Abstract
High density lipoprotein (HDL) particles are blood-borne complexes whose plasma levels have been associated with protection from cardiovascular disease (CVD). Recent studies have demonstrated the existence of distinct HDL subspecies, however, these have been difficult to isolate and characterize biochemically. Here we present the first report that employs a network-based approach to systematically infer HDL subspecies. Healthy human plasma was separated into 58 fractions using our previously published three orthogonal chromatography techniques. Similar local migration patterns among HDL proteins were captured with a novel similarity score and individual co-migration networks were constructed for each fraction. By employing a graph mining algorithm, we identified 183 overlapped cliques, among which 38 were further selected as candidate HDL subparticles. Each of these 38 subparticles had at least two literature supports. In addition, GO function enrichment analysis showed that they were enriched with fundamental biological and CVD protective functions. Furthermore, gene knockout experiments in mouse model supported the validity of these subparticles related to three apolipoproteins. Finally, analysis of an apoA-I deficient human patient’s plasma provided additional support for apoA-I related complexes. Further biochemical characterization of these putative subspecies may facilitate the mechanistic research of CVD and guide targeted therapeutics aimed at its mitigation.
Keywords: high density lipoprotein, proteomics, protein network, co-migration pattern, subspecies, apolipoprotein, maximal clique, human plasma, particle fractionation
Graphical Abstract

Introduction
Plasma high density lipoprotein (HDL) is a highly heterogeneous family of particles ranging from 7 to 13 nm in diameter. It is composed of proteins and lipids in approximately equal mass. HDL has been epidemiologically associated with protection from atherosclerotic cardiovascular disease (CVD), a leading cause of mortality around the world (1, 2). Recent proteomics studies have identified upwards of 89 distinct HDL-associated proteins (3-11). Among them, roughly 70% of the protein mass consists of apolipoprotein (apo)A-I, and another 15-20% is apoA-II. These major HDL proteins form stable complexes with phospholipids, cholesterol, triglycerides and cholesteryl esters. The remaining proteins include other classical apolipoproteins (12) such as apoC-II, C-III, E, D, M and A-IV, as well as enzymes, transfer proteins, protease inhibitors, complement factors, and even vitamin-binding proteins (13).
Plasma HDL cholesterol (HDL-C) levels are a well-known negative risk factor for the development of CVD. A widely accepted basis for the inverse relationship between human plasma HDL-C and CVD is the ability of HDL, and its major protein constituent apoA-I, to mediate reverse cholesterol transport (RCT) (14). In this process, HDL promotes cholesterol efflux from peripheral cells such as macrophage-derived foam cells in the vessel wall to transport excess cholesterol and other lipids back to the liver for catabolism. Aside from lipid-transport activities, recent studies have identified an array of additional functions that likely contribute to HDL-mediated cardiovascular protection. For example, HDL has been documented to prevent oxidative modification of LDL via the HDL-associated protein paraoxonase1 (PON1) (15). Another well recognized HDL function is its role as an anti-inflammatory regulator that may slow atherosclerosis progression (16-18). Additionally, HDL can modulate vascular tone by affecting the production of nitric oxide (NO), a key mediator of vascular smooth muscle cell contraction (19). These anti-oxidative, anti-inflammatory and pro-vasodilatory properties of HDL might have the equal importance as its well-known cholesterol efflux function in the process of protecting against the CVD development.
Recent studies have demonstrated the existence of distinct HDL subspecies that are defined by unique protein complements. Asztalos et al. used antibodies to visualize individual protein migration patterns in a native 2D gel electrophoresis system and found apoA-I in 11 distinct spots representing variously charged and sized species (20). In our previous research, we separated human lipoproteins into five individual fractions by ultracentrifugation (7) and 17 fractions by size exclusion chromatography (8) and again saw highly distinct distribution patterns for individual proteins. Additionally, there is emerging evidence that distinct protein particle compositions can result in defined and unexpected functions. The most impressive example to date is the HDL particle dubbed Trypanosome Lytic Factor (TLF) (21, 22). This particle contains apoA-I, haptoglobin-related protein (HPR) and apoL-I, and has been shown to have specific lytic activity against the protozoan Trypanosoma brucei. This activity is absolutely dependent on the presence of these proteins. Given the widely diverse functions of the HDL-associated proteins and the fact that HDL’s CVD protection is mediated through similarly diverse functions, it is reasonable to believe that many of these subspecies may play unique biological roles.
Unfortunately, our knowledge of the structure and functions of HDL subspecies is limited. So far, no existing method can directly reveal the composition of distinct HDL subspecies. Density gradient ultracentrifugation (UC) is currently the preferred method of HDL isolation because density is a major resolving factor between non-lipid and lipid-bound proteins. The underlying principle of this method is to quantitatively float the relatively light lipid-bound proteins away from the heavy non-lipid associated proteins. However, this method often involves prolonged high speed centrifugation steps and the use of high salt concentrations, which can modify the protein structure and deplete apolipoproteins from the final isolates. In addition, the high salt must also be removed for further analysis of lipoprotein fractions, a process that results in poor recoveries (23). Furthermore, Van’t Hooft et al. has shown that more than a half of apoE was disassociated with HDL proteins during ultracentrifugation (24). All of these issues can interfere with our ability to identify specific protein-protein interactions (PPIs) on lipoprotein particles. Thus, there is an urgent need to analyze the HDL proteome using alternative separation techniques.
In our previous study, we applied three novel, non-density based, orthogonal chromatographic separation techniques on human plasma in order to better characterize the structural composition and functions of HDL subspecies (9). These techniques were used to fractionate normal human plasma into phospholipid-containing fractions, and the identities of the proteins were determined using mass spectrometry (MS). The spectral counts obtained from MS analysis of those fractions correlated well with protein abundance as determined by immunological analyses (7). As such, the distribution of each protein across all the fractions, i.e., a migration pattern, was obtained. Our previous work clearly established that certain HDL proteins could co-migrate across the separation methodologies, suggesting the existence of discrete subspecies. However, our initial co-migration analysis was limited in that it only inferred static interactions by considering global distribution similarity between HDL proteins, which may not effectively reflect the diversity of compositions for distinct HDL subspecies. Moreover, one global protein network was constructed based on those static PPIs using the conventional strategy, similar to several previous large-scale proteomics studies (25-28). This single-network strategy is certainly beneficial to either uncover important principles of global protein organization, or reveal novel protein interactions and complexes. However, it simply assumes spatial-temporal co-existence among nearby network nodes and edges (proteins and interactions) so that one identified protein complex may involve multiple smaller protein complexes in reality. Indeed, in our static protein interaction network, while certain sub-networks were clearly identifiable, the majority of the proteins clustered into one single sub-network, limiting the utility of the data. Customized network construction strategies and profound graph mining algorithms are crucial for further investigation of HDL subspecies.
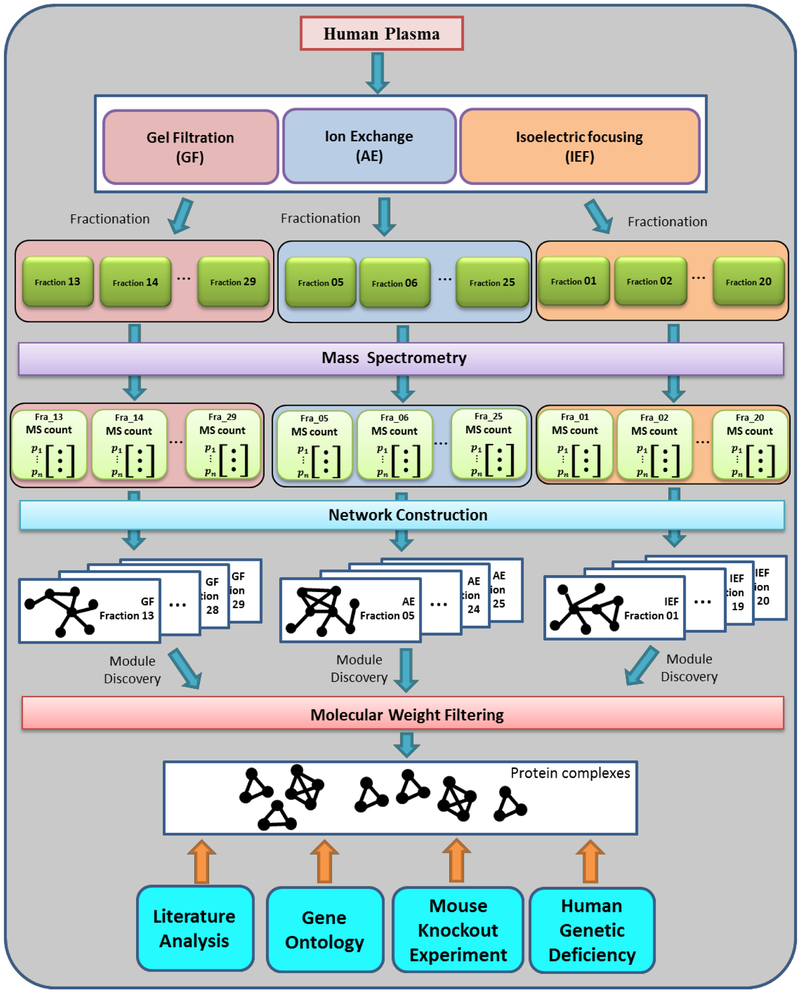
In this work, we developed a novel multi-network based computational approach to systematically identify structural HDL subspecies (Fig. 1). The organization of this article is as follows: First, we described the proteomics data upon which our analysis was based followed by the novel scoring system that quantitatively measured the similarity between two given proteins’ local distribution patterns. Next, we constructed individual local co-migration networks for all fractions and searched the HDL complex candidates in those networks. Finally, we tested those identified candidates using distinct experimental and computational approaches, including literature search analysis and Gene Ontology (GO) functional enrichment analysis, a mouse HDL study and a human genetic disease study.
Figure 1.
Schematic diagram of the systematic approach to identify HDL complexes based on three orthogonal chromatographic separation techniques. Healthy human plasmas were separated by three separation methods into an array of fractions. For each fraction, MS analysis was performed to discover the identities and spectral counts of proteins. Based on abundance profiles, individual co-migration networks were constructed with our Local S-score. A graph mining algorithm was applied to discover protein complexes. After filtering with molecular weight threshold, multiple validation approaches were utilized to test the putative HDL subspecies.
Results
Fractionation of HDL proteomics dataset.
Lipid associated proteins’ distribution dataset (see Material and Methods) was obtained in our previous study (9), where healthy human plasma was fractionated by three optimized non-density based orthogonal chromatographic separation techniques (gel filtration (GF), anion exchange (AE), and isoelectric focusing (IEF)). We first filtered an HDL-associated protein dataset from original lipid associated protein data. Many research groups have used different proteomic techniques to study HDL compositions, and each approach assessing HDL fractionation reflected specific physicochemical properties of the particles, such as density, size and electrophoretic mobility. Due to the differences in HDL separation techniques, sample preparation, and instruments, the total list of HDL proteins varies dramatically from study to study. We have compared the existing data from 16 proteomics studies published to date and compiled a list of 89 high-confidence HDL-associated proteins that were observed in at least three different studies or have independent biochemical evidence of HDL residence (see Material and Methods). Therefore, we mainly focused on these 89 high-confidence HDL-associated proteins here and removed other data.
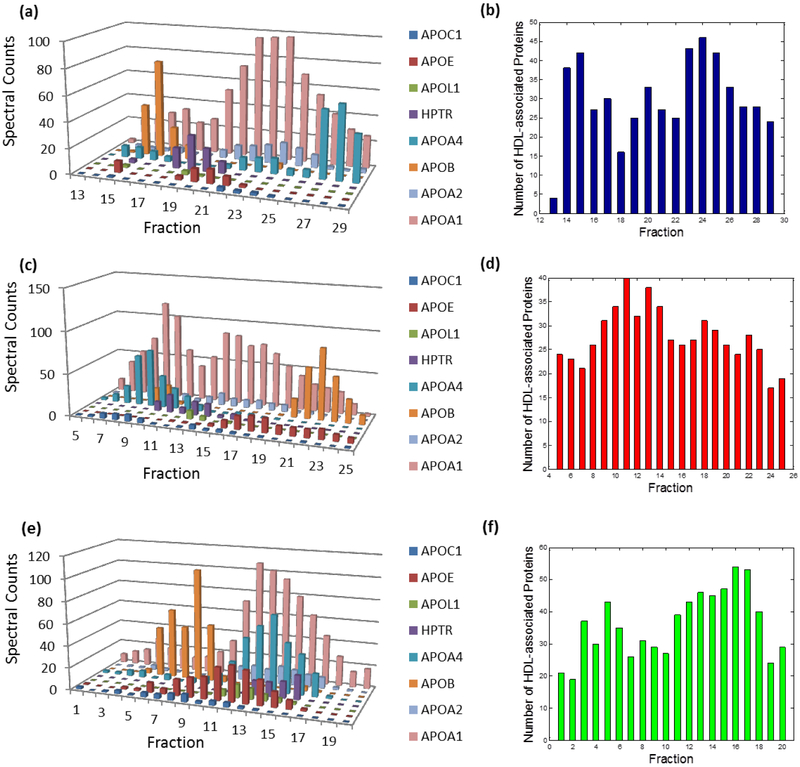
Abundance profiles of several known HDL-associated proteins from three separation techniques were presented (Fig. 2a, c and e). As expected, the most abundant protein, apoA-I, occurred in nearly every fraction while other apolipoproteins displayed relatively distinct distributions. Intriguingly, we noticed that only a portion of proteins co-occurred when we took a closer look at any individual fraction (Fig. 2b, d and f). Additionally, many proteins, except the abundant ones, only existed in certain fractions rather than across the whole spectrum. The diversity of these proteins’ distributions implicitly reflected the heterogeneity of HDL subspecies. HDL proteins co-existing within the same subparticles should travel together during the plasma separation; thus, proteins on a given subparticle are likely to be detected by MS in the same fraction. Hence, we first separated all proteins discovered in our experiments into different groups based on their co-existence in individual fractions. Consequently, the entire proteomics data pool was divided into 58 subsets based on GF (17 fractions), AE (21 fractions), and IEF (20 fractions) separation techniques. Although the total number of identified proteins in the three separation methods was 159, proteins co-existing within the individual fraction were commonly fewer than 40. It is possible that those co-existing proteins may come from more than one subspecies. But, the separation of detected proteins into subsets effectively distinguished “likely” and “unlikely” co-localized proteins. With the help of the following analyses, we were able to uncover the composition of certain subspecies.
Figure 2.
Abundance profile examples of multiple HDL-associated proteins and number of proteins detected by MS within each fraction from the separation method (a-b) GF, (b-d) AE, and (e-f) IEF.
Construction and graph analysis of local co-migration networks to identify HDL subspecies.
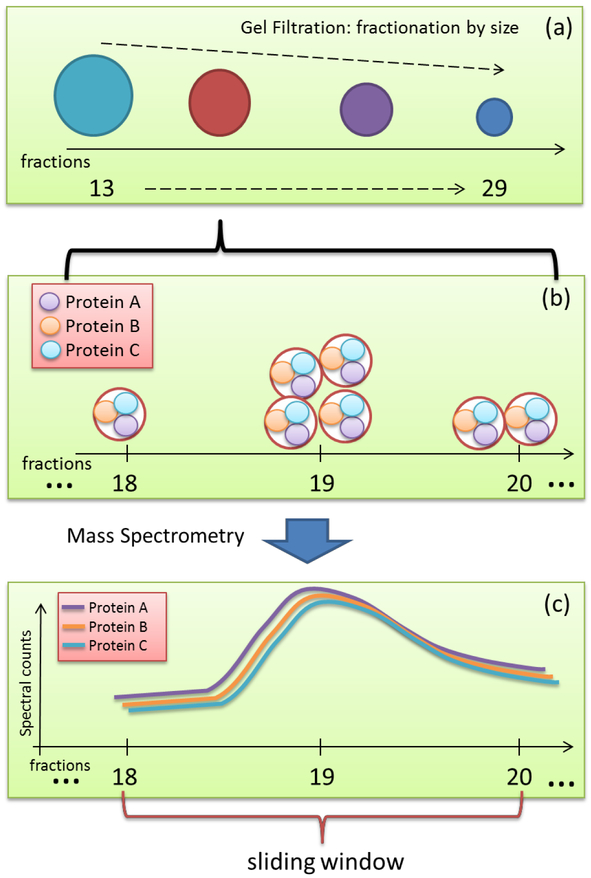
Individual co-migration networks were constructed for each fraction from the different separation techniques to represent the co-migration relationship between any pair of HDL proteins. Vertices of the network for a given fraction represented MS-detected HDL-associated proteins. Edges between vertices represented the local co-migration relationship, and each one is associated with a similarity score. Here, we developed a novel score, named local Spearman’s rank correlation coefficient Score (Local S-score) (see Material and Methods) to quantitatively measure such local similarities (Fig. 3). Conventionally, variables with a correlation coefficient larger than 0.8 are regarded as strongly correlated. As such, we chose the similarity threshold of 0.8 to transfer our weighted graphs to binary graphs. Using this multi-network strategy, we constructed a total of 58 protein local co-migration networks (17, 21, and 20 networks for GF, AE, and IEF techniques, respectively). Those 58 co-migration networks have very diverse topologies, indicating the heterogeneity of subspecies’ composition. To illustrate the overall structure of the HDL interactome, we merged the 58 co-migration networks to form a comprehensive HDL interactome map that contains 70 proteins, 1540 edges (Fig. S1).
Figure 3.
The rationale of Local S-score for capturing the local similarity of proteins’ migration patterns. (a) The mechanism of our separation methods is utilizing different characteristics of each subspecies to create their specific distributions, i.e., migration patterns. For example, in the GF approach, larger size subparticles are likely to migrate fast and fall into early fractions, while smaller size ones would be detected in the following fractions. (b) Successive neighboring fractions may contain the same type of particles. The same subparticles may distribute among multiple neighboring fractions in a specific pattern, e.g., one subparticle containing proteins A, B, and C may distribute as shown in the figure. (c) Those proteins within the same subspecies are likely to have highly similar migration patterns locally. This local similarity may be only reflected within a certain range of fractions, i.e., sliding window.
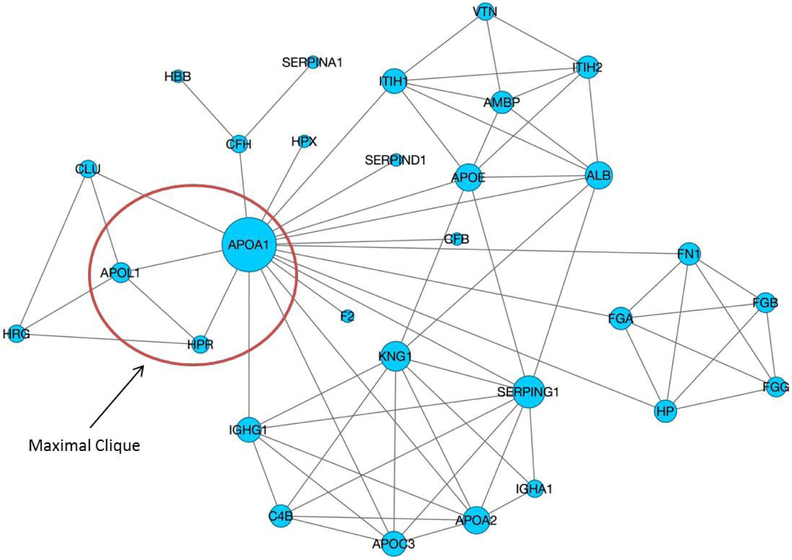
Within each co-migration network, if all proteins included in one subspecies have similar migration patterns, they tend to form a highly-connected cohesive network module. In graph theory, a clique in a network is a fully connected subset of the vertices where every two nodes in this subset are connected by an edge. A maximal clique is a clique that cannot be extended by including any more adjacent nodes and it has been applied in protein networks to discover core network elements (29). The maximal clique concept fits well with our purpose of discovering such highly-connected cohesive modules. Fig. 4 illustrates a local co-migration network constructed for 19th fraction of GF method and one of maximal cliques within this network. In this study, we identified all maximal cliques using the Bron-Kerbosch algorithm (30) from individual co-migration networks. Please note that we required a clique to contain at least three nodes as a protein complex, and dismissed all two-node cliques that represent pair-wise PPIs. Those discovered maximal cliques represented the likely protein complex candidates and were subject to further validation as follows.
Figure 4.
A local co-migration network constructed for 19th fraction of GF method. Size of the vertex reflects network degree of this vertex. Circled subnet is a maximal clique within this network, corresponding to the well-known TLF particle.
The molecular weight was applied as a filter to remove any unlikely candidate. The molecular weight of an HDL complex is usually observed to be less than 400,000 Da, including roughly 50% (by mass) HDL-associated proteins and 50% (by mass) lipids (31, 32). Accordingly, we set a conservative upper limit of molecular weight for HDL subspecies as 200,000 Da. After filtering all cliques with this molecular weight threshold, 183 candidates (Table S1) were selected for further investigation. Due to the inability to directly observe a given subspecies, we had to look for multiple lines of indirect evidences to test identified subspecies candidates. We tested those subspecies with six lines of evidences, including: literature analysis, GO functional analysis, two PPIs models based on our mouse HDL study and two PPIs models based on our human genetic deficiency disease study. Finally, we presented 38 putative HDL subspecies which are supported by at least three lines of evidences (Table 1). In the following sections, we presented independent lines of evidence that support the validity of the identified putative HDL subspecies.
Table 1.
Putative HDL subspecies which are supported by at least three lines of evidence.
| Size | Subspecies | Literature | GO Enrichment |
Mouse Matrix |
Mouse Spoke |
Human Matrix |
Human Spoke |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | APOA1 | CLU | HPR | 2 | 3.92E-03 | 1 | 1 | 3 | 2 | ||
| 3 | APOA1 | APOL1 | CLU | 2 | 3.92E-03 | 1 | 1 | 3 | 2 | ||
| 3 | APOA1 | APOL1 | HPR | 2 | 5.87E-02 | 0 | 0 | 3 | 2 | ||
| 3 | APOA1 | APOE | SERPING1 | 3 | 6.17E-03 | 3 | 2 | 0 | 0 | ||
| 3 | APOA1 | APOE | ITIH1 | 3 | 6.17E-03 | 3 | 2 | 0 | 0 | ||
| 3 | APOA1 | APOC3 | PON1 | 2 | 6.17E-03 | 3 | 2 | 3 | 2 | ||
| 3 | APOA1 | CLU | SERPIND1 | 2 | 3.92E-03 | 3 | 2 | 1 | 1 | ||
| 3 | APOA1 | APOC2 | GPLD1 | 2 | 1.38E-02 | 3 | 1 | 3 | 2 | ||
| 3 | APOA1 | APOA2 | APOC1 | 3 | 2.49E-03 | 3 | 3 | 3 | 2 | ||
| 3 | APOA1 | APOC3 | APOE | 3 | 4.05E-04 | 3 | 2 | 1 | 1 | ||
| 3 | APOA1 | APOA2 | ITIH1 | 3 | 3.92E-03 | 3 | 3 | 1 | 1 | ||
| 3 | APOA1 | APOC3 | CLU | 3 | 3.92E-03 | 3 | 2 | 3 | 2 | ||
| 3 | APOA1 | APOA2 | APOC3 | 3 | 4.05E-04 | 3 | 3 | 3 | 2 | ||
| 3 | APOA1 | APOC1 | CLU | 3 | 3.92E-03 | 3 | 2 | 3 | 2 | ||
| 3 | APOA1 | APOC2 | APOC3 | 3 | 1.62E-03 | 3 | 1 | 3 | 2 | ||
| 3 | APOA1 | APOC1 | APOE | 3 | 2.49E-03 | 3 | 2 | 1 | 1 | ||
| 3 | APOA1 | APOC2 | PON1 | 2 | 1.38E-02 | 3 | 1 | 3 | 2 | ||
| 3 | APOA1 | APOA2 | APOA4 | 3 | 4.05E-04 | 2 | 2 | 1 | 1 | ||
| 4 | ALB | APOA1 | APOE | SERPING1 | 6 | 8.95E-03 | 3 | 2 | 0 | 0 | |
| 4 | APOA1 | APOC1 | APOC3 | HP | 6 | 5.92E-03 | 4 | 2 | 3 | 2 | |
| 4 | APOA1 | AHSG | HPX | SERPIND1 | 4 | 8.84E-02 | 3 | 2 | 1 | 1 | |
| 4 | ALB | APOA1 | APOC1 | CFB | 6 | 1.56E-02 | 3 | 2 | 1 | 1 | |
| 4 | ALB | APOA1 | APOA2 | APOC1 | 6 | 3.92E-03 | 3 | 3 | 3 | 2 | |
| 4 | ALB | APOA1 | APOC1 | HPR | 4 | 1.56E-02 | 1 | 1 | 3 | 2 | |
| 4 | APOA1 | APOA2 | CLU | F2 | 4 | 6.00E-03 | 3 | 3 | 3 | 2 | |
| 4 | APOA1 | APOC1 | APOE | GSN | 6 | 3.92E-03 | 3 | 2 | 1 | 1 | |
| 4 | APOA1 | APOA2 | CFB | TTR | 6 | 6.00E-03 | 2 | 2 | 1 | 1 | |
| 5 | APOA1 | APOA2 | APOC3 | SERPING1 | IGHG1 | 10 | 1.62E-03 | 5 | 4 | 3 | 2 |
| 5 | APOA1 | APOA2 | APOA4 | IGHG1 | PON1 | 7 | 1.62E-03 | 4 | 4 | 3 | 2 |
| 5 | APOA1 | APOA2 | APOE | IGHG1 | F2 | 10 | 3.92E-03 | 3 | 3 | 1 | 1 |
| 5 | APOA1 | APOA2 | APOC3 | F2 | TTR | 10 | 1.62E-03 | 3 | 3 | 3 | 2 |
| 5 | APOA1 | APOA2 | CLU | F2 | TTR | 8 | 6.35E-03 | 3 | 3 | 3 | 2 |
| 5 | APOA2 | APOC3 | APOM | F2 | TTR | 6 | 6.35E-03 | 3 | 2 | 3 | 0 |
| 5 | APOA2 | APOM | CLU | F2 | TTR | 4 | 1.38E-02 | 3 | 2 | 3 | 0 |
| 5 | APOA1 | AHSG | HP | SERPING1 | TTR | 10 | 8.01E-02 | 3 | 2 | 1 | 1 |
| 5 | APOA1 | APOA2 | APOA4 | HPX | SERPIND1 | 7 | 1.62E-03 | 3 | 3 | 1 | 1 |
| 5 | APOA1 | APOA2 | CFB | HP | TTR | 10 | 6.35E-03 | 3 | 2 | 1 | 1 |
| 5 | APOA1 | APOC3 | CFB | CLU | TTR | 9 | 6.35E-03 | 6 | 3 | 3 | 2 |
Comparison of our local-network approach with four traditional methods on network construction and clique identification.
We first assessed the accuracy of the predicted HDL interactome network based on known HDL PPIs deposited in the Human Protein Reference Database (HPRD), where all PPIs were collected from published literature based on experiments (e.g., yeast-2-hybrid, co-IP, etc.) and manually curated to avoid errors (33). The 352 PPIs involving 64 HDL proteins represent a comprehensive collection of known HDL PPIs, and we believed it is an ideal dataset to test the coverage of our predictions in terms of pairwise interactions. All the reported PPIs related to HDL-associated proteins were used as positive instances, and all unreported ones were treated as negative instances.
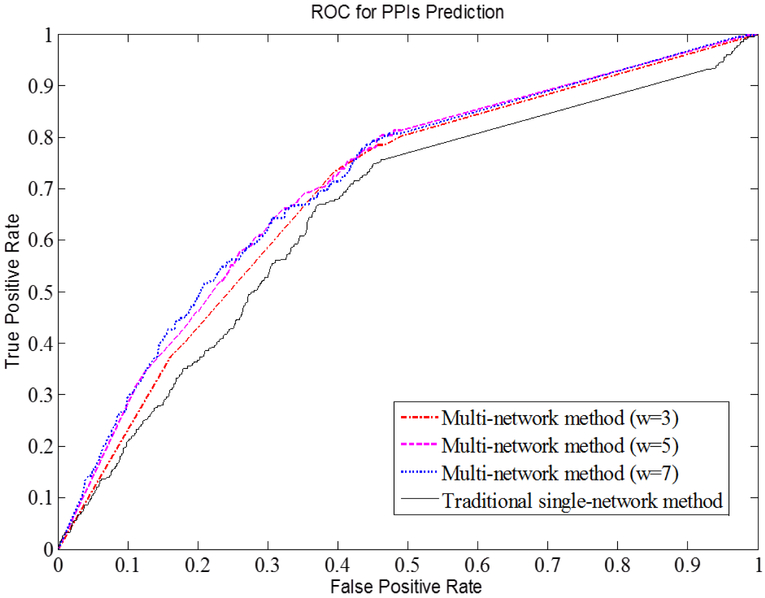
Based on this gold-standard dataset, we compared the performance of our local-network method a simpler, more widely used method (i.e., constructing a single-network with a global similarity score derived based on distribution across all fractions) in terms of PPIs prediction. We tested the PPIs prediction on the traditional model and our models with various sliding windows w. Fig. 5 shows receiver operating characteristic (ROC) curves, common graphical plots illustrating the performance of a binary classifier. For each predictive model, we calculated the area under the curve (AUC), which is equal to the probability that a given scoring system will return a higher score for a random positive PPI than the one for a random negative PPI. The higher AUC is, the better a predictive method is. Our local-network models with (w=5 or w=7) had the highest AUC of 0.71, while the AUC of the model (w=3) was 0.69. In contrast, the AUC of the traditional score was 0.65. The results indicate our local-network method has a better performance in predicting PPIs. Since the smaller sliding window requires less computational time, (w=5) sliding window becomes the preferred parameter in the following analyses.
Figure 5.
ROC curves for the traditional score and our Local S-score with various sliding windows w.
Alternatively, we conducted an analysis to construct three PPI networks from three separation methods (see Supplemental Materials, Fig. S2a-c), and only considered the overlapping edges among all networks. The consensus network only consists of five well-known PPIs with negligible coverage (5/352) of known HDL PPIs (Fig. S2d). This comparison suggests that our local-network method was tolerant of a potential artifactual lack of co-migration due to experimental perturbation in one of the separations, thus the failure of a given complex to survive one of the separations does not necessarily preclude the identification of the subspecies. Each of the separation methods has drawbacks that can potentially perturb particle migration or integrity, e.g., dilution effects in GF and salt effects in AE. The conventional method that only considers the simultaneously co-associated protein pairs among all three separation methods may yield a very limited number of PPIs.
In the third comparison, we found many PPIs that can only be discovered by our method but not a traditional method. In our previous study (9), we have applied a traditional global correlation analysis across all fractions to identify a few highly correlated protein pairs, e.g., fibrinogen alpha chain (FGA): fibrinogen beta chain (FGB), FGB: fibrinogen gamma chain (FGG) and apoA-I: apoA-II (9). The top ranked pairs discovered in our previous study were confirmed once again in this work. In addition to those pairs, our new score is able to reveal more co-migrated relations that could not be reflected in conventional correlation analysis. For example, apoA-I: apoL-I, an interaction observed in other independent studies (21, 22) and in our human apoA-I deficiency study, was discovered by our local co-migration analyses in several fractions. However, the migration patterns of apoA-I and apoL-I were not strongly correlated across all fractions, and therefore the interaction was not identified in our previous study.
Finally, we explored whether distinct HDL complexes could be uncovered from the densely-connected merged HDL interactome (Fig. S1) rather than from 58 individual local networks. We noted that the complexes identified by the graph mining algorithms in such a densely connected network often have molecular weights that are too large for HDL particles. This may be due to that HDL proteins, especially the abundant ones, are likely to participate in different subparticles and interact with distinct proteins (8, 9, 34) so that the traditional single-network strategy is likely to uncover a large protein complex that involves multiple subparticles in reality. Apparently, such diverse characteristics of HDL complexes cannot be properly represented with static links in one conglomerate protein interactome map (35).
Each of the putative HDL subspecies has at least two literature supports.
We first examined the PPIs within identified HDL complexes using the reported PPIs in HPRD database. The complexes with more reported PPIs are likely to exist in reality. Each of 38 HDL subspecies has multiple literature supports. With the HPRD database and literature review, we presented the number of PPIs supported by literature for each individual HDL subspecies in Table 1.
The most recognized HDL subparticle, the TLF particle (apoA-I, apoL-I and HPR) was discovered from several fractions in our analysis, demonstrating the effectiveness of our systematic strategy. The network in Fig.4 is one of the co-migration networks containing this TLF particle. Our Local S-score between apoL-I and HPR was up to 0.97 (p-value=0.017). In contrast, we performed co-expression based PPI prediction on STRING (36) by querying either apoL-I or HPR, and PPI between apoL-I and HPR was not presented. Also, the traditional global score between apoL-I and HPR reported in our previous work is only up to 0.72 among three separation methods, below the required correlation threshold of 0.8.
We also looked for other independent studies to support the existence of our subparticle candidates. One example is the (apoA-I, apoC-I and apoE) complex (37, 38). It has been reported that apoC-I binds free fatty acids and reduces their intracellular esterification, and its function to modulate the interaction of apoE and beta-migrating VLDL(39). At the same time, apoE and apoA-I binding to ATP-binding cassette, sub-family A member 1 (ABCA1) is essential for the HDL formation (40). ApoA-I mainly works in the cholesterol efflux process, and many studies have supported the idea that apoC-I and apoE may work together with apoA-I to help with lipid metabolism. We may infer from this prior knowledge that the complex (apoA-I, apoC-I and apoE) may exist in certain molecular processes.
Another example is complex (apoA-I, apoA-II, Complement factor B and Transthyretin), whose all pairwise PPIs were supported by literature. Transthyretin (TTR), the plasma carrier for both thyroxine and retinol, has connections to apoA-I (41, 42). TTR was shown to affect HDL biology and the development of atherosclerosis by reducing cholesterol efflux and increasing the apoA-I amyloidogenic potential (43). It was shown that TTR can cleave apoA-I, decreasing its ability to promote cholesterol efflux from cholesterol-loaded macrophages (44). Moreover, the apoA-I cleaved by TTR has reduced affinity for ABCA1, as assessed by cross-linking (45). These support the notion that this particle may exist and play a critical role in cholesterol efflux.
Diverse biological functions of HDL complexes.
Being enriched for biological functions supports the existence of the protein complex candidates. For each of the identified subspecies, to examine whether it is enriched for certain GO terms, the hypergeometric test was used to test the significance against the null hypothesis, i.e., all proteins clustered as one subparticle were picked randomly from the HDL watch list (see Material and Methods). In Table 1, we only listed the p-value corresponding to the most significant GO terms. Of the 38 subspecies candidates, 31 have significantly enriched functions after Benjamini correction (p-value <0.01). Take the case of subparticle (apoA-I, apoA-II, and apoA-IV), for example, with the most significant annotation of “regulation of intestinal cholesterol absorption” (p-value: 5.37E-04). Its individual members commonly participate in the cholesterol metabolism process (46-48), suggesting that this subparticle may exist in nature and perform a specific metabolic function.
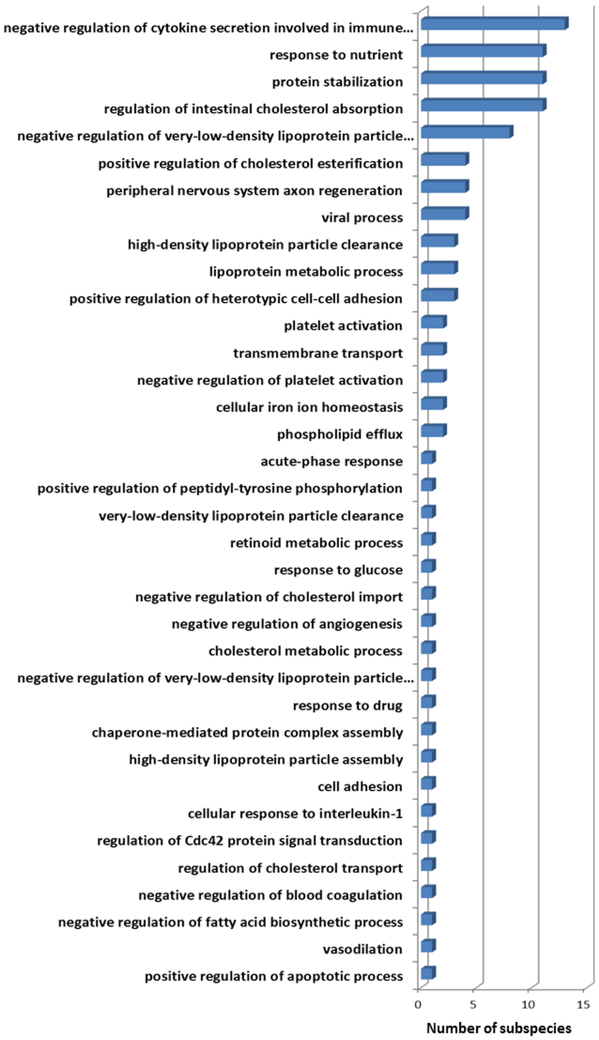
To examine the variety of GO terms enriched among the 183 subparticle candidates, we summarized the frequency of the most significant GO terms associated with individual putative subspecies in Fig 6. Several enriched GO terms of identified subparticles turn out to be the annotation of fundamental biological processes. Those functions are essential to keep the normal cellular activity, e.g., “positive regulation of heterotypic cell-cell adhesion”. On the other hand, several other enriched functions are consistent with previously known HDL functions, such as “reverse cholesterol transport”, “anti-oxidation”, and “immune response” and “hemostasis”. Although the CVD protective functions have been the major focus of various HDL studies, our results suggested that distinct HDL subspecies may be responsible for different functionalities, from fundamental biological functions to CVD-protective functions. We are aware that the current GO knowledge may be incomplete; however, since it was not our intention to discover new HDL biological functions based on the current GO knowledge, the GO enrichment analysis primarily served the purpose to validate reasonable protein clustering in that irrelevant proteins in term of GO functions should not dominate the candidate subparticles.
Figure 6.
Histogram of the most significantly enriched functions of 38 identified HDL subspecies. Distribution of the functions covers both fundamental biological functions and CVD-protective functions.
Validation of HDL subspecies using a mouse HDL study.
To study the composition of HDL subspecies, we have utilized a dataset from our mouse HDL study (49) and analyzed protein distribution patterns of mice when three major HDL-associated genes (APOA1, APOA2 and APOA4) were knocked out individually. The mouse model is an invaluable system for the study of genetic effects on lipoprotein metabolism. Their plasma contains lipoproteins that are roughly similar to those in the human in terms of protein and lipid composition (50). Plasma separation and MS analysis assays were performed for three wild type (WT) mice and three gene knockout (KO) mice in three genetic knockout experiments, respectively. Please refer to (49) for more experimental details in our mouse HDL study. Here, we hypothesized that two proteins are likely to co-exist on the same protein complex if one of them has a shifted distribution after another is absent (Fig. S3a). On the other hand, two proteins are less likely to co-exist on the same particle if one protein has no distribution change after another is ablated (Fig. S3b). To quantitatively evaluate the shifting of the proteins’ distribution patterns, we developed a lag-score (L-score) migration pattern shift analysis (see Material and Methods). Proteins with ∣L∣ ≥ 1 were identified as influenced proteins. Then, the identified mouse proteins were mapped back to human HDL-associated proteins by sequence homology.
With our L-score, we inferred 26, 16 and 4 human HDL-associated proteins from the mouse HDL study with the ablation of APOA1, APOA2 and APOA4, respectively. All influenced proteins and their corresponding L-scores were listed in Table 2. The absolute value of L-score indicates the significance level of pattern changes. Small absolute value indicates that only subtle changes were detected, while large absolute value indicates obvious change of the migration patterns. Proteins were ranked with the absolute value of L-score. The ablation of APOA1 had a significant impact on the distribution of 26 other HDL-associated proteins. The ablation of APOA2 had a smaller effect on 16 mapped HDL-associated proteins. Only apoA-II, apoC-III, HPR and Haptoglobin (HP) were identified during the protein mapping according to the APOA4 knockout experiment. Both spoke and matrix PPIs models (27) were adopted to determine the validity of the identified subspecies (see Material and Methods). Among 38 putative complexes, 37 subparticles are supported by at least one PPI with the spoke PPIs model, while 37 subspecies have at least one PPI supported by the matrix PPIs model (Table 1).
Table 2.
Human HDL-associated proteins identified in the migration pattern shifting analysis. Each column lists all the influenced proteins, ranked by absolute value of their L-scores, due to the absence of one apolipoprotein in the mouse model.
| apoA-I | apoA-II | apoA-IV | |||
|---|---|---|---|---|---|
| Influenced Proteins |
L-score | Influenced Proteins |
L-score | Influenced Proteins |
L-score |
| SAA4 | 10 | APOC1 | 12 | APOA2 | 1 |
| AZGP1 | 9 | APOC2 | 11 | HP HPR | 1 |
| PON1 | 6 | APOC3 | 2 | APOC3 | 1 |
| GPLD1 | 2 | GPLD1 | 2 | -1 | |
| APOC3 | −2 | APOA1 | 1 | ||
| AHSG | −1 | APOE | 1 | ||
| AMBP | −1 | APOM | 1 | ||
| APOC1 | −1 | AZGP1 | 1 | ||
| APOE | −1 | ITIH2 | 1 | ||
| CLU | −1 | ITIH1 | 1 | ||
| C1S | −1 | ITIH4 | 1 | ||
| C9 | −1 | AMBP | 1 | ||
| CFB | −1 | PON1 | 1 | ||
| C2 | −1 | CLU | −1 | ||
| ITIH2 | −1 | C9 | −1 | ||
| ITIH1 | −1 | CFH | −1 | ||
| LUM | −1 | ||||
| PGLYRP2 | −1 | ||||
| SERPINA1 | −1 | ||||
| SERPINA3 | −1 | ||||
| SERPING1 | −1 | ||||
| SERPINA4 | −1 | ||||
| SERPINF1 | −1 | ||||
| SERPINC1 | −1 | ||||
| SERPIND1 | −1 | ||||
| SERPINF2 | −1 | ||||
Validation of the identified subspecies with human apoA-I deficiency disease study.
To provide further information in the human system, we obtained a plasma sample from a patient with familial apoA-I deficiency. The genetic deficiency of apoA-I is extremely rare and has been reported in only 16 families throughout the world (51). Due to the rarity of this type of mutation in humans, larger-scale studies on this population would be extremely difficult and we were able to recruit only one participant for this study. It is also interesting to note that although these patients have markedly reduced plasma HDL-C levels, usually around 20-25 mg/dl, there is no clear association with an increased occurrence of cardiovascular diseases (34). Similar to the rationale of the mouse model study, the deficient proteins are likely to disrupt the formation or at least change the molecular size of the corresponding subspecies. Details of assays are described in Material and Methods. Forty-one known HDL-associated proteins were identified among a total of 103 MS detected proteins.
Migration pattern shift analysis was performed similar to that conducted in the mouse study. Of the 41 MS-detected HDL-associated proteins, 16 were determined to have a shift in distribution patterns with L-score (∣L∣ ≥ 1) (Table 3). Proteins were ranked with the absolute value of L-score. Some migration pattern comparison examples are shown in Fig. S4. ApoA-I migration patterns were first provided to show the lack of apoA-I in the patient compared to the healthy control. Another major protein, apoA-II, was detected with minor shift in our analysis (L-score=−1). Next, we found that both HPR and apoL-I cannot be detected in the lipoprotein fractions of the patient plasma, which is consistent with the TLF particle reported by Rifkin and Raper et al. (21, 22). Additionally, apoC-I and apoC-II’s migration patterns were shifted similarly (L-score=8), suggesting that they may co-exist with apoA-I on certain complexes. Indeed, subparticle (apoA-I, apoC-I and apoC-II) was discovered in our analysis (Table 1).
Table 3.
Human apoA-I deficiency influenced HDL-associated proteins. Proteins were ranked by the absolute value of their L-scores.
| Influenced Proteins | L-score |
|---|---|
| APOL1 | 10 |
| GPLD1 | 10 |
| APOC1 | 8 |
| APOC2 | 8 |
| SAA4 | 8 |
| HPR | 6 |
| APOM | 4 |
| APOC3 | 3 |
| PON1 | −3 |
| APOA2 | −1 |
| APOH | −1 |
| CLU | −1 |
| AHSG | −1 |
| ITIH4 | 1 |
| KLKB1 | −1 |
| RBP4 | −1 |
Again, both matrix and spoke PPIs models were adopted for this apoA-I deficiency study to test the identified subspecies. Thirty-three of the 37 subparticles containing apoA-I had at least one PPI directly supported by the spoke model. With the matrix model, 35 subspecies had at least one PPI being supported.
Discussion
In this study, we systematically identified and characterized structural HDL subspecies through analysis of proteins’ co-migration patterns generated by three orthogonal chromatographic separation techniques. Current studies of HDL proteomics are few and are limited in that, by only looking at pools of total HDL or very broad density subsets (i.e., HDL2 or HDL3), they do not take into account the heterogeneity of the HDL population. In contrast, our proteomic experiments fractionated the total HDL pool more extensively to enable discrimination between distinct subspecies. Greater fractionation of the total HDL population allows us to examine protein variances between particles more closely and identify specific HDL subspecies. To take full advantage of our separation techniques, a multi-network based approach was employed to study subspecies composition. This work is novel in the following respects:
First, most existing studies on HDL complexes, including our previous work, are limited by focusing on pairwise HDL PPIs. In contrast, our research aimed at systematically inferring HDL subspecies by identifying protein complexes. A major difficulty in the current attempts to study HDL subspecies is the lack of a direct experimental approach to reveal and validate the protein composition in individual subspecies. As such, we proposed an integrative approach by combining experimental fractionation, computational inference and multi-dimensional validation. In this paper, we presented 38 candidate protein complexes, which have not been published before.
Second, in order to achieve our goal of identifying protein complexes in HDL subspecies, we reanalyzed our previously published proteomics data, by deploying novel computational approaches. For example, we developed a novel Local S-score that maximally captures the migration similarities between proteins’ abundance profiles, and a multi-network strategy to construct 58 local co-migration networks for individual fractions, rather than one conglomerate protein network, for increasing the resolution for subspecies discovery. These novel approaches enable us to infer distinguishable HDL subspecies in the analyses.
Finally, multi-dimensional substantial experiments as well as computational analysis were performed to test the validity of the identified subspecies in this study. Gene knockout datasets from APOA1, APOA2, and APOA4 deficient mice were utilized. Analyses results have not been published, although the experimental details were discussed in our previous work. Furthermore, a novel human genetic deficiency study was applied for our complex validation. In this study, one apoA-I deficient patient was studied to look at the associated complexes. Additionally, literature searching and GO functional enrichment were used to test our putative complexes. The putative HDL complexes with multiple pieces of evidence from the validations are the highly confident candidates for future biochemical studies aimed at determining HDL protein physical interactions or co-localization, e.g., immunoprecipitation.
Due to the limitation of discover-type MS proteomics experiment, some low-abundance HDL-associated proteins were not found in our MS scanning, such as Phospholipid transfer protein (PLTP). A targeted MS/MS scanning may improve data acquisition quality. In spite of the missing peptides in the MS scanning, it is fortunate that our analysis pipeline is independent on the completeness of MS data. Our identified putative HDL complexes in Table 1 would not be compromised, since they may only represent subsets (subparticles) of HDL particles in nature.
Our future work will focus on associating the putative subspecies to their biological functions. Particularly, we are more interested in those low-abundance proteins that are newly identified. One idea is that apoA-I and apoA-II act as organizing scaffolds for proteins that principally mediate the classical lipid transport roles of HDL (52, 53). However, lower-abundance proteins, many of which have putative functions that differ quite significantly from lipid transport, may represent the most promising candidates for participation in distinct HDL subspecies. Indeed, this has been demonstrated in the case of the TLF particle which contains HPR and apoL-I (21, 22). This particle clearly plays a major role in innate immune function with an abundance level too low to play a meaningful role in lipid transport. Further studies will be needed to determine if some of the putative subspecies identified here have similar distinctive functions.
Overall, our work is the first report that employs MS-determined local co-migration patterns and graph pattern mining to systematically infer HDL subspecies with individual high-resolution networks. It uncovered the composition of some HDL subspecies with a novel multi-network computational approach. Our work also has significant implications in current cardiovascular disease research. A better understanding of the HDL subspecies that are either protective against or permissive to CVD will lead directly to new diagnostic tests for these species. For example, if we find that a particular subspecies is correlated with the development of CVD, we can envision screening CVD patients for this deleterious HDL subspecies in a clinical assay. Furthermore, existing therapeutics tend to boost HDL-C levels indiscriminately without regard for functions; and, since HDL comprises numerous particle populations, simply raising HDL-C may increase the wrong sorts of particles at the expense of the cardio-protective ones. Therefore, identification of CVD-protective HDL subspecies may help focus on new HDL-raising therapies that offer more specificity than those currently under exploration.
Material and Methods
Lipid-associated proteins distribution patterns dataset from three orthogonal separation chromatography techniques.
We developed three non-density based orthogonal separation chromatography techniques to fractionate normal human plasma to phospholipid-containing fractions (9): gel filtration (GF) chromatography that separates particles by molecular size; anion exchange (AE) chromatography that separates particles by charge; and isoelectric focusing (IEF) chromatography that separates particles based on the isoelectric point or the pH at which a particle has a net charge of zero. Considering the completeness of our current systematic strategy, we briefly described our published data here. Please refer to (9) for more experimental details. We recruited three healthy male blood donors and separated their plasma using all three separation techniques. Lipid associated proteins were isolated and their distributions across fractions were determined using HPLC-ESI-MS/MS. GF separated each plasma sample into 17 successive size-based fractions across which 106 lipid-associated proteins were identified. The AE method separated plasma samples into 21 fractions and identified 140 lipid-associated proteins. IEF method identified 93 proteins in 20 fractions.
Human apoA-I deficiency disease study.
An apoA-I deficient female participant (age 40) was paired with a control female participant (age 43). Both subjects have no known CVD, BMIs in the normal range and normal blood pressure. Venous blood was collected from participants after a 12-hour fast by a trained phlebotomist using BD Vacutainer® Plus Plastic Citrate Tubes containing buffered sodium citrate (0.105 M) as an anticoagulant. Cellular components were pelleted by centrifugation at ~1,590 × g for 15 minutes in a Horizon mini-E (Quest Diagnostics) at room temperature. Plasma was stored at 4°C until applied to the GF separation technique, always within 16 hours. Details of GF assays are same as in our previous work (8). Samples were never frozen. Written consent was obtained from participants in compliance with institutional regulations. Compositional analysis of lipoprotein fractions was assayed for these two subjects. HPLC-ESI-MS/MS was used to determine the distribution patterns for proteins across fractions.
HDL watch list.
A comprehensive HDL-associated proteins watch list is being continuously maintained and updated by the Davidson lab at: http://homepages.uc.edu/~davidswm/HDLproteome.html. The most up-to-date list has tracked 89 “likely” HDL proteins of 224 proteins from 16 independent studies, where likely HDL proteins are defined as those that appeared in at least three studies from three independent laboratories.
Local Spearman’s rank correlation coefficient Score.
Local Spearman’s rank correlation coefficient Score (S-score) was based on the Spearman’s rank correlation coefficient of the local migration patterns, which is more tolerant to errors in low spectral counts produced by low-abundance proteins. To reflect the local similarity, a sliding window w was utilized to restrict the abundance profiles within a certain range of fractions. Due to individual differences, we normalize the protein abundance profile by dividing the maximum of spectral counts of all fractions for any given protein in one plasma sample. Suppose we have a protein abundance profile X = (X1, … , X, … , X) where X is the normalized spectral count of protein Px in the fraction i. For each fraction i, we only considered the normalized protein spectral counts within the sliding window of w fractions , averaged from all v plasma samples into one abundance vector. The total number of fractions is w and the index j denotes the jth fraction in the abundance vector. Then, for each protein pair (Px, Py) in the fraction i, the Local S-score for a given separation method was calculated by:
where xj and yj are abundance ranks, obtained from the normalized spectral counts Xj and Yj of the protein pair (Px, Py) in the jth fraction for all plasma samples from one separation approach. Local S-score is a real number in the range of [−1, 1], Local S-score is highly effective at measuring a monotonic similarity between variables by considering the rank of normalized abundance levels and it also alleviates the bias against low-abundance proteins.
Functional enrichment analysis.
We collected function annotations of human HDL-associated proteins from the GO website. For each of the identified subparticle candidates, to examine whether it is enriched within a certain function, we used the hypergeometric test to test the null hypothesis that proteins within one subparticle were picked randomly from the 89 master HDL proteins list. The p-values were calculated as:
where n is the total number of genes on the watch list, m is the number of genes in the clique, p is the number of genes on our watch list associated with the given GO term, and q is the number of genes in this clique that were also associated with that GO term. The smaller the p-value is, the more significant the clique is enriched with the given function.
Lag-score.
We developed a lag-score (L-score), reflecting a quantitatively shifted distribution for a given protein. Although our Local S-score is also able to detect the local dissimilarities between the migration patterns for any given fraction, our L-score is able to measure the global pattern differences in term of the shifted fractions across all the fractions. The L-score was calculated based on time-lag between the normalized aggregated abundance profiles for protein Px from WT and KO mice groups. Time-lag is a concept in electronic signal processing to measure the shift between two signals. We applied this concept here to discover the migration patterns change between WT and KO groups. Assuming m WT mice and η KO mice, L-score Lx of a given protein Px between WT and KO groups was obtained by:
where , are the normalized spectral counts vector (i.e., abundance profiles) of protein Px from kth WT mouse and lth KO mouse, respectively. And , . is time-lag function to calculate signal lag in signal processing with cross-correlation. Cross-correlation is a similarity measure between two waveforms as one of them has time lag from the other. For discrete functions, the cross-correlation is an array defined as:
where f*[m] denotes the complex conjugate of f and m is the time lag. Time-lag between two signals is the distance between the center and the maximum of the cross-correlation vector. Larger L-score indicates more significant fractions shifts of protein migration pattern due to the ablation of genes. L-score is an integer, either positive or negative, reflecting distribution shifts positively or negatively.
Spoke and Matrix PPIs models.
A spoke model assumes all impacted proteins in Table 2 and 3 only interact with the absent protein (Fig. S5a). It is a direct interpretation of gene knockout experiments. In contrast, a matrix model assumes interactions among all proteins, including absent and pattern shifted proteins (Fig. S5b). We established three matrices to represent all interactions among all proteins (absent and pattern-shifted proteins). Using either model, PPIs derived from gene knockout experiments are able to serve as direct pieces of evidence that two interacting proteins may co-localize within the same subparticle. Thus, we directly utilized the number of edges (PPIs) being supported by the matrix and spoke models as the evidence in Table 1.
Supplementary Material
ACKNOWLEDGMENTS
WSD and LJL conceived and designed the research plan; SMG and WSD designed and performed the experiments; HL, XZ, JD and DS designed the algorithms and analyzed the data; all authors participated in writing the paper; WSD and LJL supervised the research.
The authors gratefully acknowledge the invaluable assistance of Huaiyu Zhang and Sheng Ren for the statistical analysis.
FUNDING
This work was supported by National Institutes of Health [grant number HL111829 to L. J. L].
Abbreviations
- HDL
high density lipoprotein
- apo
apolipoprotein
- CVD
cardiovascular disease
- CETP
cholesteryl ester transfer protein
- PLTP
phospholipid transfer protein
- RCT
reverse cholesterol transport
- TLF
trypanosome lytic factor
- UC
ultracentrifugation
- GF
gel filtration
- AE
anion exchange
- IEF
isoelectric focusing
- GO
Gene Ontology
- S-score
Spearman’s rank correlation coefficient score
- L-score
lag-score
- WT
wild-type
- KO
knock out
Footnotes
Supporting Information:
Supporting figures for alternative layouts of HDL interactome map combining all individual local networks. HDL interactome maps for GF, AE and IEF methods based on global correlation scores and HDL interactome map containing co-associated PPIs among three separation method, migration pattern shifting analysis for mouse gene knockout experiments, protein migration pattern comparison between the apoA-I deficient patient and the normal control, and spoke and matrix model for protein-protein interactions,. Description on a global score-based method. Supporting tables for full list of identified HDL subspecies.
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
References
- 1.Mahmood SS; Levy D; Vasan RS; Wang TJ, The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective. The Lancet 2014, 383, (9921), 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alwan A, Global status report on noncommunlcable diseases 2010. World Health Organization: 2011. [Google Scholar]
- 3.Rezaee F; Casetta B; Levels JH; Speijer D; Meijers JC, Proteomic analysis of high-density lipoprotein. Proteomics 2006, 6, (2), 721–30. [DOI] [PubMed] [Google Scholar]
- 4.Vaisar T; Pennathur S; Green PS; Gharib SA; Hoofnagle AN; Cheung MC; Byun J; Vuletic S; Kassim S; Singh P; Chea H; Knopp RH; Brunzell J; Geary R; Chait A; Zhao XQ; Elkon K; Marcovina S; Ridker P; Oram JF; Heinecke JW, Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest 2007, 117, (3), 746–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlsson H; Leanderson P; Tagesson C; Lindahl M, Lipoproteomics II: mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics 2005, 5, (5), 1431–45. [DOI] [PubMed] [Google Scholar]
- 6.Heller M; Stalder D; Schlappritzi E; Hayn G; Matter U; Haeberli A, Mass spectrometry-based analytical tools for the molecular protein characterization of human plasma lipoproteins. Proteomics 2005, 5, (10), 2619–30. [DOI] [PubMed] [Google Scholar]
- 7.Davidson WS; Silva RA; Chantepie S; Lagor WR; Chapman MJ; Kontush A, Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arterioscler Thromb Vasc Biol 2009, 29, (6), 870–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon SM; Deng J; Lu LJ; Davidson WS, Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography. J Proteome Res 2010, 9, (10), 5239–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon SM; Deng J; Tomann AB; Shah AS; Lu LJ; Davidson WS, Multi-dimensional co-separation analysis reveals protein-protein interactions defining plasma lipoprotein subspecies. Mol Cell Proteomics 2013, 12, (11), 3123–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riwanto M; Rohrer L; Roschitzki B; Besler C; Mocharla P; Mueller M; Perisa D; Heinrich K; Altwegg L; von Eckardstein A; Luscher TF; Landmesser U, Altered activation of endothelial anti- and proapoptotic pathways by high-density lipoprotein from patients with coronary artery disease: role of high-density lipoprotein-proteome remodeling. Circulation 2013, 127, (8), 891–904. [DOI] [PubMed] [Google Scholar]
- 11.Sreckovic I; Birner-Gruenberger R; Obrist B; Stojakovic T; Scharnagl H; Holzer M; Scholler M; Philipose S; Marsche G; Lang U; Desoye G; Wadsack C, Distinct composition of human fetal HDL attenuates its anti-oxidative capacity. Biochim Biophys Acta 2013, 1831, (4), 737–46. [DOI] [PubMed] [Google Scholar]
- 12.Link JJ; Rohatgi A; de Lemos JA, HDL cholesterol: physiology, pathophysiology, and management. Curr Probl Cardiol 2007, 32, (5), 268–314. [DOI] [PubMed] [Google Scholar]
- 13.Shah AS; Tan L; Long JL; Davidson WS, Proteomic diversity of high density lipoproteins: our emerging understanding of its importance in lipid transport and beyond. J Lipid Res 2013, 54, (10), 2575–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glomset JA, The plasma lecithins:cholesterol acyltransferase reaction. J Lipid Res 1968, 9, (2), 155–67. [PubMed] [Google Scholar]
- 15.Mackness MI; Arrol S; Durrington PN, Paraoxonase prevents accumulation of lipoperoxides in low-density lipoprotein. FEBS Lett 1991, 286, (1-2), 152–4. [DOI] [PubMed] [Google Scholar]
- 16.Nofer JR; Assmann G, Atheroprotective effects of high-density lipoprotein-associated lysosphingolipids. Trends Cardiovasc Med 2005, 15, (7), 265–71. [DOI] [PubMed] [Google Scholar]
- 17.Wadham C; Albanese N; Roberts J; Wang L; Bagley CJ; Gamble JR; Rye KA; Barter PJ; Vadas MA; Xia P, High-density lipoproteins neutralize C-reactive protein proinflammatory activity. Circulation 2004, 109, (17), 2116–22. [DOI] [PubMed] [Google Scholar]
- 18.Kulkarni S; Woollard KJ; Thomas S; Oxley D; Jackson SP, Conversion of platelets from a proaggregatory to a proinflammatory adhesive phenotype: role of PAF in spatially regulating neutrophil adhesion and spreading. Blood 2007, 110, (6), 1879–86. [DOI] [PubMed] [Google Scholar]
- 19.Kuvin JT; Harati NA; Pandian NG; Bojar RM; Khabbaz KR, Postoperative cardiac tamponade in the modern surgical era. Ann Thorac Surg 2002, 74, (4), 1148–53. [DOI] [PubMed] [Google Scholar]
- 20.Asztalos BF; Schaefer EJ, High-density lipoprotein subpopulations in pathologic conditions. Am J Cardiol 2003, 91, (7A), 12E–17E. [DOI] [PubMed] [Google Scholar]
- 21.Rifkin MR, Identification of the trypanocidal factor in normal human serum: high density lipoprotein. Proc Natl Acad Sci U S A 1978, 75, (7), 3450–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raper J; Fung R; Ghiso J; Nussenzweig V; Tomlinson S, Characterization of a novel trypanosome lytic factor from human serum. Infect Immun 1999, 67, (4), 1910–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawle A; Higgins MK; Olivant MP; Higgins JA, A rapid single-step centrifugation method for determination of HDL, LDL, and VLDL cholesterol, and TG, and identification of predominant LDL subclass. J Lipid Res 2002, 43, (2), 335–43. [PubMed] [Google Scholar]
- 24.van't Hooft F; Havel RJ, Metabolism of apolipoprotein E in plasma high density lipoproteins from normal and cholesterol-fed rats. J Biol Chem 1982, 257, (18), 10996–1001. [PubMed] [Google Scholar]
- 25.Havugimana PC; Hart GT; Nepusz T; Yang H; Turinsky AL; Li Z; Wang PI; Boutz DR; Fong V; Phanse S; Babu M; Craig SA; Hu P; Wan C; Vlasblom J; Dar VU; Bezginov A; Clark GW; Wu GC; Wodak SJ; Tillier ER; Paccanaro A; Marcotte EM; Emili A, A census of human soluble protein complexes. Cell 2012, 150, (5), 1068–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spirin V; Mirny LA, Protein complexes and functional modules in molecular networks. Proceedings of the National Academy of Sciences 2003, 100, (21), 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bader GD; Hogue CW, An automated method for finding molecular complexes in large protein interaction networks. BMC bioinformatics 2003, 4, (1), 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu LJ; Xia Y; Paccanaro A; Yu H; Gerstein M, Assessing the limits of genomic data integration for predicting protein networks. Genome research 2005, 15, (7), 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin CC; Juan HF; Hsiang JT; Hwang YC; Mori H; Huang HC, Essential core of protein-protein interaction network in Escherichia coli. J Proteome Res 2009, 8, (4), 1925–31. [DOI] [PubMed] [Google Scholar]
- 30.Bron C; Kerbosch J, Finding All Cliques of an Undirected Graph [H]. Communications of the Acm 1973, 16, (9), 575–577. [Google Scholar]
- 31.Gordon SM; Hofmann S; Askew DS; Davidson WS, High density lipoprotein: it's not just about lipid transport anymore. Trends Endocrinol Metab 2011, 22, (1), 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eisenberg S, High density lipoprotein metabolism. J Lipid Res 1984, 25, (10), 1017–58. [PubMed] [Google Scholar]
- 33.Prasad TK; Goel R; Kandasamy K; Keerthikumar S; Kumar S; Mathivanan S; Telikicherla D; Raju R; Shafreen B; Venugopal A, Human protein reference database—2009 update. Nucleic acids research 2009, 37, (suppl 1), D767–D772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos RD; Schaefer EJ; Asztalos BF; Polisecki E; Wang J; Hegele RA; Martinez LR; Miname MH; Rochitte CE; Da Luz PL; Maranhao RC, Characterization of high density lipoprotein particles in familial apolipoprotein A-I deficiency. J Lipid Res 2008, 49, (2), 349–57. [DOI] [PubMed] [Google Scholar]
- 35.Zhang M; Lu LJ, Investigating the validity of current network analysis on static conglomerate networks by protein network stratification. BMC Bioinformatics 2010, 11, 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franceschini A; Szklarczyk D; Frankild S; Kuhn M; Simonovic M; Roth A; Lin J; Minguez P; Bork P; von Mering C, STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research 2013, 41, (D1), D808–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schamaun O; Olaisen B; Mevag B; Gedde-Dahl T Jr.; Ehnholm C; Teisberg P, The two apolipoprotein loci apo A-I and apo A-IV are closely linked in man. Hum Genet 1984, 68, (2), 181–4. [DOI] [PubMed] [Google Scholar]
- 38.Ordovas JM; Schaefer EJ, Genetic determinants of plasma lipid response to dietary intervention: the role of the APOA1/C3/A4 gene cluster and the APOE gene. Br J Nutr 2000, 83 Suppl 1, S127–36. [DOI] [PubMed] [Google Scholar]
- 39.Puppione DL, Higher primates, but not New World monkeys, have a duplicate set of enhancers flanking their apoC-I genes. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 2014, 11, 45–48. [DOI] [PubMed] [Google Scholar]
- 40.Karten B; Campenot RB; Vance DE; Vance JE, Expression of ABCG1, but not ABCA1, correlates with cholesterol release by cerebellar astroglia. J Biol Chem 2006, 281, (7), 4049–57. [DOI] [PubMed] [Google Scholar]
- 41.Monaco HL, The transthyretin-retinol-binding protein complex. Biochim Biophys Acta 2000, 1482, (1-2), 65–72. [DOI] [PubMed] [Google Scholar]
- 42.Sousa MM; Berglund L; Saraiva MJ, Transthyretin in high density lipoproteins: association with apolipoprotein A-I. J Lipid Res 2000, 41, (1), 58–65. [PubMed] [Google Scholar]
- 43.Liz MA; Faro CJ; Saraiva MJ; Sousa MM, Transthyretin, a new cryptic protease. J Biol Chem 2004, 279, (20), 21431–8. [DOI] [PubMed] [Google Scholar]
- 44.Lee M; Kovanen PT; Tedeschi G; Oungre E; Franceschini G; Calabresi L, Apolipoprotein composition and particle size affect HDL degradation by chymase: effect on cellular cholesterol efflux. J Lipid Res 2003, 44, (3), 539–46. [DOI] [PubMed] [Google Scholar]
- 45.Liz MA; Gomes CM; Saraiva MJ; Sousa MM, ApoA-I cleaved by transthyretin has reduced ability to promote cholesterol efflux and increased amyloidogenicity. J Lipid Res 2007, 48, (11), 2385–95. [DOI] [PubMed] [Google Scholar]
- 46.Boucher J; Ramsamy TA; Braschi S; Sahoo D; Neville TA; Sparks DL, Apolipoprotein A-II regulates HDL stability and affects hepatic lipase association and activity. J Lipid Res 2004, 45, (5), 849–58. [DOI] [PubMed] [Google Scholar]
- 47.Duverger N; Murry-Brelier A; Latta M; Reboul S; Castro G; Mayaux JF; Fruchart JC; Taylor JM; Steinmetz A; Denefle P, Functional characterization of human recombinant apolipoprotein AIV produced in Escherichia coli. Eur J Biochem 1991, 201, (2), 373–83. [DOI] [PubMed] [Google Scholar]
- 48.de Haan W; Out R; Berbee JF; van der Hoogt CC; van Dijk KW; van Berkel TJ; Romijn JA; Jukema JW; Havekes LM; Rensen PC, Apolipoprotein CI inhibits scavenger receptor BI and increases plasma HDL levels in vivo. Biochem Biophys Res Commun 2008, 377, (4), 1294–8. [DOI] [PubMed] [Google Scholar]
- 49.Gordon S; Li H; Zhu X; Shah A; Lu LJ; Davidson WS, A Comparison of the Mouse and Human Lipoproteome: Suitability of the Mouse Model for Studies of Human Lipoproteins. Journal of proteome research 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camus MC; Chapman MJ; Forgez P; Laplaud PM, Distribution and characterization of the serum lipoproteins and apoproteins in the mouse, Mus musculus. J Lipid Res 1983, 24, (9), 1210–28. [PubMed] [Google Scholar]
- 51.Al-Sarraf A; Al-Ghofaili K; Sullivan DR; Wasan KM; Hegele R; Frohlich J, Complete Apo AI deficiency in an Iraqi Mandaean family: case studies and review of the literature. J Clin Lipidol 2010, 4, (5), 420–6. [DOI] [PubMed] [Google Scholar]
- 52.Huang Y; Wu Z; Riwanto M; Gao S; Levison BS; Gu X; Fu X; Wagner MA; Besler C; Gerstenecker G; Zhang R; Li XM; DiDonato AJ; Gogonea V; Tang WH; Smith JD; Plow EF; Fox PL; Shih DM; Lusis AJ; Fisher EA; DiDonato JA; Landmesser U; Hazen SL, Myeloperoxidase, paraoxonase-1, and HDL form a functional ternary complex. J Clin Invest 2013, 123, (9), 3815–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Philips MC, New insights into the determination of HDL structure by apolipoproteins: Thematic review series: high density lipopoprotein structure, function and metabolism. J Lipid Res 2013, 54, (8), 2034–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.