differences in the sensitivity of studied sites to radiation. Only locus 1p31.1 was significantly more resistant to radiation at 3.0 Gy compared with other chromosome loci. Nevertheless, the analysis of CNVs in our work is limited by cytogenetically visible changes. We do not exclude the possibility of occurrence of small deletions and duplications as well as more inter-locus differences that are not recognizable by the method applied.
Regarding the mechanisms of ionizing radiation-induced de novo CNVs formation, Arlt et al. [7] hypothesized that the prevalence of copy number gains is difficult to explain via non-homologous end joining (NHEJ) of DNA double strand breaks. Thus, it was suggested that irradiation-induced CNVs are more likely to occur via replication-dependent mechanisms, e.g. repair of more abundant DNA single strand breaks or base lesions. Gains of CNVs in five chromosome loci are also found after irradiation with accelerated electrons; thus, this mechanism is the most appropriate for explaining the observed effects.
According to this analysis of de novo CNVs in human chromosomes 1, 7, 9, 10 and 16 it was shown that the level of fluorescence intensities in CNVs sites negatively correlated with chromosome size after exposure to 3.0 Gy, while no correlation was observed at lower doses of irradiation. These results allow assuming that LDEA-induced CNVs are more probable to occur in small chromosomes rather than in big chromosomes. Earlier Sommer et al. [46], assessing the radiation sensitivity of chromosomes 2, 8 and 14, demonstrated inverse correlation between chromosome DNA content and number of irradiation induced aberrations. Although this contradicts results of Cigarrán et al. [47] who demonstrated, with the exception of chromosome 20, a positive correlation between the DNA content and the number of exchange-type aberrations and the number of breaks in irradiated human blood lymphocytes. No hotspots of de novo CNVs in the chromosomes were identified in offspring of accidentally irradiated parents, confirming that induced CVNs fit the random-effect model of radiation exposure [10]. The literature data thus show both random and non-random distribution of damage across the genome obtained under various irradiation conditions using different biomarkers of radiation.
Negative correlation between increase of fluorescence intensity and gene density in cells irradiated with 0.5 Gy allows to assume that CNVs are more probable to occur in chromosomes with low gene density rather that in chromosomes with high gene density. This is in agreement with results of Rapp et al. [48] research, where the authors with implementation of comet-FISH technique demonstrated that UV-A-induced fragments of gene-rich chromosome 1 can be found in only 3% whereas fragmentation of the gene-poor chromosome 8 was observed in 25% of all comets. Interestingly, Surrallés et al. [49] demonstrated that chromosomes with high gene density are preferentially repaired after influence of mutagen compared to gene-poor chromosomes. Given the limitations of our research, we can only assume that the probability of formation of LDEA-induced mutations in the CNVs sites is inversely related to the chromosomes size and gene density. Further extended studies are needed to describe the distribution of de novo CNVs in chromosomes after irradiation with accelerated electrons.
CNVs observed in current study were persistent in blood cells 72 h post-exposure with accelerated electrons. According to Arlt et al. [7] CNVs can persist in fibroblast cell line after ionizing radiation for more than 7 days. Clones of human primary fibroblast after ionizing radiation displayed an increased rate of CNVs after 20 population doublings [45]. Long-term presence indicates the stability of de novo induced CNVs and their importance as bioindicator of radiation. Analysis of persistent cytogenetic changes in human population is critical for study of biological consequences of radiation exposure [24, 50, 51].
Several previous studies described radiation-induced CNVs in TK6 cells, a human B-cell lymphoblastoid cell line of nonmalignant origin after gamma irradiations with the prevalence of gains [52], and in aneuploid A549 male non-small cell lung cancer adenocarcinoma cell line irradiated with X rays [53]. Increase of CNVs was shown in mouse thymic lymphomas induced by gamma-irradiation in vivo [54]. Current knowledge on DNA copy number alterations in papillary thyroid carcinomas and on strategies to identify radiation-specific changes in these tumors was presented by Zitzelsberger and Unger [55]. According to Arlt et al. [7] the spectrum of DNA damaging agents that lead to increased rates of CNV formation, and the mechanisms by which these agents might act, are very poorly understood.
Thus, research in this area is of particular interest, especially since radiation-induced CNVs can be transmitted to next generation [9, 10] and may play an important role in radiation-induced carcinogenesis [45].
Conclusions
In summary, we have shown that irradiation with laser-driven electron bunches can induce CNVs in human blood leukocytes in vitro. These CNVs occurred due to duplications or amplifications which according to preliminary data can inversely correlate with chromosome size and gene density. Taking in consideration our previous and current results and literature data we can assume that LDEA may induce replication stress although other mechanisms of CNV induction are not precluded. The observation that CNVs can last in cell population as stable chromosomal changes for several days after radiation exposure allows recommending them for characterization of genetic effects of accelerated electrons. These findings should be complemented with other studies with implementation of more sophisticated methods for CNV analysis.
Acknowledgements
We thank all blood donors and MSc. medical doctor Arthur Avagyan for realization of venipuncture.
Funding
This study was funded by RA MES CS (#17A-1F008), RA MES CS - BMBF (#A/G-17) and DAAD (Eastern Partnership Program).
Availability of data and materials
The data sets supporting the conclusions of this article are included within the article.
Authors’ contributions
TH and GH drafted the manuscript. TL and RA edited the manuscript. TH and AS performed the FISH and CNV analysis. TH and GH performed the statistical analysis. TH, GH, RA and TL designed the study. AHA-R did the probe amplification and labeling. AW provided locus specific BAC probes for this study. BG realized irradiation of blood on AREAL accelerator. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written consent was obtained from all donors. The study was approved by the Ethic Committee of the National Center of Bioethics (Yerevan State University, Faculty of Biology).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tigran Harutyunyan, Email: tigranharutyunyan@ysu.am.
Galina Hovhannisyan, Email: galinahovhannisyan@ysu.am.
Anzhela Sargsyan, Email: angela.sargsyan@ysu.am.
Bagrat Grigoryan, Email: grigory@asls.candle.am.
Ahmed H. Al-Rikabi, Email: Ahmed.Al-Rikabi@med.uni-jena.de
Anja Weise, Email: ANJA.WEISE@med.uni-jena.de.
Thomas Liehr, Email: Thomas.Liehr@med.uni-jena.de.
Rouben Aroutiounian, Email: genetik@ysu.am.
References
- 1.Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16(3):172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 2.Shivakumar BM, Rotti H, Vasudevan TG, Balakrishnan A, Chakrabarty S, Bhat G, et al. Copy number variations are progressively associated with the pathogenesis of colorectal cancer in ulcerative colitis. World J Gastroenterol. 2015;21(2):616–622. doi: 10.3748/wjg.v21.i2.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong Y, Pan Y, Wang R, Zhang Z, Xi Q, Liu RZ. Copy number variations in spermatogenic failure patients with chromosomal abnormalities and unexplained azoospermia. Genet Mol Res. 2015;14(4):16041–16049. doi: 10.4238/2015.December.7.17. [DOI] [PubMed] [Google Scholar]
- 4.Grayton HM, Fernandes C, Rujescu D, Collier DA. Copy number variations in neurodevelopmental disorders. Prog Neurobiol. 2012;99(1):81–91. doi: 10.1016/j.pneurobio.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39(10):1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arlt MF, Ozdemir AC, Birkeland SR, Wilson TE, Glover TW. Hydroxyurea induces de novo copy number variants in human cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17360–17365. doi: 10.1073/pnas.1109272108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW. Copy number variants are produced in response to low-dose ionizing radiation in cultured cells. Environ Mol Mutagen. 2014;55(2):103–113. doi: 10.1002/em.21840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harutyunyan T, Hovhannisyan G, Babayan N, Othman MA, Liehr T, Aroutiounian R. Influence of aflatoxin B1 on copy number variants in human leukocytes in vitro. Mol Cytogenet. 2015;8:25. doi: 10.1186/s13039-015-0131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adewoye AB, Lindsay SJ, Dubrova YE, Hurles ME. The genome-wide effects of ionizing radiation on mutation induction in the mammalian germline. Nat Commun. 2015;6:6684. doi: 10.1038/ncomms7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa EOA, Pinto IP, Gonçalves MW, da Silva JF, Oliveira LG, da Cruz AS, et al. Small de novo CNVs as biomarkers of parental exposure to low doses of ionizing radiation of caesium-137. Sci Rep. 2018;8(1):5914. doi: 10.1038/s41598-018-23813-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faure J, Glinec Y, Pukhov A, Kiselev S, Gordienko S, Lefebvre E, et al. A laser-plasma accelerator producing monoenergetic electron beams. Nature. 2004;431:541–544. doi: 10.1038/nature02963. [DOI] [PubMed] [Google Scholar]
- 12.Andreassi MG, Borghini A, Pulignani S, Baffigi F, Fulgentini L, Koester P, et al. Radiobiological effectiveness of ultrashort laser-driven Electron bunches: micronucleus frequency, telomere shortening and cell viability. Radiat Res. 2016;186(3):245–253. doi: 10.1667/RR14266.1. [DOI] [PubMed] [Google Scholar]
- 13.Malka V, Faure J, Glinec Y, Lifschitz AF. Laser-plasma accelerators: a new tool for science and for society. Plasma Phys Control Fusion. 2005;47:B481. [Google Scholar]
- 14.Malka V, Faure J, Glinec Y, Lifschitz AF. Laser-plasma accelerator: status and perspectives. Philos Trans A Math Phys Eng Sci. 2006;364:601–610. doi: 10.1098/rsta.2005.1725. [DOI] [PubMed] [Google Scholar]
- 15.Hooker SM. Developments in laser-driven plasma accelerators. Nat Photonics. 2013;7:P775–P782. [Google Scholar]
- 16.Laschinsky L, Karsch L, Leßmann E, Oppelt M, Pawelke J, Richter C, et al. Radiobiological influence of megavoltage electron pulses of ultra-high pulse dose rate on normal tissue cells. Radiat Environ Biophys. 2016;55(3):381–391. doi: 10.1007/s00411-016-0652-7. [DOI] [PubMed] [Google Scholar]
- 17.Lee WH, Nguyen PK, Fleischmann D, Wu JC. DNA damage-associated biomarkers in studying individual sensitivity to low-dose radiation from cardiovascular imaging. Eur Heart J. 2016;37(40):3075–3080. doi: 10.1093/eurheartj/ehw206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tice RR, Strauss GH. The single cell gel electrophoresis/comet assay: a potential tool for detecting radiation-induced DNA damage in humans. Stem Cells. 1995;13(1):207–214. [PubMed] [Google Scholar]
- 19.Olive PL. DNA damage and repair in individual cells: applications of the comet assay in radiobiology. Int J Radiat Biol. 1999;75(4):395–405. doi: 10.1080/095530099140311. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Xu C, Du LQ, Cao J, Liu JX, Su X, et al. Evaluation of the comet assay for assessing the dose-response relationship of DNA damage induced by ionizing radiation. Int J Mol Sci. 2013;14(11):22449–22461. doi: 10.3390/ijms141122449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olive PL. Impact of the comet assay in radiobiology. Mutat Res. 2009;681(1):13–23. doi: 10.1016/j.mrrev.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Seidel C, Lautenschläger C, Dunst J, Müller AC. Factors influencing heterogeneity of radiation-induced DNA-damage measured by the alkaline comet assay. Radiat Oncol. 2012;7:61. doi: 10.1186/1748-717X-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Löbrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, et al. GammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 2010;9(4):662–669. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- 24.Tucker JD. Low-dose ionizing radiation and chromosome translocations: a review of the major considerations for human biological dosimetry. Mutat Res. 2008;659(3):211–220. doi: 10.1016/j.mrrev.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Pajic J, Jovicic D, Ps Milovanovic A. Micronuclei as a marker for medical screening of subjects continuously occupationally exposed to low doses of ionizing radiation. Biomarkers. 2017;22(5):439–445. doi: 10.1080/1354750X.2016.1217934. [DOI] [PubMed] [Google Scholar]
- 26.Fenech M. The lymphocyte cytokinesis-block micronucleus cytome assay and its application in radiation biodosimetry. Health Phys. 2010;98(2):234–243. doi: 10.1097/HP.0b013e3181b85044. [DOI] [PubMed] [Google Scholar]
- 27.Vral A, Fenech M, Thierens H. The micronucleus assay as a biological dosimeter of in vivo ionising radiation exposure. Mutagenesis. 2011;26(1):11–17. doi: 10.1093/mutage/geq078. [DOI] [PubMed] [Google Scholar]
- 28.Qian QZ, Cao XK, Shen FH, Wang Q. Effects of ionising radiation on micronucleus formation and chromosomal aberrations in Chinese radiation workers. Radiat Prot Dosim. 2016;168(2):197–203. doi: 10.1093/rpd/ncv290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rigaud O, Fortunel NO, Vaigot P, Cadio E, Martin MT, Lundh O, et al. Exploring ultrashort high-energy electron-induced damage in human carcinoma cells. Cell Death Dis. 2010;1:e73. doi: 10.1038/cddis.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babayan N, Hovhannisyan G, Grigoryan B, Grigoryan R, Sarkisyan N, Tsakanova G, et al. Dose-rate effect of ultrashort electron beam radiation on DNA damage and repair in vitro. J Radiat Res. 2017;58(6):894–897. doi: 10.1093/jrr/rrx035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laschinsky L, Baumann M, Beyreuther E, Enghardt W, Kaluza M, Karsch L, et al. Radiobiological effectiveness of laser accelerated electrons in comparison to electron beams from a conventional linear accelerator. J Radiat Res. 2012;53:395–403. doi: 10.1269/jrr.11080. [DOI] [PubMed] [Google Scholar]
- 32.Beyreuther E, Karsch L, Laschinsky L, Leßmann E, Naumburger D, Oppelt M, et al. Radiobiological response to ultra-short pulsed megavoltage electron beams of ultra-high pulse dose rate. Int J Radiat Biol. 2015;91(8):643–652. doi: 10.3109/09553002.2015.1043755. [DOI] [PubMed] [Google Scholar]
- 33.Bangs CD, Donlon TA. Metaphase chromosome preparation from cultured peripheral blood cells. Curr Protoc Hum Genet. 2005;45(1):4.1. doi: 10.1002/0471142905.hg0401s45. [DOI] [PubMed] [Google Scholar]
- 34.Manvelyan M, Cremer FW, Lancé J, Kläs R, Kelbova C, Ramel C, et al. New cytogenetically visible copy number variant in region 8q21.2. Mol Cytogenet. 2011;4:1. doi: 10.1186/1755-8166-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mkrtchyan H, Gross M, Hinreiner S, Polytiko A, Manvelyan M, Mrasek K, et al. The human genome puzzle - the role of copy number variation in somatic mosaicism. Curr Genomics. 2010;11(6):426–431. doi: 10.2174/138920210793176047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weise A, Gross M, Mrasek K, Mkrtchyan H, Horsthemke B, Jonsrud C, et al. Parental-origin-determination fluorescence in situ hybridization distinguishes homologous human chromosomes on a single-cell level. Int J Mol Med. 2008;21:189–200. doi: 10.3892/ijmm.21.2.189. [DOI] [PubMed] [Google Scholar]
- 37.Iourov IY. Quantitative Fluorescence In Situ Hybridization (QFISH) In: Wan TSK, editor. Methods in molecular biology, Cancer cytogenetics methods and protocols. New York: Springer Science; 2017. pp. 143–149. [DOI] [PubMed] [Google Scholar]
- 38.Scherer S. Guide to Human Genome. http://www.cshlp.org/ (2010). Accessed 2 Aug 2018.
- 39.Manvelyan M, Hunstig F, Bhatt S, Mrasek K, Pellestor F, Weise A, et al. Chromosome distribution in human sperm - a 3D multicolor banding-study. Mol Cytogenet. 2008;1:25. doi: 10.1186/1755-8166-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sudmant PH, Mallick S, Nelson BJ, Hormozdiari F, Krumm N, Huddleston J, et al. Global diversity, population stratification, and selection of human copy-number variation. Science. 2015;349(6253):aab3761. doi: 10.1126/science.aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fanciulli M, Petretto E, Aitman TJ. Gene copy number variation and common human disease. Clin Genet. 2010;77(3):201–213. doi: 10.1111/j.1399-0004.2009.01342.x. [DOI] [PubMed] [Google Scholar]
- 43.Shaikh TH. Copy number variation disorders. Curr Genet Med Rep. 2017;5(4):183–190. doi: 10.1007/s40142-017-0129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arlt MF, Mulle JG, Schaibley VM, Ragland RL, Durkin SG, Warren ST, et al. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am J Hum Genet. 2009;84:339–350. doi: 10.1016/j.ajhg.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flunkert J, Maierhofer A, Dittrich M, Müller T, Horvath S, Nanda I, et al. Genetic and epigenetic changes in clonal descendants of irradiated human fibroblasts. Exp Cell Res. 2018;370(2):322–332. doi: 10.1016/j.yexcr.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 46.Sommer S, Buraczewska I, Wojewodzka M, Bouzyk E, Szumiel I, Wojcik A. The radiation sensitivity of human chromosomes 2, 8 and 14 in peripheral blood lymphocytes of seven donors. Int J Radiat Biol. 2005;81(10):741–749. doi: 10.1080/09553000500499381. [DOI] [PubMed] [Google Scholar]
- 47.Cigarrán S, Barrios L, Barquinero JF, Caballín MR, Ribas M, Egozcue J. Relationship between the DNA content of human chromosomes and their involvement in radiation-induced structural aberrations, analysed by painting. Int J Radiat Biol. 1998;74(4):449–455. doi: 10.1080/095530098141311. [DOI] [PubMed] [Google Scholar]
- 48.Rapp A, Bock C, Dittmar H, Greulich KO. UV-A breakage sensitivity of human chromosomes as measured by COMET-FISH depends on gene density and not on the chromosome size. J Photochem Photobiol B. 2000;56(2–3):109–117. doi: 10.1016/s1011-1344(00)00052-x. [DOI] [PubMed] [Google Scholar]
- 49.Surrallés J, Sebastian S, Natarajan AT. Chromosomes with high gene density are preferentially repaired in human cells. Mutagenesis. 1997;12(6):437–442. doi: 10.1093/mutage/12.6.437. [DOI] [PubMed] [Google Scholar]
- 50.Ulsh BA, Dolling J, Lavoie J, Mitchel RE, Boreham DR. Chromosome damage caused by accidental chronic whole-body gamma radiation exposure in Thailand. Dose Response. 2015;13(4):1559325815614302. doi: 10.1177/1559325815614302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hande MP, Azizova TV, Geard CR, Burak LE, Mitchell CR, Khokhryakov VF, et al. Past exposure to densely ionizing radiation leaves a unique permanent signature in the genome. Am J Hum Genet. 2003;72(5):1162–1170. doi: 10.1086/375041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kimmel RR, Agnani S, Yang Y, Jordan R, Schwartz JL. DNA copy-number instability in low-dose gamma-irradiated TK6 lymphoblastoid clones. Radiat Res. 2008;169(3):259–269. doi: 10.1667/RR1096.1. [DOI] [PubMed] [Google Scholar]
- 53.Muradyan A, Gilbertz K, Stabentheiner S, Klause S, Madle H, Meineke V, et al. Acute high-dose X-radiation-induced genomic changes in A549 cells. Radiat Res. 2011;175(6):700–707. doi: 10.1667/RR2341.1. [DOI] [PubMed] [Google Scholar]
- 54.Kang HM, Jang JJ, Langford C, Shin SH, Park SY, Chung YJ. DNA copy number alterations and expression of relevant genes in mouse thymic lymphomas induced by gamma-irradiation and N-methyl-N-nitrosourea. Cancer Genet Cytogenet. 2006;166(1):27–35. doi: 10.1016/j.cancergencyto.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Zitzelsberger H, Unger K. DNA copy number alterations in radiation-induced thyroid cancer. Clin Oncol (R Coll Radiol) 2011;23(4):289–296. doi: 10.1016/j.clon.2011.01.154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the conclusions of this article are included within the article.


