Abstract
Objectives
To explore the dynamics of faecal ESBL/AmpC shedding in dairy cattle and farmers, a study was conducted to examine changes in shedding by individual animals, as well as environmental exposure, and to study the association between antimicrobial use (AMU) and ESBL/AmpC shedding.
Methods
The study comprised a cross-sectional survey of 20 farms and a 1 year follow-up of 10 farms. Faecal samples were cultured by both direct inoculation on MacConkey agar + 1 mg/L cefotaxime (MC+) and enrichment in LB-broth + 1 mg/L cefotaxime with subsequent inoculation on MC+. Dust samples were collected using electrostatic dustfall collectors (EDCs). Human faecal samples were collected by the farmers. Presence of ESBL/AmpC genes was screened for by PCR and sequencing. Using mixed effects logistic regression, ORs were determined and population-attributable fractions (PAFs) calculated subsequently.
Results
In Phase 1, 8/20 farms were positive for ESBL/AmpC and, with 2 negative farms, were selected for Phase 2. Transient shedding of dominant allele variants was observed in the animals. EDCs and human faecal samples did not reflect what was observed in the animals. AMU was related to shedding of ESBLs in the next sampling moment [OR 14.6 (95% CI 3.0–80.0)] and the PAF of AMU was 0.36 (95% CI 0.08–0.77). Calves fed with colostrum from cows on dry-off therapy was not a risk factor [OR 1.7 (95% CI 0.7–4.9, P = 0.28)].
Conclusions
The presence of ESBL/AmpC could only be partly explained by AMU. No link was shown between shedding in cattle and humans or the environment. Interventions should focus on prevention of introduction.
Introduction
ESBL/AmpC-producing Enterobacteriaceae have been observed in humans, animals and the environment.1 The prevalence and diversity of different ESBL/AmpC variants observed in food-producing animals is increasing steadily.2,3 Although many studies have described the presence of ESBL/AmpC-producing Enterobacteriaceae, longitudinal data on faecal shedding of ESBL/AmpC-producing Enterobacteriaceae in farm animals remains limited.4–10 Recently, two prevalence studies were performed on Dutch conventional dairy farms and on Dutch organic dairy farms.7,8 Compared with organic farms, ESBL/AmpC prevalence on conventional farms was relatively high. In addition, the use of third- and fourth-generation cephalosporins on the conventional farms was associated with a higher prevalence of ESBL/AmpC-producing Escherichia coli, which is in line with findings in other countries.9,10 However, these cross-sectional studies do not allow study of the long-term dynamics of ESBL/AmpC organism shedding, either at the herd or individual animal level, which is relevant for understanding their potential for persistence on farms. Additionally, neither the association of animal age with the likelihood of ESBL/AmpC shedding, nor the potential hazard for people working or living on the farms was studied.
In order to further explore the ESBL/AmpC epidemiology on dairy farms, we studied: (i) the dynamics of faecal shedding of ESBL/AmpC in individual animals on multiple farms and its relationship with antimicrobial use (AMU); (ii) possible transmission between different age groups on the farms; (iii) the environmental contamination through slurry and dust sampling in the stables; and (iv) shedding in farmers and family members living on the farm.
Materials and methods
Ethics
All animal sampling was performed within the guidelines of the Dutch Animals Act (stb-2011-345) and the Animal Welfare Body Utrecht,11,12 meaning no additional licence was required.
For all human sampling, the Medical Research Ethics Committee of University Medical Center Utrecht granted permission to perform this research (METC protocol number 14/346/C).
Farm selection and sampling strategy
Phase 1: baseline
The study was divided into a baseline cross-sectional part (Phase 1) and a longitudinal part over a period of a year (Phase 2). In Phase 1, 20 conventional farms were selected from the clientele of the University Farm Animal Practice, which serves ∼350 farms housing 28 000 dairy cows. Farms were selected from the highest users of antimicrobials during 2012/2013. Usage was expressed in DDD per animal per year (DDDA), as described by Bos et al.,13 and farm sizes ranged from 85 to 190 cows in 2013. Farmers participated on a voluntary basis and received €500 reimbursement. Animals were categorized into four age groups, in accordance with the national registration system for AMU for dairy cattle (MediRund): Group 1 (0–8 weeks); Group 2 (>8 weeks–1 year); Group 3 (>1–2 years); and Group 4 (>2 years). Sampling was performed in March/April 2014 (T0) and sample size was based on an expected prevalence of 20% in a group of 100 dairy animals, with 95% confidence and an absolute precision of 15%. The calculated sample size for detection of at least one positive animal given the above-mentioned prevalence and confidence level was 13.14
Phase 2: longitudinal
For Phase 2, 10 farms were selected: all eight ESBL/AmpC-positive farms from Phase 1 and two additional ESBL/AmpC-negative farms. Starting in May/June 2014 (T2), the farms were sampled every 2 months until March/April 2015 (T12). At each sampling, all animals from Group 1 and all animals moving from either Group 1 to 2, or from Group 3 to 4 at consecutive samplings were included. Animals moving from Group 2 to 3 were included every 4 months. The rationale behind this sampling difference is that both young and older animals (Groups 1 and 4) are treated with antimicrobials more frequently than animals in Groups 2 and 3 and consequently are considered more likely to be ESBL/AmpC carriers. Furthermore, all animals positive for ESBL/AmpC at certain sampling events were included in the subsequent sampling. Additionally, a slurry tank sample was collected every round and electrostatic dustfall collectors (EDCs) were placed in the stables to collect dust from T4 to T12. The EDCs and their accompanying blank control EDCs (sealed in a Ziploc bag) were collected 2–3 weeks after placement. Farmers, their family members living at the farm and employees collected their own faecal samples at T4 and T12.
Metadata
Based on ESBL/AmpC status and the availability of detailed information on housing and AMU at animal level, five farms were selected to analyse the effect of antimicrobial treatment on the occurrence of ESBL in dairy cattle (farms 4, 5, 14, 15 and 19). The records on AMU contained the antimicrobial class, the administration route and the date of administration for each individual animal. For the analyses, antimicrobials were subcategorized into first- and second-choice antimicrobials, based on the risk for selecting ESBLs.15 In the Netherlands, first-choice antimicrobials are allowed to be used in empirical therapy, whereas the use of second-choice antimicrobials has to be supported with either microbiological or clinical evidence regarding treatment efficacy to justify its use.
E. coli isolation
Rectal faecal samples were collected and processed for culturing the next morning at the latest. Human faecal samples were sent by regular mail. Informed consent was given by all participants. In the case of minors, consent was provided by their parents.
All samples were cultured qualitatively (detection limit: 2 cfu/g faeces) as well as quantitatively (detection limit: 500 cfu/g) using the track dilution method as described by Baede et al.16 If a sample showed growth after direct inoculation (quantitative culture), three isolates showing typical E. coli morphology were selected for further analysis. Since E. coli may show various morphologies on MacConkey, isolates showing different morphologies were selected, if applicable. If direct inoculation was negative, but the corresponding sample after enrichment (qualitative culture) was positive, one isolate showing typical E. coli morphology was selected for further analysis.
Species identification
All isolates from faecal samples were pure-cultured on Columbia sheep blood agar (Oxoid, the Netherlands). The species was determined using MALDI-TOF (Bruker, the Netherlands).
DNA extraction
DNA from all faecal sample isolates was extracted by boiling one colony of each strain in 500 μL Tris-EDTA (TE) buffer (Sigma–Aldrich, Germany) for 10 min and subsequently spinning it down at 14 000 rpm for 1 min. The supernatant was transferred to a Micronic tube (VWR, the Netherlands) and stored at 4°C for further analysis.
DNA from dust obtained in the EDCs was extracted as described previously,17 with the modification of using 20 mL/cloth Aqua B. Braun water + 0.05% Tween 20 for homogenizing EDCs. Freeze-drying was performed for 3–4 days and samples were stored at −20°C.
DNA isolation for WGS was performed using the UltraClean® Microbial DNA Isolation Kit (MO BIO, QIAGEN, USA).
ESBL/AmpC and E. coli characterization
Isolates were screened for the presence of blaCTX-M, blaTEM, blaSHV, blaCMY and blaOXA by conventional PCR using the primers shown in Table S1 (available as Supplementary data at JAC Online). The PCR mix (20 μL) contained 5 μL of DNA lysate, 2× GoTaq Hotstart Green Master Mix (Promega Benelux B.V., the Netherlands), 0.5 μM forward and reverse primers and molecular grade water (Sigma–Aldrich). Positive PCR products were purified with ExoSAP-IT (Affymetrix, USA) and subsequently sent for sequencing (BaseClear, the Netherlands). Sequences were analysed using BioNumerics v7.5 software (Applied Maths, Belgium). ESBL/AmpC annotations as reported on www.lahey.org/studies were used as a reference. From each positive farm, a selection with the size of the square root of the number of isolates harbouring the same gene was taken, to determine clonality of the isolates by WGS on a NextSeq platform (Illumina, USA). Selected isolates were evenly distributed over the samples of interest (ordered by animal ear tag number).
Data analysis
Statistical analyses were performed using the R v3.2.2 statistical programming language.18 The OR for the presence of ESBL/AmpC with AMU was estimated with mixed effects logistic regression using the ‘glmer’ function of the ‘mle4’ package in which the farm was considered a random effect.19 Akaike’s information criterion (AIC) was used for model selection. The OR of the best model was used to calculate the population-attributable fraction (PAF) assuming that the OR is an adequate estimate of the relative risk. Fisher’s exact test was used to test proportions of ESBL/AmpC in calves fed with colostrum of dams, with or without antimicrobial dry-cow therapy.
Genome sequences were assembled with SPAdes v3.10.1.20 Core-genome alignments were made using Parsnp v1.2,21 corrected for recombination regions using Gubbins and visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).22In silico MLST typing was performed as described by T. Seemann and phylogrouping was performed using BLAST by checking the presence of chuA, yjaA and TSPE4.23,24 DNA sequences were deposited in the European Nucleotide Archive under project number PRJEB30024.
Results
Farm selection and descriptive results of ESBL/AmpC presence
AMU on the 20 farms in Phase 1 ranged from 3.56 to 9.16 DDDA in 2012, and from 3.50 to 6.91 DDDA in 2013.
At T0, in 8 out of 20 farms ESBL/AmpC-producing E. coli were isolated from either animals, slurry or both (Table 1). The number of samples taken at each farm ranged from 64 to 105 (median = 92). On five of the eight farms the prevalence was very low, with either one or two positive samples (farms 4, 5, 7, 10 and 12). On three farms the prevalence was relatively high, ranging from 11% to 27% (farms 14, 15 and 19).
Table 1.
Total number of samples screened (S) and number of positive samples (E) in age groups 1–4 and slurry for each sampling time (T0–T12) for 10 conventional dairy cattle farms selected after Phase 1 (T0), during 2014–15
| T0 Phase 1 |
T2 Phase 2 |
T4 Phase 2 |
T6 Phase 2 |
T8 Phase 2 |
T10 Phase 2 |
T12 Phase 2 |
Total |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Farma | Sample | S | E | S | E | S | E | S | E | S | E | S | E | S | E | S | E |
| 4* | Group 1 | 8 | — | 12 | — | 3 | — | 4 | 1 | 6 | — | 4 | 1 | 3 | — | 40 | 2 |
| Group 2 | 21 | 1 | 4 | — | 15 | 2 | 4 | — | 11 | — | 9 | — | 24 | — | 88 | 3 | |
| Group 3 | 32 | — | 1 | — | 8 | — | 1 | — | 6 | — | 10 | — | 23 | — | 81 | 0 | |
| Group 4 | 30 | — | 7 | 3 | 3 | — | 8 | — | 13 | — | 10 | — | 23 | — | 94 | 3 | |
| Total 1–4 | 91 | 1 | 24 | 3 | 29 | 2 | 17 | 1 | 36 | 0 | 33 | 1 | 73 | 0 | 303 | 8 | |
| Slurry | 4 | 1 | 4 | — | 4 | — | 4 | — | 4 | 1 | 4 | — | 4 | — | 28 | 2 | |
| 5* | Group 1 | 10 | — | 6 | — | 6 | 1 | 9 | — | 13 | — | 7 | — | 2 | — | 53 | 1 |
| Group 2 | 29 | — | 10 | — | 18 | — | 7 | — | 24 | — | 14 | — | 40 | — | 142 | 0 | |
| Group 3 | 29 | — | 5 | — | 8 | — | 0 | — | 25 | — | 5 | — | 41 | — | 113 | 0 | |
| Group 4 | 29 | — | 2 | — | 12 | 2 | 10 | — | 6 | — | 5 | — | 33 | — | 97 | 2 | |
| Total 1–4 | 97 | 0 | 23 | 0 | 44 | 3 | 26 | 0 | 68 | 0 | 31 | 0 | 116 | 0 | 405 | 3 | |
| Slurry | 5 | 1 | 5 | — | 5 | — | 5 | — | 5 | — | 5 | — | 5 | — | 35 | 1 | |
| 7 | Group 1 | 2 | — | 5 | — | 2 | — | 2 | — | 1 | — | 1 | — | 3 | — | 16 | 0 |
| Group 2 | 19 | — | 2 | — | 5 | — | 2 | — | 13 | — | 1 | — | 9 | — | 51 | 0 | |
| Group 3 | 22 | — | 1 | — | 1 | — | 0 | — | 5 | — | 3 | — | 19 | — | 51 | 0 | |
| Group 4 | 30 | — | 1 | — | 2 | — | 4 | — | 5 | — | 4 | — | 15 | — | 61 | 0 | |
| Total 1–4 | 73 | 0 | 9 | 0 | 10 | 0 | 8 | 0 | 24 | 0 | 9 | 0 | 46 | 0 | 179 | 0 | |
| Slurry | 5 | 1 | 3 | 1 | 1 | — | 1 | — | 3 | — | 3 | — | 2 | — | 18 | 2 | |
| 10 | Group 1 | 8 | — | 9 | — | 7 | — | 11 | — | 6 | — | 6 | — | 4 | — | 51 | 0 |
| Group 2 | 28 | — | 8 | — | 28 | 2 | 5 | — | 17 | — | 6 | — | 27 | — | 119 | 2 | |
| Group 3 | 22 | — | 2 | — | 7 | — | 6 | — | 20 | — | 3 | — | 32 | — | 92 | 0 | |
| Group 4 | 28 | — | 8 | — | 1 | — | 9 | — | 5 | — | 8 | — | 15 | — | 74 | 0 | |
| Total 1–4 | 86 | 0 | 27 | 0 | 43 | 2 | 31 | 0 | 48 | 0 | 23 | 0 | 78 | 0 | 336 | 2 | |
| Slurry | 5 | 1 | 3 | — | 3 | — | 3 | — | 3 | — | 3 | — | 3 | — | 23 | 1 | |
| 11 | Group 1 | 2 | — | 6 | — | 6 | — | 3 | — | 8 | 3 | 9 | 1 | 4 | — | 38 | 4 |
| Group 2 | 30 | — | 3 | — | 20 | — | 9 | — | 21 | 2 | 9 | — | 37 | — | 129 | 2 | |
| Group 3 | 31 | — | 8 | — | 24 | 2 | 3 | — | 22 | 14 | 16 | — | 36 | — | 140 | 16 | |
| Group 4 | 29 | — | 5 | — | 8 | — | 7 | — | 7 | 2 | 12 | — | 17 | — | 85 | 2 | |
| Total 1–4 | 92 | 0 | 22 | 0 | 58 | 2 | 22 | 0 | 58 | 21 | 46 | 1 | 94 | 0 | 392 | 24 | |
| Slurry | 3 | — | 3 | — | 3 | — | 3 | — | 3 | 2 | 3 | 1 | 3 | — | 21 | 3 | |
| 12 | Group 1 | 1 | — | 7 | 1 | 3 | 3 | 9 | 3 | 4 | 3 | 10 | — | 4 | — | 38 | 10 |
| Group 2 | 22 | — | 1 | — | 18 | 2 | 6 | 2 | 19 | — | 13 | 2 | 28 | — | 107 | 6 | |
| Group 3 | 21 | — | 3 | 3 | 9 | — | 0 | — | 20 | — | 2 | — | 15 | — | 70 | 3 | |
| Group 4 | 29 | 1 | 3 | — | 4 | 1 | 5 | 2 | 4 | — | 12 | — | 10 | — | 67 | 4 | |
| Total 1–4 | 73 | 1 | 14 | 4 | 34 | 6 | 20 | 7 | 47 | 3 | 37 | 2 | 57 | 0 | 282 | 23 | |
| Slurry | 2 | 1 | 2 | 1 | 2 | — | 2 | — | 2 | — | 2 | — | 2 | — | 14 | 2 | |
| 14* | Group 1 | 4 | — | 9 | — | 11 | — | 5 | — | 7 | — | 16 | — | 6 | — | 58 | 0 |
| Group 2 | 28 | — | 4 | — | 11 | 1 | 11 | — | 15 | — | 6 | — | 44 | — | 119 | 1 | |
| Group 3 | 29 | — | 0 | — | 0 | — | 0 | — | 9 | — | 4 | — | 32 | — | 74 | 0 | |
| Group 4 | 28 | 10 | 19 | 18 | 24 | 8 | 26 | 4 | 26 | 19 | 38 | — | 47 | — | 208 | 59 | |
| Total 1–4 | 89 | 10 | 32 | 18 | 46 | 9 | 42 | 4 | 57 | 19 | 64 | 0 | 129 | 0 | 459 | 60 | |
| Slurry | 3 | — | 2 | 1 | 2 | — | 3 | 2 | 3 | — | 3 | — | 3 | — | 19 | 3 | |
| 15* | Group 1 | 3 | 3 | 2 | 2 | 3 | 3 | 5 | 4 | 3 | 3 | 5 | 2 | 5 | 3 | 26 | 20 |
| Group 2 | 33 | 4 | 7 | 3 | 27 | 5 | 14 | 8 | 17 | 1 | 14 | 6 | 15 | 11 | 127 | 38 | |
| Group 3 | 30 | 2 | 8 | 9 | 22 | 1 | 8 | 8 | 28 | 1 | 12 | 1 | 34 | 26 | 142 | 48 | |
| Group 4 | 29 | 16 | 24 | 7 | 24 | 23 | 26 | 11 | 27 | 2 | 37 | — | 41 | 42 | 208 | 101 | |
| Total 1–4 | 95 | 25 | 41 | 21 | 76 | 32 | 53 | 31 | 75 | 7 | 68 | 9 | 95 | 82 | 503 | 207 | |
| Slurry | 2 | 1 | 2 | 2 | 2 | 2 | 2 | — | 2 | 1 | 2 | 1 | 3 | — | 15 | 7 | |
| 18 | Group 1 | 2 | — | 3 | — | 5 | — | 8 | — | 1 | — | 3 | — | 2 | — | 24 | 0 |
| Group 2 | 16 | — | 2 | — | 9 | — | 4 | — | 13 | — | 1 | — | 19 | — | 64 | 0 | |
| Group 3 | 21 | — | 3 | — | 11 | — | 0 | — | 11 | — | 4 | — | 18 | — | 68 | 0 | |
| Group 4 | 30 | — | 2 | 1 | 4 | — | 5 | — | 2 | — | 6 | — | 6 | — | 55 | 1 | |
| Total 1–4 | 69 | 0 | 10 | 1 | 29 | 0 | 17 | 0 | 27 | 0 | 14 | 0 | 45 | 0 | 211 | 1 | |
| Slurry | 2 | — | 2 | — | 1 | — | 2 | 1 | 2 | — | 2 | — | 2 | — | 13 | 1 | |
| 19* | Group 1 | 4 | — | 3 | 1 | 9 | 4 | 4 | 1 | 7 | — | 7 | — | 7 | 3 | 41 | 9 |
| Group 2 | 28 | 2 | 5 | 5 | 17 | 9 | 18 | 1 | 23 | — | 16 | — | 29 | — | 136 | 17 | |
| Group 3 | 30 | 1 | 6 | 5 | 12 | 12 | 13 | 4 | 23 | 4 | 19 | 2 | 37 | 1 | 140 | 29 | |
| Group 4 | 30 | 10 | 14 | 14 | 20 | 20 | 23 | 2 | 27 | 1 | 28 | 1 | 30 | — | 172 | 48 | |
| Total 1–4 | 92 | 13 | 28 | 25 | 58 | 45 | 58 | 8 | 80 | 5 | 70 | 3 | 103 | 4 | 489 | 103 | |
| Slurry | 4 | 2 | 4 | — | 4 | 2 | 4 | — | 4 | 2 | 4 | 2 | 4 | — | 28 | 8 | |
Eight farms (4, 5, 7, 10, 12, 14, 15 and 19) were selected because of ESBL presence. Farm 11 and Farm 18 were selected based on the absence of ESBLs in Phase 1.
Farms included for analyses of AMU are indicated with an asterisk.
In Phase 2 (T2–T12), all eight positive farms were included for follow-up. Additionally, two farms for which the ESBL status resulting from Phase 1 was still being confirmed at the start of Phase 2 were included (farms 11 and 18). All 10 farms were positive for ESBL/AmpC twice or more (Table 1) throughout the study. Most farms that showed a low prevalence at T0, were either negative or showed low numbers of positive samples during subsequent samplings. One exception was farm 11, where 36% of the samples were positive at T8. However, only a single positive sample was observed during the next sampling. Farms with a relatively high prevalence at T0 remained highly positive throughout the year, although a downward trend was observed, with the exception of the final sampling at farm 15 (T12).
ESBL/AmpC and E. coli genotyping in animals and slurry
All ESBL/AmpC genes detected throughout the study on the relatively high prevalence farms are shown in Tables S2 to S6. The general trend on all farms, but especially on the highly positive farms, is that the presence of ESBL/AmpC at a specific sampling was dominated by one or two gene variants. This is confirmed by the selection of strains that were subjected to core-genome analysis (Figure S1). The clear clustering of isolates is associated with a combination of phylogroup, sequence type and, to a lesser extent, ESBL/AmpC gene. Also, many clusters are from isolates from multiple farms, indicating a certain overlap in E. coli type present on the farms. Furthermore, the dominant gene variants at a farm usually varied between samplings, although some overlap was observed. From all animals with a positive ESBL/AmpC sample in T0–T10 (n = 340), 24 animals (7%) were positive for exactly the same ESBL/AmpC allele variant (or combination of ESBL plus narrow-spectrum β-lactamase, e.g. blaTEM-1) in an isolate at the subsequent sampling (T2–T12). In the vast majority of faecal samples a single ESBL/AmpC gene variant was detected. Farms with a high number of positive samples at a specific sampling moment also showed a higher diversity of gene variants present in one faecal sample. This was most evident in samples collected from farm 11 (T8) and farms 15 and 19 (multiple samplings). The most frequently observed ESBL/AmpC gene variants were blaCTX-M-1, blaCTX-M-14, blaCTX-M-15,blaCTX-M-32 and blaCMY-2 (Tables S2 to S6). Less frequently observed gene variants were blaCTX-M-2, blaCTX-M-3, blaTEM-52c and an inhibitor-resistant TEM, blaTEM-79. In young animals, blaTEM-79 was observed only on farm 12 at multiple sampling moments, suggesting a recurring event (Groups 1 and 2; Table S3).
ESBL/AmpC from EDCs
The ESBL/AmpC-encoding genes obtained from the dust samples differed from those in the faecal samples. The vast majority of genes observed on most farms were different variants of the narrow-spectrum β-lactamase blaTEM-1, blaOXA-1 or blaOXA-2 (Table S7). The gene blaCTX-M was observed five times on farms 4, 7, 11 and 15. In all cases the determination of the exact allele variant remained inconclusive. Farm 15, at T10, is the only potential match in presence of blaCTX-M in both faecal samples and a dust sample. On farm 5 (T8) and farm 10 (T4) a blaCMY was observed in an EDC.
ESBL/AmpC prevalence among farmers, workers and family members
At T4, 38 persons were willing to participate, representing 9 out of 10 farms in Phase 2 (except farm 12). One person from farm 10 was positive for blaCTX-M-14. This gene was not observed in any of the farm samples throughout the year. At T12, 25 persons (also representing 9 out 10 farms) were willing to participate. This time, persons from farm 11 did not participate. One person from farm 4 was positive for blaCTX-M-1. This gene was not observed at that farm at T12, but was observed at T0, T2, T8 and T10 (in combination with blaTEM-1b) in either animals or slurry. This person was negative at T4. Animal isolates from this farm carrying blaCTX-M-1 that were also included in the WGS selection showed different STs and phylogroups at each sampling (Figure S1).
Effect of antimicrobial treatment on the occurrence of ESBL/AmpC
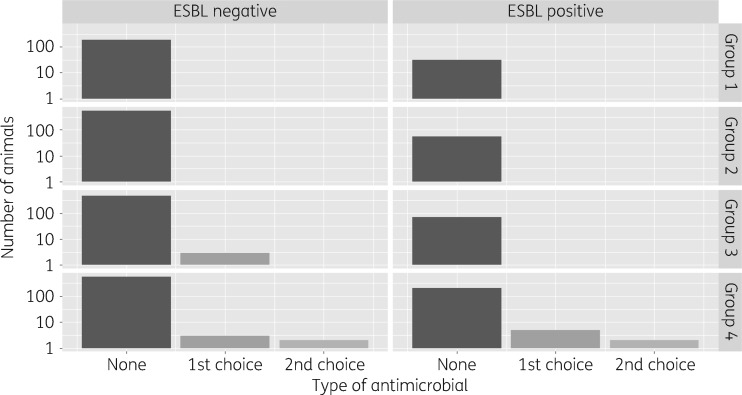
The number of animals treated with antimicrobials is shown in Figure 1. If second-choice antimicrobials were used, the same animal was also treated previously or concurrently with first-choice antimicrobials. The presence of ESBL/AmpC at a certain point in time was best explained by a model incorporating the use of antimicrobials prior to sampling. A model with total use of antimicrobials had a slightly better fit than a model with first-choice and first- plus second-choice antimicrobials distinguished (Table 2). The following ORs (95% CIs) apply for use versus non-use of antimicrobials: including use of first-choice antimicrobials only, 11.4 (1.6–87.3); use of second-choice following first-choice antimicrobials, 21.8 (1.9–299.4); and use of antimicrobials without distinction between first or second choice, 14.6 (3.0–80.4) (Table 3). The PAF estimates (95% CIs) vary between first- and second-choice antimicrobials [0.22 (0.02–0.70) and 0.22 (0.01–0.80), respectively] and for antimicrobials without this categorization 0.36 (0.08–0.77).
Figure 1.
Distribution of negative and positive samples over ‘none’, ‘first-choice’ or ‘second-choice’ antimicrobials and the different age groups on five selected Dutch conventional dairy cattle farms during 2014–15.
Table 2.
Total number of animal samples (S) and number of ESBL-positive samples (E) grouped by exposure to individual animal treatment 2 months prior to sampling, excluding exposure due to colostrum feeding of calves, for five selected Dutch conventional dairy cattle farms, for which detailed AMU data were available, during 2014–15
| No antimicrobial |
Only first-choice antimicrobial |
First- and second-choice antimicrobial |
|||||
|---|---|---|---|---|---|---|---|
| Farm | Sample | S | E | S | E | S | E |
| 4 | Group 1 | 40 | 2 | 0 | 0 | 0 | 0 |
| Group 2 | 88 | 1 | 0 | 0 | 0 | 0 | |
| Group 3 | 80 | 2 | 1 | 0 | 0 | 0 | |
| Group 4 | 89 | 2 | 2 | 0 | 3 | 1 | |
| Total 1–4 | 297 | 7 | 3 | 0 | 3 | 1 | |
| 5 | Group 1 | 53 | 1 | 0 | 0 | 0 | 0 |
| Group 2 | 141 | 0 | 1 | 0 | 0 | 0 | |
| Group 3 | 111 | 0 | 2 | 0 | 0 | 0 | |
| Group 4 | 97 | 2 | 0 | 0 | 0 | 0 | |
| Total 1–4 | 402 | 3 | 3 | 0 | 0 | 0 | |
| 14 | Group 1 | 58 | 0 | 0 | 0 | 0 | 0 |
| Group 2 | 119 | 1 | 0 | 0 | 0 | 0 | |
| Group 3 | 74 | 0 | 0 | 0 | 0 | 0 | |
| Group 4 | 207 | 56 | 1 | 1 | 0 | 0 | |
| Total 1–4 | 458 | 57 | 1 | 1 | 0 | 0 | |
| 15 | Group 1 | 26 | 20 | 0 | 0 | 0 | 0 |
| Group 2 | 127 | 37 | 0 | 0 | 0 | 0 | |
| Group 3 | 142 | 43 | 0 | 0 | 0 | 0 | |
| Group 4 | 205 | 96 | 3 | 3 | 0 | 0 | |
| Total 1–4 | 500 | 196 | 3 | 3 | 0 | 0 | |
| 19 | Group 1 | 41 | 9 | 0 | 0 | 0 | 0 |
| Group 2 | 136 | 16 | 0 | 0 | 0 | 0 | |
| Group 3 | 140 | 27 | 0 | 0 | 0 | 0 | |
| Group 4 | 169 | 46 | 2 | 1 | 1 | 1 | |
| Total 1–4 | 486 | 98 | 2 | 1 | 1 | 1 | |
Table 3.
Relationship between AMU and ESBL excretion in the 2 months prior to sampling in five selected Dutch conventional dairy cattle farms during 2014–15
| OR (95% CI) |
||||
|---|---|---|---|---|
| Model | first choice | second choice following first choice | AMU | AIC |
| Only intercept | — | — | — | 205.6 |
| Antimicrobiala | — | — | 14.6 (3.0–80.4) | 196.6 |
| First choice only plus second choice following first choiceb | 11.4 (1.6–87.3) | 21.8 (1.9–299.4) | — | 198.4 |
Parameter estimates of the fixed effects logistic regression models and AIC.
Animals treated with first-choice antimicrobial with or without being followed by second choice.
Animals treated with only first-choice antimicrobials or for animals treated with second choice following a first-choice treatment.
No effect of movement to the next age group was observed on ESBL/AmpC excretion, because all animals with positive samples had a positive sample in at least one of their previous groups.
Of 180 calves fed on colostrum from dams that were dried off without antimicrobials, 24 tested positive for ESBL/AmpC in the first sample after birth. Eighteen of 105 calves fed with colostrum of dams that were on antimicrobial dry-off therapy were positive for ESBL/AmpC, resulting in an OR of 1.7 (95% CI 0.7–4.9, P = 0.28) (Table 4).
Table 4.
Presence of ESBL/AmpC in the first sample from calves that received colostrum from a mother with or without dry-cow therapy
| ESBL/AmpC |
||||||
|---|---|---|---|---|---|---|
| negative |
positive |
Total |
||||
| N | % | N | % | N | % | |
| Dry-off therapy | ||||||
| Yes | 87 | 82.9 | 18 | 17.1 | 105 | 100 |
| No | 67 | 89.3 | 8 | 10.7 | 75 | 100 |
| Total | 154 | 85.6 | 26 | 14.4 | 180 | 100 |
Fisher’s exact P = 0.28.
Discussion
This study focused on the dynamics of ESBL/AmpC organism shedding in Dutch dairy cattle. Because it was focused on dynamics of faecal shedding, farms with relatively high AMU were included to increase the likelihood of collecting positive samples. The results at the first sampling showed that ESBL/AmpCs were not detected in most farms, or were detected in only a small proportion of the animals. Our finding of eight positive farms (40%) is in accordance with a previous Dutch cross-sectional study that also showed an on-farm slurry tank ESBL/AmpC prevalence of 41%.7 Results of Phase 2 showed that positive farms demonstrating a relatively high prevalence at the start, remained relatively highly positive throughout the following year. Likewise, farms with a low prevalence at the start generally showed either a low number of positive samples or were negative during the subsequent samplings. However, due to purposeful sampling at age group movements and previously positive animals, these observed high prevalences are biased towards higher values.
The ESBL/AmpC gene variants found indicate the transient character of ESBL/AmpC shedding in dairy cattle. On a specific farm, peak prevalence seemed to be dominated by one or two ESBL/AmpC variants, suggesting an introduction of ESBL/AmpC variants that spread among the animals and that subsequently disappear. Only 7% of the animals found positive showed the same ESBL/AmpC gene at the subsequent sampling. A similar dynamic of faecal shedding has also been observed in Dutch veal calves.25 Although veal and dairy cattle industries are very different with respect to herd composition, farm management and AMU, transient shedding is found in both livestock sectors. Also, the ESBL/AmpC variants observed are similar, which may be explained by the movements of calves from dairy to veal farms.
Remarkably, no overlap was found between ESBL/AmpC-encoding genes from faecal samples compared with dust samples (Tables S2–S7), even on highly positive farms. This is in contrast to what has been observed in Dutch pig farming.17 The lack of overlap suggests exposure through air may not play a major role in the spread of ESBL/AmpC-producing E. coli in dairy cattle within stables.
Longitudinal studies on dairy cattle have also been performed in the UK.4–6 Unfortunately, since all focused on one farm and applied different approaches/goals, comparing prevalence between studies is difficult. Nonetheless, these studies showed a high occurrence of blaCTX-M, which is consistent with our findings. The aforementioned studies also focused on diversity in ESBL/AmpC-producing E. coli based on PFGE typing. This diversity was high. Given the high abundance of specific dominant variants within a limited time span, as observed in our study, the high prevalence in some of the herds most likely resulted from a point-source introduction, possibly followed by clonal dispersion. However, since STs also varied in multiple cases (Figure S1), horizontal transfer or parallel introduction of other ESBL/AmpC-producing E. coli are likely to have occurred as well.
This study found no difference in ESBL/AmpC shedding between calves fed colostrum from cows on antimicrobial dry-off therapy and calves fed colostrum from cows without antimicrobial dry-off therapy. This has also been shown on dairy and beef cattle farms in the Bavarian area of Germany.26 In the UK, feeding waste milk containing antimicrobials was shown to be a risk factor, leading to higher bacterial cell counts of ESBL/AmpC-producing E. coli and increased persistence.4 One of the reasons for this difference could be the type and/or concentration of antimicrobials present in the colostrum/waste milk. In the paper by Brunton et al.,4 most of the waste milk samples contained cefquinome residues. In the Netherlands this is a third-choice antimicrobial,15 meaning it cannot be administered to animals unless first- or second-choice antimicrobials (e.g. penicillins or penicillins with extended spectrum) have proven to be ineffective and there is no alternative for the third-choice antimicrobial of interest. The usage of third-/fourth-generation cephalosporins in dairy cattle in general, not limited to dry-off therapy, has been identified as a risk factor in other studies.9,10 Except for one animal (which was negative for ESBL/AmpC), all treated animals in the present study were treated with first- or second-choice antimicrobials.
Our results show a tendency for stronger associations of ESBL/AmpC presence with second-choice antimicrobials than with first-choice ones. This classification is made based on the ability to select for antimicrobial resistance in general, but our study focused only on ESBL/AmpC. However, co-selection due to MDR may also play a role. In the present study we have not screened for resistance determinants belonging to other antimicrobial classes. AMU could only explain 22%–36% of the ESBL/AmpC-positive samples. This, however, only explains selection, not introduction. Others have shown that the floor-cleaning method may also be relevant in the spread of ESBL/AmpC and using a teat sealant for dry-cow therapy could have a protective effect.7 The occurrence of different ESBL/AmpC genes throughout the study period on farms suggests the introduction and circulation of different strains on these farms. Factors leading to introduction of ESBL/AmpC and subsequent spread throughout the farm have not been assessed in the present study. Movement of, for example, animals, silage and equipment into the stable from outside, or pest control should be taken into account. Given our observation that most farms showed a low prevalence with only one or a few animals positive for ESBL/AmpC, it should be noted that not all introduction events may lead to extensive spread throughout the herd.
Finally, human faecal samples were analysed at two points in time. At both sampling moments only one person was found positive for ESBLs and the ESBL gene of interest could not be directly linked to ESBLs identified on the farm at the time of sampling. The blaCTX-M-1 gene found in one human at T12 was observed on the farm earlier, suggesting it was obtained in a stable. Despite this one overlapping gene and the fact that the number of included farms is small, these positive human samples are thought to be a reflection of the prevalence within the general Dutch human population living in livestock-dense areas.27 This is in contrast to other Dutch studies on either pig or poultry farms, in which a direct link in ESBL shedding between animals and farmers was shown.28,29 This finding correlates with the results from the dust samples in the present study. Since dust may be a vehicle for human exposure to ESBL/AmpC, the negative dust samples may explain the absence of a correlation between positive animals and positive humans.
In conclusion, this study has shown that, in general, animals carrying ESBL/AmpC-producing E. coli on the selected Dutch dairy farms are either absent or at a low prevalence. However, a limited proportion of farms demonstrated highly dynamic ESBL/AmpC shedding in their animals, which could only be partly explained by AMU. Multiple point-source introductions are likely to have occurred. Interventions should focus on prevention of introduction. The observed human ESBL/AmpC shedding is considered to be a reflection of that in the general Dutch population.
Supplementary Material
Acknowledgements
We thank A. Aanen, G. D. Greve, S. L. de Wind, S. Chaïbi and L. Van der Graaf-Van Bloois for their technical assistance throughout the project. We also acknowledge H. Voogd, J. Klaver, F. Beekman and D. Mevius of the External Advisory Board for their constructive support and comments. Finally, we would like to thank all participating farmers for their willingness to participate, their assistance in sampling and providing all data used this study.
Funding
This project was funded by the organization of the Dutch dairy supply chain ‘ZuivelNL’, reference 348998.
Transparency declarations
None to declare.
References
- 1. Ewers C, Bethe A, Semmler T. et al. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect 2012; 18: 646–55. [DOI] [PubMed] [Google Scholar]
- 2. Rao L, Lv L, Zeng Z. et al. Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003–2012. Vet Microbiol 2014; 172: 534–41. [DOI] [PubMed] [Google Scholar]
- 3. Hordijk J, Wagenaar JA, van de Giessen A. et al. Increasing prevalence and diversity of ESBL/AmpC-type β-lactamase genes in Escherichia coli isolated from veal calves from 1997 to 2010. J Antimicrob Chemother 2013; 68: 1970–3. [DOI] [PubMed] [Google Scholar]
- 4. Brunton LA, Reeves HE, Snow LC. et al. A longitudinal field trial assessing the impact of feeding waste milk containing antibiotic residues on the prevalence of ESBL-producing Escherichia coli in calves. Prev Vet Med 2014; 117: 403–12. [DOI] [PubMed] [Google Scholar]
- 5. Horton RA, Duncan D, Randall LP. et al. Longitudinal study of CTX-M ESBL-producing E. coli strains on a UK dairy farm. Res Vet Sci 2016; 109: 107–13. [DOI] [PubMed] [Google Scholar]
- 6. Liebana E, Batchelor M, Hopkins KL. et al. Longitudinal farm study of extended-spectrum β-lactamase-mediated resistance. J Clin Microbiol 2006; 44: 1630–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gonggrijp MA, Santman-Berends IMGA, Heuvelink AE. et al. Prevalence and risk factors for extended-spectrum β-lactamase- and AmpC-producing Escherichia coli in dairy farms. J Dairy Sci 2016; 99: 9001–13. [DOI] [PubMed] [Google Scholar]
- 8. Santman-Berends IMGA, Gonggrijp MA, Hage JJ. et al. Prevalence and risk factors for extended-spectrum β-lactamase or AmpC-producing Escherichia coli in organic dairy herds in the Netherlands. J Dairy Sci 2017; 100: 562–71. [DOI] [PubMed] [Google Scholar]
- 9. Snow LC, Warner RG, Cheney T. et al. Risk factors associated with extended spectrum β-lactamase Escherichia coli (CTX-M) on dairy farms in North West England and North Wales. Prev Vet Med 2012; 106: 225–34. [DOI] [PubMed] [Google Scholar]
- 10. Tragesser LA, Wittum TE, Funk JA. et al. Association between ceftiofur use and isolation of Escherichia coli with reduced susceptibility to ceftriaxone from fecal samples of dairy cows. Am J Vet Res 2006; 67: 1696–700. [DOI] [PubMed] [Google Scholar]
- 11.Anon. Dutch Animals Act 2011. https://zoek.officielebekendmakingen.nl/stb-2011-345.html.
- 12.Anon. Animal Welfare Body Utrecht 2017. http://www.ivd-utrecht.nl/en/.
- 13. Bos MEH, Taverne FJ, van Geijlswijk IM. et al. Consumption of antimicrobials in pigs, veal calves, and broilers in the Netherlands: quantitative results of nationwide collection of data in 2011. PLoS One 2013; 8: e77525.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dhand NK, Khatkar MS.. Statulator: An Online Statistical Calculator. Sample Size Calculator for Estimating a Single Proportion 2014. http://statulator.com/SampleSize/ss1P.html.
- 15.Working Group on Veterinary Antimicrobial Policy. Directive on Classification of Veterinary Antimicrobials [in Dutch]. Royal Netherlands Veterinary Association. http://wvab.knmvd.nl/richtlijn-WVAB.
- 16. Baede VO, Wagenaar JA, Broens EM. et al. Longitudinal study of extended-spectrum-β-lactamase- and AmpC-producing Enterobacteriaceae in household dogs. Antimicrob Agents Chemother 2015; 59: 3117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dohmen W, Schmitt H, Bonten M, Heederik D.. Air exposure as a possible route for ESBL in pig farmers. Environ Res 2017; 155: 359–64. [DOI] [PubMed] [Google Scholar]
- 18.R Development Core Team. R: A Language and Environment for Statistical Computing Vienna, Austria, 2008. https://www.R-project.org.
- 19. Bates D, Machler M, Bolker BM, Walker SC.. Fitting linear mixed-effects models using Ime4. J Stat Softw 2015; 67: 1–48. [Google Scholar]
- 20. Bankevich A, Nurk S, Antipov D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Treangen TJ, Ondov BD, Koren S. et al. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 2014; 15: 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Croucher NJ, Page AJ, Connor TR. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jolley KA, Maiden MCJ.. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 2010; 11: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seemann T. MLST. https://github.com/tseemann/mlst.
- 25. Hordijk J, Mevius DJ, Kant A. et al. Within-farm dynamics of ESBL/AmpC-producing Escherichia coli in veal calves: a longitudinal approach. J Antimicrob Chemother 2013; 68: 2468–76. [DOI] [PubMed] [Google Scholar]
- 26. Schmid A, Hörmansdorfer S, Messelhäusser U. et al. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli on Bavarian dairy and beef cattle farms. Appl Environ Microbiol 2013; 79: 3027–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wielders CCH, van Hoek AH, Hengeveld PD. et al. Extended-spectrum β-lactamase- and pAmpC-producing Enterobacteriaceae among the general population in a livestock-dense area. Clin Microbiol Infect 2016; 23: 120.e1–8. [DOI] [PubMed] [Google Scholar]
- 28. Dierikx C, van der Goot J, Fabri T. et al. Extended-spectrum-β-lactamase- and AmpC-β-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J Antimicrob Chemother 2013; 68: 60–7. [DOI] [PubMed] [Google Scholar]
- 29. Dohmen W, Bonten MJM, Bos MEH. et al. Carriage of extended-spectrum β-lactamases in pig farmers is associated with occurrence in pigs. Clin Microbiol Infect 2015; 21: 917–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.