Abstract
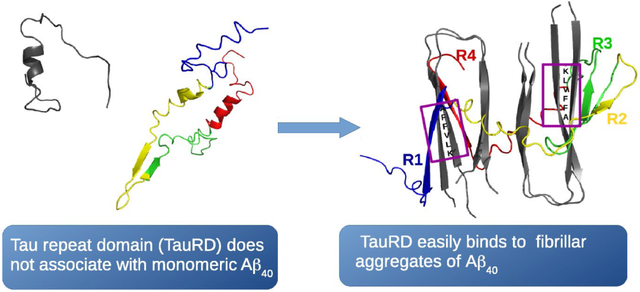
One of the hallmarks of Alzheimer’s disease is the formation of aggregates of the tau protein, a process that can be facilitated by the presence of fibrils formed by the Amyloid β peptide (Aβ). However, the mechanism that triggers tau aggregation is still a matter of debate. The effect of Aβ40 fibrils on the aggregation of the repeat domain of tau (TauRD) is investigated here by employing coarse-grained molecular dynamics simulations. The results indicate that the repeat domain of tau has high affinity for Aβ40 fibrils, with the 261GSTENLK267 fragment of tau driving TauRD towards the 16KLVFFA21 fragment in Aβ40. Monomeric Aβ40, in which the 16KLVFFA21 fragment is rarely found in an extended conformation (as in the fibril), has low affinity for the TauRD, indicating that the ability of Aβ40 fibrils to bind to the TauRD depends on the 16KLVFFA21 fragment of Aβ adopting an extended conformation.
Graphical Abstract
Introduction
The brain of patients with Alzheimer’s disease (AD)1 presents extracellular plaques formed by fibrils of the Amyloid β peptide (Aβ) and intracellular tangles of a different fibrillar material, often referred to as paired helical filaments (PHFs), formed by the tau protein2. A number of studies have indicated that these two events are deeply connected3-6. Unlike Aβ, which aggregates easily and forms fibrils both in vitro and in vivo, tau is highly soluble and does not aggregate normally7-8. The widely accepted “amyloid hypothesis”9 states that Aβ fibrils appear early in the disease, and they facilitate the aggregation of tau. According to this hypothesis, it is through the tau aggregates that Aβ exerts its toxicity, causing neuronal degeneration and death9-10.
Many studies have shown that Aβ fibrils and pre-fibrillar amyloids can facilitate tau aggregation3-4, 11-13. Aβ and tau can form mixed aggregates11-12, and this interaction might promote aggregation-prone conformations in tau. Aβ can also precipitate the formation of the PHFs of tau by a more complex mechanism, in which Aβ facilitates hyperphosphorylation of tau, which in its turn can lead to fibrillization of tau3, 11, 14. Hence, it is clear that interaction with Aβ can trigger aggregation of tau. However, the details of this interaction – critical for the design of drugs aimed to prevent it – are not well understood.
In addition, it is not clear which species of Aβ (soluble, fibrillar or pre-fibrillar) or which isoform (Aβ40 or Aβ42) is responsible for Aβ-induced aggregation of tau. While some studies have indicated that in soluble form, only Aβ42, and not Aβ40, can trigger tau hyperphosphorylation and aggregation14, others have shown that in their fibrillar form both Aβ40 or Aβ42 can produce the same effect3, 11. Intriguingly, another study has shown that pre-fibrillar aggregates of Aβ42, but not monomers or fibrils, can facilitate the formation of tau trimers that can subsequently associate into PHF13. The contradictory results regarding the Aβ species implicated in tau aggregation might arise from the fact that Aβ42 aggregates at a much faster rate than Aβ4015. Therefore, experiments using soluble Aβ42 might have Aβ42 aggregates within minutes of mixing, while samples containing Aβ40 will take longer to develop aggregates and will contain mostly monomeric Aβ40. Another source of contradiction is the fact that Aβ fibrils, or smaller aggregates, prepared by different protocols can have different morphologies16. Hence, two different samples of Aβ42 fibrils, each with a particular morphology, might interact very differently with tau and produce a very different result. Therefore, it is important to understand the structural principles governing the interaction between Aβ and tau. With this goal in mind, in this article, the effect of Aβ40 fibrils and monomers on the aggregation of the repeat domain of tau is investigated using molecular dynamics (MD) simulations with the coarse-grained united-residue (UNRES) force field17-21.
Methods
The UNRES model.
Detailed descriptions of the UNRES model and its parametrization are available in references 17-20, 22-23. The implementation used here to simulate multichain complexes was reported in reference 22. Here, for completeness, we give a short description of the model. In the UNRES model,17-20, 22-23 a polypeptide chain is represented as a sequence of α-carbon (Cα) atoms with united peptide groups (p) located halfway between the consecutive Cα atoms and united side chains (SC) attached to the Cα atoms. The force field has been derived as the potential of mean force (PMF) of a system of polypeptide chain(s) in water, where all degrees of freedom except the coordinates of the Cα atoms and SC centers have been averaged out17, 20. The effective energy function contains local and site-site interactions, as well as multibody terms, which have been obtained by decomposing the PMF into factors corresponding to clusters of interactions within and between coarse-grained sites17. The SC-SC interaction potentials implicitly include the contribution from solvation17, 20. The force field was calibrated to reproduce the structure and thermodynamics of small model proteins18-19, 23. In this study, the parameters derived from the study of the WW domain from the Formin-binding protein 28 (PDB ID: 1E0L)19 were used.
Simulation setup.
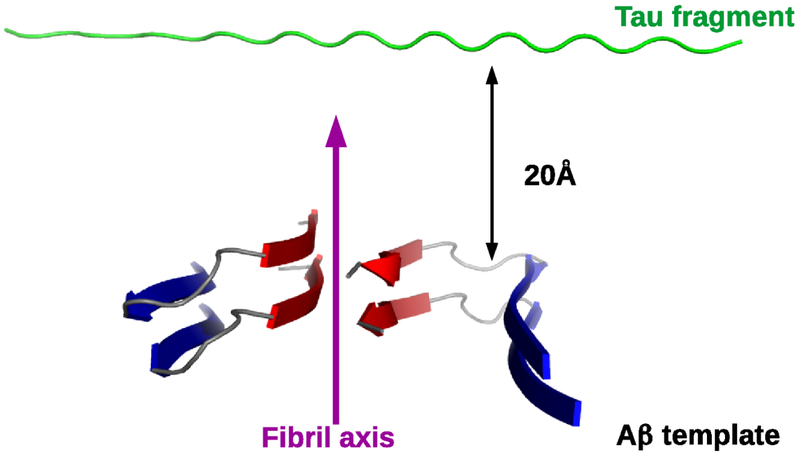
In simulations that included an Aβ40 template, the template was modeled using the coordinates derived by Petkova et al.24 (Fig. 1). Our previous study on this system25 showed that a large fibril can be mimicked by using only two layers. Therefore, the template used in this work consisted of two layers, resulting in a total of 4 chains. To give the template the stability of a long fibril, distance restraints were applied to the Cα carbons as in reference 45. The free monomer was initially placed at a 20 Å distance from the edge of the template in a random coil conformation (Fig. 1). When simulations included two free monomers, the second monomer was placed 20 Å away from the first, and 20 Å from the template. In simulations that included only free monomers, the chains were initially in an extended conformation, parallel to each other, and separated by 20 Å distance. In all simulations the system was enclosed in a spherical volume, as in reference 25, the size of which was set to result in a concentration of 3 mM.
Figure 1:
Simulation Setup. A two-layers (4 chains) fibril, based on the model of Petkova et al24 for Aβ40 fibrils, was used as a template to simulate the interaction between an Aβ40 fibril and the repeat fragment from tau protein. The N-terminal and C-terminal strands of the Aβ40 are colored in blue and red, respectively. Distance restraints were used to stabilize the template, but no restraints were applied on the monomer. The monomer was initially placed at a 20 Å distance from the template in a completely unfolded conformation. The tau fragment (green color) was either Tau3RD, Tau4RD, R1, R2, R3 or R4.
Molecular dynamics (MD) simulations.
Simulations were carried out using replica exchange molecular dynamics (REMD). For each system, a total of 120 trajectories were simulated with temperatures ranging from 280 K to 320 K. Each trajectory was 72×105 steps long, which was equivalent to 3.6×105 UNRES molecular time units (mtu),26 which is approximately 17.6 μs. Based on previous benchmarks26, a time step of 0.05 mtu was used, with a variable-time-step algorithm27. Temperature exchanges were attempted every 20,000 steps. Between exchanges, the temperature was held constant with the Berendsen thermostat28.
When averages such as β-content, radius of gyration or number of hydrogen bonds are reported, they are obtained by averaging over conformations from the last 22 × 105 steps and using only conformations from simulations at 300 K. When the number of trajectories showing dimers is reported, the values are obtained using all conformations.
To calculate the fraction of trajectories that resulted in binding of the tau fragment (either one or several repeats) to the template, we used 3 amino acids long β-sheet as the cutoff. In other words, if the monomer formed a β-sheet with the template that was at least 3-amino acids long, then it was considered to be bound to the template.
The binding propensity of Tau3RD and Tau4RD along the Aβ40 sequence, which are shown in Figure 2a, have been averaged over all the chains (i.e, divided by 4). This was done because for some trajectories, the tau fragment binds across the surface of the template, and often to the same region of Aβ40 (as in Fig. 2b), resulting in double counting of the binding at that region. Therefore, to obtain a measure of binding propensity that was independent of the number of exposed chains, the results were averaged over all the chains in the template.
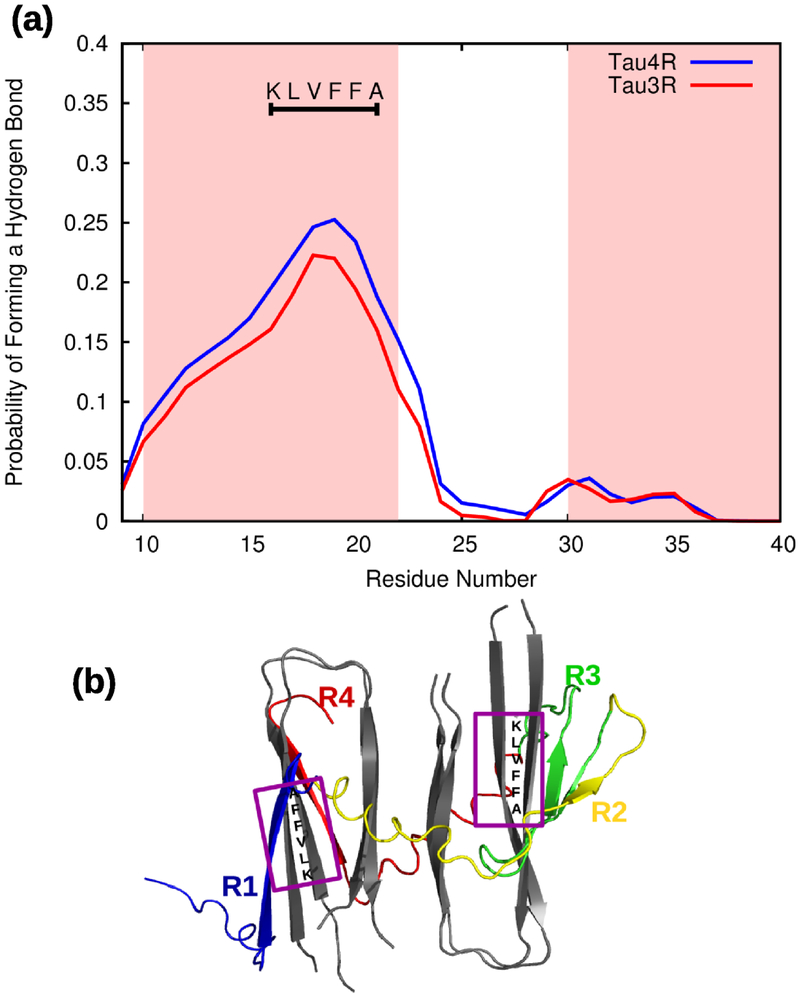
Figure 2.
Simulations of Tau3RD or Tau4RD and an Aβ40 fibril template. (a) The probability for each residue in the Aβ peptides of forming a hydrogen bond with any of the residues in the Tau monomer. The two regions along the Aβ peptides that adopt a β-sheet conformation are shaded in pink. Hydrogen bonding occurs mainly along the N-terminal strand of the Aβ peptides. The region with the highest probability, residues 16-21, is indicated by a horizontal black line, as well as the sequence along this fragment. (b) Representative snapshot showing the binding of Tau4RD and the Aβ40 template (gray). Different colors are used for each of the tau repeats: R1 in blue, R2 in yellow, R3 in green and R4 in red. A purple box indicates the KLVFFA fragment on Aβ40.
Results and Discussion
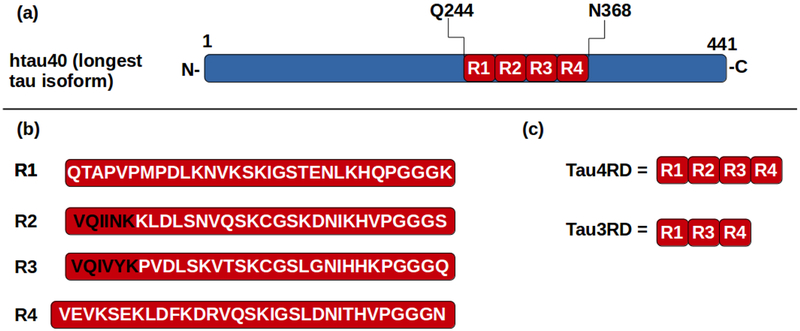
Six different tau isoforms exist29-30 in the human brain. The longest tau isoform, htau40, is 441 residues long (Fig. 3a). The C-terminal half of tau contains what is known as the repeat domain (Fig. 3b). Three of the six tau isoforms contain four repeats (R1 to R4), but the other three contain only three of the repeats (R1, R3 and R4). Both three- and four-repeat isoforms are found in tau filaments from AD brains31. The repeat domain of tau (TauRD) forms the core of tau fibrils32 and promotes β-sheet formation33-36. Therefore, we focused our study on this region, more specifically on the three-repeat (Tau3RD) and the four-repeat (Tau4RD) tau fragments (Fig. 3c). For this purpose, we carried out MD simulations of a 4-chains (2 layers) Aβ40 template24 interacting with Tau3RD or Tau4RD (Fig. 1).
Figure 3.
(a) Representation of the longest tau isoform, htau40, showing the location of the repeat domain (R1 to R4). (b) The sequence of each of the four repeats, R1, R2, R3 and R4. The two fragments known to form fibrils by themselves34, 36 and to be crucial for tau fibrillization35 are shown in black. (c) Composition of Tau3RD and Tau4RD fragments.
Results show that Tau3RD and Tau4RD have high affinity for the Aβ40 template.
The tau monomer forms hydrogen bonds with the template in 69% of Tau4RD trajectories and in 67% of Tau3RD ones. By tracking the regions of Aβ40 participated in the hydrogen bonds formed with Tau3RD or Tau4RD, and averaging over the four chains in the template, a binding propensity along the Aβ40 sequence was calculated (Fig. 2a). The results show that the residues in Aβ40 responsible for the binding to Tau3RD and Tau4RD are primarily residues along the N-terminal half of Aβ40 (see Fig. 2a and 2b), with the highest binding propensity seen along the KLVFFA fragment (residues 16-21) of Aβ40.
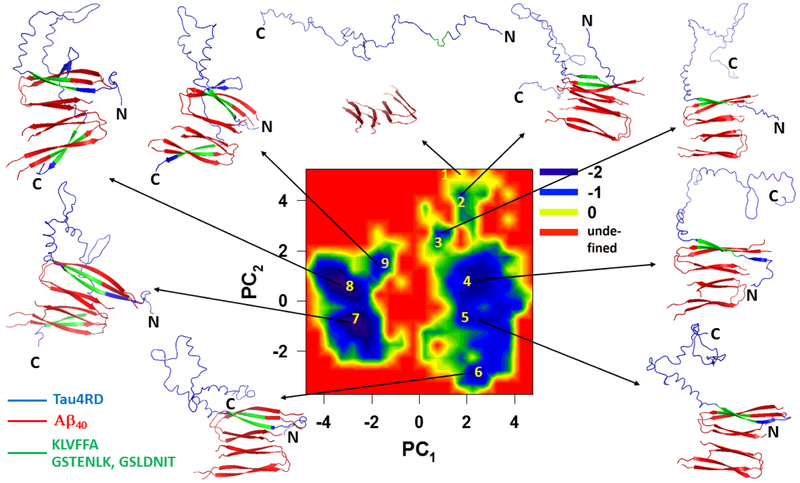
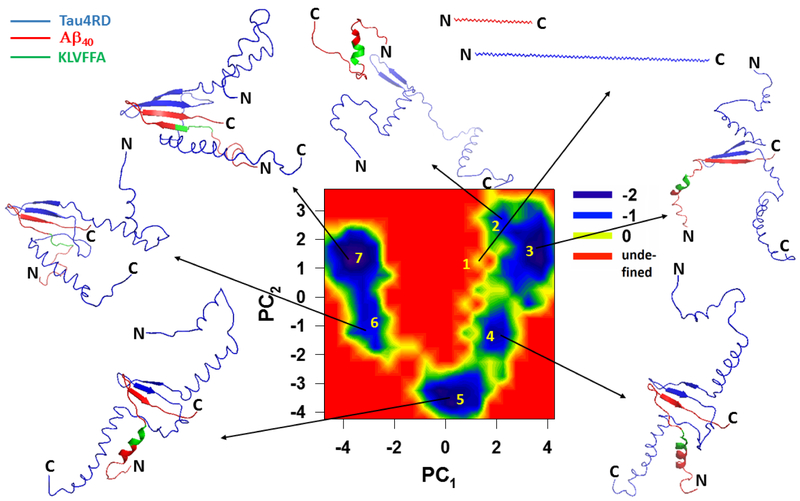
In order to describe, in detail, the pathway along which the tau monomer binds to the fibril template, we built the free-energy landscape (FEL) along the principal components (PCs) (Fig. 4) obtained from principal component analysis (PCA) of a representative trajectory. PCs typically capture most of the total displacement from the average protein structure with the first few PCs during a simulation37-39. It appears that, at an early stage in the trajectory, the GSTENLK fragment of Tau4RD (residues 18-24 of the R1 repeat) drives the tau protein towards the KLVFFA fragment (residues 16-21) of one of the Aβ40 chains, and binds this fragment forming a β-sheet (representative structures of minima 2-6). The R2, R3 and R4 repeats do not interact with the Aβ40 template during this time interval. Only after finishing formation of the β-strand and binding of the R1 repeat with the Aβ40 template, does the R4 repeat start moving towards the second semifilament of the Aβ40 template, and the GSLDNIT fragment (residues 19-25 of R4 repeat) binds to the KLVFFA fragment of one of the chains with anti-parallel orientation and forms a β-strand (see representative structures of minima 7-9 in Fig. 4 and Fig. 2b). The highest affinity is seen along a region of Aβ40 containing the KLVFFA sequence (Fig. 2a). Interestingly, the KLVFFA fragment has been shown to be critical for the formation of Aβ fibrils40-41; it is also capable of forming amyloid fibrils by itself and to sequester small compounds into these fibrils42. Our simulations show that this same region is critical for interaction with Tau3RD and Tau4RD.
Figure 4.
Free-energy landscape (in kcal/mol) along the first two PCs, μ (PC1, PC2) = - kBT ln P (PC1, PC2), with representative structures at the minima (labeled 1 to 9) for the trajectory of tau protein binding to the Aβ40 template.
It is interesting to note that Tau4RD and Tau3RD show a considerable amount of α-helical structure throughout the simulations (see representative structures of minima in Fig. 4). These helices are quite flexible and often unfold or convert into β-sheets. However, it is evident that the presence of the Aβ40 template can offset the β-sheet-to-α-helix balance in the tau fragments by stabilizing β-sheets with the template.
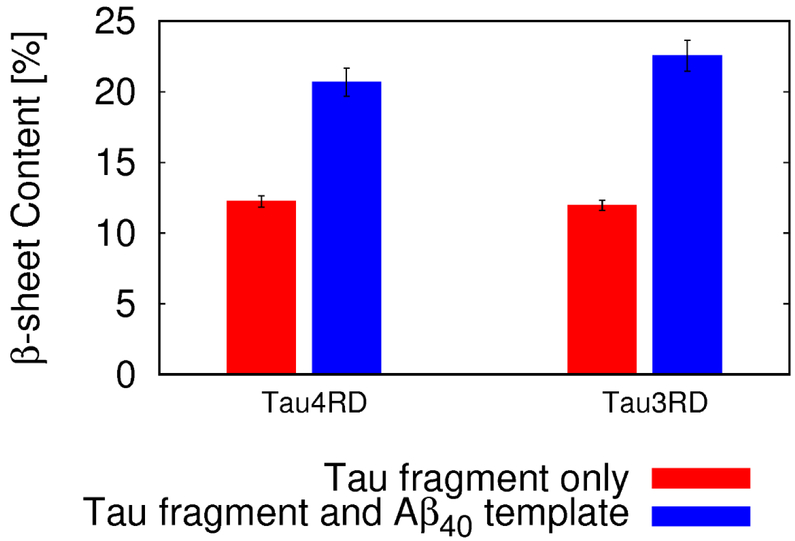
Tau3RD and Tau4RD exhibit higher β-sheet content in the presence of the Aβ40 template.
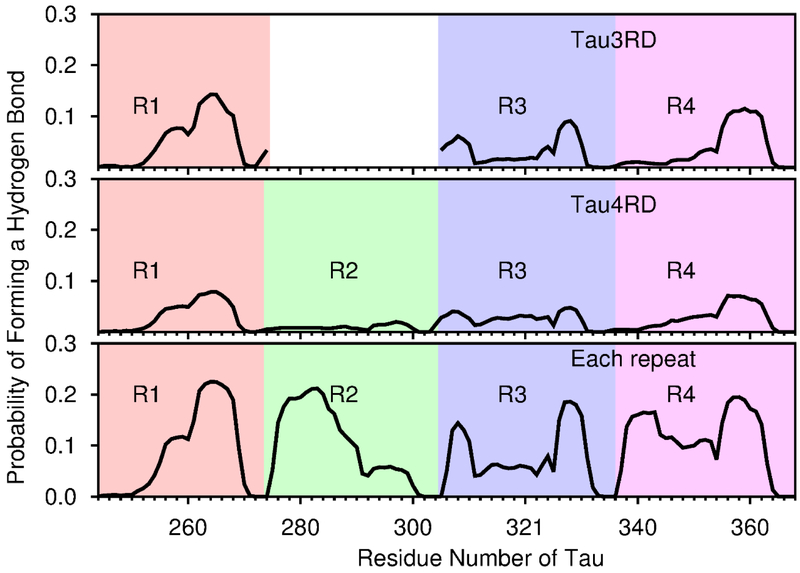
To further understand the effect of the Aβ40 template on the secondary structure of the tau fragments, simulations of Tau3RD or Tau4RD, without the Aβ40 template, were carried out, and compared with those with the Aβ40 template. These comparisons show that the Aβ40 template promotes conformations with higher β content along Tau3RD and Tau4RD (Fig. 5). The increase in β content is primarily due to β-sheets between Aβ40 and the R1 or R4 repeats of tau. This can be seen when evaluating the probability of each residue on the tau repeat to form a hydrogen bond with the Aβ40 template (Fig. 6, top and middle panels). Areas with higher probabilities (i.e., areas that often form hydrogen bonds with the Aβ40 template) are those that are frequently involved in β sheets with the Aβ40 template. Therefore, our simulations indicate that binding occurs more often through hydrogen bonds involving residues along repeats R1 and R4 (Fig. 6, top and middle panels).
Figure 5:
The β-sheet content (in %) of Tau3RD and Tau4RD fragments in the absence (red) or presence (blue) of the Aβ40 template. The errors were obtained from fluctuations between the different trajectories.
Figure 6.
The probability for each residue in Tau3RD (top), Tau4RD (middle), or any of the repeats independently (bottom), of forming a hydrogen bond with any of the residues from the Aβ chains. The results from simulations of each of the repeats independently are shown next to each other to facilitate comparison with the upper panels. The regions corresponding to each of the repeats is indicated by different background colors, pink (R1), green (R2), blue (R3) and purple (R4). Numbering of residues corresponds to the longest Tau isoform, with R1=244-274, R2=275-305, R3=306-336 and R3=337-368. For top plot (Tau3RD), the region corresponding to the second repeat is empty because this region does not exist in Tau3RD.
Interactions between tau repeats hinder their ability to bind to the Aβ40 template.
The lower tendency of repeats R2 and R3 to form hydrogen bonds with the Aβ peptides could be due to their sequence having low affinity with Aβ or to interactions between the repeats (R2 and R3 with each other or with the other repeats) competing with the tau-Aβ interactions. To investigate this question, simulations of each of the repeats (R1, R2, R3 or R4) interacting with an Aβ template were carried out. The tendency of each of the repeats to form hydrogen bonds with the template is shown in Fig. 6, bottom panel. These results indicate that, when the other repeats are absent, each of the four repeats has similar affinity with the peptides in the Aβ40 template. Moreover, their tendency to form hydrogen bonds with the template is higher when they interact with the template by themselves than when the other repeats are also present (Fig. 6). Therefore, interactions between the repeats must be hindering binding to Aβ. Indeed, the fragment 306VQIVYK311, the regions with the highest β-structure potential in tau33, is often seen forming a β-hairpin in our simulations (Fig. S1a-c). This β-hairpin is present in simulations of monomers, as well as in simulations with the template, and it forms in early stage of the simulations (Fig. S2 and S3), indicating that its formation and stabilization is due to intramolecular contacts and does not depend on interactions with the Aβ40 template. This suggests that the presence of the β-hairpin might hamper the ability of the regions involved in it to bind to the Aβ40 template.
The presence of the Aβ40 template does not facilitate dimerization of TauRD.
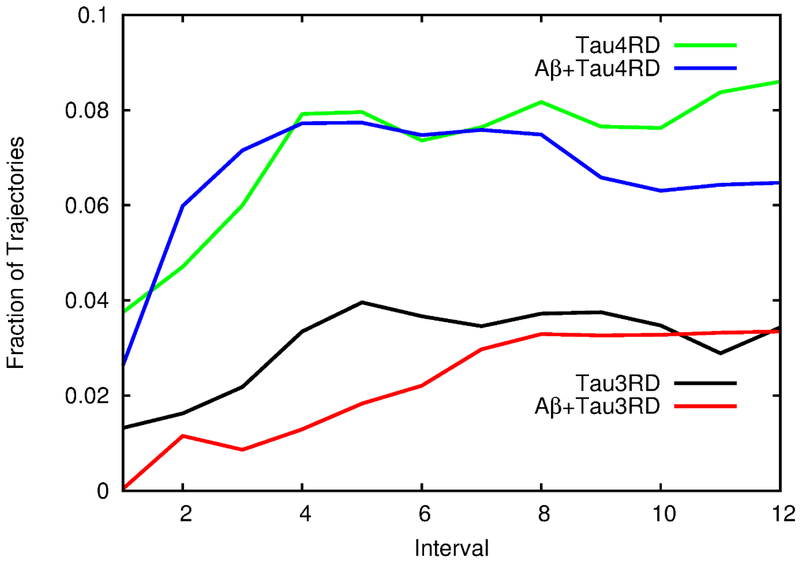
The fact that, upon interacting with the Aβ template, Tau3RD and Tau4RD show an increase in their β-sheet content suggest that the presence of Aβ might make tau more prone to aggregation. To investigate this, MD simulations of dimers of Tau3RD or Tau4RD with or without an Aβ template were carried out. The MD simulations were partitioned into twelve consecutive intervals, and the average number of trajectories in which a β-sheet had formed between the TauRD monomers were calculated. The results show different behaviors for Tau3RD and Tau4RD. Consistent with aggregation experiments8, Tau4RD shows a higher tendency to form homodimers than Tau3RD (Fig. 7). Contrary to our expectations, the presence of the template does not seem to affect the formation of dimers by Tau3RD or Tau4RD (Fig. 7).
Figure 7:
The effect of the Aβ40 template on dimer formation by Tau3RD or Tau4RD. The trajectories were divided into 12 equal intervals, and the fraction of trajectories in which an intermolecular β-sheet was formed between the two tau monomers (either Tau3RD or Tau4RD) is shown at each interval. Values are shown for Tau4RD in the absence (yellow) or presence (blue) of the template and for Tau3RD in the absence (black) or presence (red) of the template.
Although our simulations do not show a higher rate of dimerization in the presence of the Aβ template, it is possible that this is an effect that cannot be observed within the time scale of our simulations, or that it cannot be seen on the Tau3RD or Tau4RD fragments, but that the complete tau protein needs to be simulated. Another possibility is that a larger number of monomers need to attach to the Aβ template for aggregation to be observed. It should also be mentioned that the only experimental evidence of Aβ alone been capable of triggering tau aggregation is a study where tau peptides were subjected to several rounds of incubation with decreasing levels of Aβ aggregates before the presence of tau trimers (not PHFs) was detected13. Therefore, it is possible that the increase in β-sheet content on tau observed in our simulations (Fig. 5) might result, after hours of incubation and several remixing steps, in tau oligomerization. Aβ-induced aggregation of tau was also studied through MD simulations by Qi et al.43. Their simulations indicated that an Aβ42 template causes Tau3RD and Tau4RD fragments to adopt more extended conformations, which would presumably facilitate their assembly into fibrils. This “stretching-and-packing”43 mechanism was reflected in their simulations in higher radii of gyration when the fragments interacted with the Aβ42 template, i.e., the presence of the template caused the tau fragments to stretch out. Our simulations did not show this effect. While the radii of gyration that we obtain (Fig. S4) are more consistent with a disordered peptide44, we do not see any significant difference between the simulations of a free monomer and those of a monomer and a template. The discrepancies between our results and those of Qi et al.43 might be attributed to differences in the force fields or effective simulation times, but they can also arise from the fact that our simulations were carried out with an Aβ40 template instead of Aβ42.
TauRD’s binding affinity to Aβ40 is largely reduced when Aβ40 is a non-aggregated form.
Many experimental studies have indicated that Aβ42, but not Aβ40, can induce tau aggregation14, 45. However, these studies often use the Aβ peptides in a non-aggregated state14. Because Aβ42 aggregates at a faster rate than Aβ40, the studies might be evaluating the effect of different aggregation states. This possibility was pointed out in a study by Rank et al11, where the authors found that soluble Aβ42, but not Aβ40, could form mixed aggregates with tau. However, if pre-aggregated Aβ40 was used, the effect on tau aggregation was similar to that of non-aggregated Aβ42.
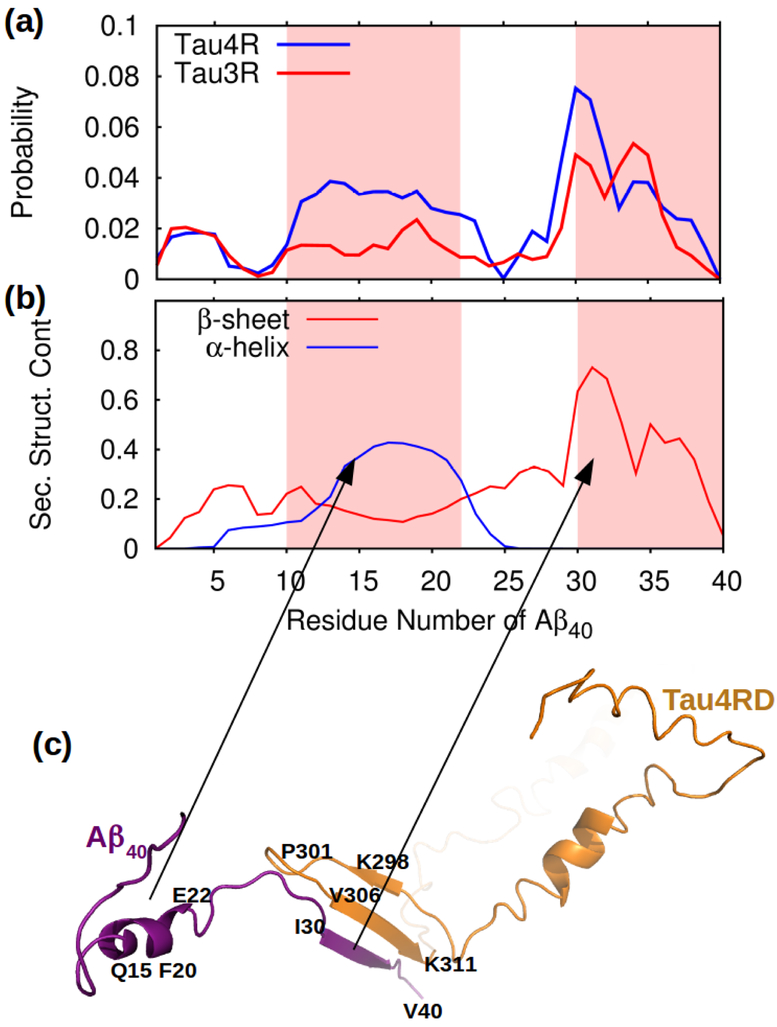
To see if, as in Rank et al.11 experiments, the ability of Aβ40 to bind to Tau3RD and Tau4RD fragments was only seen when Aβ40 was in the fibrillar conformation, simulations in which the tau fragments interacted with an Aβ40 monomer (i.e. non-aggregated) were carried out. The number of hydrogen bonds formed between the Tau4RD or Tau3RD fragments and the Aβ40 monomer (Fig. 8a) is considerably lower than it was for the Aβ40 template (Fig. 2a). This indicates that Tau4RD and Tau3RD have little affinity for non-aggregated Aβ40. Similar simulations carried out with Aβ42 show that TauRD’s affinity for Aβ42 monomers is also low (Fig. S5). Taken together, our results indicate that neither Aβ40 or Aβ42 monomers can form aggregates in complex with tau. Most likely, when the mix contains Aβ42, tau binds to the Aβ42 that has already started to form aggregates.
Figure 8.
Simulations of dimerization between Aβ40 and TauRD (a) The probability that each of the residues along the Aβ40 sequence would have formed a hydrogen bond with any of the residues in Tau4RD (blue) or Tau3RD (red). The regions that in the fibrils are in a β-strand conformation are shaded in pink. (b) β-sheet and α-helix content for along the Aβ40 sequence. (c) Example of a dimer formed between Tau4RD (orange) and Aβ40 (purple) illustrating the higher tendency to form hydrogen bonds along the C-terminal region of Aβ40.
When Aβ40 and Tau4RD, or Tau3RD, do form dimers, it is mostly through intermolecular β-sheets involving residues along the C-terminal portion of Aβ40 (Fig. 8a). The reason for Tau4RD and Tau3RD associating with the C-terminal half of Aβ40, instead of the N-terminal half [as was the case with the Aβ40 template (Fig. 2a)], can be found in the secondary structure of the Aβ40 monomers. The 16KLVFFA21 region of Aβ40, which in the fibril is part of the N-terminal β-strand, and that so efficiently attracts Tau4RD and Tau3RD (Fig. 2a), is more often found in an α-helical conformation in the monomeric state (Fig. 8b). Due to this change in secondary structure, the tau fragments bind to the C-terminal strand instead, which is found in a β-sheet conformation most of the time (see example in Fig. 8c), but to which they seem to have lower affinity. Hence, Aβ40 or Aβ42 monomers are rarely found in complex with Tau3RD or Tau4RD. As an illustrative example, we built the FEL along the principal components for one of the MD trajectories to describe, in detail, the pathway along which the TauRD and Aβ40 monomers form heterodimers (Fig. 9). It should be noted that for this trajectory, after certain time, the 16KLVFFA21 fragment loses the helical secondary structure (representative structure of minimum 6) and adopts the β-stand conformation (representative structure of minimum 7), however the dimer does not change the conformation (Fig. 9).
Figure 9.
Free-energy landscape (in kcal/mol) along the first two PCs with representative structures at the minima (labeled 1 to 7) for the trajectory of tau protein binding to the Aβ40 monomer.
Conclusions
To summarize, MD trajectories generated with the UNRES force field were used to study the effect of Aβ40 fibrils on the aggregation of Tau4RD and Tau3RD, two fragments from the repeat domain of the tau protein. Our results indicate that both Tau3RD and Tau4RD have higher affinity for the Aβ40 template than for themselves. Aβ40 fibrils act as sticky surfaces onto which the tau fragments easily bind forming intermolecular β-sheets. More importantly, the affinity between Aβ and the tau fragments was not observed when Aβ40 or Aβ42 were in a non-aggregated form. Put together, these findings might explain why soluble Aβ42, but not Aβ40, can trigger tau aggregation11, 14. The difference might simply be caused by the fact that Aβ42 aggregates at a much faster rate than Aβ40, and the experiments are comparing the binding of tau to β-rich aggregates of Aβ42 with its binding to monomeric Aβ40. The results also suggest that other Aβ40 fibril polymorphs may sequester Tau3RD and Tau4RD, as long as the 16KLVFFA21 fragment of Aβ40 adopts an extended conformation.
Certain limitations of our model deserve some attention. For example, inspection of the different trajectories shows that two chains of Tau4RD or Tau 3RD can form hydrogen bonds with the Aβ40 template simultaneously, one on each side of it, and still interact with each other. This is something that could not happen on real fibrils, with typical lengths of at least tens of nm46. However, this could happen with pre-fibrillar aggregates that exhibit spherical or disk-shaped morphology47-48. NMR and deuterium exchange data indicate that these aggregates have abundant β-sheet conformation, with the C-terminal region buried in the interior of the structure, and the N-terminal region more exposed to solvent47-48. Therefore, the propensity of tau fragments to bind to the N-terminal region of Aβ, when this region is in an extended conformation, indicates that the tau fragments could have high affinity for Aβ’s pre-fibrillar aggregates, and thus the aggregates might also be able to trap monomeric tau, preventing it from performing its function, and stabilizing its β-sheet content. Moreover, due to the spherical morphology of the aggregates, and because they are formed by as little as 10 Aβ chains46-47, several tau fragments could bind to each Aβ aggregate and interact with each other, which might eventually facilitate oligomerization.
Our MD simulations also indicate that the tau fragments have affinity for extended conformations, and thus they might be prone to interacting with other amyloids. Indeed, oligomeric α-synuclein, another protein known to form amyloid fibrils, can also induce tau aggregation13. This suggests that Aβ and α-synuclein might not be the only amyloidogenic proteins capable of facilitating tau aggregation
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM-14312) and the National Science Foundation (MCB10-19767). This research was conducted by using the resources of: (a) our 588- processor Beowulf cluster at the Baker Laboratory of Chemistry and Chemical Biology, Cornell University, (b) the supercomputer resources at the Interdisciplinary Center of Mathematical and Computer Modeling (ICM) and (c) the Informatics Center of the Metropolitan Academic Network (IC MAN) in Gdańsk.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the http://pubs.acs.org
Formation of β-hairpin in Tau3RD and Tau4RD; Histograms of radius of gyration for Tau4RD and Tau3RD in the presence and absence of the Aβ40 template; TauRD’s binding affinity to non-aggregated Aβ42 monomers.
The authors declare no competing financial interest.
References
- 1.Alzheimer A; Stelzmann RA; Schnitzlein HN; Murtagh FR An English translation of Alzheimer's 1907 paper, "Uber eine eigenartige Erkankung der Hirnrinde". Clin. Anat. 1995, 8, 429–431. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe DJ The molecular pathology of Alzheimer's disease. Neuron 1991, 6, 487–498. [DOI] [PubMed] [Google Scholar]
- 3.Busciglio J; Lorenzo A; Yeh J; Yankner BA β-Amyloid fibrils induce tau phosphorylation and loss of microtubule binding. Neuron 1995, 14, 879–888. [DOI] [PubMed] [Google Scholar]
- 4.Gotz J; Chen F; van Dorpe J; Nitsch RM Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Aβ42 fibrils. Science 2001, 293, 1491–1495. [DOI] [PubMed] [Google Scholar]
- 5.LaFerla FM Pathways linking Aβ and tau pathologies. Biochem. Soc. Trans. 2010, 38, 993–995. [DOI] [PubMed] [Google Scholar]
- 6.Flores-Rodriguez P; Ontiveros-Torres MA; Cardenas-Aguayo MC; Luna-Arias JP; Meraz-Rios MA; Viramontes-Pintos A; Harrington CR; Wischik CM; Mena R; Floran-Garduno B; et al. The relationship between truncation and phosphorylation at the C-terminus of tau protein in the paired helical filaments of Alzheimer's disease. Front. Neurosci. 2015, 9, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weingarten MD; Lockwood AH; Hwo SY; Kirschner MW A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. U. S. A. 1975, 72, 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barghorn S; Mandelkow E Toward a unified scheme for the aggregation of tau into Alzheimer paired helical filaments. Biochemistry 2002, 41, 14885–14896. [DOI] [PubMed] [Google Scholar]
- 9.Hardy J; Selkoe DJ The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [DOI] [PubMed] [Google Scholar]
- 10.Masters CL; Selkoe DJ Biochemistry of amyloid β-protein and amyloid deposits in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rank KB; Pauley AM; Bhattacharya K; Wang Z; Evans DB; Fleck TJ; Johnston JA; Sharma SK Direct interaction of soluble human recombinant tau protein with Aβ 1–42 results in tau aggregation and hyperphosphorylation by tau protein kinase II. FEBS Lett. 2002, 514, 263–268. [DOI] [PubMed] [Google Scholar]
- 12.Guo JP; Arai T; Miklossy J; McGeer PL Aβ and tau form soluble complexes that may promote self aggregation of both into the insoluble forms observed in Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 1953–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lasagna-Reeves CA; Castillo-Carranza DL; Guerrero-Muoz MJ; Jackson GR; Kayed R Preparation and characterization of neurotoxic tau oligomers. Biochemistry 2010, 49, 10039–10041. [DOI] [PubMed] [Google Scholar]
- 14.Hu X; Li X; Zhao M; Gottesdiener A; Luo W; Paul S Tau pathogenesis is promoted by Aβ1–42 but not Aβ1–40. Mol. Neurodegener. 2014, 9, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stine WB Jr.; Dahlgren KN; Krafft GA; LaDu MJ In vitro characterization of conditions for amyloid-β peptide oligomerization and fibrillogenesis. J. Biol. Chem. 2003, 278, 11612–11622. [DOI] [PubMed] [Google Scholar]
- 16.Tycko R Amyloid polymorphism: structural basis and neurobiological relevance. Neuron 2015, 86, 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liwo A; Czaplewski C; Pillardy J; Scheraga HA Cumulant-based expressions for the multibody terms for the correlation between local and electrostatic interactions in the united-residue force field. J. Chem. Phys. 2001, 115, 2323–2347. [Google Scholar]
- 18.Ołdziej S; Ła̧giewka J; Liwo A; Czaplewski C; Chinchio M; Nanias M; Scheraga HA Optimization of the UNRES Force Field by Hierarchical Design of the Potential-Energy Landscape. 3. Use of Many Proteins in Optimization. J. Phys. Chem. B 2004, 108, 16950–16959. [Google Scholar]
- 19.Liwo A; Khalili M; Czaplewski C; Kalinowski S; Ołdziej S; Wachucik K; Scheraga HA Modification and optimization of the united-residue (UNRES) potential energy function for canonical simulations. I. Temperature dependence of the effective energy function and tests of the optimization method with single training proteins. J. Phys. Chem. B 2007, 111, 260–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liwo A; Czaplewski C; Ołdziej S; Rojas AV; Kazmierkiewicz R; Makowski M; Murarka RK; Scheraga HA Simulation of Protein Structure and Dynamics with the Coarse-Grained UNRES Force Field In Coarse-Graining of Condensed Phase and Biomolecular Systems, Voth G, Ed. CRC Press: 2008; pp 107–122. [Google Scholar]
- 21.Maisuradze GG; Senet P; Czaplewski C; Liwo A; Scheraga HA Investigation of protein folding by coarse-grained molecular dynamics with the UNRES force field. J. Phys. Chem. A 2010, 114, 4471–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojas AV; Liwo A; Scheraga HA Molecular dynamics with the United-residue force field: ab initio folding simulations of multichain proteins. J. Phys. Chem. B 2007, 111, 293–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y; Xiao Y; Liwo A; Scheraga HA Exploring the parameter space of the coarse-grained UNRES force field by random search: selecting a transferable medium-resolution force field. J. Comput. Chem. 2009, 30, 2127–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petkova AT; Yau WM; Tycko R Experimental constraints on quaternary structure in Alzheimer's β-amyloid fibrils. Biochemistry 2006, 45, 498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rojas A; Liwo A; Browne D; Scheraga HA Mechanism of fiber assembly: treatment of Aβ peptide aggregation with a coarse-grained united-residue force field. J. Mol. Biol. 2010, 404, 537–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khalili M; Liwo A; Jagielska A; Scheraga HA Molecular dynamics with the united-residue model of polypeptide chains. II. Langevin and Berendsen-bath dynamics and tests on model α-helical systems. J. Phys. Chem. B 2005, 109, 13798–13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalili M; Liwo A; Rakowski F; Grochowski P; Scheraga HA Molecular dynamics with the united-residue model of polypeptide chains. I. Lagrange equations of motion and tests of numerical stability in the microcanonical mode. J. Phys. Chem. B 2005, 109, 13785–13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berendsen HJC; Postma JPM; Gunsteren WFV; DiNola A; Haak JR Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar]
- 29.Goedert M; Spillantini MG; Jakes R; Rutherford D; Crowther RA Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer's disease. Neuron 1989, 3, 519–526. [DOI] [PubMed] [Google Scholar]
- 30.Goedert M; Jakes R Expression of separate isoforms of human tau protein: correlation with the tau pattern in brain and effects on tubulin polymerization. EMBO J. 1990, 9, 4225–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goedert M; Spillantini MG A century of Alzheimer's disease. Science 2006, 314, 777–781. [DOI] [PubMed] [Google Scholar]
- 32.Wegmann S; Medalsy ID; Mandelkow E; Müller DJ The fuzzy coat of pathological human Tau fibrils is a two-layered polyelectrolyte brush. Proc. Natl. Acad. Sci. U. S.A. 2013, 110, E313–E321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Bergen M; Friedhoff P; Biernat J; Heberle J; Mandelkow EM; Mandelkow E Assembly of τ protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming β structure. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 5129–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goux WJ; Kopplin L; Nguyen AD; Leak K; Rutkofsky M; Shanmuganandam VD; Sharma D; Inouye H; Kirschner DA The formation of straight and twisted filaments from short tau peptides. J. Biol. Chem. 2004, 279, 26868–26875. [DOI] [PubMed] [Google Scholar]
- 35.Li W; Lee VM Characterization of two VQIXXK motifs for tau fibrillization in vitro. Biochemistry 2006, 45, 15692–15701. [DOI] [PubMed] [Google Scholar]
- 36.Wiltzius JJ; Landau M; Nelson R; Sawaya MR; Apostol MI; Goldschmidt L; Soriaga AB; Cascio D; Rajashankar K; Eisenberg D Molecular mechanisms for protein-encoded inheritance. Nat. Struct. Mol. Biol. 2009, 16, 973–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maisuradze GG; Liwo A; Scheraga HA Principal component analysis for protein folding dynamics. J. Mol. Biol. 2009, 385, 312–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maisuradze GG; Liwo A; Scheraga HA How adequate are one- and two-dimensional free energy landscapes for protein folding dynamics? Phys. Rev. Lett. 2009, 102, 238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maisuradze GG; Liwo A; Scheraga HA Relation between free energy landscapes of proteins and dynamics. J. Chem. Theory Comput. 2010, 6, 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tjernberg LO; Callaway DJ; Tjernberg A; Hahne S; Lilliehook C; Terenius L; Thyberg J; Nordstedt C A molecular model of Alzheimer amyloid β-peptide fibril formation. J. Biol. Chem. 1999, 274, 12619–12625. [DOI] [PubMed] [Google Scholar]
- 41.Rojas A; Maisuradze N; Kachlishvili K; Scheraga HA; Maisuradze GG Elucidating Important Sites and the Mechanism for Amyloid Fibril Formation by Coarse-Grained Molecular Dynamics. ACS Chem. Neurosci. 2017, 8, 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landau M; Sawaya MR; Faull KF; Laganowsky A; Jiang L; Sievers SA; Liu J; Barrio JR; Eisenberg D Towards a pharmacophore for amyloid. PLoS Biol. 2011, 9, e1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi R; Luo Y; Wei G; Nussinov R; Ma B Aβ “stretching-and-packing” cross-seeding mechanism can trigger tau protein aggregation. J. Phys. Chem. Lett 2015, 6, 3276–3282. [Google Scholar]
- 44.Mylonas E; Hascher A; Bernado P; Blackledge M; Mandelkow E; Svergun DI Domain conformation of tau protein studied by solution small-angle X-ray scattering. Biochemistry 2008, 47, 10345–10353. [DOI] [PubMed] [Google Scholar]
- 45.McGowan E; Pickford F; Kim J; Onstead L; Eriksen J; Yu C; Skipper L; Murphy MP; Beard J; Das P; et al. Aβ42 is essential for parenchymal and vascular amyloid deposition in mice. Neuron 2005, 47, 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dubnovitsky A; Sandberg A; Rahman MM; Benilova I; Lendel C; Hard T Amyloid-β protofibrils: size, morphology and synaptotoxicity of an engineered mimic. PLoS One 2013, 8, e66101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chimon S; Shaibat MA; Jones CR; Calero DC; Aizezi B; Ishii Y Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer's β-amyloid. Nat. Struct. Mol. Biol. 2007, 14, 1157–1164. [DOI] [PubMed] [Google Scholar]
- 48.Ahmed M; Davis J; Aucoin D; Sato T; Ahuja S; Aimoto S; Elliott JI; Van Nostrand WE; Smith SO Structural conversion of neurotoxic amyloid-β1-42 oligomers to fibrils. Nat. Struct. Mol. Biol. 2010, 17, 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.