Abstract
Objectives
Drug-resistant TB remains a public health challenge. Rifamycins are among the most potent anti-TB drugs. They are known to target the RpoB subunit of RNA polymerase; however, our understanding of how rifamycin resistance is genetically encoded remains incomplete. Here we investigated rpoB genetic diversity and cross-resistance between the two rifamycin drugs rifampicin and rifabutin.
Methods
We performed WGS of 1003 Mycobacterium tuberculosis clinical isolates and determined MICs of both rifamycin agents on 7H10 agar using the indirect proportion method. We generated rpoB mutants in a laboratory strain and measured their antibiotic susceptibility using the alamarBlue reduction assay.
Results
Of the 1003 isolates, 766 were rifampicin resistant and 210 (27%) of these were rifabutin susceptible; 102/210 isolates had the rpoB mutation D435V (Escherichia coli D516V). Isolates with discordant resistance were 17.2 times more likely to harbour a D435V mutation than those resistant to both agents (OR 17.2, 95% CI 10.5–27.9, P value <10−40). Compared with WT, the D435V in vitro mutant had an increased IC50 of both rifamycins; however, in both cases to a lesser degree than the S450L (E. coli S531L) mutation.
Conclusions
The observation that the rpoB D435V mutation produces an increase in the IC50 of both drugs contrasts with findings from previous smaller studies that suggested that isolates with the D435V mutation remain rifabutin susceptible despite being rifampicin resistant. Our finding thus suggests that the recommended critical testing concentration for rifabutin should be revised.
Introduction
Rifampicin resistance (RR) is the hallmark of MDR in TB. By the WHO’s most recent estimates, 4.1% of new TB cases are RR/MDR and this rate is 19% among retreatment cases.1,2 The estimated global incidence of RR/MDR was 600 000 in 2016, a figure that increased from 480 000 the year prior.1,2 Timely and accurate diagnosis of RR remains a major challenge and, together with challenges related to the duration and complexity of treatment regimens, progress in RR/MDR control has been limited. There is, however, increasing adoption of molecular diagnostic tests that rapidly detect genetic mutations in the RNA polymerase β subunit gene (rpoB) as a proxy for resistance to this drug class and MDR-TB.2 There are over 25 different resistance mutations detected by the latest generation, one such test being the Xpert MTB/RIF Ultra, and the results are currently summarized as binary, i.e. rifampicin resistance ‘detected’ versus ‘non-detected’.3 At the same time, it is well recognized that genetic mutations can have a range of effects on the resistance phenotype and that these effects can vary by bacterial lineage or genetic background.4,5 This variability may extend to differential susceptibility to other drugs from the same class.6 Given the complexity and side effects associated with MDR-TB therapy compared with susceptible TB, identifying subtypes of Mycobacterium tuberculosis (MTB) isolates that are apparently resistant, but may continue to be treatable with a higher dose of a first-line agent or a related drug from the same class is highly desirable.
WGS of MTB isolates is generating an abundance of information on the bacterial pathogen. To date, most research utilizing WGS data has attempted to recapitulate the results of phenotypic culture-based drug susceptibility testing (DST) in the hope of WGS potentially replacing this expensive and time-consuming approach.7–9 However, the power of WGS lies in potentially going beyond DST to allow the personalization of antibiotic therapy based on genotypic information. One such mutation, rpoB D435V, thought to encode about 30% of rifampicin resistance, has been highlighted as associated with rifabutin susceptibility at the CLSI-recommended critical cut-off point of 0.5 mg/L.10–12 The drug rifabutin is considered as effective as rifampicin in the treatment of drug-susceptible TB. Rifabutin is also substituted for rifampicin in HIV patients with TB due to rifampicin’s known interaction with components of ART.13 A few small or uncontrolled studies of rifabutin in patients with RR/MDR have suggested potential efficacy, but there have been no prospective trials.14–16 Most of the data have stemmed from comparisons between rifampicin and rifabutin DST and rpoB sequencing of MTB clinical isolates.17–22 Constructing rpoB mutant alleles in isogenic laboratory strains can directly measure the effect of mutations like D435V on rifamycin cross-resistance, but reports published to date have been conflicting. Williams et al.23 introduced the rpoB D435V mutation through plasmid transduction into the H37Rv lab strain and measured a step up in MIC of rifampicin, but not of rifabutin. Gill and Garcia,24 on the other hand, measured a large step up in the rifamycin concentration needed to inhibit 50% of ex vivo transcription with the introduction of the rpoB D435V mutation for both rifampicin and rifabutin. Notably the D435V mutation has been consistently found to have lower effects on the rifampicin MIC than S450L in several reports.4 To try to add clarity to the debate, we report on the largest collection of clinical MTB isolates for which rifampicin and rifabutin resistance were quantified with MICs and compared with WGS results. We describe the diversity of rpoB variants seen, assess the degree of rifampicin/rifabutin cross-resistance and construct rpoB mutants in a laboratory strain to study the in vitro effect of D435V on rifampicin and rifabutin MICs.
Methods
Patient cohort/sample collection
MTB sputum-based culture isolates from Peru were selected to overrepresent MDR from samples collected for clinical care and then archived at the Massachusetts State Laboratory (n = 496)7 or sampled from a longitudinal cohort of patients with TB from Lima, Peru (n = 567).25
Culture and drug resistance/MIC determination
Löwenstein–Jensen (LJ) culture was initially performed from sputum specimens using standard NALC-NaOH (where NALC stands for N-acetyl-l-cysteine) decontamination and isolates were frozen/archived. Cryopreserved isolates were transported to the National Jewish Health (NJH) laboratory, Colorado for regrowth and MIC determination. Each isolate was thawed and subcultured on LJ medium and then underwent rifampicin and rifabutin MIC determination on 7H10 agar medium using the indirect proportion method in a staged fashion as follows. Isolates were first tested at three low concentrations that include the WHO-recommended critical concentration. If the isolate was resistant at the critical concentration, then testing at six higher concentrations was also performed. The testing concentrations deviated from the traditional doubling to better detect intermediate-level MICs that are between the critical concentration and within theoretically achievable levels in patient sera (i.e. the Cmax) based on available pharmacodynamics data.26 The drug concentrations (mg/L) were as follows: rifampicin low: 0.25, 0.5 and 1; rifampicin high: 2, 3, 5, 8, 10 and 50; rifabutin low: 0.125, 0.25 and 0.50; and rifabutin high: 0.60, 0.75, 0.875, 1.0, 1.5 and 2.5.
DNA extraction and WGS
DNA from patient isolates was extracted from cryopreserved cultures as follows. Each isolate was thawed and subcultured on LJ medium and a big loopful of colony growth was lysed with lysozyme and proteinase K to obtain DNA using CTAB/chloroform (where CTAB stands for cetyl trimethylammonium bromide) extraction and ethanol precipitation. DNA was sheared into ∼250 bp fragments using a Covaris sonicator (Covaris, Inc.) and prepared using the TruSeq Whole-Genome Sequencing DNA Sample Preparation Kit (Illumina, Inc.) and sequenced on an Illumina HiSeq 2500 sequencer with paired-end reads of length 125 bp.
Variant calling and phylogeny construction
We aligned the Illumina reads to the reference MTB isolate H37Rv using Stampy 1.0.2327 and variants were called by Platypus 0.5.228 using default parameters. Genome coverage was assessed using SAMtools 0.1.1829 and FastQC30 and read-mapping taxonomy was assessed using Kraken.31 Strains that (i) failed sequencing at a coverage of less than 95% of rpoB sites at 10× or more reads for each site or (ii) had a mapping percentage of less than 90% to MTB complex were excluded. Variants were filtered if they had a quality of <15, purity of <0.4 or did not meet the PASS filter designation by Platypus. TB genetic lineage was called by constructing a neighbour-joining phylogeny using MEGA-532 including lineage-representative MTB isolates from Sekizuka et al.33 and confirmed by the SNP barcode method described in Coll et al.34
Statistical analysis
The Fisher exact test was used for testing proportions and the Wilcoxon rank sum test was used for comparing MIC distributions using R version 3.2.3. The significance threshold was set at <0.01. A mutation was determined to be homoplasic if it was found in isolates that belonged to more than one sublineage.
Construction and susceptibility testing of rpoB mutants
Using the laboratory Beijing-type MTB strain HN878 we obtained rpoB mutants by isolating individual colonies from cultures selected on solid 7H10 medium (Middlebrook 7H10 medium/0.5% glycerol/10% OADC/0.05% Tween 80) containing rifampicin at 2 mg/L, as previously described.35 The rifampicin resistance-determining region (RRDR) of rpoB (codons 426–454) was sequenced for each colony and the causal mutation was identified. We determined the MIC for the parental HN878 strain, as well as D435V and S450L mutants, of both rifampicin and rifabutin using the alamarBlue reduction method. Briefly, strains were grown to mid-logarithmic phase (OD600 0.8–1.0) in 7H9 medium (7H9 salts/0.2% glycerol/10% OADC/0.05% Tween 80) and subcultured to OD600 0.006 in fresh 7H9 medium. This diluted stock was then combined 1:1 with 2× drug-containing medium to a final volume of 200 μL in 96-well plates. The concentrations tested were 32, 8, 2, 0.5, 0.125, 0.03, 0.008 and 0.002 mg/L rifampicin and 0.5, 0.125, 0.03, 0.008, 0.002, 0.0005, 0.00012 and 0.00003 mg/L rifabutin. Antibiotics were dissolved in DMSO and, for the no-drug conditions, DMSO was added at an equal volume. Each condition was generated in three independent triplicate wells. Strains were cultured for 4 days with shaking at 37°C and then a 1/10 volume of alamarBlue (Bio-Rad) was added. After an additional 4 days of culture, the reduction of alamarBlue was quantified by measurement of OD570. Normalized growth was calculated for each replicate by subtracting the OD570 of the cell-free well from each condition and scaling by the background-subtracted OD570 measurement of a no-drug well. Because of a relatively high background OD seen at low drug concentrations with the alamarBlue reduction assay we report the fold increase in IC50 rather than the MIC.
Sequence data
All sequence data are available on NCBI under Bioproject number PRJEB26000.
Results
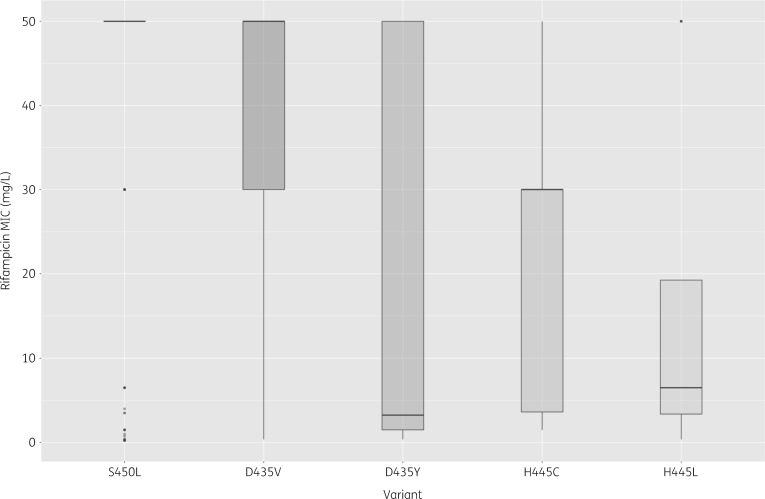
Of the total number of isolates, 60 had sequencing data that failed quality criteria and were excluded. Of the remaining 1003 isolates, 87 (9%) isolates belonged to the Beijing (L2) lineage (86 L2.2 and 1 L2.1) and those remaining were Euro-American (125 L4, 307 L4.1, 461 L4.3, 10 L4.8, 7 L4.7, 5 L4.6.1 and 1 L4.5). Of the total, 766 were RR (MIC >1 mg/L) and, of these, 210 (27%) displayed a rifabutin MIC ≤0.5 mg/L. Table 1 compares the rifampicin and rifabutin MICs for the isolates. The median rifabutin MIC for isolates that were rifampicin susceptible (MIC ≤1 mg/L, n = 236) was ≤0.125 mg/L (IQR ≤0.125 to ≤0.125 mg/L). We identified 94 different genetic variants in the rpoB gene or upstream intergenic region that occurred in one or more RR isolate (Table S1, available as Supplementary data at JAC Online). The gene body contained 74 unique non-synonymous single or double nucleotide substitutions and 8 indels at 51 different codons. The three most common rpoB RRDR mutations occurred at codons S450L, D435V and H445Y in 439, 131 and 21 RR isolates, respectively. Several non-RRDR rpoB mutations were homoplasic, i.e. occurred in isolates belonging to more than one TB sublineage. There were five such non-synonymous mutations that occurred in five or more isolates. Four co-occurred with one or more RRDR mutation, but the overlap was not complete for two: E250G and V170F. Only the mutation I491F was homoplasic and occurred in isolates with no RRDR mutation found. Five other nearby mutations (P454R, I480V, I488L, I491S and L494P) occurred in one to three RR isolates, each for a total of eight isolates. Although these variants did not display any homoplasy, there were no co-occurring RRDR mutations in these isolates to explain RR (Table S1).
Table 1.
Rifampicin versus rifabutin MICs for the 1003 clinical isolates
| Rifampicin | Rifabutin |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.125 | 0.250 | 0.500 | 0.600 | 0.750 | 0.875 | 1.0 | 1.5 | 2.5 | >2.5 | |
| ≤0.250 | 131 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 0.500 | 63 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.0 | 40 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2.0 | 29 | 1 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3.0 | 8 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| 5.0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8.0 | 12 | 2 | 1 | 3 | 2 | 0 | 0 | 1 | 0 | 1 |
| 10.0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| 50.0 | 37 | 11 | 3 | 9 | 0 | 0 | 1 | 6 | 3 | 2 |
| >50 | 44 | 27 | 15 | 17 | 9 | 10 | 15 | 36 | 83 | 351 |
All MICs are in mg/L and were determined using 7H10 agar proportions as detailed in the Methods section.
Of the 210 isolates with rifabutin susceptibility and RR, labelled rifamycin ‘discordant isolates’, 102 isolates carried the rpoB mutation D435V. There was no clonality among the isolates harbouring D435V; the isolates belonged to four sublineages (2.2, 4, 4.1 and 4.3) and the minimum isolate pairwise SNP distance among them was 19 SNPs. Three additional variants (S450L, H445Y and D435Y) were each observed in >5% of the discordant isolates (Table 2). Overall, the discordant isolates carried 31 different non-synonymous rpoB variants (Table 2). We tested all the observed variants for association with the discordant versus the concordant (rifampicin resistant and rifabutin resistant) phenotype; significantly associated mutations are shown in bold in Table 2. Discordant isolates were more likely to carry a D435V, D435Y, H445L or H445C mutation than those resistant to both agents. On the other hand, concordant isolates were more likely to carry an S450L or an H445Y mutation. We tested whether the mutations associated with discordance resulted in lower rifampicin MICs as compared with S450L and this was the case for all four mutations (P value <10−15 in each case) (Figure 1). We also tested whether there was a difference in rifabutin MIC observed for isolates from the Beijing (2.2) versus the Euro-American (4, 4.1, 4.3) sublineages with D435V mutations and observed the median MIC for the Beijing isolates to be 0.375 mg/L (IQR 0.188–1.5 mg/L), significantly higher than the rifabutin MIC for the Euro-American pool of isolates [median MIC ≤0.125 mg/L (IQR ≤0.125–0.188 mg/L)] (P value 6 × 10−4). Overall 67/210 isolates with the D435V mutation and rifamycin discordance had a rifabutin MIC of 0.25 or 0.5 mg/L, i.e. greater than the median WT rifabutin MIC of ≤0.125 mg/L observed among rifampicin-susceptible isolates (n = 236) and among the subset without any rpoABC mutations (n = 103).
Table 2.
rpoB variant alleles found in isolates with discordant rifamycin resistance (rifabutin MIC <0.5 mg/L and rifampicin MIC >1 mg/L) versus concordant rifamycin resistance
| H37Rv position | Nucleotide change | Amino acid change | Count for discordant isolates (N = 210), n (%) | Count for concordant isolates (N = 556), n (%) | Rifampicin MIC (median) | Rifampicin MIC (IQR) | Rifabutin MIC (median) | Rifabutin MIC (IQR) | OR | 95% CI | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 761110 | A>T | D435V | 102 (49) | 29 (5) | >50 | 30–50 | ≤0.125 | ≤0.125–0.375 | 17.2 | 10.5–27.9 | <0.001 |
| 761155 | C>T | S450L | 19 (9) | 420 (76) | >50 | 50–50 | >2.5 | 2 to >2.5 | 0.03 | 0.02–0.05 | <0.001 |
| 761889 | G>C | V695L | 18 (9) | 51 (9) | >50 | 1.5–50 | 0.9375 | ≤0.125–2 | 0.9 | 0.5–1.6 | 0.9 |
| 761109 | G>T | D435Y | 12 (6) | 3 (<1) | 3.25 | 1.5–50 | ≤0.125 | ≤0.125–0.328 | 11.2 | 3.1–40.1 | <0.001 |
| 761140 | A>T | H445L | 12 (6) | 0 | 6.5 | 2.125–6.5 | ≤0.125 | ≤0.125 to ≤0.125 | undefined | 7.65–undefined | <0.001 |
| 761139 | CA>TG | H445C | 7 (3) | 0 | 30 | 17–30 | ≤0.125 | ≤0.125 to ≤0.125 | undefined | 3.88–undefined | <0.001 |
| 761161 | T>C | L452P | 5 (2) | 4 (1) | 4 | 1.5–40 | ≤0.125 | ≤0.125–0.9 | 3.4 | 0.9–12.7 | 0.07 |
| 760555 | A>G | E250G | 4 (2) | 9 (2) | 1.5 | ≤0.25–50 | ≤0.125 | ≤0.125 to >2.5 | 1.2 | 0.4–3.9 | 0.76 |
| 761277 | A>T | I491F | 4 (2) | 2 (<1) | 3.25 | 1.75–5.875 | 0.25 | ≤0.125–0.506 | 5.4 | 1.0–29.6 | 0.05 |
| 761095 | T>G | L430R | 4 (2) | 1 (<1) | >50 | 30–50 | 0.375 | 0.188–0.375 | 10.8 | 1.2–97.2 | 0.02 |
| 761128 | C>T | S441L | 3 (1) | 0 | 30 | 30–30 | ≤0.125 | ≤0.125–0.156 | — | 1.09–undefined | 0.02 |
| 761880 | G>A | A692T a | 2 (1) | 45 (8) | >50 | >50 to >50 | 2 | 1.25 to >2.5 | 0.1 | 0.03–0.5 | <0.001 |
| 761154 | TC>CA | S450Q | 2 (1) | 3 (<1) | >50 | >50 to >50 | 0.55 | 0.375–0.935 | 1.8 | 0.3–10.7 | 0.62 |
| 761101 | A>C | Q432P | 2 (1) | 1 (<1) | >50 | >50 to >50 | 0.375 | 0.281–0.525 | 5.4 | 0.5–59.3 | 0.19 |
| 761149 | G>A | R448Q | 2 (1) | 1 (<1) | >50 | >50 to >50 | 0.375 | 0.375–0.812 | 5.4 | 0.5–59.3 | 0.19 |
| 761109 | G>A | D435N | 2 (1) | 0 | >50 | >50 to >50 | 0.375 | 0.375–0.375 | — | 0.5–undefined | 0.08 |
| 761148 | CG>AA | R448K | 2 (1) | 0 | >50 | >50 to >50 | 0.1875 | 0.188–0.188 | — | 0.5–undefined | 0.08 |
| 761139 | C>T | H445Y | 1 (<1) | 20 (4) | >50 | >50 to >50 | >2.5 | >2.5 to >2.5 | 0.1 | 0.02–0.96 | 0.01 |
| 761139 | C>G | H445D | 1 (<1) | 7 (1) | >50 | 45 to >50 | >2.5 | >2.5 to >2.5 | 0.4 | 0.05–3.1 | 0.46 |
| 760425 | G>A | E207K | 1 (<1) | 6 (1) | >50 | >50 to >50 | >2.5 | >2.5 to >2.5 | 0.4 | 0.05–3.6 | 0.68 |
| 760645 | C>T | P280L | 1 (<1) | 0 | 30 | — | ≤0.125 | — | — | — | 0.28 |
| 761083 | delGCACCA | 426 | 1 (<1) | 0 | 6.5 | — | ≤0.125 | — | — | — | 0.28 |
| 761103 | insTTC | 433 | 1 (<1) | 0 | 30 | — | 0.1875 | — | — | — | 0.28 |
| 761108 | GGA>AGG | MD434- 435IG | 1 (<1) | 0 | >50 | — | ≤0.125 | — | — | — | 0.28 |
| 761109 | GA>TT | D435F | 1 (<1) | 0 | 6.5 | — | ≤0.125 | — | — | — | 0.28 |
| 761115 | delAAC | 437 | 1 (<1) | 0 | >50 | — | ≤0.125 | — | — | — | 0.28 |
| 761139 | CA>GG | H445G | 1 (<1) | 0 | >50 | — | ≤0.125 | — | — | — | 0.28 |
| 761139 | CA>TC | H445S | 1 (<1) | 0 | 1.5 | — | ≤0.125 | — | — | — | 0.28 |
| 761167 | C>T | P454L | 1 (<1) | 0 | ≤0.25 | — | ≤0.125 | — | — | — | 0.28 |
| 761248 | A>C | E481A | 1 (<1) | 0 | 30 | — | 0.375 | — | — | — | 0.28 |
| 761905 | C>T | P700L | 1 (<1) | 0 | >50 | — | ≤0.125 | — | — | — | 0.28 |
All MICs are in mg/L.
A695T occurred only in lineage 4.3 isolates and always with S450L (46 isolates) or D435V (1 isolate) and thus this association is likely confounded; A695T was not observed in rifampicin-susceptible isolates. See Table S1 for more information on non-RRDR variants.
Figure 1.
Rifampicin MIC is significantly lower for the four mutations found to be associated with rifampicin/rifabutin discordance relative to S450L (associated with concordant resistance).
We investigated whether concurrent mutations in genes that encode the β′ and α subunits of MTB DNA-directed RNA polymerase (rpoC and rpoA, respectively) associate with rifamycin discordance. We observed 212 rpoC and 25 rpoA variants in the 1003 isolates. Twenty-eight rpoC and two rpoA variants were observed to be homoplasic (Table S2). After excluding rpoC G594E and the synonymous variant rpoC A542A, which were very common among 4.3 and 4.1 isolates, respectively (Table S2), and among both rifamycin-susceptible and -resistant isolates, we calculated the proportion of concordant susceptible, discordant and concordant resistant isolates that harboured one of the remaining homoplasic rpoC or rpoA variants (Table S3). Only 9 of the 236 concordant susceptible and 9 of the 210 discordant isolates harboured an rpoA/C homoplasic variant, with no difference in proportions among the two (Fisher OR 1.13, P value 0.81). However, 220 of the 556 concordant resistant isolates harboured an rpoA/C homoplasic variant, significantly higher than among the discordant isolates (Fisher OR 14.5, P value <10−15).
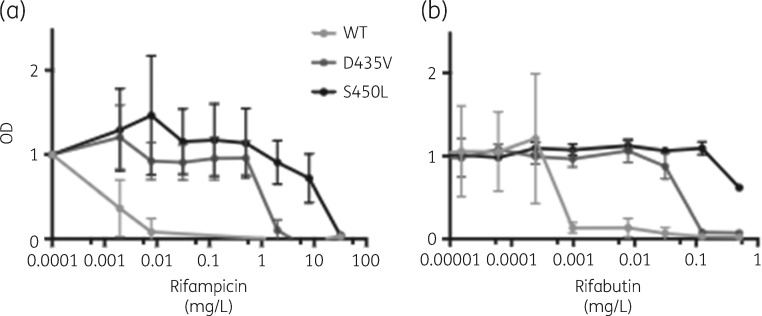
As D435V was the most common mutation associated with the apparent lack of cross-resistance to rifabutin we selected a D435V and an S450L mutant in the laboratory strain HN878 by identifying spontaneous mutants resistant to rifampicin. Compared with the isogenic parental HN878 strain, the D435V mutation resulted in an increase in MIC of both rifampicin and rifabutin; however, in both cases this increase was less than what was observed for the S450L mutant (Figure 2). For rifabutin and D435V the IC50 was 62.5× that of WT, as compared with >250× for S450L; for rifampicin, D435V and S450L mutants had IC50s that were 1000× and 16 000× that of WT, respectively. The D435V mutant was almost completely inhibited at 0.5 mg/L rifabutin, while S450L mutants retained substantial capacity for growth at that drug concentration.
Figure 2.
The rpoB D435V mutation increases the MIC of both rifampicin and rifabutin. Measurement of antibiotic resistance by alamarBlue reduction assay with varying concentrations of rifampicin (a) and rifabutin (b). OD570 is normalized with the growth of an antibiotic-free control set to 1. Each data point represents the mean of three independent cultures with the standard deviation.
Discussion
In this study we observed RR isolates to have considerable rpoB genetic diversity. Several recent reports have challenged the notion that RR can only be caused by mutations within the RRDR; for example, the I491F mutation was found to be a common cause of RR in Swaziland.36 Homoplasy is one method of measuring variants under positive selection and hence of relevance to the resistance phenotype.37 Although our study does not establish causation, we found several mutations outside of the RRDR, several in the vicinity of I491F, in RR isolates that have no other clearly causative mutation. This highlights the complexity of resistance even to the first drug rifampicin and emphasizes the need to look beyond the RRDR, especially when patient treatment response is not as expected.
We also find discordance between rifampicin and rifabutin to be relatively common, occurring in 27% of RR isolates. Rifabutin resistance among rifampicin-susceptible isolates was very rare and occurred in only one isolate. Again, there was considerable rpoB variant diversity among the discordant isolates, but the D435V mutation was by far the most common and occurred in nearly half of the discordant isolates (102/210, 49%). Homoplasic rpoA and rpoC mutations were rare among rifamycin-discordant isolates, occurring in 4% of such isolates. This proportion was similar among rifamycin-concordant susceptible isolates. Four rpoB mutations were significantly associated with discordance and two were significantly associated with concordance. Interestingly, all four of the mutations significantly associated with discordance were also associated with lower MICs of rifampicin itself and not just of rifabutin. This was also confirmed by determining MICs for the two isogenic strains harbouring D435V and S450L, with D435V found to raise both rifampicin and rifabutin MICs, albeit to a lower extent than S450L. The effect of D435V on rifampicin MIC does appear more profound than on rifabutin, increasing the IC50 by 1000-fold versus 62.5-fold, respectively, but we found D435V to increase IC50 more than has been previously reported in clinical isolates, estimated at 2–4-fold greater than that of WT MIC in one study that examined clinical isolates from the atypical Beijing lineage.10 As HN878 belongs to the typical Beijing lineage we cannot exclude that the observed difference in inhibitory concentration may be a result of interaction between the D435V mutation and the genetic background. In the clinical isolate pool examined, we did observe a higher rifabutin MIC for Beijing isolates than Euro-American isolates, providing some evidence for interactions between the genetic background and resistance mutation. Finally, it is important to note that we phenotyped the in vitro rpoB mutants with the alamarBlue microtitre assay. Although MICs determined using colorimetric microtitre assays and those determined using agar proportions are not identical they have been found to correlate highly, with AUC >0.95 in prior studies.38,39
Overall our finding that the D435V variant results in a large step up in rifabutin inhibitory concentration, especially in HN878 and East Asian-Beijing 2.2 isolates, suggests that rifabutin may have lower efficacy against such isolates and that the rifabutin critical testing concentration, currently recommended at 0.5 mg/L, may be too high. Rifamycin killing of MTB is concentration dependent, but achieving an adequate rifabutin peak serum concentration (Cmax ≥0.45 mg/L40,41) has proven challenging even when treating rifamycin-susceptible TB with currently recommended doses.26,41,42 Thus, even small increases in rifabutin MIC may have significant consequences on treatment efficacy.26,42 Ideally more retrospective data can be collected on the rifabutin treatment response of patients whose MTB isolates harbour mutations associated with discordance as there may not be equipoise to conduct a prospective rifabutin trial in such patients. Alternatively, trials in which rifabutin is added to an otherwise adequate MDR regimen in patients with such isolates can be considered.
Supplementary Material
Acknowledgements
We thank the Peruvian team for their patient care and for providing the patient isolates that enabled this research.
Funding
Funded by the National Institute of Allergy and Infectious Diseases (NIAID) as a Center for Excellence in Translational Research (U19-AI109755) and also by National Institutes of Health (NIH) Big Data to Knowledge (BD2K) Initiative K01 (MRF ES026835). The funding sources had no role in any aspect of the study, manuscript or decision to submit for publication.
Transparency declarations
None to declare.
References
- 1. Global Tuberculosis Report 2016. Geneva, Switzerland: WHO, 2016. http://apps.who.int/medicinedocs/documents/s23098en/s23098en.pdf. [Google Scholar]
- 2. Global Tuberculosis Report 2017. Geneva, Switzerland: WHO, 2017. http://apps.who.int/medicinedocs/documents/s23360en/s23360en.pdf. [Google Scholar]
- 3. Chakravorty S, Simmons AM, Rowneki M. et al. The new Xpert MTB/RIF Ultra: improving detection of Mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. mBio 2017; 8: e00812-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nebenzahl-Guimaraes H, Jacobson KR, Farhat MR. et al. Systematic review of allelic exchange experiments aimed at identifying mutations that confer drug resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 2014; 69: 331–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Safi H, Lingaraju S, Amin A. et al. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-d-arabinose biosynthetic and utilization pathway genes. Nat Genet 2013; 45: 1190–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farhat MR, Jacobson KR, Franke MF. et al. Gyrase mutations are associated with variable levels of fluoroquinolone resistance in Mycobacterium tuberculosis. J Clin Microbiol 2016; 54: 727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farhat MR, Sultana R, Iartchouk O. et al. Genetic determinants of drug resistance in Mycobacterium tuberculosis and their diagnostic value. Am J Respir Crit Care Med 2016; 194: 621–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bradley P, Gordon NC, Walker TM. et al. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun 2015; 6: 10063.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walker TM, Kohl TA, Omar SV. et al. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis 2015; 15: 1193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sirgel FA, Warren RM, Böttger EC. et al. The rationale for using rifabutin in the treatment of MDR and XDR tuberculosis outbreaks. PLoS One 2013; 8: e59414.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes: Approved Standard M24-A. NCCLS, Wayne, PA, USA, 2003. [PubMed] [Google Scholar]
- 12. Dheda K, Limberis JD, Pietersen E. et al. Outcomes, infectiousness, and transmission dynamics of patients with extensively drug-resistant tuberculosis and home-discharged patients with programmatically incurable tuberculosis: a prospective cohort study. Lancet Respir Med 2017; 5: 269–81. [DOI] [PubMed] [Google Scholar]
- 13. Nahid P, Dorman SE, Alipanah N. et al. Official American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America clinical practice guidelines: treatment of drug-susceptible tuberculosis. Clin Infect Dis 2016; 63: e147–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jo K-W, Ji W, Hong Y. et al. The efficacy of rifabutin for rifabutin-susceptible, multidrug-resistant tuberculosis. Respir Med 2013; 107: 292–7. [DOI] [PubMed] [Google Scholar]
- 15. Grassi C, Peona V.. Use of rifabutin in the treatment of pulmonary tuberculosis. Clin Infect Dis 1996; 22 Suppl 1: S50–4. [DOI] [PubMed] [Google Scholar]
- 16. Lee H, Ahn S, Hwang NY. et al. Treatment outcomes of rifabutin-containing regimens for rifabutin-sensitive multidrug-resistant pulmonary tuberculosis. Int J Infect Dis 2017; 65: 135–41. [DOI] [PubMed] [Google Scholar]
- 17. Jamieson FB, Guthrie JL, Neemuchwala A. et al. Profiling of rpoB mutations and MICs for rifampin and rifabutin in Mycobacterium tuberculosis. J Clin Microbiol 2014; 52: 2157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berrada ZL, Lin S-YG, Rodwell TC. et al. Rifabutin and rifampin resistance levels and associated rpoB mutations in clinical isolates of Mycobacterium tuberculosis complex. Diagn Microbiol Infect Dis 2016; 85: 177–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chien HP, Yu MC, Ong TF. et al. In vitro activity of rifabutin and rifampin against clinical isolates of Mycobacterium tuberculosis in Taiwan. J Formos Med Assoc 2000; 99: 408–11. [PubMed] [Google Scholar]
- 20. Dickinson JM, Mitchison DA.. In vitro activity of new rifamycins against rifampicin-resistant M. tuberculosis and MAIS-complex mycobacteria. Tubercle 1987; 68: 177–82. [DOI] [PubMed] [Google Scholar]
- 21. Senol G, Erbaycu A, Ozsöz A.. Incidence of cross resistance between rifampicin and rifabutin in Mycobacterium tuberculosis strains in Izmir, Turkey. J Chemother 2005; 17: 380–4. [DOI] [PubMed] [Google Scholar]
- 22. Uzun M, Erturan Z, Anğ O.. Investigation of cross-resistance between rifampin and rifabutin in Mycobacterium tuberculosis complex strains. Int J Tuberc Lung Dis 2002; 6: 164–5. [PubMed] [Google Scholar]
- 23. Williams DL, Spring L, Collins L. et al. Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 1998; 42: 1853–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gill SK, Garcia GA.. Rifamycin inhibition of WT and Rif-resistant Mycobacterium tuberculosis and Escherichia coli RNA polymerases in vitro. Tuberculosis (Edinb) 2011; 91: 361–9. [DOI] [PubMed] [Google Scholar]
- 25. Zelner J, Murray M, Becerra M. et al. Protective effects of household-based TB interventions are robust to neighbourhood-level variation in exposure risk in Lima, Peru: a model-based analysis. Int J Epidemiol 2018; 47: 185–92. [DOI] [PubMed] [Google Scholar]
- 26. Alsultan A, Peloquin CA.. Therapeutic drug monitoring in the treatment of tuberculosis: an update. Drugs 2014; 74: 839–54. [DOI] [PubMed] [Google Scholar]
- 27. Lunter G, Goodson M.. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 2011; 21: 936–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rimmer A, Phan H, Mathieson I. et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet 2014; 46: 912–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H, Handsaker B, Wysoker A. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babraham Bioinformatics. FastQC: A Quality Control Tool for High Throughput Sequence Data https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 31. Wood DE, Salzberg SL.. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 2014; 15: R46.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tamura K, Peterson D, Peterson N. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28: 2731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sekizuka T, Yamashita A, Murase Y. et al. TGS-TB: total genotyping solution for Mycobacterium tuberculosis using short-read whole-genome sequencing. PLoS One 2015; 10: e0142951.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Coll F, McNerney R, Guerra-Assunção JA. et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun 2014; 5: 4812.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ford CB, Lin PL, Chase MR. et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet 2011; 43: 482–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sanchez-Padilla E, Merker M, Beckert P. et al. Detection of drug-resistant tuberculosis by Xpert MTB/RIF in Swaziland. N Engl J Med 2015; 372: 1181–2. [DOI] [PubMed] [Google Scholar]
- 37. Farhat MR, Shapiro BJ, Kieser KJ. et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet 2013; 45: 1183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Palomino J-C, Martin A, Camacho M. et al. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2002; 46: 2720–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rancoita PMV, Cugnata F, Gibertoni Cruz AL. et al. Validating a 14-drug microtiter plate containing bedaquiline and delamanid for large-scale research susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother 2018; 62: e00344-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weiner M, Benator D, Burman W. et al. Association between acquired rifamycin resistance and the pharmacokinetics of rifabutin and isoniazid among patients with HIV and tuberculosis. Clin Infect Dis 2005; 40: 1481–91. [DOI] [PubMed] [Google Scholar]
- 41. Naiker S, Connolly C, Wiesner L. et al. Randomized pharmacokinetic evaluation of different rifabutin doses in African HIV-infected tuberculosis patients on lopinavir/ritonavir-based antiretroviral therapy. BMC Pharmacol Toxicol 2014; 15: 61.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boulanger C, Hollender E, Farrell K. et al. Pharmacokinetic evaluation of rifabutin in combination with lopinavir-ritonavir in patients with HIV infection and active tuberculosis. Clin Infect Dis 2009; 49: 1305–11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.