Abstract
Over the past decade a suite of new mass spectrometry-based proteomics methods has been developed that now enables the conformational properties of proteins and protein-ligand complexes to be studied in complex biological mixtures, from cell lysates to intact cells. Highlighted here are seven of the techniques in this new toolbox. These techniques include chemical cross-linking (XL-MS), hydroxyl radical footprinting (HRF), Drug Affinity Responsive Target Stability (DARTS), Limited Proteolysis (LiP), Pulse Proteolysis (PP), Stability of Proteins from Rates of Oxidation (SPROX) and Thermal Proteome Profiling (TPP). The above techniques all rely on conventional bottom-up proteomics strategies for peptide sequencing and protein identification. However, they have required the development of unconventional proteomic data analysis strategies. Discussed here are the current technical challenges associated with these different data analysis strategies as well as the relative analytical capabilities of the different techniques. The new biophysical capabilities that the above techniques bring to bear on proteomic research are also highlighted in the context of several different application areas in which these techniques have been used including the study of protein ligand binding interactions (e.g., protein target discovery studies and protein interaction network analyses) and the characterization of biological states.
Keywords: thermodynamics, protein folding, proteomics, mass spectrometry
Graphical Abstract

1. INTRODUCTION
Over the past decade a new toolbox of mass spectrometry-based techniques has been established for probing the conformational properties of proteins on the proteomic scale. These techniques have involved different combinations of protease digestion, chemical modification, protein precipitation, chemical denaturation, and thermal denaturation strategies with quantitative mass spectrometry-based proteomics platforms. This new toolbox of proteomics techniques has enabled the study of conformational properties of proteins and protein-ligand binding interactions on the proteomic scale using a whole cell approach.
For many years, protein structure and protein-ligand binding interactions have been studied using purified systems. Such studies have revealed a vast amount of information on protein folding, structure, and protein-ligand binding interactions. However, the environmental differences between dilute solutions and the densely packed cellular environment raises the question as to whether these in vitro studies provide physiologically relevant structural and functional information.1 Macromolecular crowding in the cell, for example, plays a major role on protein interactions exerting influence on conformational distributions, protein stabilization, specificity of interactions, and diffusion of molecules.2–4 It is also believed that even weak/non-specific interactions of proteins with various components of the densely packed cellular environment (e.g., other proteins and small molecules) can alter the different conformational states that proteins populate in the cell.5, 6 These interactions are not present in the purified protein systems that have been extensively used to study conformational properties of proteins using traditional biophysical methods (i.e. circular dichroism, fluorescence, nuclear magnetic resonance) or even using a more recent method, hydrogen-deuterium exchange coupled with mass spectrometry (HDX-MS) and other mass spectrometry-based methods. The growing number of studies documenting the effects of the cellular environment on protein structure and function underscores the importance of using a whole cell approach to study proteins.5, 7, 8
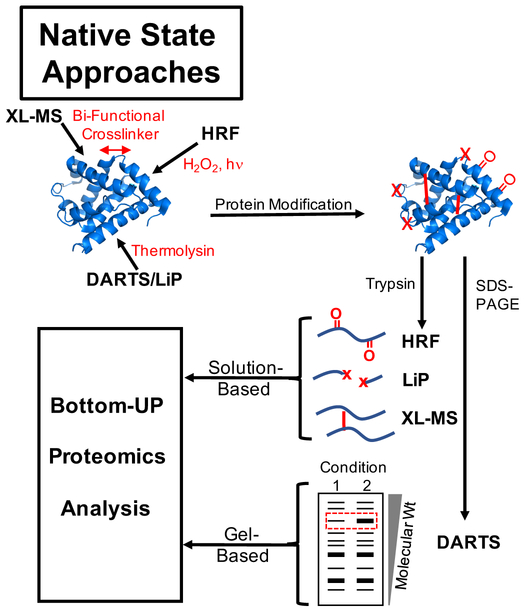
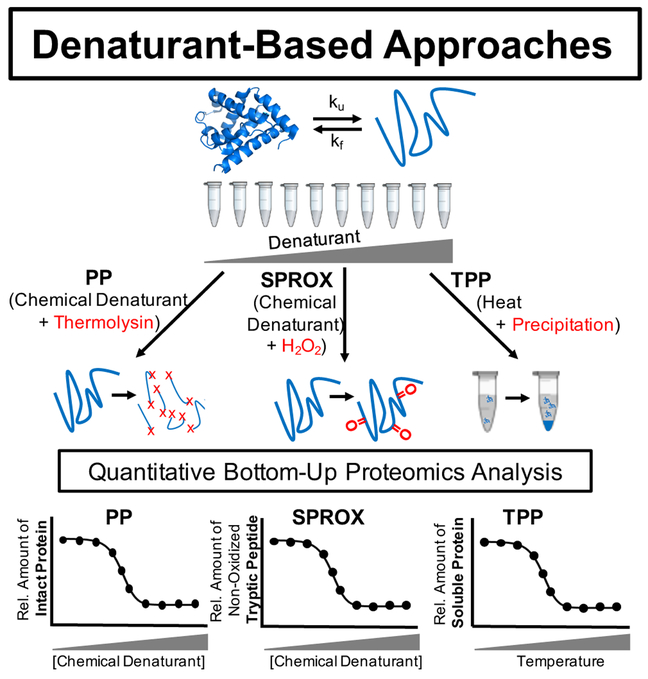
Highlighted in this Perspective are a series of seven experimental methods for characterizing the conformational properties of proteins on the proteomic scale using a whole cell approach. The methods, which are divided here into two different families, have created a new field of proteomics research that we coin proteome-wide structural biology. One family of methods driving this new field is comprised of limited proteolysis and covalent labelling strategies that involve the use of either a low concentration of or no protein denaturant (Figure 1). Included in this family of methods are the chemical cross-linking (XL-MS),9 hydroxyl radical footprinting,10, 11 Drug Affinity Responsive Target Stability (DARTS),12 and Limited Proteolysis (LiP)13, 14 techniques. These techniques all probe the local structural features of proteins and protein interactions of proteins. The second family of methods driving this new field of proteome-wide structural biology includes the Pulse Proteolysis (PP),15–18 Stability of Proteins from Rates of Oxidation (SPROX),19, 20 and Thermal Proteome Profiling (TPP)21 techniques (Figure 2). The techniques in this second family of methods utilize a protein denaturant, and probe the global unfolding/refolding reactions of proteins.
Figure 1.
Schematic representation of experimental workflows utilized in native state approaches highlighted here including the XL-MS, HRF, DARTS, and LiP techniques.
Figure 2.
Schematic representation of experimental workflows utilized in the SPROX, PP, and TPP techniques that utilize denaturant to probe the more global unfolding/refolding properties of proteins.
Common to all the techniques highlighted here is their exploitation of conventional bottom-up proteomics methods. However, their unique experimental workflows have required the development of unconventional proteomic data analysis strategies. These strategies and the different data analysis challenges associated with each technique are discussed here, as are the new biophysical capabilities that the above techniques bring to proteomic research. The goal of this perspective is to educate the reader on the differences and similarities between the different approaches and to introduce the new field of proteome-wide structural biology that these techniques have created. Thus, also highlighted are representative studies illustrating the advantages, disadvantages and existing challenges of the different techniques, and several different application areas including the study of protein-ligand binding interactions (e.g., protein target discovery studies and protein interaction network analyses) and the characterization of biological states, such as those associated with normal biological processes and different diseases.
2. EXPERIMENTAL WORKFLOWS
Described below are the different combinations of protease digestion, chemical modification, protein precipitation, chemical denaturation, thermal denaturation, and bottom-up proteomics strategies that are employed in the experimental workflows of the seven different methods covered here. Summarized in Table 1 are several important technical features associated with each experimental workflow. Summarized in Table 2 are the different types of biophysical information that each technique generates and the applications in which they have so far been used.
Table 1.
Summary of the technical features of the seven experimental methods highlighted in this Perspective.
| Techniques | Buffer Conditions | In/ex-vivo | Readout Target | Proteome Coverage (# Proteins) |
|---|---|---|---|---|
| XL-MS | Native | Both | Cross-Linked Peptides | 2111 3202 3163 |
| HRF | Oxidized Peptides | 13914 | ||
| DARTS | Ex-vivo | Intact Proteins | 5005 | |
| UP | Cleaved Peptides | 10006 | ||
| PP | Denaturing | Intact Proteins or Cleaved Peptides | 9007 | |
| SPROX | Methionine-Containing Peptides | 10008 | ||
| TPP | Both | Soluble Proteins | 50009 |
Pseudomonas Aeruginosa cells from reference112.
Escherichia coli cells from reference38.
Mouse heart tissue from reference110.
Vero cells from reference 42.
U87-MG cells (human glioblastoma) from reference 44.
Yeast cells from reference 13.
Escherichia coli cells from reference 18.
MDA-MB-231 cells from reference67.
K562 cells from reference 51.
Table 2.
Biophysical information, applications, and limitations of the seven techniques highlighted in this perspective.
| Techniques | Biophysical Information | Applications | Limitations |
|---|---|---|---|
| XL-MS | ▪ Protein structure modeling constraints: distance constraints (XL-MS, HRF), and solvent accessible surface area (HRF) | ▪ Protein interaction network analysis | ▪ Requires advanced MS method for peptide identification |
| HRF | ▪ Ligand binding determination ▪ Biological State Analysis |
▪ Requires advanced searching strategy for the complex data generated ▪ Difficult to enrich for oxidized peptides |
|
| DARTS | ▪ Protease susceptibility | ▪ Drug target discovery | ▪ Target protein cannot be resistant |
| UP | ▪ Biological state analysis | to proteolysis in native condition ▪ Need to separate the intact protein or proteolyzed protein fragments from digested product |
|
| PP | ▪ Protein folding free energies (AGf values), binding free energies (AAGf values), and dissociation constants (Kd values) | ▪ Drug target discovery | ▪ Need to separate the intact protein from digested product |
| SPROX | ▪ Drug target discovery ▪ Biological state analysis |
▪ Requires detection of methionine-containing peptides | |
| TPP | ▪ Protein melting temperature (Tm) | ▪ Drug target discovery ▪ Protein Interaction network Analysis | ▪ No general relationship between protein Tm shift and drug binding affinity |
Native State Approaches
XL-MS.
XL-MS has been applied to a number of purified proteins and protein complexes over the last several decades,22, 23 and it was one of the first covalent labelling approaches for protein conformational analyses to be used on the proteomic scale.24–26 The experimental workflow in XL-MS experiments (Figure 1) involves treating the protein sample with a chemical crosslinking reagent containing two or more protein-reactive functional groups (e.g., thiol reactive maleimides, carboxyl group reactive diazoacetateesters, and/or amine reactive N-hydroxysuccinimide (NHS) esters) that are connected by a linker region. Ultimately, the sites of inter- and intra-protein crosslinks are defined using the cross-linked peptides detected in a bottom-up proteomics analysis of the sample. Cross-linkers containing various reactive groups and linker lengths have been developed and used to achieve spatial information on proteins and protein-protein complexes.27–31 In recent years, there have been an increasing number of in vivo XL-MS applications studying protein interactions in the native cellular environment.27–32 Several recently developed XL-MS approaches, including the protein interaction (PIR) technology,33 the incorporation of photo-reactive cross-linkers into proteins during translation in cell culture,32 and the use of disuccinimdyl dibutyric acid (DBSU)34 have greatly improved the detection and identification of cross-linked peptides in proteome-wide XL-MS experiments.9, 35–38
HRF.
Another covalent labelling method for studying protein interactions is hydroxyl-radical footprinting (HRF). This method utilizes hydroxyl radicals to oxidatively modify solvent accessible sites in proteins (Figure 1). The changes in solvent accessibility resulting from ligand binding or from a structural change induced by a different biological state can be used to identify interaction sites and regions of conformational change. Multiple methods have been used to generate hydroxyl radicals including synchrotron radiation,39 laser photolysis of hydrogen peroxide,40 and UVirradiation.41 Two laser-based methods for generating hydroxyl radicals, fast photochemical oxidation of proteins (FPOP)11, 42 and nanosecond laser photolysis,43 have proven especially useful in HRF applications to proteins in intact cells. Ultimately, the sites of modification, which can include the side chains of 17 of the 20 amino acids, are identified using a bottom-up proteomics analysis.
DARTS.
The DARTS approach is a limited proteolysis-based strategy that was originally developed to identify protein targets of small molecules.12 It is based on the premise that drug binding induces conformational changes in proteins that either increase or decrease the susceptibility of the protein to proteolytic digestion with a non-specific protease (e.g., thermolysin or proteinase K). In the original DARTS workflow, proteins with different cleavage patterns in the presence and absence of ligand were identified using a gel-based proteomics readout (Figure 1).12 More recently several gel-free LC MS/MS based proteomics approaches have been proposed and successfully demonstrated with DARTS.44, 45 The LC-MS/MS approaches in DARTS (whether they are gel-based or gel-fee) are protein centered. That is, the bottom-up proteomics data generated in the LC-MS/MS analyses is used to generate quantitative information about the proteins to which they map.
LiP.
The LiP experiment46 is fundamentally similar to DARTS. As in DARTS, the LiP experiment involves treating the protein samples under study (e.g., a cell lysate in the presence and in the absence of a test ligand) with a non-specific protease (e.g., thermolysin, or proteinase K) under solution conditions in which the proteins in the sample are in their native state. The non-specific proteolysis reaction is quenched, and the differential cleavage pattern observed between the protein samples is determined using bottom-up proteomics methods. In contrast to DARTS, the LC-MS/MS readout in the bottom-up proteomics analysis is peptide centered. That is, the differential cleavage patterns are directly ascertained from the tryptic (and semi-tryptic) peptides identified and quantified in the bottom-up proteomics analysis (Figure 1). Spectral counting,46 SRM,46 and SILAC14 have all been employed in LiP experiments to quantify the relative amounts of fully (and semi-) tryptic peptides generated in the test protein samples.
Denaturation-Based Approaches
PP.
PP is also a limited proteolysis-based approach. However, it is conceptually different than the DARTS and LiP techniques. In PP, the chemical denaturant dependence of a non-specific proteolytic digestion reaction is used to evaluate the thermodynamic properties of a protein’s folding reaction.47 The PP protocol involves incubating the protein sample in a series of buffers containing increasing concentrations of urea and then treating the samples with a non-specific protease (e.g., thermolysin) to selectively digest the unfolded protein population in each urea-containing buffer (Figure 2). Proteome-wide applications of PP have utilized several different mass spectrometry and/or gel-based readouts to quantify the relative amount of intact protein in each protein sample.15–18 These readouts have included: i) the use of a two-dimensional gel electrophoresis strategy for the differential analyses of gel band intensities;15 ii) the use of a fractionation strategy prior to the use of one-dimensional gel electrophoresis;16 iii) the use of a SILAC quantitation strategy using one-dimensional gel electrophoresis;17 and iv) the use of a filter-assisted sample preparation (FASP) protocol in combination with tandem mass tags (TMT) labels.18 An important step in all of the above PP workflows is the separation of the digestion products (i.e., protein fragments) from the intact proteins in the denaturant-containing protein samples, which must be must be done prior to subjecting them to bottom-up proteomics analysis. Also, as in DARTS, the quantitative bottom-up proteomics analysis in PP is protein centered.
SPROX.
SPROX is a covalent labelling technique that involves protein oxidation like HRF. However, the oxidation reaction conditions in SPROX are significantly milder than those used in HRF. The SPROX technique utilizes a chemical denaturant in much the same way as PP. However, in SPROX, it is the chemical denaturant dependence of a highly selective methionine oxidation reaction involving hydrogen peroxide that is used to report on the thermodynamic properties of a protein’s folding reaction.48 The SPROX protocol involves equilibrating the protein sample in a series of buffers containing increasing concentrations of a chemical denaturant (e.g., GdmCl or Urea) and then treating the samples with hydrogen peroxide using reaction conditions under which the primary site of oxidation is the thioether group in the side chain of methionine residues that become solvent exposed as proteins in the denaturant-containing buffers are unfolded (Figure 2). Proteome wide applications of SPROX have utilized either isobaric mass tags48 or SILAC20 to quantify the extent of methionine oxidation as a function of chemical denaturant. This is accomplished using standard bottom-up proteomics methods and ultimately measuring the relative quantity of un-oxidized methionine-containing peptides (or oxidized methionine-containing peptides) in the different denaturant-containing protein samples. To facilitate the detection and quantitation of methionine-containing peptides in quantitative LC-MS/MS analyses of SPROX samples, a methionine-containing peptide enrichment strategy has been incorporated into the SPROX protocol.49
TPP.
In TPP experiments the temperature dependence of a protein aggregation reaction is used to report on the thermal denaturation properties of proteins.50, 51 The TPP protocol involves incubating the protein sample at a series of different temperatures for a given time (typically 3 minutes) (Figure 2). After cooling the samples to room temperature, the proteins that are unfolded at a given temperature aggregate, and the aggregated proteins at each temperature are separated from the soluble (folded) proteins in an ultracentrifugation step. The soluble proteins recovered at each temperature are quantified in a quantitative bottom-up proteomics analysis using isobaric mass tags.51 The aggregated proteins that are removed from solution during the ultracentrifugation step can also be quantified in a quantitative bottom-up proteomics analysis also using isobaric mass tags.52 The TPP approach has also been combined with a differential ingel electrophoresis (DIGE) readout to identify differentially stabilized proteins in two different samples (e.g., cells incubated with and without drug).53 The quantitative bottom-up proteomics strategies used in TPP experiments are protein centered (i.e., data collected on the peptides in the LC-MS/MS readout is used to quantify the precipitated protein).
3. DATA ACQUISITION and ANALYSIS
The above techniques all rely on standard bottom-up proteomics methods for peptide and protein identification and quantitation. Therefore, all of the current limitations and challenges associated with using bottom-up proteomics methods are at play in each technique. For example, peptide and protein coverages in the different techniques are all limited by the speed and sensitivity of existing mass spectrometry instrumentation. Fundamentally, the protein centered readouts in TPP, PP, and DARTS are similar to those used in conventional bottom-up proteomics analyses. However, as discussed below the SPROX, LiP, HRF, and XL-MS techniques have additional limitations and challenges associated with their peptide centered readouts. All of the techniques have unique data analysis strategies with different challenges, which are also highlighted below.
Peptide Detection
The SPROX, LiP, HRF and XL-MS techniques all rely on the detection of specific peptides in the bottom-up proteomics experiment. The SPROX technique requires the detection of methionine-containing peptides. The LiP and HRF techniques require detection of specific peptide(s) that include the sites of proteolysis or oxidation respectively, and the XL-MS technique requires detection of the specific peptides that are crosslinked. On the one hand, this means that the SPROX, LiP, HRF, and XL-MS techniques can provide site-specific information about the location of a detected conformational change in a protein. On the other hand, this makes the bottom-up proteomics analysis in these experiments especially challenging because the specific peptides that need to be detected typically represent only a fraction of the peptides present in the bottom-up proteomics samples generated in these techniques. Thus, the detection of these specific peptides is difficult using shot-gun proteomics methods, and ultimately the scope of these techniques can be limited by high false negative rates.
Strategies to enrich for the methionine-containing peptides in SPROX and the cross-linked peptides in XL-MS have been developed and found to significantly increase the scope of these techniques.9, 49, 54 Enrichment strategies for the proteolyzed peptides in LiP and the chemically modified peptides in HRF would also help expand the scope of these techniques and lower false negative rates. However, no such enrichment strategies for the proteolyzed and oxidized peptides generated in LiP and FPOP, respectively, have been reported, to date. In a recent nanosecond laser photolysis-based HRF experiment, immunoprecipitation was used as an enrichment strategy. However, Western blot analysis showed a decrease in protein detected after the pull down. This observation was consistent with a decrease in binding affinity upon oxidation, and it highlights the difficulty in using enrichment strategies with HRF methods.43
Peptide Identification
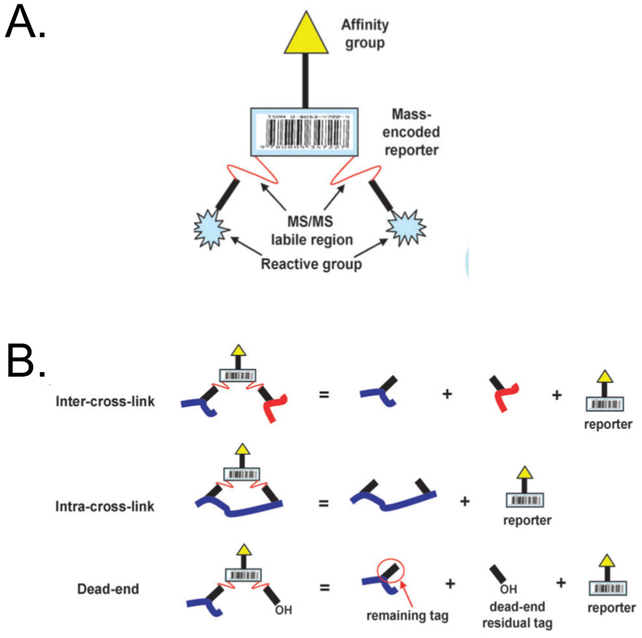
The peptide centered readouts in XL-MS and HRF present a formidable challenge from a bioinformatics perspective. The crosslinked peptides generated in XL-MS and the chemically modified peptides generated in HRF can be difficult to identify using conventional bottom-up proteomics methods for data acquisition and analysis. The PIR technology developed for XL-MS experiments not only helps overcome the “needle in a haystack” problem with the low abundance of cross-linked peptides, but it also facilitates data analysis (Figure 3). Using an MS method termed ReACT, precursor ions are subject to a low energy fragmentation event, that releases the two cross-linked peptides as well as a reporter ion of specific mass.55 In the resulting MS2 spectra, the summation of the mass of the reporter ion plus the two released peptides are searched in real time. The released peptides are then fragmented further via MS3 analysis. For each MS2 spectra, two MS3 spectra are recorded, one for each of the cross-linked peptides. Database searching, is then performed on the resulting MS3 spectra for identification of cross-linked peptides.56 For other types of cross-links, specialized software has been developed to identify cross-linked peptides.56 These include StavroX57 and Xlink Analyzer.58
Figure 3.
PIR structure. (A) Conceptual design of cross protein interaction reporters (PIRs). (B) Examples of fragmentation patterns of PIR-labeled peptides. Adapted with permission from Tang and Bruce, A new cross-linking strategy: protein interaction reporter (PIR) technology for protein-protein interaction studies, Mol Biosyst, 6(6), pages 939–947. Copyright 2010 the Royal Society of Chemistry.
In HRF experiments, many different amino acids can be modified and several residues can undergo multiple modification types. This not only increases the complexity of the data but also increases the search space for peptide identification. Several platforms have been used for data analysis of in vitro data including commonly used software such as Mascot,59 Proteome Discoverer60 and Byonic.61 Other platforms, such as ProtMap MS,62 were specifically developed for analyzing HRF data. For IC-FPOP, the search space is further increased owing to the large number of peptides present in the cell lysate. A multi-level strategy with stringent filters is typically used to reduce search times and limit false positives.42 In this strategy, unmodified peptides are searched in the first level. In subsequent levels, different modifications are searched (i.e., +16 modifications in level two and +14 modifications in level 3). Using the multi-level strategy reduces the data search time for IC-FPOP while still searching the entire complement of possible modifications.
Peptide Quantitation
The peptide centered readouts in the SPROX and LiP techniques create a unique challenge for the quantitative data analysis strategies used with these techniques. In both SPROX and LiP experiments, the quantitative LC-MS/MS data from a single peptide is used for hit selection. This is in contrast to more conventional, quantitative bottom-up proteomics analyses (e.g., protein expression level analyses) as well as the TPP and PP techniques, in which the quantitative LC-MS/MS data from multiple peptides derived from the same protein can be used for the analysis). Using the quantitative LC-MS/MS data from a single peptide for hit selection in SPROX and LiP, makes these techniques especially vulnerable to the pitfalls associated with quantitative bottom-up proteomics analyses (e.g., isomass peptide interferences in isobaric mass tagging strategies). This can adversely impact the false positive and false negative rates of these techniques. To help differentiate false positive from true positives in LiP and SPROX experiments, the number of biological and technical replicates can be increased, and/or consistent hit behavior can be required between wild-type and oxidized methionine-containing peptide pairs in SPROX and between tryptic and semi-tryptic peptide pairs in LiP.
Data Analysis
The quantitative data analysis strategies used for hit selection (e.g., the identification of protein conformational changes) in DARTS, LiPs, HRF, and XL-MS require evaluating the relative amount of a given peptide in two different samples (e.g., one in the presence of ligand and one not). The differential amounts of specific peptides (e.g., those that are non-specifically digested with a protease in DARTS and LiPs and those that are chemically modified in HRF and XL-MS) directly report on the different conformational properties of the proteins to which they map (e.g., more or less solvent exposed in the DARTS, LiPs and HRF experiments and crosslinked or not crosslinked in the XL-MS experiment). The quantitative proteomics strategies employed in the DARTS, LiPs, HRF, and XL-MS (e.g., spectral counting, stable isotope labelling, and label-free methods) are fundamentally very similar to those used in conventional bottom-up proteomics, although they have some unique challenges (see above).
The quantitative data analysis strategies used for hit selection in SPROX, TPP, and PP are notably different from conventional bottom-up proteomics methods. In traditional quantitative bottom-up proteomics analyses the expected change in protein concentration from sample to sample is unknown. However, the expected data structure in SPROX, TPP, and PP experiments is known. The protein denaturation curves generated using these techniques are sigmoidal (see Figure 2) because the protein folding reactions on which they report are cooperative. Ultimately, the chemical denaturant concentration (in SPROX and PP) or the temperature (in TPP) at the transition midpoint (see Figure 2) is extracted from the denaturation curves generated in SPROX, TPP, and PP. In SPROX and PP this chemical denaturant concentration can be used to calculate thermodynamic parameters such as a protein folding free energy or the dissociation constant of a protein-ligand complex.47, 48, 63–65
The expected structure of SPROX, TPP, and PP data facilitates the determination of transition midpoints. For example, because the general structure of the data is known, outliers in the data can be more easily identified and dealt with than in conventional bottom-up proteomics experiments. It also means that the precision of the data points defining the protein denaturation curves generated in these techniques can be increased using normalizations based on global analyses of the data. For example, in SPROX the isobaric mass tag data generated on the methionine-containing peptides can be subject to a normalization based on the non-methionine-containing peptides, which are not expected to change from denaturant sample to denaturant sample.48, 66 “Super denaturation curves” generated using all the data from all the proteins in a sample have also been used to normalize the denaturation curves generated for individual proteins in TPP.51
Proteome Coverage
The peptides identified in the bottom-up proteomic readouts employed for all the techniques highlighted here ultimately report on the conformational properties of the proteins to which they map. Thus, a high number of identified peptides and proteins is required for in-depth proteome-wide structural information. TPP is perhaps the most comprehensive of the methods (see Table 1) since the proteomic coverage is only limited by the usual constraints of bottom-up proteomics whereas other methods have more unique limitations. As described above, SPROX relies on the detection of methionine-containing peptides, LiP relies on the detection of tryptic (or semi-tryptic) peptides covering the specific site of proteolytic cleavage, HRF relies on the detection of tryptic peptides containing the site of modification, and XL-MS relies on the detection of the specific cross-linked peptides in the bottom-up proteomics experiment. No one technique affords full proteome coverage. To date, there have been only a few direct comparisons of the proteomic coverages obtained using the above techniques.14, 67, 68 Table 1 includes a “rough” comparison of the proteomic coverages that can be obtained using the different techniques. The comparison is “rough” because the cell lines from which the samples were derived are different, as are the mass spectrometry instrumentation and methods used to acquire the data.
Due to the unique nature of the bottom-up proteomic readouts in each technique, even the techniques that probe similar conformational properties in proteins are expected to the provide some complimentary information. For example, while the SPROX, TPP, and PP techniques are expected to generate some overlapping hits in protein target discovery experiments, each technique is also likely to assay some unique proteins. The same is also likely to be true of the DARTS and LiP techniques.
4. EX VIVO vs. IN VIVO
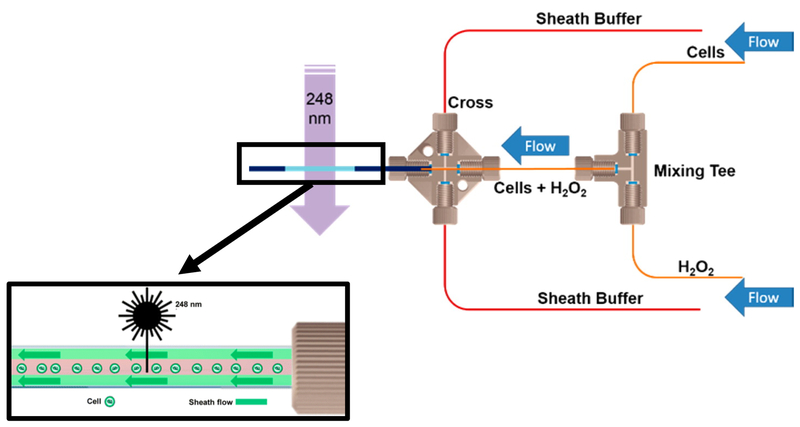
All the techniques highlighted here can be applied to unpurified protein mixtures such as cell lysates. However, only the TPP, HRF, and XL-MS techniques have so far been applied to protein in cells.10, 27, 51 In-Cell FPOP (IC-FPOP) capitalizes on the fact that hydrogen peroxide readily crosses cellular membranes. This permeation is not limited to just the plasma membrane but extends to the organelle membranes as well leading to modifications of proteins in various cellular compartments such as the cytoplasm, nucleus, and mitochondria amongst others.42 Cell viability assays have demonstrated that IC-FPOP probes live cells. For example, it has been shown that in the time frame of the IC-FPOP experiment, greater than 70% of Vero cells were still viable in the presence 20 mM hydrogen peroxide.11 Also, critical to the success of IC-FPOP experiments was the development of a single cell flow system (Figure 4) to limit cell clumping and provide equal exposure of cells to laser irradiation.42 By optimizing the laser frequency and flow rate in IC-FPOP experiments it has been possible to detect oxidative modifications on over 1300 proteins within the cell. Another attractive feature of IC-FPOP is that the protein modification reactions can be performed on a very fast (i.e., microsecond) time scale. ICFPOP currently shows great promise for in vivo applications as well using transparent animals such as C. elegans where laser irradiation can penetrate the organism. Currently, the main hurdles are hydrogen peroxide diffusion in the animal leading to a limited number of modified proteins.
Figure 4.
IC-FPOP flow cell schematic. Optimal conditions were observed with a 10:1 sheath buffer to cellular analyte ratio capillary in dark blue, window for laser light in light blue. Single cell flow after exit from the cross is depicted in the inset. Adapted with permission from Rinas et al., Development of a Microflow System for In-Cell Footprinting Coupled with Mass Spectrometry, Anal. Chem., 88(20), pages 10052–10058. Copyright 2016 American Chemical Society.
XL-MS on whole cells has also been demonstrated using both exogenous crosslinkers (e.g., PIR technology)37, 38, 69, 70 and photoreactive cross-linkers36, 71 incorporated into proteins during protein translation in cell culture. Critical to the successful use of exogenous cross-linkers is cross-linker solubility and cellular penetration, both of which are dependent on the size of the cross-linker. Cross linkers with a lower molecular weight tend to have better solubility and cellular penetration, which are crucial for in-cell studies. The penetration of a given cross-linker can also vary depending on the cellular surface, which ultimately increases the variability of cross-linker concentrations in the cell.9 This becomes a limitation of XL-MS where not all proteins are equally able to be crosslinked due to cross-linker concentration variability. However, like IC-FPOP, XL-MS experiments on whole cells, especially those involving photo-reactive cross-linkers,30 have the advantage that they can be performed on a relatively fast time-scale, creating the possibility to probe more transient protein-protein interactions.
The TPP approach is also amenable to in vivo analyses. TPP is especially attractive for in vivo analyses of proteins in intact cells because it does not involve the introduction of any exogenous chemical reagents, as is the case in IC-FPOP and XL-MS experiments. The TPP approach has been used in a number of in vivo applications including studies to identify the protein targets of drugs,51, 72–75 and even more recently to investigate protein-protein interactions.76 In TPP, the test cells are typically heated for 3 min at the desired temperatures, cooled for 3 min at 4 C, lysed, and the cell lysate subjected to a centrifugation step that takes about 20 min. While the folding properties of the proteins in the cells are mostly captured during the initial heating and cooling step, the time required for these steps is relatively long (i.e., on the order of minutes) compared to that required to capture the folding properties in IC-FPOP and XL-MS experiments (i.e., on the order of microseconds). This has the potential to compromise the ability of TPP to capture more transient protein-protein interactions present in the cell.
Whole cell analysis is advantageous because it provides an opportunity to interrogate the quinary structure of proteins that can be perturbed once the plasma membrane is disturbed. Quinary structure resulting from transient protein-protein interactions has been shown to be important for many cellular functions and tools that can analyze these structures are greatly needed.5 Although it has yet to be determined whether TPP, IC-FPOP, or XL-MS methods can effectively probe quinary structure, their demonstrated capabilities in whole cell analysis provides the possibility. Two of the techniques, IC-FPOP and XL-MS methods with photo-activated cross linkers, have an added advantage that they can be used on relatively short time-scales (microsecond timescale). This makes these methods especially well-suited for studying weak, transient interactions like those found in quinary structures.
5. Biophysical Information
Thermodynamic Parameters
The DARTS and LiP techniques provide largely qualitative information about the thermodynamic properties of the protein conformational changes induced by ligands. For example, regions of protein structure that become more or less susceptible to proteolyitic digestion in the presence of ligand are generally interpreted as being more or less stable, respectively. The XL-MS and HRF techniques can provide similarly qualitative information based on the differential reactivity of specific regions of protein structure in the presence and absence of ligand. However, the XL-MS and HRF techniques can provide more quantitative information about the structural properties of proteins (see below).
The SPROX, PP, and TPP techniques provide the most quantitative information about the conformational properties of proteins and protein-ligand complexes. The transition midpoints extracted from the protein denaturation curves generated using these techniques (Figure 2) can be used to rank the relative stabilities of different proteins and the relative stability of a given protein under two different biological conditions (e.g., in the presence and absence of ligand) or in two different biological states (e.g., normal cells and cancer cells). The SPROX and PP techniques have the advantage over the TPP technique, that they can be used to evaluate thermodynamic parameters associated with protein folding and ligand binding interactions including protein folding free energies (ΔGf values), binding free energies (ΔΔGf values), and dissociation constants (Kd values).47, 48, 63–65 The evaluation of these thermodynamic parameters using SPROX and PP is possible because i) the chemical denaturation of proteins is generally reversible, which means the protein samples in the denaturant-containing buffers are truly at equilibrium and ii) the relationship between the folding free energy of a protein and chemical denaturant is well-established.77 This is in contrast to the TPP experiment where i) the thermal denaturation of proteins is not generally reversible so the thermally unfolded proteins are not truly at equilibrium and ii) there is no general relationship between a protein’s melting temperature and its ΔGf value.51 This also complicates the evaluation of ligand binding affinities using TPP. Even the magnitude of a protein’s Tm shift upon ligand binding does not always correlate with the ligand binding affinity.51
The different readouts (i.e., peptide- vs. protein-centered) in SPROX, TPP, and PP also mean that the thermodynamic parameters generated using each technique actually reports are associated with different conformational properties. For example, the thermodynamic parameters generated in SPROX are generally those associated with the specific structural domains to which the detected methionine-containing peptides map. This is in contrast to the PP and TPP techniques where the thermodynamic parameters generated in these techniques are those defined by the aggregate biophysical properties of the entire protein.
Structural Modeling
Information gained both from XL-MS and HRF can be used as constraints in molecular modeling and structural predictions. In XL-MS, cross-linkers, which have a defined length, provide quantitative information on the proximity of proteins and this data can provide distance constraints for molecular modeling. One caveat, however, is that structural constraints are not completely translational to topology since the length of the cross-linker represents the maximum distance whereas the actual interaction may exist closer than the cross-linker length.33 An additional caveat is that the conformational flexibility of proteins may bias the generation of cross-linked peptides for one conformational state. In spite of these caveats, XL-MS constraints have been used to aid modeling studies of the binding interface of proteins in vitro.78 XL-MS constraints have also been used for de novo modeling of proteins. Kahraman et al. constructed a partial de novo full-length model of human IgBPI using 65 intra-protein crosslinks combined with distance restraint data.79 Five models with the lowest RMSD in respect to the N-terminal domain of the template structure of mouse IgBPI were chosen as the best models for IgBPI.
To date, HRF methods have mainly provided qualitative information about the structural properties of different protein conformational states. However, the use of HRF data as constraints for molecular modeling is an emerging field. One challenge in such an HRF application is that the varying reactivity of residues with hydroxyl radicals can limit the amount of information obtained from the experiment. For example, highly reactive residues may be more highly modified relative to their more solvent accessible but less reactive neighbors. To overcome this challenge, Huang et al. developed an algorithm to correlate the measured footprinting rate to a protection factor based on residue reactivity with hydroxyl radicals.80 This protection factor provides a structural view based solely on solvent accessibility providing more quantitative information on the structure. This type of normalization has facilitated the use of HRF data as constraints for molecular modeling. Xie et al. demonstrated the ability to use HRF data to distinguish molecular models of high and low accuracy.81 Aprahamian et al. recently demonstrated that atomic resolution models of proteins can be obtained when HRF-derived protection factors are used as a score term in Rosetta to predict tertiary structures.82
6. NEW APPLICATION AREAS
Applications of traditional bottom-up proteomics methods have largely focused on protein expression level analyses. Indeed, mass spectrometry-based protein expression level analysis have been widely used over the past two decades to characterize a range of different biological states83–97 and drug activities,98–100 as evidenced by the tens of thousands of publications in this area over the last two decades. While differential protein expression profiling studies can provide some insight into the cellular pathways and potential protein players associated with a biological state or activity of a therapeutic agent, the biological significance of proteins with altered expression levels in different biological states or in response to drug treatments is often dubious because a protein’s expression level is not directly tied to its function. Functionally relevant proteins with the same expression levels in different biological states also go undetected using the current paradigm of expression level profiling to characterize such states.
The seven techniques highlighted in this perspective create a new paradigm for characterizing biological states and drug action. This new paradigm, which is based on protein conformational analyses, has the potential to uncover protein biomarkers and therapeutic targets of disease that are more biologically significant than those currently generated using protein expression level analyses. The close connection between protein folding stability and function was recently illustrated in several SPROX studies including one on the thermodynamic effects of phosphorylation on the proteins in an MCF-7 cell lysates101 and another on the proteins in cancer cell lysates.102 Highlighted below are several application areas where these new proteomics methods are beginning to making an impact.
Protein Target Discovery
A major driving force for the development of many of the techniques described above has been the need for better methods for protein target discovery. The utility of DARTS, SPROX, PP, LiPs, and TPP for identifying the protein targets of drugs and other small molecules ligands, such as enzyme co-factors, has been well-established in proof-of-principle experiments using a number of different model systems12, 48, 49, 51, 63, 65, 103–105 These techniques are also being used in more and more studies to identify the protein targets of potential therapeutic agents with less well understood mechanisms of action. For example, The PP and SPROX techniques were both used to assay over 1,000 proteins in a MDA-MB-231 cell lysate grown under hypoxic conditions for interactions with manassantin A, a natural product which has been shown to have anti-cancer activity in cell-based assays but has a currently unknown mode-of-action.67 This work identified a total of 28 protein hits with manassantin A-induced thermodynamic stability changes. Of particular note, was that two hits (filamin A and elongation factor 1-alpha) were identified with manassantin A-induced stability changes using both experimental approaches.
The ability to corroborate the hits obtained using one technique with another technique is useful for differentiating true positives from false positives. In this regard, the SPROX and PP approaches are especially useful, as they report on the same chemical-denaturant-induced equilibrium unfolding properties of proteins albeit with different proteomic-readouts. Along these lines, the DART and LiP approaches are expected to be similarly useful for corroborating the hits obtained using one technique with the other. Both approaches rely on a similar limited proteolysis reaction to identify hits, however, the proteomic-readouts employed in each technique are operationally different.
Biological States
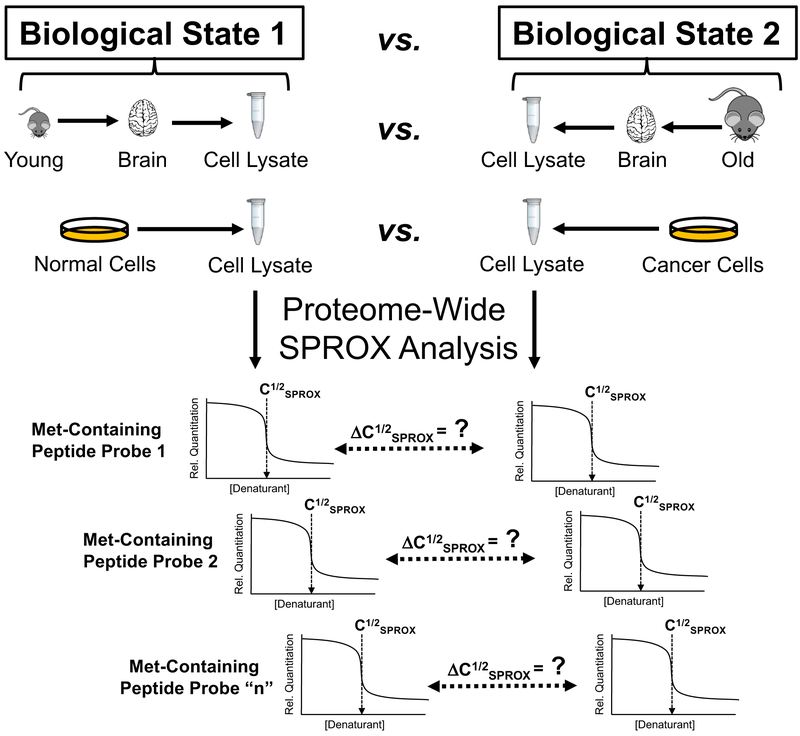
The characterization of biological states including those associated with normal biological processes (e.g., aging) and disease (e.g., cancer) is not only fundamentally important but it can facilitate the discovery of novel biomarkers that can be exploited in drug therapies and disease diagnoses. The SPROX methodology was one of the first techniques highlighted here to be applied to the analysis of biological states. Summarized in Figure 5 is the experimental workflow used in these SPROX experiments on biological states, which included those associated with a mouse model of aging and cell culture models of cancer. For example, the SILAC-SPROX technique was used to assay ~800–1000 proteins for changes in their protein folding behavior in five different cell line models of breast cancer including the MCF-10A, MCF-7, MDA-MB-231, BT474, and MDA-MB-468 cell lines.14, 102, 106 Between 10–40% of the proteins assayed in different comparative analyses displayed thermodynamic stability changes in the cell lysates from the different cell lines. The “hit” rates were all significantly higher than the false-positive rate of peptide hit discovery of ~3% established for SILAC-SPROX.104 The thermodynamic analyses enabled the benign MCF-10A breast cancer cell line to be differentiated from the MCF-7, MDA-MB-231, BT474, and MDA-MB-468 breast cancer cell lines. The protein hits with altered stabilities in the different breast cancer cell lines encompassed those with a wide range of functions and protein expression levels, and they included a significant fraction (~50%) with similar expression levels in the cell line comparisons. One MCF-7 cell line specific protein hit, calpain small subunit 1 (CAPNS1), was shown to have greater thermodynamic stability and increased catalytic activity in the MCF-7 cell line, despite no significant change in its expression level.102
Figure 5.
Schematic representation of the experimental workflow employed in several recent applications of the SPROX methodology to the characterization of biological states including those associated with a mouse model of aging110 and cell culture models of cancer.102, 106
A number of the identified protein hits in the above breast cancer cell-line comparisons were known from other biochemical studies to play a role in tumorigenesis and cancer progression. This not only substantiated the biological significance of the protein hits identified using the SILAC-SPROX approach, but it also helped elucidate the molecular basis for their dysregulation and/or dysfunction in cancer. In some cases, the hit proteins created novel molecular signatures of breast cancer and provided additional insight into the molecular basis of the disease. For example, the hit proteins in the MCF-7 vs. MDA-MA-231 comparison were enriched in hydrolases adding to the growing evidence that hydrolases are important for cell growth and invasiveness.107–109 Significantly, such information could not be gleaned from protein expression level data.102
In another biological state analysis, the SPROX methodology was also used to profile the thermodynamic stability of over 800 proteins in brain cell lysates from mice, aged 6- (n=7) and 18-months (n=9).110 The biological variability of the protein stability measurements was low and within the experimental error of SPROX (~0.1 – 0.2 M GdmCl). In this work, a total of 83 protein hits were detected with age-related stability differences in the brain samples. Remarkably, the large majority of the brain protein hits were destabilized in the old mice, and the hits were enriched in proteins that have slow turnover rates. Of particular significance was that a large fraction of the peptide hits (32 of 89) mapped to proteins known to have post-translational modifications, specifically carbonylations or oxidations, related to aging or a neurodegenerative disease with aging as a risk factor. Furthermore, 70% of the hits have been previously linked to ageing or age-related diseases. These results help validate the use of thermodynamic stability measurements to capture relevant age-related proteomic changes and establish a new biophysical link between these proteins and aging.
Protein Interaction Network Analysis
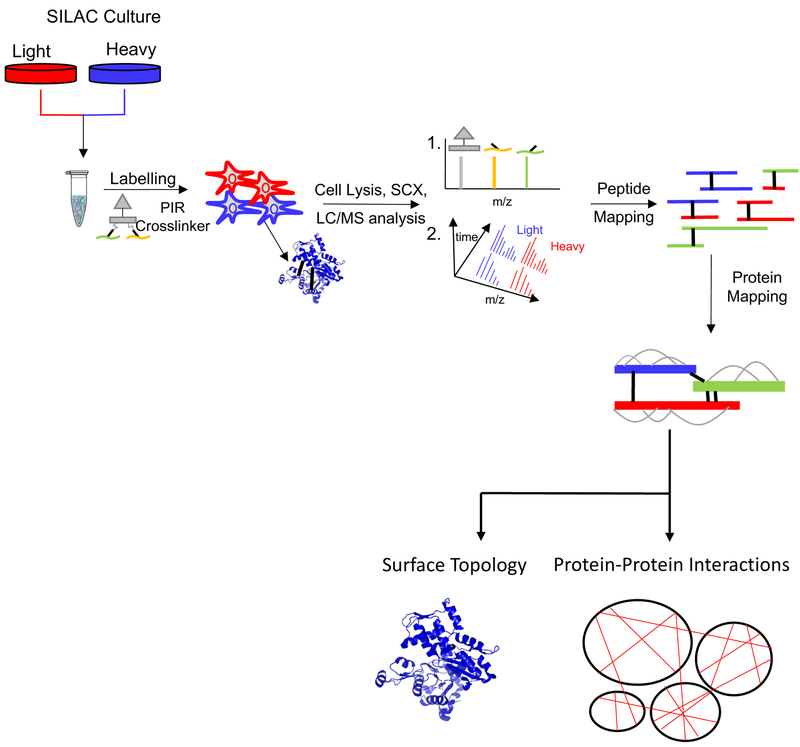
The XL-MS experiment is especially well-suited for the analysis of protein-protein-protein interactions in cells.38, 69, 111, 112 For a more quantitative approach, Chavez et al. coupled the PIR technology with SILAC (stable isotope labeling by amino acids in cell culture) to perform quantitative XL-MS (qXL-MS) for studying large scale protein structural changes and interactions in cells (Figure 6).113 The addition of SILAC allows for a comparison between a drug resistance cancer cell line and a drug sensitive parental cell line. This technique allows quantification of crosslinked peptide pairs either by MS1 based or targeted MS2 (parallel reaction monitoring (PRM)) based methods (Figure 6). In a recent report, this qXL-MS approach was employed to study the inter- and intra-molecular protein-protein interactions of Hsp90 upon treatment with a known Hsp90 inhibitor, 17-N-allylamino-17-demthoxygeldanamycin (17-AAG).69 Interestingly, there were differences in the conformational properties of the 17-AAG/HS90B complex formed in vitro and in vivo based on the comparison of in vivo and in vitro crosslinking data.
Figure 6.
Experimental workflow for in-cell XL-MS. Cells are cultured in isotopically light/heavy SILAC media followed by addition of PIR crosslinker to the cells in 1:1 mixture of light/heavy cells. The cells are lysed and the crosslinked peptides are enriched via strong cation exchange (SCX). LC/MS analysis by ReACt is used to identify cross-linked peptide pairs followed by MS1 based quantification of light/heavycrosslinked peptides. The selected crosslinked peptides are analyzed by targeted PRM. The resulting data is used to map the crosslinked peptides and further map the proteins that have been crosslinked. The crosslinked data is then complied into a protein interaction network and the crosslinks are analyzed in the context of existing structural information of the proteins.
While cell culture studies of protein-interaction networks can provide structural information on proteins within their native environment where cellular crowding effects protein structure and interactions, they do not provide significant information on organ level disease states. In a recent study, XL-MS was applied to proteins in mouse heart tissue samples to identify protein-protein interactions and to provide insight into how protein complexes exist in the context of the mouse heart.111 This study revealed insights into multiple conformational states of sarcomere proteins as well as interactions among oxidative phosphorylation complexes suggesting supercomplex assembly. This shows that the complex cellular environment plays a critical role in protein conformations and interactions that are not observed with purified proteins and complexes.
7. CONCLUSION
This perspective highlights seven new mass spectrometry-based technologies used to study protein structure on the proteomic scale. These methods provide information on protein conformations and interactions in the context of their native environment which is crowded with macromolecules. Although there are similarities between the methods, including the use of bottom-up proteomic strategies for data analysis, each method has its own unique data analysis challenges. The methods also provide different types of information from qualitative structural data, quantitative thermodynamic information, to structural constraints for molecular modeling. In some cases, using two methods in combination can provide complementary information. In other cases, the use of multiple methods can help increase the proteomic coverage and capture different types of conformational changes (e.g., more global versus more local effects on protein folding and stability). These methods provide access to new application areas for mass spectrometry including in vivo interaction networks and protein target discovery that is not solely reliant on protein expression changes. As these techniques are further developed and utilized, their applications will be further extended.
Acknowledgments
This work was supported in part by a grant from the National Institutes of General Medical Sciences at the National Institutes of Health 2R01GM084174–08 (to M.C.F.) and a grant from the National Science Foundation MCB1701692 (to L.M.J.)
REFERENCES
- 1.Gershenson A; Gierasch LM, Protein folding in the cell: challenges and progress. Curr Opin Struct Biol 2011, 21, (1), 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gierasch LM; Gershenson A, Post-reductionist protein science, or putting Humpty Dumpty back together again. Nat Chem Biol 2009, 5, (11), 774–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong J; Gierasch LM, Macromolecular crowding remodels the energy landscape of a protein by favoring a more compact unfolded state. J Am Chem Soc 2010, 132, (30), 10445–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christiansen A; Wang Q; Samiotakis A; Cheung MS; Wittung-Stafshede P, Factors defining effects of macromolecular crowding on protein stability: an in vitro/in silico case study using cytochrome c. Biochemistry 2010, 49,(31), 6519–30. [DOI] [PubMed] [Google Scholar]
- 5.Wirth AJ; Gruebele M, Quinary protein structure and the consequences of crowding in living cells: leaving the test-tube behind. Bioessays 2013, 35, (11), 984–93. [DOI] [PubMed] [Google Scholar]
- 6.Ando T; Skolnick J, Crowding and hydrodynamic interactions likely dominate in vivo macromolecular motion. Proc Natl Acad Sci U S A 2010, 107, (43), 18457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis RJ, Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr Opin Struct Biol 2001, 11, (1), 114–9. [DOI] [PubMed] [Google Scholar]
- 8.McConkey EH, Molecular evolution, intracellular organization, and the quinary structure of proteins. Proc Natl Acad Sci U S A 1982, 79, (10), 3236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang X; Bruce JE, A new cross-linking strategy: protein interaction reporter (PIR) technology for protein-protein interaction studies. Mol Biosyst 2010, 6, (6), 939–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chea EE; Jones LM, Analyzing the structure of macromolecules in their native cellular environment using hydroxyl radical footprinting. Analyst 2018, 143, (4), 798–807. [DOI] [PubMed] [Google Scholar]
- 11.Espino JA; Mali VS; Jones LM, In Cell Footprinting Coupled with Mass Spectrometry for the Structural Analysis of Proteins in Live Cells. Anal Chem 2015, 87, (15), 7971–8. [DOI] [PubMed] [Google Scholar]
- 12.Lomenick B; Hao R; Jonai N; Chin RM; Aghajan M; Warburton S; Wang JN; Wu RP; Gomez F; Loo JA; Wohlschlegel JA; Vondriska TM; Pelletier J; Herschman HR; Clardy J; Clarke CF; Huang J, Target identification using drug affinity responsive target stability (DARTS). Proc Nat Acad Sci U S A 2009, 106, (51), 21984–21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Y; De Franceschi G; Kahraman A; Soste M; Melnik A; Boersema PJ; de Laureto PP; Nikolaev Y; Oliveira AP; Picotti P, Global analysis of protein structural changes in complex proteomes. Nat Biotechnol 2014, 32, (10), 1036–44. [DOI] [PubMed] [Google Scholar]
- 14.Liu F; Fitzgerald MC, Large-Scale Analysis of Breast Cancer-Related Conformational Changes in Proteins Using Limited Proteolysis. J Proteome Res 2016, 15, (12), 4666–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu PF; Kihara D; Park C, Energetics-based discovery of protein-ligand interactions on a proteomic scale. J Mol Biol 2011, 408, (1), 147–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang Y; Schlebach JP; Verheul RA; Park C, Simplified proteomics approach to discover protein-ligand interactions. Protein Sci 2012, 21, (9), 1280–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adhikari J; Fitzgerald MC, SILAC-pulse proteolysis: A mass spectrometry-based method for discovery and cross-validation in proteome-wide studies of ligand binding. J Am Soc Mass Spectrom 2014, 25, (12), 2073–83. [DOI] [PubMed] [Google Scholar]
- 18.Zeng L; Shin WH; Zhu X; Park SH; Park C; Tao WA; Kihara D, Discovery of Nicotinamide Adenine Dinucleotide Binding Proteins in the Escherichia coli Proteome Using a Combined Energetic- and Structural-Bioinformatics-Based Approach. J Proteome Res 2017, 16, (2), 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dearmond PD; Xu Y; Strickland EC; Daniels KG; Fitzgerald MC, Thermodynamic analysis of protein-ligand interactions in complex biological mixtures using a shotgun proteomics approach. J Proteome Res 2011, 10, (11), 4948–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran DT; Adhikari J; Fitzgerald MC, Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC)-Based Strategy for Proteome-Wide Thermodynamic Analysis of Protein-Ligand Binding Interactions. Mol Cell Proteomics 2014, 13, (7), 1800–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savitski MM; Reinhard FB; Franken H; Werner T; Savitski MF; Eberhard D; Martinez Molina D; Jafari R; Dovega RB; Klaeger S; Kuster B; Nordlund P; Bantscheff M; Drewes G, Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 2014, 346, (6205), 1255784. [DOI] [PubMed] [Google Scholar]
- 22.Huang BX; Kim HY; Dass C, Probing three-dimensional structure of bovine serum albumin by chemical cross-linking and mass spectrometry. J Am Soc Mass Spectrom 2004, 15, (8), 1237–47. [DOI] [PubMed] [Google Scholar]
- 23.Schulz DM; Ihling C; Clore GM; Sinz A, Mapping the topology and determination of a low-resolution three-dimensional structure of the calmodulin-melittin complex by chemical cross-linking and high-resolution FTICRMS: direct demonstration of multiple binding modes. Biochemistry 2004, 43, (16), 4703–15. [DOI] [PubMed] [Google Scholar]
- 24.Fancy DA, Elucidation of protein-protein interactions using chemical cross-linking or label transfer techniques. Curr Opin Chem Biol 2000, 4, (1), 28–33. [DOI] [PubMed] [Google Scholar]
- 25.Singh P; Panchaud A; Goodlett DR, Chemical cross-linking and mass spectrometry as a low-resolution protein structure determination technique. Anal Chem 2010, 82, (7), 2636–42. [DOI] [PubMed] [Google Scholar]
- 26.Vasilescu J; Guo X; Kast J, Identification of protein-protein interactions using in vivo cross-linking and mass spectrometry. Proteomics 2004, 4, (12), 3845–54. [DOI] [PubMed] [Google Scholar]
- 27.Bruce JE, In vivo protein complex topologies: sights through a cross-linking lens. Proteomics 2012, 12, (10), 1565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novak P; Havlicek V; Derrick PJ; Beran KA; Bashir S; Giannakopulos AE, Monitoring conformational changes in protein complexes using chemical cross-linking and Fourier transform ion cyclotron resonance mass spectrometry: the effect of calcium binding on the calmodulin-melittin complex. Eur J Mass Spectrom (Chichester) 2007, 13, (4), 281–90. [DOI] [PubMed] [Google Scholar]
- 29.Sinz A, Chemical cross-linking and FTICR mass spectrometry for protein structure characterization. Anal Bioanal Chem 2005, 381, (1), 44–7. [DOI] [PubMed] [Google Scholar]
- 30.Sinz A, Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom Rev 2006, 25, (4), 663–82. [DOI] [PubMed] [Google Scholar]
- 31.Sinz A, Investigation of protein-protein interactions in living cells by chemical crosslinking and mass spectrometry. Anal Bioanal Chem 2010, 397, (8), 3433–3440. [DOI] [PubMed] [Google Scholar]
- 32.Sinz A, Divide and conquer: cleavable cross-linkers to study protein conformation and protein-protein interactions. Anal Bioanal Chem 2017, 409, (1), 33–44. [DOI] [PubMed] [Google Scholar]
- 33.Tang X; Munske GR; Siems WF; Bruce JE, Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal Chem 2005, 77, (1), 311–8. [DOI] [PubMed] [Google Scholar]
- 34.Sinz A, Cross-Linking/Mass Spectrometry for Studying Protein Structures and Protein-Protein Interactions: Where Are We Now and Where Should We Go from Here? Angew Chem Int Ed Engl 2018. [DOI] [PubMed] [Google Scholar]
- 35.Chavez JD; Eng JK; Schweppe DK; Cilia M; Rivera K; Zhong X; Wu X; Allen T; Khurgel M; Kumar A; Lampropoulos A; Larsson M; Maity S; Morozov Y; Pathmasiri W; Perez-Neut M; Pineyro-Ruiz C; Polina E; Post S; Rider M; Tokmina-Roszyk D; Tyson K; Vieira Parrine Sant’Ana D; Bruce JE, A General Method for Targeted Quantitative Cross-Linking Mass Spectrometry. PLoS One 2016, 11, (12), e0167547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piotrowski C; Ihling CH; Sinz A, Extending the cross-linking/mass spectrometry strategy: Facile incorporation of photo-activatable amino acids into the model protein calmodulin in Escherichia coli cells. Methods 2015, 89, 121–7. [DOI] [PubMed] [Google Scholar]
- 37.Schweppe DK; Chavez JD; Lee CF; Caudal A; Kruse SE; Stuppard R; Marcinek DJ; Shadel GS; Tian R; Bruce JE, Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc Natl Acad Sci U S A 2017, 114, (7), 1732–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong X; Navare AT; Chavez JD; Eng JK; Schweppe DK; Bruce JE, Large-Scale and Targeted Quantitative Cross-Linking MS Using Isotope-Labeled Protein Interaction Reporter (PIR) Cross-Linkers. J Proteome Res 2017, 16, (2), 720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chance MR; Sclavi B; Woodson SA; Brenowitz M, Examining the conformational dynamics of macromolecules with time-resolved synchrotron X-ray ‘footprinting’. Structure 1997, 5, (7), 865–9. [DOI] [PubMed] [Google Scholar]
- 40.Hambly DM; Gross ML, Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am Soc Mass Spectrom 2005, 16, (12), 2057–63. [DOI] [PubMed] [Google Scholar]
- 41.Sharp JS; Becker JM; Hettich RL, Analysis of protein solvent accessible surfaces by photochemical oxidation and mass spectrometry. Anal Chem 2004, 76, (3), 672–83. [DOI] [PubMed] [Google Scholar]
- 42.Rinas A; Mali VS; Espino JA; Jones LM, Development of a Microflow System for In-Cell Footprinting Coupled with Mass Spectrometry. Anal Chem 2016, 88, (20), 10052–10058. [DOI] [PubMed] [Google Scholar]
- 43.Zhu Y; Serra A; Guo T; Park JE; Zhong Q; Sze SK, Application of Nanosecond Laser Photolysis Protein Footprinting to Study EGFR Activation by EGF in Cells. J Proteome Res 2017, 16, (6), 2282–2293. [DOI] [PubMed] [Google Scholar]
- 44.Kim D; Hwang HY; Kim JY; Lee JY; Yoo JS; Marko-Varga G; Kwon HJ, FK506, an Immunosuppressive Drug, Induces Autophagy by Binding to the VATPase Catalytic Subunit A in Neuronal Cells. J Proteome Res 2017, 16, (1), 55–64. [DOI] [PubMed] [Google Scholar]
- 45.Lomenick B; Olsen RW; Huang J, Identification of Direct Protein Targets of Small Molecules. Acs Chemical Biology 2011, 6, (1), 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng YH; De Franceschi G; Kahraman A; Soste M; Melnik A; Boersema PJ; de Laureto PP; Nikolaev Y; Oliveira AP; Picotti P, Global analysis of protein structural changes in complex proteomes. Nat Biotechnol 2014, 32, (10), 1036–+. [DOI] [PubMed] [Google Scholar]
- 47.Park C; Marqusee S, Pulse proteolysis: a simple method for quantitative determination of protein stability and ligand binding. Nat Methods 2005, 2, (3), 207–12. [DOI] [PubMed] [Google Scholar]
- 48.West GM; Tucker CL; Xu T; Park SK; Han X; Yates JR; Fitzgerald MC, Quantitative proteomics approach for identifying protein–drug interactions in complex mixtures using protein stability measurements. Proc Natl Sci U S A 2010, 107, (20), 9078–9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeArmond PD; Xu Y; Strickland EC; Daniels KG; Fitzgerald MC, Thermodynamic analysis of protein–ligand interactions in complex biological mixtures using a shotgun proteomics approach. J Proteome Res 2011, 10, (11), 4948–4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molina DM; Jafari R; Ignatushchenko M; Seki T; Larsson EA; Dan C; Sreekumar L; Cao YH; Nordlund P, Monitoring Drug Target Engagement in Cells and Tissues Using the Cellular Thermal Shift Assay. Science 2013, 341, (6141), 84–87. [DOI] [PubMed] [Google Scholar]
- 51.Savitski MM; Reinhard FBM; Franken H; Werner T; Savitski MF; Eberhard D; Molina DM; Jafari R; Dovega RB; Klaeger S; Kuster B; Nordlund P; Bantscheff M; Drewes G, Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 2014, 346, (6205), 55–+. [DOI] [PubMed] [Google Scholar]
- 52.Peng H; Guo H; Pogoutse O; Wan C; Hu LZ; Ni Z; Emili A, An Unbiased Chemical Proteomics Method Identifies FabI as the Primary Target of 6-OH-BDE-47. Environmental Science & Technology 2016, 50, (20), 11329–11336. [DOI] [PubMed] [Google Scholar]
- 53.Park H; Ha J; Koo JY; Park J; Park SB, Label-free target identification using in-gel fluorescence difference via thermal stability shift. Chem Sci 2017, 8, (2), 1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haupl B; Ihling CH; Sinz A, Combining affinity enrichment, cross-linking with photo-amino acids, and mass spectrometry for probing protein kinase D2 interactions. Proteomics 2017. [DOI] [PubMed] [Google Scholar]
- 55.Weisbrod CR; Chavez JD; Eng JK; Yang L; Zheng C; Bruce JE, In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy. J Proteome Res 2013, 12, (4), 1569–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eng JK; Hoopmann MR; Jahan TA; Egertson JD; Noble WS; MacCoss MJ, A deeper look into Comet--implementation and features. J Am Soc Mass Spectrom 2015, 26, (11), 1865–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gotze M; Pettelkau J; Schaks S; Bosse K; Ihling CH; Krauth F; Fritzsche R; Kuhn U; Sinz A, StavroX--a software for analyzing crosslinked products in protein interaction studies. J Am Soc Mass Spectrom 2012, 23, (1), 76–87. [DOI] [PubMed] [Google Scholar]
- 58.Kosinski J; von Appen A; Ori A; Karius K; Muller CW; Beck M, Xlink Analyzer: software for analysis and visualization of cross-linking data in the context of three-dimensional structures. J Struct Biol 2015, 189, (3), 177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gau BC; Chen J; Gross ML, Fast photochemical oxidation of proteins for comparing solvent-accessibility changes accompanying protein folding: data processing and application to barstar. Biochim Biophys Acta 2013, 1834, (6), 1230–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rinas A; Espino JA; Jones LM, An efficient quantitation strategy for hydroxyl radical-mediated protein footprinting using Proteome Discoverer. Anal Bioanal Chem 2016, 408, (11), 3021–31. [DOI] [PubMed] [Google Scholar]
- 61.Bern M; Finney G; Hoopmann MR; Merrihew G; Toth MJ; MacCoss MJ, Deconvolution of mixture spectra from ion-trap data-independent-acquisition tandem mass spectrometry. Anal Chem 2010, 82, (3), 833–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaur P; Kiselar JG; Chance MR, Integrated algorithms for high-throughput examination of covalently labeled biomolecules by structural mass spectrometry. Anal Chem 2009, 81, (19), 8141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Geer MA; Fitzgerald MC, Characterization of the Saccharomyces cerevisiae ATP-Interactome using the iTRAQ-SPROX Technique. J Am Soc Mass Spectrom 2016, 27, (2), 233–43. [DOI] [PubMed] [Google Scholar]
- 64.West GM; Tang L; Fitzgerald MC, Thermodynamic analysis of protein stability and ligand binding using a chemical modification-and mass spectrometry-based strategy. Anal Chem 2008, 80, (11), 4175–4185. [DOI] [PubMed] [Google Scholar]
- 65.Xu Y; Wallace MA; Fitzgerald MC, Thermodynamic Analysis of the Geldanamycin-Hsp90 Interaction in a Whole Cell Lysate Using a Mass Spectrometry-Based Proteomics Approach. J Am Soc Mass Spectrom 2016, 27, (10), 1670–6. [DOI] [PubMed] [Google Scholar]
- 66.Strickland EC; Geer MA; Tran DT; Adhikari J; West GM; DeArmond PD; Xu Y; Fitzgerald MC, Thermodynamic analysis of protein-ligand binding interactions in complex biological mixtures using the stability of proteins from rates of oxidation. Nat Protocols 2013, 8, (1), 148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Geer Wallace MA; Kwon DY; Weitzel DH; Lee CT; Stephenson TN; Chi JT; Mook RA Jr.; Dewhirst MW; Hong J; Fitzgerald MC, Discovery of Manassantin A Protein Targets Using Large-Scale Protein Folding and Stability Measurements. J Proteome Res 2016, 15, (8), 2688–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ogburn RN; Jin L; Meng H; Fitzgerald MC, Discovery of Tamoxifen and NDesmethyl Tamoxifen Protein Targets in MCF-7 Cells Using Large-Scale Protein Folding and Stability Measurements. J Proteome Res 2017, 16, (11), 4073–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chavez JD; Schweppe DK; Eng JK; Bruce JE, In Vivo Conformational Dynamics of Hsp90 and Its Interactors. Cell Chem Biol 2016, 23, (6), 716–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang L; Zheng C; Weisbrod CR; Tang X; Munske GR; Hoopmann MR; Eng JK; Bruce JE, In vivo application of photocleavable protein interaction reporter technology. J Proteome Res 2012, 11, (2), 1027–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sinz A, Crosslinking Mass Spectrometry Goes In-Tissue. Cell Syst 2018, 6, (1), 10–12. [DOI] [PubMed] [Google Scholar]
- 72.Becher I; Werner T; Doce C; Zaal EA; Togel I; Khan CA; Rueger A; Muelbaier M; Salzer E; Berkers CR; Fitzpatrick PF; Bantscheff M; Savitski MM, Thermal profiling reveals phenylalanine hydroxylase as an off-target of panobinostat. Nat Chem Biol 2016, 12, (11), 908–+. [DOI] [PubMed] [Google Scholar]
- 73.Page BDG; Valerie NCK; Wright RHG; Wallner O; Isaksson R; Carter M; Rudd SG; Loseva O; Jemth AS; Almlof I; Font-Mateu J; Llona-Minguez S; Baranczewski P; Jeppsson F; Homan E; Almqvist H; Axelsson H; Regmi S; Gustavsson AL; Lundback T; Scobie M; Stromberg K; Stenmark P; Beato M; Helleday T, Targeted NUDT5 inhibitors block hormone signaling in breast cancer cells. Nat Commun 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reinhard FBM; Eberhard D; Werner T; Franken H; Childs D; Doce C; Savitski MF; Huber W; Bantscheff M; Savitski MM; Drewes G, Thermal proteome profiling monitors ligand interactions with cellular membrane proteins. Nat Methods 2015, 12, (12), 1129–+. [DOI] [PubMed] [Google Scholar]
- 75.Vartanian S; Ma TP; Lee J; Haverty PM; Kirkpatrick DS; Yu KB; Stokoe D, Application of Mass Spectrometry Profiling to Establish Brusatol as an Inhibitor of Global Protein Synthesis. Mol Cell Proteomics 2016, 15, (4), 1220–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan CSH; Go KD; Bisteau X; Dai L; Yong CH; Prabhu N; Ozturk MB; Lim YT; Sreekumar L; Lengqvist J; Tergaonkar V; Kaldis P; Sobota RM; Nordlund P, Thermal proximity coaggregation for system-wide profiling of protein complex dynamics in cells. Science 2018, 359, (6380), 1170–1177. [DOI] [PubMed] [Google Scholar]
- 77.Myers JK; Pace CN; Scholtz JM, Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci 1995, 4, (10), 2138–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Herzog F; Kahraman A; Boehringer D; Mak R; Bracher A; Walzthoeni T; Leitner A; Beck M; Hartl FU; Ban N; Malmstrom L; Aebersold R, Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 2012, 337, (6100), 1348–52. [DOI] [PubMed] [Google Scholar]
- 79.Kahraman A; Herzog F; Leitner A; Rosenberger G; Aebersold R; Malmstrom L, Cross-link guided molecular modeling with ROSETTA. PLoS One 2013, 8, (9), e73411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang W; Ravikumar KM; Chance MR; Yang S, Quantitative mapping of protein structure by hydroxyl radical footprinting-mediated structural mass spectrometry: a protection factor analysis. Biophys J 2015, 108, (1), 107–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xie B; Sood A; Woods RJ; Sharp JS, Quantitative Protein Topography Measurements by High Resolution Hydroxyl Radical Protein Footprinting Enable Accurate Molecular Model Selection. Sci Rep 2017, 7, (1), 4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aprahamian ML; Chea EE; Jones LM; Lindert S, Rosetta Protein Structure Prediction from Hydroxyl Radical Protein Footprinting Mass Spectrometry Data. Anal Chem 2018, 90, (12), 7721–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Walther TC; Mann M, Mass spectrometry-based proteomics in cell biology. J Cell Biol 2010, 190, (4), 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oppermann FS; Gnad F; Olsen JV; Hornberger R; Greff Z; Keri G; Mann M; Daub H, Large-scale Proteomics Analysis of the Human Kinome. Mol Cell Proteomics 2009, 8, (7), 1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alizadeh AA; Eisen MB; Davis RE; Ma C; Lossos IS; Rosenwald A; Boldrick JG; Sabet H; Tran T; Yu X; Powell JI; Yang LM; Marti GE; Moore T; Hudson J; Lu LS; Lewis DB; Tibshirani R; Sherlock G; Chan WC; Greiner TC; Weisenburger DD; Armitage JO; Warnke R; Levy R; Wilson W; Grever MR; Byrd JC; Botstein D; Brown PO; Staudt LM, Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, (6769), 503–511. [DOI] [PubMed] [Google Scholar]
- 86.Golub TR; Slonim DK; Tamayo P; Huard C; Gaasenbeek M; Mesirov JP; Coller H; Loh ML; Downing JR; Caligiuri MA; Bloomfield CD; Lander ES, Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 1999, 286, (5439), 531–537. [DOI] [PubMed] [Google Scholar]
- 87.Lu J; Getz G; Miska EA; Alvarez-Saavedra E; Lamb J; Peck D; Sweet-Cordero A; Ebet BL; Mak RH; Ferrando AA; Downing JR; Jacks T; Horvitz HR; Golub TR, MicroRNA expression profiles classify human cancers. Nature 2005, 435, (7043), 834–838. [DOI] [PubMed] [Google Scholar]
- 88.Perou CM; Sorlie T; Eisen MB; van de Rijn M; Jeffrey SS; Rees CA; Pollack JR; Ross DT; Johnsen H; Akslen LA; Fluge O; Pergamenschikov A; Williams C; Zhu SX; Lonning PE; Borresen-Dale AL; Brown PO; Botstein D, Molecular portraits of human breast tumours. Nature 2000, 406, (6797), 747–752. [DOI] [PubMed] [Google Scholar]
- 89.Sorlie T; Perou CM; Tibshirani R; Aas T; Geisler S; Johnsen H; Hastie T; Eisen MB; van de Rijn M; Jeffrey SS; Thorsen T; Quist H; Matese JC; Brown PO; Botstein D; Lonning PE; Borresen-Dale AL, Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Sci U S A 2001, 98, (19), 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sorlie T; Tibshirani R; Parker J; Hastie T; Marron JS; Nobel A; Deng S; Johnsen H; Pesich R; Geisler S; Demeter J; Perou CM; Lonning PE; Brown PO; Borresen-Dale AL; Botstein D, Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Sci U S A 2003, 100, (14), 8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van’t Veer LJ; Dai HY; van de Vijver MJ; He YDD; Hart AAM; Mao M; Peterse HL; van der Kooy K; Marton MJ; Witteveen AT; Schreiber GJ; Kerkhoven RM; Roberts C; Linsley PS; Bernards R; Friend SH, Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, (6871), 530–536. [DOI] [PubMed] [Google Scholar]
- 92.Volinia S; Calin GA; Liu CG; Ambs S; Cimmino A; Petrocca F; Visone R; Iorio M; Roldo C; Ferracin M; Prueitt RL; Yanaihara N; Lanza G; Scarpa A; Vecchione A; Negrini M; Harris CC; Croce CM, A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Sci U S A 2006, 103, (7), 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Adam BL; Qu YS; Davis JW; Ward MD; Clements MA; Cazares LH; Semmes OJ; Schellhammer PF; Yasui Y; Feng ZD; Wright GL, Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res 2002, 62, (13), 3609–3614. [PubMed] [Google Scholar]
- 94.Hanash S, Disease proteomics. Nature 2003, 422, (6928), 226–232. [DOI] [PubMed] [Google Scholar]
- 95.Li JN; Zhang Z; Rosenzweig J; Wang YY; Chan DW, Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clinical Chemistry 2002, 48, (8), 1296–1304. [PubMed] [Google Scholar]
- 96.Rikova K; Guo A; Zeng Q; Possemato A; Yu J; Haack H; Nardone J; Lee K; Reeves C; Li Y; Hu Y; Tan ZP; Stokes M; Sullivan L; Mitchell J; Wetzel R; MacNeill J; Ren JM; Yuan J; Bakalarski CE; Villen J; Kornhauser JM; Smith B; Li D; Zhou X; Gygi SP; Gu TL; Polakiewicz RD; Rush J; Comb MJ, Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007, 131, (6), 1190–1203. [DOI] [PubMed] [Google Scholar]
- 97.Yanagisawa K; Shyr Y; Xu BGJ; Massion PP; Larsen PH; White BC; Roberts JR; Edgerton M; Gonzalez A; Nadaf S; Moore JH; Caprioli RM; Carbone DP, Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet 2003, 362, (9382), 433–439. [DOI] [PubMed] [Google Scholar]
- 98.Godl K; Wissing J; Kurtenbach A; Habenberger P; Blencke S; Gutbrod H; Salassidis K; Stein-Gerlach M; Missio A; Cotten M; Daub H, An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc Natl Sci U S A 2003, 100, (26), 15434–15439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Graves PR; Kwiek JJ; Fadden P; Ray R; Hardeman K; Coley AM; Foley M; Haystead TAJ, Discovery of novel targets of quinoline drugs in the human purine binding proteome. Mol Pharmacol 2002, 62, (6), 1364–1372. [DOI] [PubMed] [Google Scholar]
- 100.Oda Y; Owa T; Sato T; Boucher B; Daniels S; Yamanaka H; Shinohara Y; Yokoi A; Kuromitsu J; Nagasu T, Quantitative chemical proteomics for identifying candidate drug targets. Anal Chem 2003, 75, (9), 2159–2165. [DOI] [PubMed] [Google Scholar]
- 101.Meng H; Fitzgerald MC, Proteome-Wide Characterization of Phosphorylation-Induced Conformational Changes in Breast Cancer. J Proteome Res 2018, 17, (3), 1129–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Adhikari J; West GM; Fitzgerald MC, Global Analysis of Protein Folding Thermodynamics for Disease State Characterization. J Proteome Res 2015, 14, (5), 2287–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Adhikari J; Fitzgerald MC, SILAC-Pulse Proteolysis: A Mass Spectrometry-Based Method for Discovery and Cross-Validation in Proteome-Wide Studies of Ligand Binding. J Am Soc Mass Spectrom 2014, 25, (12), 2073–2083. [DOI] [PubMed] [Google Scholar]
- 104.Tran DT; Adhikari J; Fitzgerald MC, StableIsotope Labeling with Amino Acids in Cell Culture (SILAC)-based strategy for proteome-wide thermodynamic analysis of protein-ligand binding interactions. Mol Cell Proteomics 2014, 13, (7), 1800–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Trindade RV; Pinto AF; Santos DS; Bizarro CV, Pulse Proteolysis and Precipitation for Target Identification. J Proteome Res 2016, 15, (7), 2236–45. [DOI] [PubMed] [Google Scholar]
- 106.Liu F; Meng H; Fitzgerald MC, Large-Scale Analysis of Breast Cancer-Related Conformational Changes in Proteins Using SILAC-SPROX. J Proteome Res 2017, 16, (9), 3277–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jessani N; Liu YS; Humphrey M; Cravatt BF, Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Sci U S A 2002, 99, (16), 10335–10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Paulick MG; Bogyo M, Application of activity-based probes to the study of enzymes involved in cancer progression. Curr Opin Genet Dev 2008, 18, (1), 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shields DJ; Niessen S; Murphy EA; Mielgo A; Desgrosellier JS; Lau SKM; Barnes LA; Lesperance J; Bouvet M; Tarin D; Cravatt BF; Cheresh DA, RBBP9: A tumor-associated serine hydrolase activity required for pancreatic neoplasia. Proc Natl Sci U S A 2010, 107, (5), 2189–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Roberts JH; Liu F; Karnuta JM; Fitzgerald MC, Discovery of Age-Related Protein Folding Stability Differences in the Mouse Brain Proteome. J Proteome Res 2016, 15, (12), 4731–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chavez JD; Lee CF; Caudal A; Keller A; Tian R; Bruce JE, Chemical Crosslinking Mass Spectrometry Analysis of Protein Conformations and Supercomplexes in Heart Tissue. Cell Syst 2018, 6, (1), 136–141 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Navare AT; Chavez JD; Zheng C; Weisbrod CR; Eng JK; Siehnel R; Singh PK; Manoil C; Bruce JE, Probing the protein interaction network of Pseudomonas aeruginosa cells by chemical cross-linking mass spectrometry. Structure 2015, 23, (4), 762–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chavez JD; Schweppe DK; Eng JK; Zheng C; Taipale A; Zhang Y; Takara K; Bruce JE, Quantitative interactome analysis reveals a chemoresistant edgotype. Nat Commun 2015, 6, 7928. [DOI] [PMC free article] [PubMed] [Google Scholar]