Abstract
Skeletal muscle has an extraordinary capacity to regenerate after injury and trauma. The muscle repair mechanism is a complex process orchestrated by multiple steps. In neuromuscular disorders such as Duchenne muscular dystrophy (DMD), the pathological consequences of the lack of dystrophin and the loss of the dystrophin-associated protein complex are dramatic, with a progressive cascade of events, such as continual influx of inflammation, repeated cycles of degeneration and impaired regeneration. Thus, muscle regeneration is a hallmark of the disease and careful monitoring of regenerative processes with robust markers should provide useful information to the field. Since decades, several indices of regeneration such as centronucleation and fibre size have been commonly used. In the present review, we discuss the impaired regenerative process in DMD, the common and new indices of regeneration and their associated methodologies. We notably highlight the regenerative marker embryonic myosin as a robust indicator of muscle regeneration. We also describe new quantitative methodologies offering the possibility of using a panel of translational regenerative biomarkers to obtain a more complete view of the regeneration processes. Upregulation of utrophin, an autosomal and functional paralogue of dystrophin, is one of the most promising therapeutic strategies as it targets the primary cause of the disease and is applicable to all DMD patients regardless their genetic defects. As utrophin is a regeneration associated protein increased in dystrophic muscle, we discuss the correlation of utrophin levels after drug treatment with regeneration markers. The recent advances in technologies and complementary markers of muscle regeneration described in this review, provide an unprecedented opportunity to develop more robust utrophin DMD based strategies for all DMD patients.
Keywords: DMD, regeneration, biomarkers, utrophin, embryonic myosin, methodologies, muscle repair, degeneration
Introduction
Muscles represent 30–40% of the body mass, play key roles in regulating metabolism and energy homeostasis in the organism and the musculoskeletal system is essential for coordinated movements, postural maintenance and independent living. This tissue is susceptible for various injuries in daily life, such as mechanical trauma, ischemia, thermal stress or neurological damage, and in various pathogenic conditions such as the Duchenne muscular dystrophin (DMD) (Guiraud et al., 2015). The innate capacity of skeletal muscle to regenerate is a complex and highly orchestrated mechanism involving several cell types, sequential and overlapping stages from the inflammatory response and the invasion of macrophages in different waves; the activation, mobilisation and differentiation of the satellite cells as major contributor to muscle regeneration, and finally the maturation of newly formed myofibres (Ciciliot and Schiaffino, 2010). Thereafter, we will discuss the impaired regenerative process as a hallmark of DMD, the common and new indices of regeneration (Baghdadi and Tajbakhsh, 2018; Guiraud et al., 2019) and the need to use panels of regenerative biomarkers to monitor on greater details the disease and the efficacy of treatments as well. We will focus on utrophin therapies for DMD (Guiraud et al., 2018) as utrophin and regeneration are deeply connected, highlighting recent methodologies to quantify utrophin (Janghra et al., 2016), new insights and fundamental questions to address. We have performed a PubMed literature search of articles published in the period January 1986–January 2019 on muscle regeneration and associated biomarkers in DMD.
Duchenne Muscular Dystrophin
DMD is a lethal X-linked recessive disorder affecting 1 in 5000 boys (Guiraud et al., 2015). The disease is characterized by a progressive muscle wasting leading to loss of ambulation by 8–12 years of age and premature death at 20–30 years due to respiratory and cardiac complications. At the molecular level, DMD is caused by mutations in the dystrophin gene leading to the absence of the protein. Dystrophin provides an essential mechanical link between the extracellular matrix and the actin cytoskeleton through the dystrophin-associated protein complex (DAPC) to maintain the strength, flexibility and stability in skeletal muscles. Absence of dystrophin and subsequent loss of the DAPC leads to progressive defects including perturbation of the calcium homeostasis, activation of proteases and pro-inflammatory cytokines, mitochondrial and satellite cells dysfunction. The lack of dystrophin also alters the morphology of neuromuscular junction and neurons in dystrophic mice (Pratt et al., 2015) and DMD is associated with neurodevelopmental disorders as evident by cognitive and behavioral abnormalities found in patients. At the cellular level, dystrophic muscle show evidence of necrosis, inflammation and fibrosis, undergo repeated bouts of degeneration and regeneration (Figure 1) with impaired vascular adaptation and suffer from contraction-induced injury resulting in muscle wasting, fatty accumulation and premature death. Although considerable progress has been made in gene-based, cell-based and pharmacological strategies (Barthelemy and Wein, 2018), there is currently no effective treatment for DMD. Clinical heterogeneity in DMD (Desguerre et al., 2009) has posed significant challenges to applying meaningful outcome measures to clinical trials and one of the major hurdles in developing treatment for DMD is the lack of translational readout to predict the benefit of experimental medicinal products. Therefore, the development of new methods to monitor disease progression will greatly help in gaining new insights about the pathology and will provide complementary and essential information to predict benefits from novel treatments for DMD, notably in the case of dystrophin-independent therapeutic avenues.
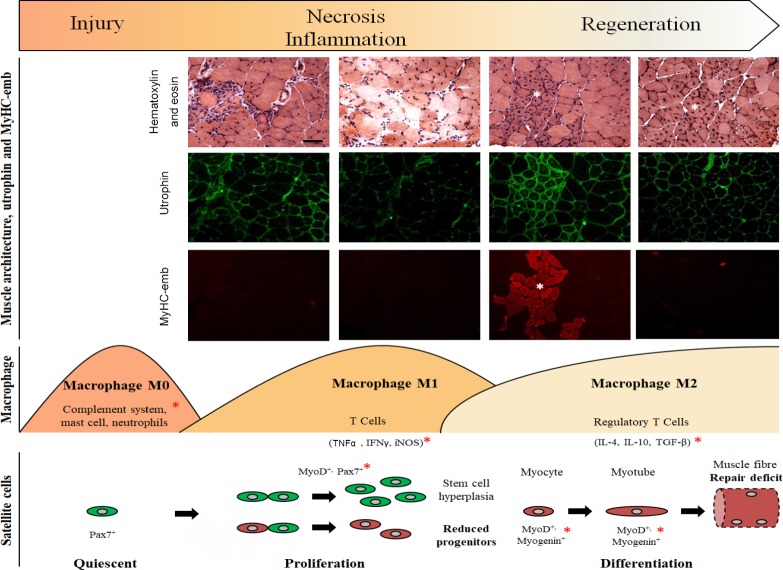
Figure 1.
Different stages of myogenesis in regenerative dystrophic muscle.
The lack of dystrophin and subsequent loss of the DAPC result in membrane fragility leading to architectural changes of the muscle fibre from the initial necrosis to the inflammatory stage and the impaired muscle regeneration (Guiraud et al., 2015). Common signs of muscle regeneration (white asterisk) are centronucleated myofibres, clusters of small fibres and presence of MyHC-emb. Utrophin expression is increased at the sarcolemma in dystrophic muscle as part of the repair process. Three different waves of immune cells occur from the first wave with the complement system, the mast cells and the neutrophils, to the pro-inflammatory M1 macrophage secreting a variety of cytokines. The recruitment of various immune cells regulates the activation, proliferation and differentiation of muscle satellite cells. Their self-renewal and commitment are governed by a gene regulatory network (Baghdadi and Tajbakhsh, 2018). The stem cell pool is maintained by symmetric satellite cell expansion and the myogenic progenitors, essential for the regeneration process, are generated by asymmetric cell divisions. In DMD, the satellite cell polarity is impaired resulting in an increased number of satellite cells and a reduction of the muscle progenitors causing a repair deficit and an impaired regeneration (Dumont et al., 2015). In addition to usual indices of regeneration, study these cytokines and transcription factors (red asterisk) will provide a more complete view on the regenerative processes, notably after drug treatment. Hematoxylin-eosin pictures and utrophin/MyHC-emb immunofluorescence images are derived from mdx skeletal muscles. Scale bar: 100 µm. IL: Interleukin; TGF-β: transforming growth factor beta; TNFα: tumour necrosis factor alpha; IFNγ: interferon gamma; iNOS: inducible nitric oxide synthase; DAPC: dystrophin-associated protein complex; DMD: Duchenne muscular dystrophin; MyHC-emb: embryonic myosin.
Regenerative Biomarkers for Duchenne Muscular Dystrophin
The pathologic processes in dystrophic muscles include marked degeneration and regeneration of myofibres. Histological examination of dystrophic skeletal muscles revealed excessive fibre size variation, large rounded hypertrophic fibres, fibres with central nuclei, as well as hypercontracted fibres and clusters off small regenerating myofibres. These features offer valuable indices of muscle regeneration, a hallmark of the disease (Figure 1). Since decades, centronucleation is used as a marker of regeneration (Treat-NMD SOP DMD_M.1.2.001) but is limited by variables which may influence the proportion of centronucleated fibres. Another commonly used indicator of regeneration is the morphological change in size of nascent muscle cells. Nevertheless, as the orientation of the sectioning angle could be misleading, the use of more robust morphometric parameters as the minimal Feret’s diameter and its associated variance coefficient is recommended (Treat-NMD SOP DMD_M.1.2.001). Force production in response to a single action potential (peak twitch tension) or to a maximal activation following a series of stimuli (tetanic tension) is also a valuable methodology to evaluate muscle regeneration for preclinical studies (Treat-NMD SOP DMD_M.1.2.002, Treat-NMD SOP DMD_M.2.2.005). Recently, we revisited the presence of developmental myosin such as embryonic myosin (MyHC-emb) and neonatal in dystrophic muscles as a meaningful indicator of muscle damage (Schiaffino et al., 1986) which correlates with functional motor score in DMD and Becker muscular dystrophy (BMD) (Janghra et al., 2016). Our study demonstrated that MyHC-emb is a robust marker of regeneration at different ages and in different muscles of the mdx mice (Guiraud et al., 2019), the most commonly used animal model for DMD. Restoration of dystrophin significantly reduced MyHC-emb levels and our results provide translational support for the use of developmental myosin as a disease biomarker in DMD clinical trials. Nevertheless, there is no single marker that unequivocally identifies a regenerating fibre and MyHC-emb suffers from limitations as it may be occasionally present in non-regenerating denervated myofibres and is expressed at different levels during the regeneration stages between muscle fibres. Therefore, knowing that muscles, in animal models as well as patients are affected to different degrees, it is essential to use a panel of complementary biomarkers to obtain a more complete view on regeneration.
In recent years, several crucial regulators of muscle regeneration have been described (Baghdadi and Tajbakhsh, 2018). Gene array studies highlighted interesting regeneration-associated genes and pathways such as Notch-Delta, Bmp15 and Nrg3 (Turk et al., 2005). Long non-coding RNAs with a role in embryonic stem cell maintenance and some evolutionary conserved microRNAs (miR) acting as post-transcriptional regulators as miR-489 or miR-206 were also described to have an essential role during skeletal muscle regeneration and could lead to interesting complementary indicators of the regenerative process. Furthermore, muscle regeneration is modulated by inflammation (Yang and Hu, 2018) and therefore these mechanisms need to be considered to gain a deeper view on regenerative processes (Figure 1). The complement system, first sensor of the muscle injury, is impaired in dystrophic muscle illustrating a deficit in immunity in DMD. The different secreted factors, such as tumour necrosis factor alpha or transforming growth factor beta, released during muscle repair and the different macrophage waves, guiding and playing central roles in the regulation of the muscle regeneration, could be also informative and offer, in conjunction with the usual regenerative indicators, a more complete view on the regeneration processes (Figure 1).
Satellite cells are essential for muscle regeneration. While the progressive decline of compensatory regeneration in DMD has been historically attributed to the functional exhaustion of the satellite cells, recent critical insights have been provided. Dumont et al. (2015) demonstrated that dystrophin is highly expressed in satellite cells and plays an essential role in the cell polarity establishment regulating the generation of myogenic progenitors. In DMD, the loss of polarity in dystrophin-deficient satellite cells results in the inability to establish the cell polarity, the abrogation of asymmetric satellite stem-cell divisions and the failure to enter the myogenic program lead to impaired regeneration. Consequently, dystrophic muscles show a satellite cell hyperplasia and a reduction in progenitors leading to repair deficit (Figure 1). Monitoring the ability of the satellite cells to enter the myogenic program and optimizing strategies to ensure satellite cell delivery are therefore important.
Utrophin, a Regeneration Associated Protein
Utrophin is a promising candidate to compensate for the lack of dystrophin in all DMD patients independent of their mutation (Guiraud et al., 2018). The autosomal paralogue to dystrophin, utrophin, has structurally similar N-terminal, cysteine-rich and C-terminal domains and despite subtle differences, shares many binding partners such as such as β-dystroglycan and F-actin (Ervasti, 2007). Animal model studies with transgenic mdx mice overexpressing utrophin demonstrated that the continuous localization of utrophin along the sarcolemma prevents signs of dystrophy in a dose dependent manner. Even a low increase of utrophin is significantly beneficial and a high level of utrophin not toxic in a broad range of murine tissues. Several therapeutics strategies resulting in increased utrophin, from the modulation of the utrophin promoter using small drugs to the stabilisation of the utrophin protein using different agents and utrophin gene therapy were previously reviewed (Guiraud et al., 2018). These studies demonstrate that utrophin act as an efficient surrogate to compensate for the lack of dystrophin and that utrophin overexpression is a promising therapeutic avenue for all DMD patients. In transgenic mdx mice overexpressing utrophin, all signs of regeneration are suppressed: the index of centronucleation, the MyHC-emb levels and the muscle function are similar to wild-type animals. Therefore, it will be interesting to elucidate the potential roles and benefits of utrophin in immunity, in macrophage regulation and to determine if, similarly to dystrophin, utrophin can restore the stem cell polarity and the generation of myogenic progenitors.
Ubiquitously expressed utrophin is found abundantly in lung, kidney, liver, spleen, and brain with lower levels in adult in skeletal and cardiac muscles (Fisher et al., 2001). In early developing muscles, utrophin is expressed at the sarcolemma and progressively replaced by dystrophin towards birth. In adult muscle, utrophin is limited to the neuromuscular and myotendinous junctions. In mdx as in DMD and BMD muscles, utrophin expression is up-regulated at the sarcolemma as part of the repair process (Figure 1). As utrophin based strategies aim to maintain utrophin in larger non-regenerative muscle fibres, distinguishing the initial regeneration-associated utrophin signal in cluster of small regenerating myofibres from the drug related utrophin signal is important for the determination of the real impact of the therapy. The sarcolemmal localisation and homogeneity of the utrophin signal across the whole muscle in correlation with the regenerative process are critical. Therefore, the development of accurate utrophin and regeneration quantification is required to study utrophin expression at the individual fibre level. Recently, some progress has been made in this direction with the development of automated, robust and reproducible staining and imaging protocols to quantify sarcolemmal utrophin levels, numbers of regenerating fibres and fibre size at the muscle fibre level in DMD and BMD patients (Janghra et al., 2016). Such tools are invaluable for assessing utrophin based drugs activity and were recently used with success in a clinical trial (NCT02858362).
The levels of regeneration and utrophin vary between muscle types, and are age dependent in both animals and DMD patients (Kleopa et al., 2006; Guiraud et al., 2018), emphasizing the complexity of quantifying utrophin in dynamic and dystrophic muscles. Therefore, beyond the development of robust, automated and quantifiable imaging solution at the muscle fibre level, several fundamental questions about utrophin still need to be addressed. Utrophin was previously described as increased in dystrophic tissue independently from regeneration (Weir et al., 2004), indicating that other mechanisms such as stabilisation of the utrophin protein at the muscle membrane occur in dystrophic muscle. Therefore, the determination of the half-life of utrophin protein as well as the turnover of this protein at the dystrophic muscle membrane will provide essential information. Another interesting fundamental question is to understand why the utrophin levels in DMD patients are higher (Ervasti, 2015) than in the mdx model whereas the regeneration processes are no longer an active process at the age of 5–12 years in patients (Turk et al., 2005). Utrophin levels were also described as significantly higher in DMD compared to BMD patients (Janghra et al., 2016). Consequently, it will be important to understand why these higher utrophin levels are not beneficial in DMD? Understanding the triggers behind these differences is essential and the answers may certainly guide us towards developing more robust utrophin drugs for DMD patients.
Concluding Remarks
The muscle regeneration, impaired in DMD, is a hallmark of the disease. Several indicators are commonly used to quantify regeneration and recently, revisited regenerative markers such as MyHC-emb and new quantitative methodologies emerged to offer this new possibility to use panel of translational regenerative biomarkers in order to obtain a more complete view on the regeneration processes. This is particularly critical to assess utrophin strategies for DMD as utrophin is a regeneration-associated protein. In correlation with inflammation and satellite cells markers linked to the regenerative processes, these indices of regeneration and recent advances in technologies provide an unprecedented opportunity to develop more robust utrophin DMD based strategies.
Footnotes
Conflicts of interest: None declared.
Financial support: None.
Copyright license agreement: The Copyright License Agreement has been signed by both authors before publication.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
C-Editors: Zhao M, Li JY; T-Editor: Jia Y
References
- 1.Barthelemy F, Wein N. Personalized gene and cell therapy for Duchenne Muscular Dystrophy. Neuromuscul Disord. 2018;28:803–824. doi: 10.1016/j.nmd.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Baghdadi MB, Tajbakhsh S. Regulation and phylogeny of skeletal muscle regeneration. Dev Biol. 2018;433:200–209. doi: 10.1016/j.ydbio.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 3.Ciciliot S, Schiaffino S. Regeneration of mammalian skeletal muscle. Basic mechanisms and clinical implications. Curr Pharm Des. 2010;16:906–914. doi: 10.2174/138161210790883453. [DOI] [PubMed] [Google Scholar]
- 4.Desguerre I, Christov C, Mayer M, Zeller R, Becane HM, Bastuji-Garin S, Leturcq F, Chiron C, Chelly J, Gherardi RK. Clinical heterogeneity of duchenne muscular dystrophy (DMD): definition of sub-phenotypes and predictive criteria by long-term follow-up. PLoS One. 2009;4:e4347. doi: 10.1371/journal.pone.0004347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dumont NA, Wang YX, von Maltzahn J, Pasut A, Bentzinger CF, Brun CE, Rudnicki MA. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat Med. 2015;21:1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ervasti JM. Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim Biophys Acta. 2007;1772:108–117. doi: 10.1016/j.bbadis.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Ervasti JM. Dystrophin and utrophin quantitation using full-length recombinant protein as standard. Measuring Dystrophin in Dystrophinopathy Patients and Interpreting the Data, Scientific Workshop. 2015 [Google Scholar]
- 8.Fisher R, Tinsley JM, Phelps SR, Squire SE, Townsend ER, Martin JE, Davies KE. Non-toxic ubiquitous over-expression of utrophin in the mdx mouse. Neuromuscul Disord. 2001;11:713–721. doi: 10.1016/s0960-8966(01)00220-6. [DOI] [PubMed] [Google Scholar]
- 9.Guiraud S, Aartsma-Rus A, Vieira NM, Davies KE, van Ommen GJ, Kunkel LM. The pathogenesis and therapy of muscular dystrophies. Annu Rev Genomics Hum Genet. 2015;16:281–308. doi: 10.1146/annurev-genom-090314-025003. [DOI] [PubMed] [Google Scholar]
- 10.Guiraud S, David R, Davies KE. The potential of utrophin modulators for the treatment of Duchenne muscular dystrophy. Expert Opin Orphan Drugs. 2018;6:179–192. [Google Scholar]
- 11.Guiraud S, Edwards B, Squire SE, Moir L, Berg A, Babbs A, Ramadan N, Wood MJ, Davies KE. Embryonic myosin is a regeneration marker to monitor utrophin based therapies for DMD. Hum Mol Genet. 2019;28:307–319. doi: 10.1093/hmg/ddy353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janghra N, Morgan JE, Sewry CA, Wilson FX, Davies KE, Muntoni F, Tinsley J. Correlation of utrophin levels with the dystrophin protein complex and muscle fibre regeneration in duchenne and becker muscular dystrophy muscle biopsies. PLoS One. 2016;11:e0150818. doi: 10.1371/journal.pone.0150818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleopa KA, Drousiotou A, Mavrikiou E, Ormiston A, Kyriakides T. Naturally occurring utrophin correlates with disease severity in Duchenne muscular dystrophy. Hum Mol Genet. 2006;15:1623–1628. doi: 10.1093/hmg/ddl083. [DOI] [PubMed] [Google Scholar]
- 14.Pratt SJP, Valencia AP, Le GK, Shah SB, Lovering RM. Pre- and postsynaptic changes in the neuromuscular junction in dystrophic mice. Front Physiol. 2015;6:252. doi: 10.3389/fphys.2015.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiaffino S, Gorza L, Dones I, Cornelio F, Sartore S. Fetal myosin immunoreactivity in human dystrophic muscle. Muscle Nerve. 1986;9:51–58. doi: 10.1002/mus.880090108. [DOI] [PubMed] [Google Scholar]
- 16.Turk R, Sterrenburg E, de Meijer EJ, van Ommen GJ, den Dunnen JT, ‘t Hoen PA. Muscle regeneration in dystrophin-deficient mdx mice studied by gene expression profiling. BMC Genomics. 2005;6:98. doi: 10.1186/1471-2164-6-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weir AP, Morgan JE, Davies KE. A-utrophin up-regulation in mdx skeletal muscle is independent of regeneration. Neuromuscul Disord. 2004;14:19–23. doi: 10.1016/j.nmd.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Yang W, Hu P. Skeletal muscle regeneration is modulated by inflammation. J Orthop Translat. 2018;13:25–32. doi: 10.1016/j.jot.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]